Abstract
Since its establishment in 2008, the US Tox21 inter-agency collaboration has made great progress in developing and evaluating cellular models for the evaluation of environmental chemicals as a proof of principle. Currently, the program has entered its production phase (Tox21 Phase II) focusing initially on the areas of modulation of nuclear receptors and stress response pathways. During Tox21 Phase II, the set of chemicals to be tested has been expanded to nearly 10,000 (10K) compounds and a fully automated screening platform has been implemented. The Tox21 robotic system combined with informatics efforts is capable of screening and profiling the collection of 10K environmental chemicals in triplicate in a week. In this article, we describe the Tox21 screening process, compound library preparation, data processing, and robotic system validation.
Keywords: 10K compound library, in vitro assays, quantitative high-throughput screening, robotic platform, Tox21 collaboration, toxicity testing
Introduction
Toxicity assessment of environmental chemicals has traditionally relied on animal-based toxicological methods, but such studies are low-throughput, costly, and inconsistently predictive of human toxicity. To potentially overcome these limitations and to rapidly and efficiently evaluate the thousands of environmental chemicals with little or no prior toxicological information, the National Toxicology Program (NTP) [1] proposed a roadmap for toxicology testing in the 21st century promoting the advancement of toxicology from a mainly observational science at the level of disease-specific and animal-based models to a predictive science focused upon a broad inclusion of target-specific mechanistic approaches [1]. In 2005, the National Research Council (NRC) [2] was commissioned by the U.S. Environmental Protection Agency [3] and the National Institutes of Environmental Health Sciences to develop a long-range vision strategic plan for toxicity testing [4]. This plan emphasized the use of new tools in molecular toxicology, computational sciences, and information technology to address the challenge of evaluating the thousands of untested chemicals present in the environment [2], stating that traditional toxicity testing methods are inadequate due to the high cost and the vast number of animals that would be needed to characterize these chemicals. Both the NTP and NRC visions rely on the use of chemical profiling strategies to study critical cellular responses to xenobiotics using primarily high-throughput screening (HTS)-based in vitro human cell models. The use of cellular targets or ‘toxicity’ pathway perturbations as new, discrete toxicological endpoints constitutes the first step to identify mechanisms of toxicity for each xenobiotic and to provide predictive potential. Mechanistic findings would be used to prioritize chemicals for more in-depth, targeted in vitro or in vivo testing leading to the development and refinement of predictive toxicological models [5].
To accomplish this vision, the Tox21 collaboration was formed between the NTP, the EPA National Center for Computational Toxicology (NCCT), and the NIH Chemical Genomics Center (NCGC) in 2008 [4] with the addition of the Food and Drug Administration (FDA) in 2010. Each organization brings a different set of complementary skills including expertise in experimental toxicology, computational toxicology, and HTS technologies. During Tox21 Phase I (proof of principle), over 75 biochemical- and cell-based assays were successfully used to screen the initial Tox21 collection of approximately 2800 compounds in a quantitative high-throughput screening (qHTS) platform using 1536-well plate format. A comprehensive list and thorough description of the Phase I assays can be found elsewhere [6–8]. Briefly, these assays interrogated different aspects of cellular physiology including overall cellular health (cytotoxicity and apoptosis induction, DNA damage), perturbation of cell signaling pathways [e.g. ARE/Nrf2, antioxidant response element/NF-E2 related factor 2; CREB, cAMP response element binding; hypoxia-inducible factor 1 alpha (HIF-1α)], inflammatory response induction [e.g. nuclear factor kappa B (NFκB); tumor necrosis factor alpha (TNFα); interleukin-8 (IL-8)], nuclear receptor modulation (e.g. androgen receptor (AR) ; estrogen receptor (ER)) as well as specific isolated cellular targets including enzyme inhibition, membrane transport inhibition, hERG (human ether-a-go-go) channel inhibition, receptor binding and specific protein–protein interaction disruption [6–8].
The use of qHTS to produce high quality and reliable data greatly reduces the frequency of false positives and false negatives, which is critical in computational modeling, low dose extrapolation, and risk assessment. In 2010, Phase II (production phase) of Tox21 was started, with the initial focus on nuclear receptor and stress response pathway assays. Most assays are multiplexed with cell viability measurements to distinguish between true positive responses and those due to increased cytotoxicity (e.g. nuclear receptor antagonism). The Tox21 Phase II library contains approximately 10,000 (10K) compounds. In 2011, a dedicated Tox21 robotic system was installed and made fully operational, which is capable of screening the Tox21 ‘10K’ compound library in 15-point concentration-responses in triplicate each week.
Screening process within the Tox21
To ensure that the Tox21 Phase II is successful, a thorough screening process has been implemented (Figure. 1). The assays nominated by the researchers from government, private, academic, and non-governmental organizations are reviewed by the Tox21 Pathways/Assays working group, one of four working groups (Chemical Selection, Pathways/Assays, Informatics, and Targeted Testing) in Tox21 collaboration, and approved by the Tox21 leadership [6,9]. The assays are selected based on their biological and toxicological relevance [10] and adaptability to miniaturization and automated screening [7]. At the NCGC, the selected assay is optimized for treatment duration and signal window and validated, using appropriate positive controls in the original assay format (e.g. 96-well or 384-well plate). The validated assay is then miniaturized into a 1536-well plate format and characterized in terms of optimal cell density per well, potency and efficacy of positive controls, and other statistical parameters including signal to background ratio (S/B) larger than three folds, coefficient of variation (CV) less than 10%, and Z′ factor larger than 0.5 [11]. Once these milestones have been achieved, the assay is ready for online validation in the robotic platform. In online validation, the assay is screened three times against the Tox21 validation library [1280 compounds from library of pharmacological active compounds (LOPAC) and 88 additional compounds selected by the Tox21 Chemical Selection Group] in the qHTS format. The data generated from the robotic online validations are evaluated for reproducibility, positive control activity consistency (EC50/IC50), S/B, CV, and Z′ factor and submitted to the Tox21 Pathways/Assays group for approval. Robotic online screenings against the Tox21 10K library are run three times on separate days in the qHTS format across 15 concentrations ranging from 1.2 nM to 92 μM, followed by data processing that includes data normalization and curve fitting (see Data Analysis section for details). All the results are first shared by the four Tox21 partners, and then are made publicly accessible through various databases including the National Center for Biotechnology Information’s PubChem [12], NTP’s Chemical Effects in Biological Systems [13], EPA’s Aggregated Computational Toxicology Resource [14], and the NCATS Tox21 Chemical Browser [15], to encourage independent evaluations of Tox21 findings.
Figure 1. The Tox21 screening process.
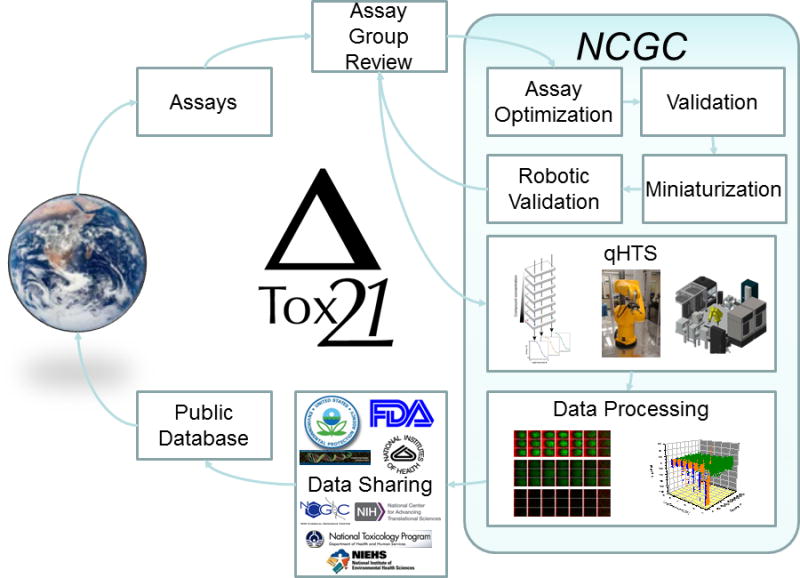
After the nominated assays are reviewed and approved by the Tox21 Pathways/Assays group and the Tox21 leadership, the selected assay is optimized, validated, and miniaturized into a 1536-well plate format, followed by the robotic validation. Robotic qHTS against the Tox21 10K library is then run, followed by data processing. The screening results are first shared by EPA, NCGC, NTP and FDA, and then are made publicly accessible.
Abbreviations: qHTS: quantitative high-throughput screening; NCGC: NIH Chemical Genomics Center.
Preparation of the Tox21 10K compound library
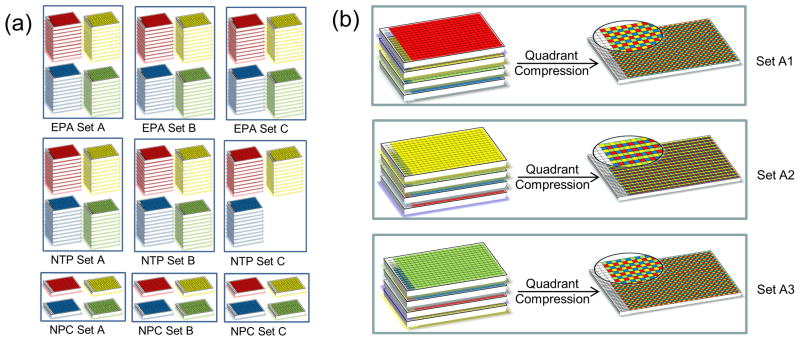
The Tox21 chemical library represents a wide range of compounds with structural diversity in relation to toxicology and commonly used in the environment (pesticides, industrial, food-use, drugs, among others). Three Tox21 partners – EPA [16], NTP [17], and NCGC [18] - each contributed over 3000 physical compounds primarily procured from commercial sources. The library totals over 10,000 plated sample solutions representing more than 8300 unique chemical entities. Main criteria for selection of the Tox21 compounds included but were not limited to known or perceived environmental hazards or exposure concerns, physicochemical properties indicating suitability for HTS (MW, volatility, solubility, logP), commercial availability and cost. In addition, the Tox21 Chemical Selection Group selected 88 diverse compounds from the Tox21 Phase I library of approximately 2800 compounds to serve as internal controls to validate assay reproducibility and examine positional plate effects: these were delivered as duplicates in all screening plates. Each Tox21 partner created three distinct sets of 384-well plates that were labeled ‘Set A’, ‘Set B’, and ‘Set C’ (Figure. 2a). EPA and NTP created ten copies of each set. NTP created two sets containing four 384-well plates each and one set containing three 384-well plates. EPA created three sets containing four 384-well plates each. NCGC provided three sets comprising the NCGC Pharmaceutical Collection (NPC, prepared as described in [18]). Upon receipt at NCGC, the plates were stored in a −80°C freezer until compound plating was initiated, thus maintaining sample integrity to the greatest extent possible. A structure–data file (SDF) containing the sample structure, sample Tox21 ID, sample plate set, plate barcode, sample well position, sample volume, sample concentration, sample supplier name, and sample supplier ID was stored electronically. NCGC assigned each sample a unique identifier and recorded the location of each sample in an Oracle database using ActivityBase [19].
Figure 2.
(a) The EPA, NTP, and NPC collections were divided into three distinct sets of unique compounds called Set A, Set B, and Set C. The EPA and NTP sets contained 10 copies of different 384-well plates called Quadrants. The NPC sets contained one copy of each quadrant. (b) The 384-well plates from each quadrant of every set are arranged in three different orders and compressed to 1536-well compound plates achieving three different plating configurations of the same compounds used for triplicate screening.
Abbreviations: EPA: Environmental Protection Agency’s; NTP: National Toxicology Program; NPC: NCGC Pharmaceutical Collection.
Unlike traditional HTS aimed at the selection of one or more starting points for drug development, where a certain level of noise and the associated false positives and negatives is expected and largely tolerated, the present screens aim at building predictive toxicology models and prioritizing chemicals for in-depth toxicological testing. Thus, the accuracy of activity assignment (i.e. whether a chemical is considered active or inactive in an assay) is of paramount importance. Therefore, to further improve the reliability of actives identification (i.e. to minimize the occurrence of false positives and false negatives), every compound is tested in triplicate in the robotic screens. As some issues associated with poor reproducibility are related to the occurrence of ‘edge effects’, also referred to as positional effects, one solution to the problem of a chemical exhibiting false activity (or lack thereof) due to its particular location in the microtiter plate is to perform each of the three replicate screens where the location of each chemical is varied from test to test. To accomplish this, a plate from each quadrant of Set A was selected and labeled ‘Set A1’, for example. Then a plate from each quadrant of the same set was selected and arranged in a different order than Set A1 and labeled ‘Set A2’. Finally, a plate from each quadrant of the same set was selected and arranged in an order different from Set A1 and Set A2 and labeled ‘Set A3’ (Figure. 2b). This was repeated for each set of the compounds. The subsets were allowed to thaw overnight in a desiccator at room temperature the day before compound plating.
An integrated process of serial dilution and final compression was specifically designed to achieve maximum precision of library preparation in combination with minimum exposure of the stock solutions to the environment: serial dilution of the 384-well plates and compression to 1536-well compound plates were simultaneously achieved on an Evolution P3 (EP3) system equipped with a 384-tip head and PlateStak Automated Microplate Handler (PerkinElmer Life and Analytical Sciences, Shelton, CT, USA). To begin, 37 μL of DMSO was dispensed into fourteen 384-well plates. The library plates were briefly centrifuged and their seals were removed. The compounds were serial diluted by aspirating 30 μL from the highest concentration 384-well plate and dispensing it into the next plate in the series, which contained 37 μL dimethyl sulfoxide (DMSO) followed by ten cycles of mixing. This was repeated until all 15 384-well plates were prepared thus achieving a √5-fold dilution for 15 concentration points. The EP3 then automatically transferred 5 μL to each well of quadrant one of six copies in 1536-well compound plates. The process was repeated for the remaining quadrants of the subset until a total of 90 1536-well compound plates were generated per plating event in four hours [19]. The plates were centrifuged briefly and heat sealed. Of the six copies made, one 15-point dilution series was covered with system-specific metal lids after seal removal and placed on the Tox21 screening robot, while the remaining five copies were stored in a −80°C freezer. The plate barcodes scanned during the preparation of dilution series were used for plate registration in ActivityBase and then automatically transferred to an in-house Oracle database for data analysis [19]. The barcodes were also loaded into an Oracle based plate management system that tracks the screening lifespan of every compound plate. After a preset time, the system sends e-mail alerts to the Tox21 screening group of impending plate expiration (currently set at four months) and the need to place a fresh screening copy on the robot.
The Tox21 robot
The Tox21 robotic system consists of a high-precision robotic arm (Stäubli, Duncan, SC, USA) surrounded by workstations that include compound plate storage units, assay plate incubators, liquid handlers, and readers (Figure 3). The six-axis robotic arm with a specially-designed gripper and barcode reader [20] is used to transport plates to different stations with a high degree of accuracy and precision. Assay plates are stored in two climate-controlled rotating incubators that can hold 1080 plates. Temperature, humidity, and CO2 content can be controlled independently in each incubator, allowing the system to run different assays in parallel. Compound storage includes two rotating compound carousels capable of holding up to approximately 1,500,000 compound samples in 972 plates (~100,000 compounds in 15-dose titrations). Any plate in either the assay incubator or compound carrousel can be accessed by the robotic arm at any time. Compound transfer to assay plates can be achieved by either using a pintool station [20] or by either of the two acoustic, noncontact liquid dispensers. As currently configured, the pin tool performs direct transfer of 23 nL of a dimethyl sulfoxide (DMSO) solution from a 1536-well compound plate to a 1536-well assay plate. The ATS-100 (EDC Biosystems, Fremont, CA, USA), an acoustic dispenser, is capable of nanoliter transfers by using acoustic energy to eject liquid sample from either 384-well or 1536-well compound plate to 1536-well assay plate without any physical contact, which can eliminate compound cross contamination. For delivery of common assay reagents, the system uses two Bioraptr FRD workstations (Beckman Coulter, Indianapolis, IN, USA) that allow for liquid transfer of 0.2–10 μL of up to four different reagents simultaneously into a 1,536 well plate, as well as a Multidrop Combi reagent dispenser (Thermo Fisher, Waltham, MA, USA) capable of high speed dispense of one reagent using eight tips. The system currently integrates two different types of detectors, the ViewLux (PerkinElmer, MA, USA) and the Envision (PerkinElmer), that cover almost the entire spectrum of fluorescence, luminescence, and absorbance measurements required for HTS. The above components are integrated by Wako director software that possesses a wide range of control features designed to ensure a reliable and efficient system operation. Overall, the system is capable of storing compound collections and assay plates, and performing assay steps including liquid transfers and diverse measurements in a fully integrated manner allowing hands-free assay completion. This robotic system is the only one currently in operation in a governmental setting that is capable of running qHTS screenings in a 1536-well plate format exclusively dedicated to toxicological studies. Moreover, through an open assay nomination process, any public health stakeholder (e.g. government, private, academic, non-governmental organizations) can propose toxicological relevant and mechanism based assays to be used to screen the Phase II Tox21 library using the robot.
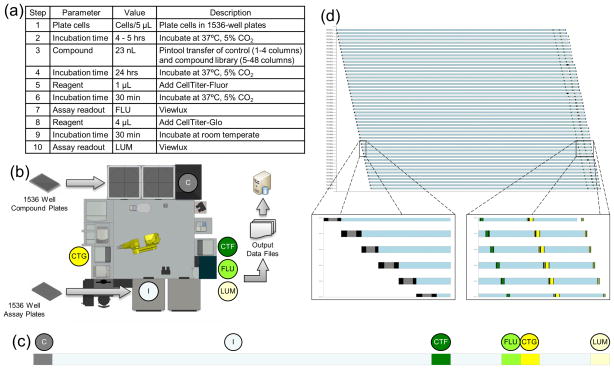
Figure 3. HTS system workflow.
The assay protocol (a) is run on the Tox21 robotic system (b), represented graphically as a Gantt chart element (c). The entire run is represented by a combined Gantt Chart (d), with each element of the Y-axis representing the steps each individual assay plate undergoes while completing each step of the protocol, with the X-axis representing time.
Abbreviations: C: compound addition; I: incubation; CTF: CellTiter-Fluor addition; CTG: CellTiter-Glo addition; FLU: fluorescence reading; LUM: luminescence reading.
To conduct a screen, a combination of physical components (robot, plates, and reagents, as dictated by the assay protocol, and as described above) and software programs is required. To program the screen, assay plates act as the input to the HTS system with associated control and compound plates as required. Each assay plate is considered to be an assay object. Assay method (or protocol) with detailed description provided in the table (Figure 3a) is the set of steps that each assay object will go through, with each step associated with some peripheral device (dispenser, plate reader, among others) or an incubation period (Figure 3b and 3c). A group of assay objects running the same method is considered to be a screen. At the start of a screen, all assay objects are placed in a first-in, first-out (FIFO) Wait Queue. A Wait Queue is a queue of objects waiting for a lock, which represents control of a peripheral device or location on the screening system. When the lock is unlocked, the objects acquire the lock in the order of the queue. Objects still in the queue are in the blocked state until the lock is released. Each step or a series of linked steps in a method has an associated lock, with this type of synchronization being called a Mutex (mutual exclusion). It is important to note that this type of scheduling is not time-based but rather resource-based; assay objects perform each step of their associated method in the order in which they are queued. In this manner, the robot can execute a wide range of biological assay protocols at maximum speed, while at the same time ensuring that steps associated with critical timing features are followed precisely. Within the current system, the scheduling software provides an event that can be captured for every protocol step for every assay object currently running, giving a complete real time history (Figure 3d). We have utilized this feature to create an external application written in LabView (National Instruments, Austin, TX, USA) to communicate with the scheduler to capture these events. This LabView application is also configured to communicate with other external applications to both send and receive data; examples include sending an alarm email message to the robot operator, performing a basic QC analysis of the screening data and sending the results to a user-accessible database, which can then be accessed by a web based application to actively monitor assays both current and previous runs through a Gantt Chart based display.
Data processing and analyses
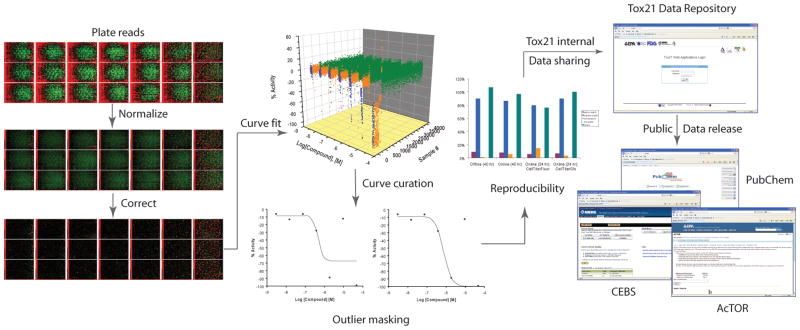
Gross assay performance is assessed initially using quality metrics from each plate and also visually. For each plate, CV, S/B, and Z′-factor are calculated and examined during the execution of the primary screen. ‘Failed plates’ identified by abnormally poor values are inspected visually and, if necessary, excluded from further data analysis. Analysis of compound concentration–response data was performed as previously described [21]. Briefly, raw plate reads for each titration point were first normalized relative to the positive control compound (100% for agonist mode and -100% for antagonist mode assays) and DMSO-only wells (0%) placed in the first four columns of each plate as follows: % Activity = [(Vcompound − VDMSO)/(Vpos − VDMSO)] × 100, where Vcompound denotes the compound well values, Vpos denotes the median value of the positive control wells, and VDMSO denotes the median values of the DMSO-only wells, and then corrected using compound-free control plates (i.e. DMSO-only plates) at the beginning and end of the compound plate stack to remove background patterns and subtle abnormalities such as tip effects or blotting from cell dispenses [22]. Concentration–response titration points for each compound were fitted to a four-parameter Hill equation [23] yielding concentrations of half-maximal activity (AC50) and maximal response (efficacy) values[24]. Compounds were designated as Class 1–4 according to the type of concentration–response curve observed [21,25]. Curve classes are heuristic measures of data confidence, classifying concentration–responses on the basis of efficacy, the number of data points observed above background activity, and the quality of fit. The most problematic concentration responses are automatically assigned curve class 5 based on considerations like the direction of activity (observing alternately both increases and decreases in signal over a short concentration range) and unusually large signal at low sample concentrations (activity at zero concentration is estimated to be < 3SD of control). Class 5 curves and other cases where an inconsistency between the highest compound activity and the curve class assigned is identified, for example, a compound with a positive response is assigned a negative curve class, are manually inspected to correct the curve class, if necessary, normally by masking or unmasking data points improperly masked by the automated curve fitting process to adjust the curve fit (Figure 4).
Figure 4. Tox21 data pipeline.
After the qHTS data come off the robot, the raw plate reads are normalized, corrected, and pivoted to form concentration response curves (CRCs). The CRCs are fit to the Hill equation and classified into different curve classes based on potency, efficacy and quality of curve fit. The curve fits are further inspected manually to correct any misfit curves. The ‘clean’ curve fitting results are then assessed for activity reproducibility to determine the final assay performance. After the initial data parsing and assessment at the NCGC are complete, the qHTS data are shared with the Tox21 partners through an access controlled site – the Tox21 data repository, where the data are further scrutinized for quality and utility. The data will then be released to the public domain in a number of public databases, including PubChem [27], CEBS [13] and ACToR [14].
After manual curation, the ‘clean’ curve fitting results from the triplicate runs as well as the Tox21-88 duplicated compounds are assessed for activity reproducibility to determine the final assay performance. Each assay is assigned a score and grade based on the reproducibility of the data [21]. As soon as the initial data parsing and assessment at the NCGC are complete, the concentration response data, curve fitting results as well as the raw plate reads, assay conditions, and sample mapping information are shared with the Tox21 partners through a suite of databases and software tools custom built by the NCGC for the Tox21 program [26]. Within the first six months of data generation, the assay data are only made available to the Tox21 partners through the aforementioned accessed controlled site where the data are further scrutinized for quality and utility. The data will then be released to the public domain in a number of public databases including PubChem [27], CEBS [13] and ACToR [14]. The complete Tox21 data pipeline is shown in Figure 4. The high quality concentration response data generated on a wide spectrum of pathways and phenotypic toxicity endpoints provide a valuable resource for predictive toxicity modeling. These data can not only serve as in vitro signatures that could be used to predict in vivo toxicity endpoints [28,29] and to prioritize chemicals for more in depth toxicity testing [30] that help to fulfill the goals of the Tox21 program, but also provide rich training data sets for the quantitative structure–activity relationship (QSAR) modeling community to build more robust models [31,32].
Validation of Tox21 robotic system
To validate the Tox21 robotic system, a well characterized cell viability assay (CellTiter-Glo® luminescence cell viability assay, Promega, Madison, WI, USA) was run in parallel using either the robotic system (termed online run) or similar standalone equipment (offline run). This assay determines the viability based on intracellular adenosine triphosphate (ATP) content, which is directly linked to metabolically active cells. The assay is conducted by a single addition of the CellTiter-Glo reagent which lyses the cells and generates a luminescent signal proportional to the amount of ATP present. In the online run every experimental step was handled by the robotic system, with the exception of cell culture and cell dispensing into the 1536-well plates while in the offline run every single step was performed using standalone equipment operated by a scientist. For both methods, the HepG2 cells were used to screen against Tox21 validation library in the qHTS format with a concentration range of 2.9 nM–46 μM in triplicates. Experimental details have been described previously [33]. The cell viability assay performed equally well in both methods with CVs of 8.95 ± 2.73 and 8.26 ± 1.62, Z′ factors of 0.80 ± 0.13 and 0.84 ± 0.04 and S/B ratios of 63.94 ± 10.70 and 53.27 ± 2.30 for the online and offline runs, respectively (Mean ± SD). The IC50 values from the tetraoctyl ammonium bromide positive control dose–response curves run in every plate were slightly different 0.98 ± 0.11 μM for the online run and 3.43 ± 0.80 μM for the offline run. Overall, the results from the two runs were very similar with only 0.36% of the tested compounds yielding a different outcome when assessed for reproducibility.
In cell-based assays it is necessary to determine cell viability in addition to the particular toxicological pathway(s) that may be affected by a given compound. To increase screening throughput and efficiency and to decrease the cost from a parallel viability run, we explored the feasibility of a multiplexed assay, where two distinct outputs from a single well in the single run can be measured. To determine the compatibility of the multiplexed viability output, we chose two viability assays, CellTiter-Fluor (Promega) and CellTiter-Glo viability assays, with different readouts to compare directly in concurrent wells of a 1536 well plate (Figure 3a). CellTiter-Fluor (Promega) is a non lytic fluorescence based assay that determines relative cell viability in a given population of cells by measuring protease activity of a conserved and constitutive protease that is only active in viable intact cells. The Tox21 validation library was screened in HepG2 cells in a concentration range of 2.9 nM–46 μM in the CellTiter-Fluor viability assay and the CellTiter-Glo viability assay on the same wells in the plates. After 24 hours of treatment, the CellTiter-Fluor reagent was first added into the assay plates and the fluorescence was determined in Viewlux plate reader (Perkin Elmer), followed by addition of CellTiter-Glo reagent and the luminescence was measured in Viewlux plate reader (Figure 3a). Both assays performed well with CV of 9.9 ± 0.57, Z′ factor of 0.75 ± 0.05 and S/B of 5.2 ± 0 for the CellTiter-Fluor assay, and CV of 4.99 ± 0.86, Z′ factor of 0.88 ± 0.03 and S/B of 30.2 ± 0.35 for the CellTiter-Glo viability assay, respectively. These findings support that the two assays can be multiplexed in the 1536-well plates. A reproducibility comparison of the compounds tested was performed based on the results of the two different viability assays. When examined for mismatches – a situation where one assay run shows a positive response for an effect on viability and the other assay shows a negative response to the same compound—resulted in a very low mismatch rate of less than 0.5% for CellTiter-Fluor assay and less than 0.1% for CellTiter-Glo assay. Active compounds showed high overlap (~98%) between the two assays with an active match of 1.8% in triplicate runs in CellTiter-Fluor assay and 1.97% in triplicate runs in CellTiter-Glo assay. The remaining inactive compounds were inactive in both assays. Taken together these findings support the utility of multiplexing viability assays with different readouts for qHTS on a robotic platform, and indicate that both assays are able to determine compounds that influence cell viability in a qHTS format.
Concluding remarks
The Tox21 collaboration is a multiagency effort among the EPA, FDA, NTP and NCGC to advance in vitro toxicological testing in the 21st century. After completion of proof of principle in Tox21 Phase I, the focus of Tox21 Phase II is on developing and evaluating a battery of in vitro assays with target-specific and mechanism-based readouts adoptable to a high throughput and high-content screening platform. All of the cell-based pathway/target assays can be run multiplexed together with a cell viability readout in the same assay well to differentiate true actives from cytotoxic compounds. During Phase II, robotic online validation with the Tox21 validation library, run in triplicate, is included for each new assay to be run on the Tox21 robot. If, when tested online, the assay performs as it did during offline optimization, it can be moved forward to robotic screening. To evaluate the screening reproducibility, each assay is screened against the Tox21 10K compound collection on three separate days, with the same compound contained in three different well locations.
The Tox21 Pathways/Assays group has selected approximately 40 assays that will be run during the initial stage of Phase II mostly using fluorescence and luminesnce as endpoints. New assays will be evaluated periodically and included for screening in the future based on assay feasibility and target relevance for toxicology. The robot platform is flexible enough to be upgraded to include new technologies, such as high-throughput flow cytometry and high-content screening analysis, depending on the program needs. These information rich approaches can be coupled with the real time data analysis and automated liquid handling to efficiently run confirmatory and mechanistic follow up studies simultaneously with the primary screen. The high quality data generated from the primary screening is instrumental to identifying mechanisms of compound action, prioritizing substances for further toxicological evaluation in vivo and developing predictive models for biological response. All the assay results are then released to public databases, which provide rich data sets to researchers for further data mining, generation of new hypothesis and building reliable QSAR models [32]. The predictive computational models generated from these high quality data sets will shed light on the potential of using in vitro assays as an alternative approach for assessing the toxicity of environmental chemicals.
Highlights.
The Tox21 effort is to advance in vitro toxicological testing in the 21st century.
Tox21 chemical library contains approximately 10,000 environmental chemicals.
A battery of in vitro assays will be validated and screened in a qHTS platform.
Tox21 robotic system is capable of screening the Tox21 chemical library in triplicates in a week.
Acknowledgments
We gratefully acknowledge Danielle VanLeer and Tongan Zhao for developing the compound plate tracking system. This work was supported by the Intramural Research Programs of the National Toxicology Program (Interagency agreement #Y2-ES-7020-01), the National Institute of Environmental Health Sciences, and the U.S. Environmental Protection Agency.
The views expressed in this paper are those of the authors and do not necessarily reflect the statements, opinions, views, conclusions, or policies of the National Institute of Environmental Health Sciences (NIEHS), National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH), U.S. Environmental Protection Agency, or the United States government. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Conflicts of interest
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.NTP. A National Toxicology Program for the 21st Century: A Roadmap for the Future 2004 [Google Scholar]
- 2.NRC. Toxicity Testing in the 21st Century: A Vision and a Strategy. The National Academies Press; 2007. [Google Scholar]
- 3.EPA. [[accessed 2 April 2013]]; http://www.epa.gov/ncct. [RE1]
- 4.Collins FS, et al. Toxicology. Transforming environmental health protection. Science. 2008;319:906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krewski D, et al. Toxicity testing in the 21st century: a vision and a strategy. Journal of toxicology and environmental health. Part B, Critical reviews. 2010;13:51–138. doi: 10.1080/10937404.2010.483176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tice RR, et al. Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect. 2013 doi: 10.1289/ehp.1205784. doi:10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shukla SJ, et al. The Future of Toxicity Testing: A Focus on In vitro Methods Using a Quantitative High Throughput Screening Platform. Drug Discov Today. 2010;15:997–1007. doi: 10.1016/j.drudis.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Attene-Ramos M, et al. High Throughput Screening. In: Wexler P, editor. Encyclopedia of Toxicology. Elsevier; 2013. [Google Scholar]
- 9.Kavlock RJ, et al. Toxicity testing in the 21st century: implications for human health risk assessment. Risk analysis : an official publication of the Society for Risk Analysis. 2009;29:485–487. doi: 10.1111/j.1539-6924.2008.01168.x. discussion 492–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Judson R, et al. Perspectives on validation of high-throughput assays supporting 21st century toxicity testing. ALTEX. 2013;30:51–56. doi: 10.14573/altex.2013.1.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang JH, et al. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 12.Bolton E, et al. Annual Reports in Computational Chemistry. Vol. 4. American Chemical Society; 2008. PubChem: Integrated Platform of Small Molecules and Biological Activities. [Google Scholar]
- 13. [[accessed 2 April 2013]];CEBS. http://tools.niehs.nih.gov/cebs3/ui/
- 14.ACToR. [[accessed 2 April 2013]]; http://actor.epa.gov.
- 15. [[accessed 2 April 2013]];NCATS Tox21 Chemical Browser. http://www.ncats.nih.gov/research/reengineering/tox21/tox21.html.
- 16. [[accessed 2 April 2013]];EPA. http://www.epa.gov/ncct/dsstox/sdf_tox21s.html.
- 17.NTP. [[accessed 2 April 2013]]; http://www.epa.gov/ncct/dsstox. [RE3]
- 18.Huang R, et al. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Science translational medicine. 2011;3:80ps16. doi: 10.1126/scitranslmed.3001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yasgar A, et al. Compound Management for Quantitative High-Throughput Screening. JALA. 2008;13:79–89. doi: 10.1016/j.jala.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michael S, et al. A robotic platform for quantitative high-throughput screening. Assay Drug Dev Technol. 2008;6:637–657. doi: 10.1089/adt.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang R, et al. Chemical genomics profiling of environmental chemical modulation of human nuclear receptors. Environ Health Perspect. 2011;119:1142–1148. doi: 10.1289/ehp.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Southall NT, et al. Enabling the Large Scale Analysis of Quantitative High Throughput Screening Data. In: Seethala R, Zhang L, editors. Handbook of Drug Screening. Taylor and Francis; 2009. pp. 442–463. [Google Scholar]
- 23.Hill AV. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol (London) 1910;40:4–7. [Google Scholar]
- 24.Wang Y, et al. A grid algorithm for high throughput fitting of dose-response curve data. Curr Chem Genomics. 2011;4:57–66. doi: 10.2174/1875397301004010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inglese J, et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci USA. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. [[accessed 2 April 2013]];Tox21 Applications. http://tripod.nih.gov/tox.
- 27. [[accessed 2 April 2013]];PubChem. http://pubchem.ncbi.nlm.nih.gov/ [RE4]
- 28.Sipes NS, et al. Predictive models of prenatal developmental toxicity from ToxCast high-throughput screening data. Toxicol Sci. 2011;124:109–127. doi: 10.1093/toxsci/kfr220. [DOI] [PubMed] [Google Scholar]
- 29.Martin MT, et al. Predictive model of rat reproductive toxicity from ToxCast high throughput screening. Biol Reprod. 2011;85:327–339. doi: 10.1095/biolreprod.111.090977. [DOI] [PubMed] [Google Scholar]
- 30.Judson RS, et al. In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Environ Health Perspect. 2010;118:485–492. doi: 10.1289/ehp.0901392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang R, et al. Weighted feature significance: a simple, interpretable model of compound toxicity based on the statistical enrichment of structural features. Toxicol sci. 2009;112:385–393. doi: 10.1093/toxsci/kfp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun H, et al. Paradigm shift in toxicity testing and modeling. AAPS J. 2012;14:473–480. doi: 10.1208/s12248-012-9358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia M, et al. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ Health Perspect. 2008;116:284–291. doi: 10.1289/ehp.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]