Abstract
The widespread adoption of high-throughput next-generation sequencing (NGS) technology among the Australian life science research community is highlighting an urgent need to up-skill biologists in tools required for handling and analysing their NGS data. There is currently a shortage of cutting-edge bioinformatics training courses in Australia as a consequence of a scarcity of skilled trainers with time and funding to develop and deliver training courses. To address this, a consortium of Australian research organizations, including Bioplatforms Australia, the Commonwealth Scientific and Industrial Research Organisation and the Australian Bioinformatics Network, have been collaborating with EMBL-EBI training team. A group of Australian bioinformaticians attended the train-the-trainer workshop to improve training skills in developing and delivering bioinformatics workshop curriculum. A 2-day NGS workshop was jointly developed to provide hands-on knowledge and understanding of typical NGS data analysis workflows. The road show–style workshop was successfully delivered at five geographically distant venues in Australia using the newly established Australian NeCTAR Research Cloud. We highlight the challenges we had to overcome at different stages from design to delivery, including the establishment of an Australian bioinformatics training network and the computing infrastructure and resource development. A virtual machine image, workshop materials and scripts for configuring a machine with workshop contents have all been made available under a Creative Commons Attribution 3.0 Unported License. This means participants continue to have convenient access to an environment they had become familiar and bioinformatics trainers are able to access and reuse these resources.
Keywords: training, next-generation sequencing, NGS, cloud, workshop
INTRODUCTION
Decreasing DNA sequencing costs, and increased sequencing throughput, is driving the application of next-generation sequencing (NGS) to new areas of life science research by even small-to-medium sized laboratories. As well as being used in the assembly of genomes (de novo and resequencing), NGS is now routinely used to further our understanding of transcriptional regulation and DNA/protein interactions through RNA-Seq and ChIP-seq, respectively.
Although NGS data can now be acquired with ease, the same cannot be said for the downstream analysis of NGS data. NGS data sets are growing in size and are being applied to ever increasingly large and complex biological questions. In addition, most NGS data analysis tools are developed for use at the Linux command line and have large computational requirements. Many laboratories are finding it increasingly difficult to move forward on the analysis of valuable data sets, owing to a lack of competency with these capabilities. While workflow-based softwares such as Galaxy [1], Yabi [2] and Taverna [3] are starting to provide a mechanism through which turn-key NGS data analysis pipelines can be delivered, many applications of NGS are not yet routine and thus not easily incorporated into such workflows. As such, there is a need for hands-on NGS training, particularly among the traditional wet-lab biologist, as they struggle to handle the ‘data deluge’ and communicate effectively with their bioinformatic colleagues.
Nationally, Australia has a shortage of cutting-edge bioinformatics training courses. This is primarily owing to the scarcity of skilled trainers with time and funding devoted to developing and delivering such courses, as well as the lack of specialized bioinformatics training facilities in which to host them. Australia is a large sparsely populated country with eight geographically distant capital cities, which are home to almost two-third (14.7 million) of the population (22.3 million) [4]. Therefore, the establishment of a coherent Australian bioinformatics training network (BTN) capable of delivering quality hands-on bioinformatics training nationwide is challenging. In this article, we discuss the key issues and solutions we have implemented to deliver a hands-on NGS road show workshop.
AUSTRALIAN BIOINFORMATICS TRAINING NETWORK INITIATIVE
A consortium of Australian research organizations, including Bioplatforms Australia (BPA), the Commonwealth Scientific and Industrial Research Organisation (CSIRO) and the Australian Bioinformatics Network (ABN) initiated a collaboration with EMBL-EBI training team to establish a NGS bioinformatics training platform to address the increasing local demand for NGS bioinformatics training. The concept is simple: develop a national network of bioinformatics trainers, capable of travelling interstate to deliver road show–style hands-on bioinformatics workshops. To this end, eight Australian bioinformaticians attended a train-the-trainer workshop, developed by EMBL-EBI. This workshop was designed to improve skills for delivering successful hands-on training and covered topics on effective teaching methods, workshop planning and curriculum development. Subsequently, a 2-day hands-on NGS workshop was jointly developed and delivered at Sydney and Melbourne with the assistance of EBI faculty and then independently in Brisbane, Adelaide and Canberra.
International collaborative approach to providing bioinformatics training courses within Australia
Australia lacks large bioinformatics research hubs, rather bioinformatics expertise is found within smaller centres located around the country; few of these bioinformatics centres have dedicated funding and people resources to provide training outreach to the Australian research community at the scale that is currently required. In contrast, several large international research centres and networks such as The Netherlands Bioinformatics Centre, EMBL, Harvard Medical School, Michigan State University, Nowgen and the European Cooperation in Science and Technology initiative SeqAhead are producing and delivering training courses that cover the latest cutting-edge technologies and applications. Similarly to our needs to collaborate and share experiences, more global efforts to get together and establish an open and collaborative platform to share training efforts are maturing. For example, the BTN [5] is currently evolving into the Global Organisation for Bioinformatics Learning, Education & Training (GOBLET; http://mygoblet.org/) and will be a rich source of training material in the future. In addition, Software Carpentry (http://software-carpentry.org/) is establishing itself as a global network of instructors focused on increasing the productivity of researchers by teaching computing skills and creating open access material. To begin utilizing this wealth of training material and expertise, we have collaborated with the EMBL-EBI training team to develop and deliver NGS training courses within Australia.
EMBL-EBI ‘train-the-trainer’ workshop
To create a critical mass of bioinformaticians and engender a cohesive bioinformatics community within Australia, three research organizations, BPA, CSIRO and EMBL-Australia, have formed a partnership to catalyse the ABN (http://australianbioinformatics.net). From within this bioinformatics community, eight potential trainers based in capital cities around Australia, and members of the ABN, were funded to travel to EMBL-EBI to attend an EMBL-EBI NGS hands-on workshop and a User Training Trainer course. Because most of the people involved in developing and delivering training workshops are not specifically employed for that role, it is important to clarify our use of the terms ‘trainer’ and ‘trainee’. We simply use them to describe the role of the people within the workshop learning environment. The User Training Trainer course is a ‘train-the-trainer’ type course aimed to cover the andragogical aspects of training. It specifically targets hands-on (i.e. sitting at a computer letting their fingers do the learning) training in bioinformatics. It covered the planning, delivering and assessment of bioinformatics end-user training as well as the development of trainer’s skills in communicating and understanding of learning preferences. Jointly the EMBL-EBI training team and the Australian trainers, using the current EMBL-EBI training materials, devised a modular 2-day hands-on NGS training workshop tailored for the Australian scientific community.
To maximize the success of the workshop, three experienced EMBL-EBI trainers were funded to attend and teach the first two workshops run in Melbourne and Sydney. This allowed the Australian trainers to focus on the requirements and logistics of delivering a road show–style workshop. Following this, a peer-review model was used where trainers observed each other, and through feedback, learnt from each other.
NGS HANDS-ON WORKSHOP
One of the major outcomes from the User Training Trainer course was the importance to perform a training needs analysis. From this, we identified training needs in the areas of ChIP-Seq, RNA-Seq and de novo genome assembly. These were based on the experience of the Australian bioinformatics trainers and interactions with Australian research scientists at a grassroots level. Therefore, a course that focused primarily on these areas together with a background in data quality control and read alignment was found to be of most benefit to the Australian genomics research community. The aims of the workshop were to provide the following:
- Hands-on experience in the analysis of Illumina NGS data, including the following:
- Knowledge of the various sequencing technologies and data formats.
- Hands-on experience with common analytical workflows for ChIP-Seq, RNA-Seq data and de novo genome assemblies using computer exercises.
- An overview of the software tools used in the course including discussions on parameters and limitations. The following tools are those at the core of the workshop:
- FastQC and FASTX-Toolkit for read QC.
- Bowtie for aligning reads to a reference.
- Samtools for manipulating SAM/BAM files.
- IGV for visualizing read alignment in BAM files.
- UCSC tools and BEDTools for manipulating BED and BigWig files.
- MACS for analysing ChIP-Seq data.
- Ensembl Genome Browser for visualizing ChIP-Seq–derived BigWig files.
- PeakAnalyzer for automated peak splitting and functional annotation of ChIP-Seq data.
- MEME and TOMTOM for motif discovery/analysis.
- Tophat and Cufflinks for the analysis of RNA-Seq data.
- DAVID for the functional annotation of differentially expressed genes.
- Velvet for the de novo assembly of a bacterial genome.
- AMOS Hawkeye for visualizing an assembly in the context of paired-end reads.
A 2-day programme, trying to adhere to rules of developing short bioinformatics training courses [6], consisting of a set of modular topics (Table 1) was developed.
Table 1:
Overview of the modules developed and their key learning objectives
| Module | Key learning objectives |
|---|---|
| Data quality |
|
| Read alignment |
|
| ChIP-Seq |
|
| RNA-Seq |
|
| de novo genome assembly |
|
TARGET AUDIENCE
The interactive 2-day hands-on course was designed primarily for bench biologists at the level of advanced PhD students or early career postdoctoral researchers with little or no bioinformatics experience. However, to maximize the potential impact of the workshop, the target audience was expected to have an interest in the analysis of NGS data or have data in-hand to analyse. The overarching aim was to increase research productivity by empowering bench scientists to independently look at their NGS data and enable them to engage bioinformaticians in constructive conversations about the analyses.
Course attendance was by application only, and selection was based on a set of criteria aimed at homogenizing the group of trainees selected to attend. Applicants were required to describe their current research interests, previous bioinformatics experience and their expectations from attending the course. Applicants were selected such that ability levels were similar and their expectations matched what the course could realistically deliver. Preference was given to applicants that were local to the workshop or in regional areas that were less likely to be hosts of a future event and to those with NGS data in hand. In our experience, it is important that all participants are of a similar ability because a course containing a mixture of both novices and those more experienced with bioinformatics analysis is difficult to run. Novices can be left behind, experienced bioinformaticians can be held back by a slower pace and trainers’ time can be monopolized by advanced students wishing to discuss more complex topics not covered in the course or by novices not being able to keep up.
CONTENT AND LEARNING OBJECTIVES
Content was planned around a 2-day workshop because an event of this duration more easily fits into the weekly schedule of a life scientist. It also allows for the possibility of running back-to-back workshops in two different cities within a single week. However, the course modules (Table 1) can be delivered individually if required.
To maximize the amount of hands-on time, we limited the lecture style components of this course to 10–20-min presentations. These presentations were not designed to provide a detailed view of the subject matter but instead to provide a brief overview of the basic concepts with sufficient context for the following hands-on session. Participants work independently using online training tutorials written to emphasize important concepts and steps within the analysis process (open access, https://github.com/nathanhaigh/ngs_workshop). Monitoring of the students understanding was done through the inclusion of specific questions that were required to be answered throughout the tutorials. Interactions between participants and trainers were actively encouraged by having group participation activities throughout the course. For example, early in the workshop, trainees were asked to form small groups and provided with the elements of commonly used workflows for RNA-Seq, ChIP-Seq and de novo assembly. They were asked to discuss and piece together the elements into what they thought were appropriate workflows. Not only did this encourage interactions, it introduced and allowed the trainees to discuss some basic terminology and concepts. In addition, the workshop was fully catered to provide additional opportunities for networking away from the computers.
Generally we had four to six trainers per workshop and recruited additional bioinformaticians from the local area. Including local bioinformaticians was not only helpful on the day but provides trainees with a local point-of-contact for post-workshop advice and networking opportunities. In addition, it provides the local bioinformaticians with an insight into the running of the workshop so they may later choose to run a workshop themselves with the materials developed.
Trainees were provided with a workshop manual in both electronic and hardcopy. The manual contains workshop-related information, tutorials and questions for the five modules (Table 1) as well as post-workshop information about access to computational resources, workshop materials and data sets. Trainers were also provided with a ‘Trainer’s Manual’ containing the answers to the questions posed in the tutorials. This trainer’s manual was made available to the trainees on completion of the workshop.
FEEDBACK SURVEY
An important aspect of any training is to obtain feedback to continually improve and evolve the content and running of the workshop over time. To facilitate this, we used SurveyMonkey to develop a post-workshop questionnaire containing 25 questions to evaluate the course content, quality and clarity of presentations, relevance of content and usefulness of individual modules, workshop organization, catering and ideas for future workshops. Anonymized survey results for the first four workshops are available (Supplementary 1–4).
GOING MOBILE
The successful delivery of any hands-on NGS training is not only dependant on the workshop content and materials, but also on access to computing infrastructure capable of meeting its compute intensive requirements. In our experience, finding a suitable venue was the biggest obstacle to successfully delivering a road show–style workshop. To overcome this, we considered the following options: (i) request trainees to supply their own computers; (ii) use a host institute computing suite and (iii) provide a remote computer onto which trainees can connect. Each of these options has their pros and cons as we shall now discuss.
Trainee provided computers
Asking trainees to provide their own computers for a hands-on session is appealing as it opens the possibility of delivering road show–style workshops. It also ensures the trainees are using a computer they are already familiar with and will continue to have access to once they return to their ‘day jobs’. Trainees will be able to bookmark resource sites as they learn and install any necessary software and plug-ins, thus enabling them to use their setup when they resume their daily routine. This is an important consideration that will increase the chances that the trainee will continue his/her learning following the workshop. However, this option has some strong pitfalls.
Problems that are commonplace with this type of setup are: (i) it is unreasonable to ask inexperienced trainees to configure their own computers for an NGS workshop; (ii) the host institution may not allow foreign computers onto their network; (iii) trainees may not have administrator level access to their computers for installing any prerequisite software and (iv) distributing example data sets and other workshop-related content can be time-consuming. This can often lead to much chaos in the first hour or two on the first day of training and recovering from these poor first impressions is difficult, if not impossible.
Even if these issues are successfully addressed, there is likely to be a heterogeneous pool of hardware specifications and operating systems. In terms of an NGS workshop, this means that some computers will not meet the minimum hardware requirements and different operating systems will need to be supported by the trainers for the duration of the workshop.
Host institute computing suite
A host institute computing suite has the advantage of providing a homogeneous pool of computers in terms of hardware and software and are already on the institute’s network. Private companies exist that are able to provide modern training facilities and computing hardware that meet most workshop requirements but their costs are generally prohibitive for academic/non-profit institutions. Universities and other research institutions are potential hosts and generally have computing suites available. The biggest issue with using these venues has been the lack of availability because they are heavily used for teaching during term and are in high demand out of term. As such, rooms need to be booked several months ahead of time.
We also found it difficult in getting the required level of support for running workshops out of these facilities. Firstly, most institutions’ computer suites use the Microsoft (MS) Windows OS and there is a reluctance to install virtualization software or configure a dual boot system to support the Linux OS needed for the workshop. Some institutions already use virtualization for commissioning computers in these suites, but we have encountered resistance to allowing a ‘foreign’ OS on the host institute’s network.
For this to be in anyway a viable option, a strong link needs to exist between the trainers and a person on-the-ground at the host institution capable of addressing the workshop’s computer hardware requirements. The absence of such a link means it is almost impossible to host a workshop from that venue.
Providing remote computing resources
This option provides the most flexibility because a central resource can be managed remotely by the geographically distributed network of trainers. Road show–style workshops become a real option, as local computer hardware requirements are minimized, as they simply act as terminals for connecting to the remote resource. Common setups used for providing remote computing resources are as follows: (i) provide a single machine for each of the trainees to log into or (ii) provide each trainee with his/her own machine to log into.
Although a single machine is easier to maintain, consideration needs to be given to the performance of the system when multiple (20–30) concurrent users are logged in. This is particularly important in an NGS workshop where several gigabytes (GB) of memory and multiple CPUs are required per user.
Providing each trainee with his/her own machine to log into allows discrete resources to be allocated to each trainee so their processes do not impact those of another trainee. Rather than providing physical hardware for each user to log into, virtualization technology allows machines to be created virtually. These virtual machines (VMs) may or may not exist on the same physical hardware and can be configured so each VM has its own dedicated resources. Creating and managing the 20–30 machines (physical or virtual) required for a training session requires additional time and skills to administer. For example, developing and maintaining OS images, deploying and managing VMs for and during the workshop. We found parallel versions of secure shell (SSH)-based tools (parallel-SSH) to be particularly useful for administering VMs during the workshop.
An important aspect to consider with this setup regards the actual interaction of the trainees with the remote machine(s). Forcing trainees, with little or no command line experience, to use the Linux command line via SSH is another way to overwhelm and possibly alienate trainees. Therefore, a remote desktop-like connection is preferable as it provides a better stepping stone into the world of Linux. Exposing trainees, unfamiliar with working on remote computers, to the concepts of ‘here’ (local) and ‘there’ (remote) can cause confusion, particularly if this is done at the start of the workshop when other new concepts are also being introduced. We have found that exposing trainees to the fact they are working remotely on the second day of a workshop is less problematic, causes less confusion and is ultimately beneficial to their understanding of common bioinformatic practices.
THE NeCTAR RESEARCH CLOUD
Given the multi-institutional and geographically distributed nature of the current collaboration together with the long-term goal of building a strong network of Australian bioinformatic trainers, we felt that the use of a remote computing setup provided the most flexibility. The development of this NGS workshop coincided with the rolling out of the first node of the National eResearch Collaboration Tools and Resources (NeCTAR) Research Cloud. NeCTAR is an Australian government project to build new infrastructure specifically for the needs of Australian researchers. Australian researchers are able to launch a VM on NeCTAR Research Cloud resources via Australian Access Federation credentials free-of-charge, simply by using their institutional username and password.
Not only does the NeCTAR Research Cloud offer all the advantages of remote computing, but is also independent of any one institution and so is more easily accessible to a multi-institutional and global collaboration.
NGS training platform
Most institutions, in which bioinformaticians work, run MS Windows or Mac OS X on desktop machines and have servers running a Linux OS. Linux is the operating system of choice for most bioinformaticians. There are many reasons for this but most stem from the fact that it is a free open-source OS, which allows software developers greater freedom and flexibility in the development of their tools. As such, most bioinformatic tools are specifically developed on Linux. The majority of the world’s high-performance computing facilities run on Linux/UNIX OSs and so those seriously looking to analyse their own NGS data need to begin to familiarizing themselves with a Linux operating system. However, this presents problems for new bioinformatics groups because the computing infrastructure they require is often not supported by the institution’s IT group, which tend to be MS Windows based. It also presents training problems, as a suitable Linux OS is unavailable for the purposes of training. We address this issue by providing NeCTAR Research Cloud–based VMs for use in the training sessions.
To limit the time required to develop an OS, which contained all the required tools for deploying cloud-based VMs for the NGS workshop, we opted to use a modified version of Cloud BioLinux [7] built on top of Ubuntu 12.04 LTS. A complete build of Cloud BioLinux installs a plethora of bioinformatic tools and data, which exceeds the 10 GB primary disk space available to default nodes of the NeCTAR Research Cloud. Therefore, we customized the build scripts to our requirements and to exclude tools that were deemed unlikely to be used (https://github.com/nathanhaigh/cloudbiolinux).
Rather than forcing trainees to use a terminal, we opted to use NX technology to provide the trainees with a remote desktop-like GUI. NX technology operates in a server–client model to optimize bandwidth usage and minimize lag. The NX client creates a local cache of data to reduce lag and thus provides a more responsive session than other remote desktop protocols like VNC. We configured the FreeNX server on the VM to accept connections for any user that is able to authenticate via SSH. From the client computer’s perspective, all that is required is the installation and configuration of the free NoMachine NX Client (available for several operating systems). Configuration of the client is simple and only requires the IP address and a username/password combination for the remote machine.
In the NoMachine NX Client, it is also possible to configure a ‘fullscreen’ mode, which does not contain any window decorations, thus ‘hiding’ the local operating system from the user. This is an appealing option when one does not want to expose too many new concepts to trainees at the start of the workshop. To simplify the process of configuring the NX Client software, we opted to set up default username/password combinations for a trainee user account as well as a sudoer (administrative) account.
An image of the OS is available in the NeCTAR Research Cloud dashboard. The image is also publically available for download as a Virtual Disk Image (VDI) for use with VirtualBox.
DEPLOYING THE NGS WORKSHOP CONTENT
We developed a BASH script to facilitate the deployment of the NGS workshop content onto a Linux machine. The script downloads all the workshop materials and configures the file system ready for use by the trainee. This provides greater flexibility in the development and deployment of future workshop content which can be maintained independently of the OS. The script is freely available, configurable via command line arguments and will work on any Linux/UNIX-like OS.
Due to primary disk space limitations (10 GB) on default nodes of the NeCTAR Research Cloud, the script places all data into/mnt/NGS_workshop/data/and a working directory is created as/mnt/NGS_workshop/working_dir/. To simplify life for trainees, a series of directories and symbolic links (symlinks) are created in the working directory to provide structure to the way data and results are accessed and created. Convenient symlinks are placed on the desktop and in the home directory of the trainee user.
In addition to deploying the workshop content other workshop-specific customizations may also be desirable. For instance, setting the appropriate time zone for the location where the workshop is being delivered and providing desktop links to online resources. We found dropcanvas (http://dropcanvas.com/) to be enormously useful in providing a convenient way to disseminate electronic copies of workshop materials to the trainees before, during and after the workshop. As such, we provided a desktop link to a read-only dropcanvas for each of the NGS workshops we delivered.
A complete working example that demonstrates the above mentioned features is available in https://github.com/nathanhaigh/ngs_workshop/tree/master/workshop_deployment/examples/. When instantiating the 20–30 NGSTrainingV1.2 VMs on the NeCTAR Research Cloud, this example script can be passed as a post-creation customization script, thereby creating VMs that are workshop ready.
Client computer configuration
Following the instantiation of the required number of NGS workshop VMs, attention then turns to the configuration of the computers at the hosting institution, hereafter called the client computers. By default, it is possible to connect to the VMs in one of two ways: (i) using the command line via an SSH client or (ii) using NX technology via an NX Client. Because trainees are not expected to be knowledgeable with the command line, we briefly describe the process for configuring the NX Client to provide a remote desktop-like GUI (full details are available in the NGS workshop handout under the ‘Remote Desktop with the NoMachine NX Client’ heading: https://github.com/downloads/nathanhaigh/ngs_workshop/trainee_handout_latest.pdf).
Ahead of running a workshop, the following needs to be confirmed: (i) the NoMachine NX Client has been installed on the client computers by an administrator and (ii) connections to the VMs, through TCP port 22, are allowed through the hosting institution’s firewall(s). Some institutions can take up to 2 weeks to have the appropriate firewall rules implemented, so instantiation of the VMs may be required well ahead of time.
Open access to workshop resources
We aim to foster and encourage the continued growth of bioinformatics training collaborations both nationally through the ABN and internationally through the BTN and the GOBLET. To this end, we have released the workshop resources under a Creative Commons Attribution 3.0 Unported License via github (https://github.com/nathanhaigh/ngs_workshop/).
Operating system
The operating system is available to Australian researchers as a snapshot on the NeCTAR Research Cloud (named NGSTrainingV1.2). It is also available in VDI format from NeCTAR Research Cloud storage (https://swift.rc.nectar.org.au:8888/v1/AUTH_33065ff5c34a4652aa2fefb292b3195a/VMs/NGSTrainingV1.2.vdi). The VDI image file is publically accessible and is suitable for use with VirtualBox (http://www.virtualbox.org/), a free virtualization software released under a GPLv2 licence.
Handout materials
The NGS workshop handout materials have been written in LaTeX, a plain text markup language for document typesetting. Documentation in LaTeX has several advantages over commonly used binary files, such as MS Word and PowerPoint files. Plain text files are easily version controlled to track changes as the documents evolve over time. The use of a web-accessible version control repository, such as github, means the code is available for contributions from across the globe.
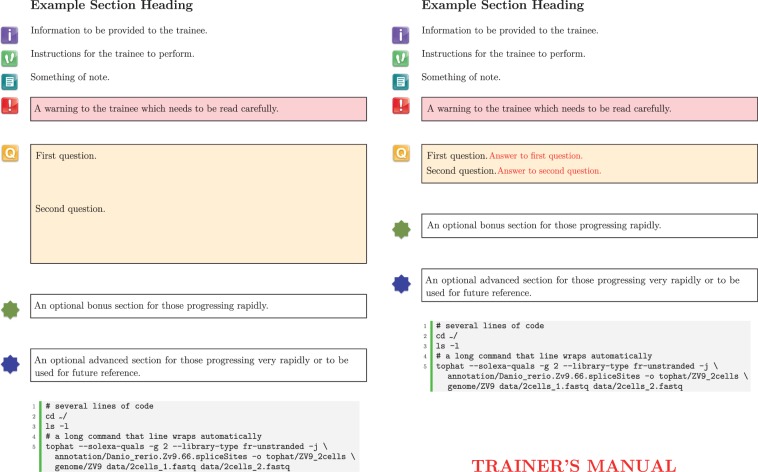
We provide a LaTeX style file (btp.sty), which defines several environments (Table 2) to make the styling and writing of the documentation straightforward, even for those with little or no LaTeX experience. The example.tex file provides example usage of these environments and generates the output seen in Figure 1. Some effort has been made to provide consistent styling of code within the lstlisting environment and provide a feature (Adobe Reader supports this, other PDF viewers may not) whereby the whole block of code can be copy-and-pasted into a terminal. This ensures less time is wasted debugging trainee’s typos and frees up time for the trainees to consider what tasks the commands are performing. However, this has a disadvantage: trainees can have the tendency to rush through executing commands without thinking about the commands they execute.
Table 2:
Details of the LaTeX environments defined to make styling of the workshop handouts consistent and easier
| Environment name | Example usage | Styling notes |
|---|---|---|
| Information | \begin{information} Information to be provided to the trainee. \end{information} | A purple information icon is placed in the left margin aligned to the top of the text within the environment. |
| Steps | \begin{steps} Instructions for the trainee to perform. \end{steps} | A green footprint icon is placed in the left margin aligned to the top of the text within the environment. |
| Note | \begin{note} Something of note. \end{note} | A turquoise note icon is placed in the left margin aligned to the top of the text within the environment. |
| Warning | \begin{warning} A warning to the trainee, which needs to be read carefully. \end{warning} | A red exclamation icon is placed in the left margin aligned to the top of the text within that environment. The text is emphasized by being placed in a red shaded box. |
| Questions | \begin{questions} One or more questions to pose to the trainee. \end{questions} | A yellow question icon is placed in the left margin aligned to the top of the text within that environment. The text is emphasized by being placed in a yellow-shaded box. Paragraph spacing is set to 2 cm, to allow sufficient space in which answer can be written. However, this is only the case if the trainermanual option is used when loading the btp package. |
| Answer | \begin{questions} First question. \begin{answer} Answer to first question. \end{answer} Second question. \begin{answer} Answer to second question. \end{answer} \end{questions} | Text within the answer environment is coloured red. However, it is only visible if the trainermanual option is used when loading the btp package. |
| Bonus | \begin{bonus} An optional bonus section for those progressing rapidly. \end{bonus} | A green star icon is placed in the left margin aligned to the top of the text within the environment. The text is emphasized by being bounded by a box with a black outline. |
| Advanced | \begin{advanced} An optional advanced section for those progressing very rapidly or to be used for future reference. \end{advanced} | A blue star icon is placed in the left margin aligned to the top of the text within the environment. The text is emphasized by being bounded by a box with a black outline. |
| Lstlisting | \begin{lstlisting} cufflinks—help \end{lstlisting} | Text within this environment is formatted as computer code and is intended to be executed at a Linux terminal. The text is styled using a monospaced font on a grey background. Line numbers are provided and separated from the code by a vertical green bar. Long commands are automatically split over multiple lines with the line continuation character ‘\’ inserted where required. When viewing the resulting PDF, with Adobe Reader, the whole text contents of this environment can be copied-and-pasted verbatim into a terminal. |
Figure 1:
Both trainee (left) and trainer (right) handouts are maintained as a single LaTeX document. The difference seen in styling is achieved simply by using the trainermanual option when loading the btp package.
The trainee- and trainer-specific content can be maintained in the same LaTeX document. To change the output styling, simply use the trainermanual option when loading the bpt package (\usepackage[trainermanual]{btp}). This option applies trainer-specific styling and includes trainer-specific content in the output. These include the following: (i) the front cover appears in red, (ii) the words ‘TRAINER’S MANUAL’ appear in large red letters in the header and footer of the front cover and in the footer of most other pages and (iii) the text contents of the answer environments become visible.
NGS workshop content deployment script
The NGS workshop deployment script is available via the workshop’s github repository (https://github.com/nathanhaigh/ngs_workshop/raw/master/workshop_deployment/NGS_workshop_deployment.sh). This script retrieves a copy of the publicly available data sets from NeCTAR cloud storage. This was to improve network bandwidth and thus content deployment times for the Australian hosted workshops.
Using this script as a template, it is possible to rework it for a completely different workshop that fetches data from any location referenced by a URL. A globally accessible repository of such scripts could make it far easier for trainers to deploy workshop-specific content in an automated fashion.
LOOKING FORWARD
Using Cloud BioLinux as the base OS provided a convenient, open-source starting point for a Bioinformatics Training Platform. However, it should be noted that it is a huge monolithic install containing many tools that will never be used in a focused workshop environment. The size of Cloud BioLinux adds significant overheads in the time required to instantiate VMs from the large NGSTrainingV1.2 image (approximately 9 GB) we have created. Instead, a system capable of provisioning a VM containing a minimal set of software, data and file system layout would be advantageous. Puppet (http://puppetlabs.com) is IT automation software for provisioning and configuring software. Using plain text files and Puppet’s declarative configuration language, the desired state of a system can be defined. These so called Puppet manifests could be used to declare what tools, data and resources are required for a given workshop. They could be version controlled, maintained and made publicly available for the bioinformatics training community to use. We are currently investigating its use as part of a Bioinformatics Training Platform.
Recently, to complement our two-day workshop, we trialled an introduction to the command line course developed by Software Carpentry before the NGS workshop. We found it invaluable in getting the participants oriented and comfortable with the command line before undertaking the bioinformatics analysis modules and a useful addition to the platform.
The community approach that we have taken to address a lack of bioinformatics training in Australia is a model that can easily be adapted by others facing similar bioinformatics challenges. Indeed all the necessary material, software and the operating system are freely available. This consortium is continuing to develop bioinformatics training modules in collaboration with international training programs to improve the bioinformatics skills of Australian researchers. To support the introduction of future courses, the trainer base will be expanded through recruitment of bioinformaticians that are committed to training and have the support of their host institutions to contribute time to this program for the greater benefit of the Australian research community.
The success of this NGS bioinformatics training program has been the result of the Australian bioinformatics community and Australian research organizations unifying to address the lack of training opportunities in Australia and importantly, the receptiveness, openness and ongoing mentoring provided by the EMBL-EBI training team in the UK.
SUPPLEMENTARY DATA
Supplementary data are available online at http://bib.oxfordjournals.org/.
Key points.
The need for bioinformatics training is increasing.
Cloud computing resources can offload computational requirements from hosting institutions.
Providing open access resources facilitates and maximizes reuse by the bioinformatics training communities.
We provide a virtual machine image and workshop deployment scripts to expedite NGS workshop deployment by bioinformatics trainers.
Acknowledgements
We would like to thank all those who have provided much assistance during the course of running the workshops. In particular the Australian Bioinformatics Network (David Lovell), Staff at University of NSW (Marc Wilkins and Simone Li), Monash eResearch (Paul Bonnington), University of Queensland (Scott Beatson), CSIRO (Ondrej Hlinka), University of Adelaide (Stephen Bent, Bastien Llamas and Arther Ng), Australian Centre for Plant Functional Genomics (Ute Baumann), NeCTAR (Glenn Moloney) and EMBL-EBI (Cath Brooksbank).
Biographies
Nathan S. Watson-Haigh is a bioinformatician with expertise and interest in systems biology approaches, de novo genome assembly, NGS data analysis, cloud computing and workshop development and delivery.
Catherine A. Shang is a project manager working within large Australian collaborative systems biology research consortiums. Catherine is actively involved in establishing collaborative bioinformatics networks and developing and delivering open access bioinformatics training courses for the benefit of the Australian research community.
Matthias Haimel is a bioinformatician working with different types of next-generation sequencing data, with a focus on assembling and analysing genomes. During the past 4 years, Matthias has been involved in developing and delivering next-generation sequencing courses either at the EBI or abroad.
Myrto Kostadima is a PhD student at the European Bioinformatics Institute (EBI) with interest in transcription and the mechanisms of transcriptional regulation in humans. Myrto has been involved in organizing and delivering courses on next-generation sequencing for the past 3 years either at the EBI or abroad.
Remco Loos is a postdoc working on understanding the mechanisms behind pluripotency and self-renewal in stem cells through next-generation sequencing, including Ribonucleic acid-sequencing (RNA-seq) and ChIP-seq, as well as sequencing approaches to epigenetic features (DNA methylation, histone modification, nucleosome positioning). In addition, Remco has been involved in developing and delivering training in next-generation sequencing analysis for the past 3 years.
Nandan Deshpande is a bioinformatician with focus on systems biology–based data analysis and experience in genome assembly and de novo transcriptomics.
Konsta Duesing Leads a team of bioinformaticians and statisticians engaged in genomics, epigenomics and metagenomics research. Konsta has developed and delivered training courses in NGS analysis and has a strong interest in systems approaches to biology and health. He is currently developing methods for the multiple ‘omics’ data space.
Xi Li is a bioinformatician working on genome annotation, NGS data processing and web-based biodata analysis platforms, also with particular interests in developing computational algorithms for the study of gene regulation as well as NGS training workshops.
Annette McGrath is the Bioinformatics Core Leader at the Commonwealth Scientific and Industrial Research Organisation. She leads a team of bioinformaticians involved in genomics research, bioinformatics infrastructure and tool development and provision and with a strong interest in training and development.
Sean McWilliam is a bioinformatician with a focus on genome annotation, genome assembly and variant analysis.
Simon Michnowicz assists researchers with high-performance computing requirements use the National Computational Infrastructure, hosted at Australian National University. He has a background in computational Proteomics.
Paula Moolhuijzen is a bioinformatician experienced in the analysis of NGS genome, transcriptome and metagenomic data, with a particular interest is the development high-throughput workflows for bioinformatics analyses.
Steve Quenette is the deputy director of the Monash eResearch Centre and project sponsor of the NeCTAR Research Cloud node at Monash University, with expertise in research platforms and productivity, and a research interest in computational science.
Jerico Nico De Leon Revote is a developer involved in research collaborations, 3D immersive visualizations and the NeCTAR Australian national research cloud.
Sonika Tyagi is a bioinformatician working in the genomics area with main focus on high-throughput sequence analysis. She is involved in developing computational methods and protocols for transcriptomics, post-transcription gene regulation analysis, exome and variation analysis.
Maria V. Schneider is the Head of Training and Outreach for The Genome Analysis Centre (TGAC) where she is responsible for the strategic coordination of the in-house and external TGAC Training and Outreach activities. Before this, she was the User Training Coordinator at EMBL-EBI.
FUNDING
Bioplatforms Australia is funded by the Australian government through the National Collaborative Research Infrastructure Strategy and the 2009 Super Science Initiative. NeCTAR is an Australian Government project conducted as part of the Super Science initiative and financed by the Education Investment Fund.
References
- 1.Giardine B, Riemer C, Hardison RC, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–5. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter AA, Macgregor AB, Szabo TO, et al. Yabi: an online research environment for grid, high performance and cloud computing. Source Code Biol Med. 2012;7:1. doi: 10.1186/1751-0473-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hull D, Wolstencroft K, Stevens R, et al. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W279–32. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Australian Bureau of Statistics. Regional Population Growth, Australia (cat. no. 3218.0.), 2011.
- 5.Schneider MV, Walter P, Blatter MC, et al. Bioinformatics Training Network (BTN): a community resource for bioinformatics trainers. Brief Bioinform. 2012;13:383–9. doi: 10.1093/bib/bbr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Via A, De Las Rivas J, Attwood TK, et al. Ten simple rules for developing a short bioinformatics training course. PLoS Comput Biol. 2011;7:e1002245. doi: 10.1371/journal.pcbi.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krampis K, et al. Cloud BioLinux: pre-configured and on-demand bioinformatics computing for the genomics community. BMC Bioinformatics. 2012;13:42. doi: 10.1186/1471-2105-13-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.