Abstract
With the advent of YouTube channels in bioinformatics, open platforms for problem solving in bioinformatics, active web forums in computing analyses and online resources for learning to code or use a bioinformatics tool, the more traditional continuing education bioinformatics training programs have had to adapt. Bioinformatics training programs that solely rely on traditional didactic methods are being superseded by these newer resources. Yet such face-to-face instruction is still invaluable in the learning continuum. Bioinformatics.ca, which hosts the Canadian Bioinformatics Workshops, has blended more traditional learning styles with current online and social learning styles. Here we share our growing experiences over the past 12 years and look toward what the future holds for bioinformatics training programs.
Keywords: continuing education, bioinformatics, online learning, massive open online courses (MOOCs)
EXTERNAL INFLUENCES IN EDUCATION
Computer technology and the World Wide Web have forever shifted the way in which our society functions and behaves. The English lexicon now includes vocabulary based on computers and the Internet landscape, like Google [1] and tweet [2], as well as terms more familiar in bioinformatics and computational space, like cloud computing [2] and BitTorrent [1]. Within the realm of education and learning, computer technology, the World Wide Web and its associated social media tools have also penetrated longstanding traditions and models. Wikipedia (which in 2005 was one of the most searched Google terms [3]) has replaced print edition encyclopedias [4], and PubMed Central, publisher’s websites and online academic libraries have supplanted excursions to the stacks at the library. Notebook and tablet computers, for the most part, substitute for handwritten class notes, which now take the form of footnotes entered directly into the lecturer’s presentation (which has been made available and downloaded before class). Skype, chat rooms and forum posts too have made face-to-face group sessions less relevant. The explosion of video sharing sites like YouTube have allowed educators to flip the classroom [5], so that acquiring new concepts is done online at home, and homework problems are worked through in the classroom. More recently, the creation of massive open online courses (MOOCs) such as iTunes University and others [6] have further pushed delivery strategies in education. The impact of these emergent technologies on education throughout the world is so profound that since 2004, the New Media Consortium (nmc.org) and the EDUCAUSE Learning Initiative (educase.edu) have annually published the Horizon Report (nmc.org/horizon), which forecasts the technologies most likely to impact on education [7].
Despite computation and the use of web-based tools and databases being inherent in the definition of bioinformatics, advances in computational technology and web tools have also impacted continuing education in bioinformatics. Here we retrospectively review how one continuing education series in bioinformatics, the Canadian Bioinformatics Workshops (CBWs), has learned to evolve its training program to better integrate these advances and realities in bioinformatics education or risk becoming out-of-date.
BIOINFORMATICS CONTINUING EDUCATION BEFORE THE INTERNET
Bioinformatics.ca first introduced the CBWs, its continuing education programs in bioinformatics, in 1999. The initial workshops introduced life science and computational researchers alike to the fields of bioinformatics, genomics, proteomics and tool development. In lieu of graduate programs, undergraduate coursework (at least in Canada; [8]) and even textbooks, these workshops were at the time, a way of addressing the need for a skilled computational science workforce in Canada [9]. Participants of these first workshops were graduate- and postgraduate-level learners with an interest and/or need in gaining both an understanding and introductory skill set in bioinformatics, its tools and databases, and typically had no prior exposure to bioinformatics in their formal education.
These workshops were hands-on and intensive, involving marked assignments and group work presentations. Students were supplied with laptop computers preconfigured with the required open source software and hooked up to the external web as well as a classroom server with in-house copies of the various biological databases (e.g. GenBank [10], RCSB Protein Data Bank (PDB) [11], etc.). As early as 2000, students were instructed to prepare for the workshop by reviewing online tutorials in UNIX [12] and Perl [13].
Following the traditional postsecondary pedagogy, the workshop schedule was divided between face-to-face didactic instruction and extended periods of hands-on exploration and skill development, both within the classroom setting. As a precursor to online lecture notes, students were provided with binders of lecture and laboratory material at the beginning of the workshop so that notes could be written in the margins. Unlike most classroom environments, individual learning benefitted from a high faculty to student ratio of 1:5, as well as from pairing of students identifying as ‘life scientists’ with students identifying as ‘computational’.
Content introduced students to the fundamentals of the computational molecular biology world and included presentations in ‘Using Biological Databases’ and ‘Submitting and Retrieving DNA sequences’, ‘Sequence Searching Tools’ and ‘BLAST’, as well as ‘Molecular Evolution’, ‘Phylogeny’ and ‘Multiple Sequence Alignment’. Students were also exposed to ‘UNIX’ and ‘Perl’ skills, and learned how to download and install a local instance of BLAST from the National Center for Biotechnology Information (NCBI) [14]. Workshop content and classroom exercises were supported by a private workshop wiki and webpages, adding HTML to the skill set. Demand for these introductory continuing education workshops was high and sustained for several years after their introduction.
A SHIFT IN BIOINFORMATICS CONTINUING EDUCATION NEEDS
Approximately 5 years later, however, this initial series of CBWs began to experience a decline in interest and enrollment. Any number of reasons may have contributed to this decline, but the most obvious retrospectively was a shift in need. The need for such introductory workshops in bioinformatics at the continuing education level was likely impacted by the rapidly evolving computer and web worlds. Between 1998 and 2005, the number of webpages in the Google index soared from 25 million [15] to over 8 billion webpages [16], making any postings of bioinformatics training material widely accessible to a global audience. Indeed, the CBWs began sharing and posting all of its workshop content and materials in 2004 (bioinformatics.ca/workshops/2004). Similarly, popular tools like BLAST began creating and linking tutorials and other learning materials to their bioinformatic tools [17], and Nature Genetics published a special issue entitled ‘A User’s Guide to the Human Genome’ in 2002 [18]. Over this same timeframe, the number of freely accessible, online tools, databases and resource materials for the bioinformatics and life science communities as tracked in the Bioinformatics Links Directory (bioinformatics.ca/links_directory) jumped from 385 links in 2002 to 700+ links in 2005 [19].
Further shifts in need occurred with the introduction of online academic ‘courseware’ at the university level throughout the 2000’s. A recent review of online bioinformatics courseware by Searls highlights the rapid growth and maturity of this content for the motivated self-learner [20]. This online courseware improved greatly with the creation of YouTube in 2005 [21], used by early online educators like Khan Academy (khanacademy.org), and again with the emphasis on the ‘structured presentation and high production quality’ rigorously enforced in online course curators like Coursera (coursera.org) and Udacity (udacity.com) [20], who more naturally mimic the face-to-face classroom environment by using videoed lectures linked with assignments and discussion forums. NCBI began posting tutorials and other instructional video content on YouTube in 2010 [22]; such content created by a tool’s own developers is invaluable because it can be readily trusted. As new bioinformatics content becomes available online, such as the recent collection of bioinformatics articles at PLoS Computational Biology that ‘can be used as a reference or tutorial’ [23] and the start of a WikiBook in bioinformatics [24], continuing education needs in bioinformatics will continue to shift in response.
ADAPTATION TO CHANGING CONTINUING EDUCATION NEEDS AND LEARNING ENVIRONMENT
Given this changing landscape, the CBWs decided to overhaul its workshops and methods. Over the course of 2007, input from bioinformatics experts, CBW faculty members and life science researchers from diverse fields was sought to determine new strategies and growth opportunities for continuing education curricula in bioinformatics. The series of advanced-topic, niche-focused workshops first presented in 2008 are a product of these discussions. Similar to the participants in the initial workshops, participants in this current series of workshops are graduate- and postgraduate-level learners with a need for bioinformatics skills. However, current participants typically already have extensive exposure to introductory bioinformatics concepts, tools and databases through their formal education, their research environment or through self-learning on the internet, and are seeking more specific focused tool- or data-centric analysis training.
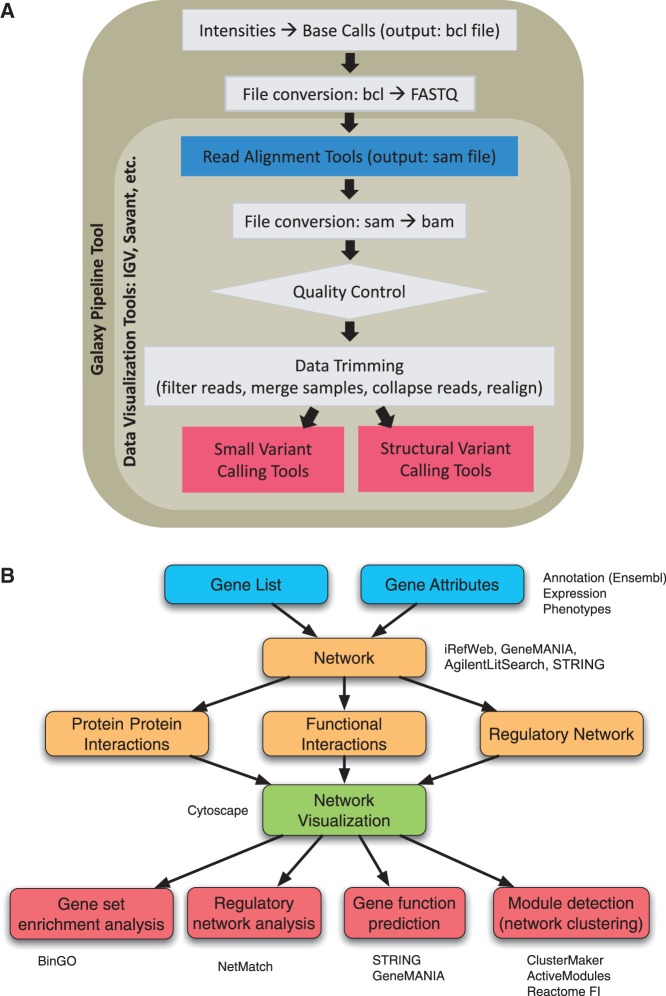
In response to the explosion of accessible introductory bioinformatics course work and materials, the CBW workshops thus changed from the introductory overview style to a focused niche-topic–specific style. In this new style, lectures and hands-on exercises follow workflows typically encountered by researchers in that topic so that researchers gain an understanding of how to progress through analysis of their data (detailed course objectives and outlines for each of the niche-topic workshops can be found at bioinformatics.ca/workshops). The workflow also serves as a diagrammatic representation of the course outline and is referred back to throughout a workshop. Lead faculty for a given workshop develop and revise a workshop’s workflow based on recent research, newly available tools and their experiences. In class, workflows are accompanied by (i) lectures introducing the background context and analysis considerations for a given step, reviewing the available tools and discussing the analysis process; and (ii) hands-on exercises that work through the execution of a given analysis step or tool functionality. For example, the highly successful ‘Informatics on High-throughput Sequencing Data’ workshop follows a workflow commonly performed by users of next-generation sequencers: read alignment, file conversion, quality control and data trimming, variant calling and data visualization (Figure 1A). Another good example of the CBW workflow occurs in the ‘Pathway and Network Analysis of –omics Data’ workshop, which follows a workflow typically encountered during analysis of a gene list (Figure 1B) [25]. These niches topic areas are reviewed on an annual basis and new workshops regularly introduced to stay current of continuing education needs in bioinformatics. Recent additions in 2013 include ‘Informatics for RNA-seq Analysis’ and ‘Flow Cytometry Data Analysis using R’ (bioinformatics.ca/workshops).
Figure 1:
Bioinformatics.ca workshops follow a workflow model. (A) The ‘Informatics for High-throughput Sequencing Data’ workshop follows a workflow commonly performed by users of next-generation sequencers. (B) The ‘Pathways and Network Analysis of –omics Data’ workshop follows a workflow decision tree encountered during gene list analysis.
Rather than move all workshops to an online delivery format as exemplified in MOOCs like Coursera, Udacity and others, however, CBW decided that there was still value and a need for live hands-on exercises and face-to-face instruction with expert lecturers to complement such open learning. As Searls states, ‘one great risk to the proposition of online bioinformatics education is that students never really get to grips with applying newfound computational or analytic skills to real biological data and actual problems in the full context of the scientific establishment’ [20], although similar arguments can be raised for isolated classroom learning as well. CBW chose to retain its split lecture and hands-on exercise schedule (∼30 min of lecture for every hour of hands-on activity), and instead altered supporting aspects of its workshops in reflection of the changing landscape. The choice to retain a mixed model of online learning and face-to-face instruction was made because audience feedback valued the opportunity for networking and one-on-one instruction on difficult complex informatic topics. While such feedback may be biased to the type of participants seen in CBW workshops, it speaks to the need to offer other learning models to aid those individuals who prefer to learn outside of a strict online environment. Faculty sentiment is that this mixed model allows the participant to investigate learning the necessary bioinformatics skills on their own pre-workshop and attempt their application to their own research data, but then allows the participant to reinforce or gain detailed tool-usage techniques and analysis strategies specific to their own data set in the face-to-face learning environment.
In response to the changing learning landscape, CBW changed various supporting aspects of its workshops. Students, for example, are no longer offered a preconfigured laptop, but are instead given detailed instructions for creating the informatic tool environment necessary for a given workshop on their own personal laptop (students without sufficient computer capacity on their laptop or without access to a personal laptop are supplied with a preconfigured laptop). While in-class troubleshooting is more challenging given the diversity in computer operating systems brought to the classroom, students are often able to directly apply their knowledge to their own data set located on the same computer, and students leave the workshop with a working and familiar tool environment. Other continuing education programs in bioinformatics make use of virtual machine copies of the classroom computational environment (personal communication with European Bioinformatics Institute and Australia Bioinformatics Network). Given the advanced-topic nature of most of the new workshops, students are assigned pre-workshop online tutorials in UNIX or R as before, although these online tutorials have matured greatly and become more sophisticated. Workshop content and classroom exercises are still supported by a dedicated workshop wiki, but most personal laptops have sufficient capacity to operate as a personal server eliminating the need for a dedicated workshop server. Where necessary, this in-house server has been replaced with an Amazon Web Services Elastic Compute Cloud (EC2) (aws.amazon.com/ec2) configured with all of the databases and tools required of the computationally intensive data-heavy workshops like ‘Bioinformatics for Cancer Genomics’ and ‘Informatics on High-throughput Sequencing Data’. At the conclusion of each workshop, an Amazon Machine Image (AMI) is created and deposited for the community of EC2 users (Table 1). AMIs capture the configuration of tools within an EC2 instance (such as those used for a given workshop), allowing other EC2 users to use this configuration within their own EC2 instance; workshop AMIs become freely accessible once logged in to an Amazon EC2 instance. On return to their home research environments, workshop participants may launch their own EC2 instance and load in the preconfigured workshop AMI, rapidly recreating the workshop compute environment.
Table 1:
The AMIs created and made available for various computationally intensive CBWs held in 2012
| CBW title | AMI |
|---|---|
| CBW Cancer Genomics 2012 | ami-0c64c565 |
| CBW HT-seq 2012 | ami-d21bbbbb |
Users having logged into an Amazon EC2 instance may access these AMIs through the community AMIs, rapidly recreating the workshop compute environment.
Reacting to the emergence of YouTube educational video content, CBW in 2009 also began recording and producing voice-over-presentation videos of its lectures, which are then made freely available on the bioinformatics.ca website (bioinformatics.ca/workshops). The online video content and accompanying presentation material is accessed by between 7% and 15% of bioinformatics.ca site users (depending on the workshop, as per Google Analytics) and serves to engage new participants seeking content details, to support past participants requiring a reminder of their session and/or to teach forthcoming participants skills in topics not covered within a given workshop (such as R statistical skills). With the current emphasis on channels at YouTube, the CBW is also working toward developing its own channel on YouTube for its workshop videos.
New technologies and teaching strategies to aid the CBW continuing education bioinformatics workshops are often suggested by past participants through the feedback surveys or by faculty during the annual workshop review meeting. Where practical, feasible and useful to the learning process, new technologies and teaching strategies are then piloted within the next series of workshops. If successful within a pilot workshop as based on faculty experience and participant feedback, the new technology or strategy is then introduced into the other workshops. Often this is an iterative process to achieve maximal impact on learning. Within the new niche-topic–specific workshops for example, the evening of the first day for all 2-day workshops was left as an open learning session for participants to practice and apply their new skills to their own data. However, the lack of structure to this evening session resulted in only a handful of participants interested in staying for the session and rated poorly in feedback and faculty experience. These evening sessions were subsequently revised to include an integrated assignment that reinforced the day’s skill set using a different data set, with the option for the participants to use their own data set; attendance and feedback on the integrated assignment improved immensely. New technologies and teaching strategies suggested and being explored for 2013 include instant messaging during a workshop to capture audience questions and an expanded 3-day workshop model to provide more hands-on practical time.
THE FUTURE OF BIOINFORMATICS CONTINUING EDUCATION INVOLVES KEEPING PACE WITH NEEDS AND THE LEARNING ENVIRONMENT
Survey feedback from each of the CBW workshops often indicates the desire for students to continue their interaction and learning, and to seek bioinformatics input beyond the classroom environment. While CBW Alumni have group space on Facebook, such social media tools do not adequately address the need. Support for continued bioinformatics skills development and advice, while burdensome for the CBW faculty when they return to their own research environments, is being better addressed by new entrants in online forums, such as SEQanswers (SEQanswers.com) launched in 2007 to ‘enable scientists to collaboratively advance genomics and high-throughput sequencing (HTS) technologies’ [26], and BioStar (biostars.org) launched more recently in 2010 to ‘allow researchers to pose questions and offer solutions to bioinformatics-related problems’ [27]. BioStar in particular seems most amenable to meeting the CBW’s needs for continued learning and advice beyond the classroom because ‘questions and answers are rated by members of the site and can be edited by any member, in a manner analogous to a wiki’ [27], allowing for the best and most trusted advice to float to the top.
One limitation to the current niche-topic–specific workshops is that many of these workshops require a basic working knowledge of the command line, especially those workshops that rely on R Statistical software (r-project.org). While the CBW offers an introductory ‘Exploratory Analysis of Biological Data using R’ workshop, many participants cannot accommodate multiple workshops in a single year. Workshops dependent on UNIX and R skills list completion of online tutorials as a prerequisite to attending the workshop. Many participants, however, neglect completing these tutorials in advance of the workshop or if they do, they do not grasp a working knowledge of the syntax and architecture. Other mechanisms for ensuring these skills in advance of a workshop are required and additional online resources may serve this purpose. Some basic coding skills may be gained through such sites as Software Carpentry (software-carpentry.org), although tutorial exercises are not used to reinforce skills. One new online bioinformatics-related education site CBW will be watching is Rosalind (Rosalind.info) launched in 2012. Designed to aid coders to develop and improve their coding skills with direct application to biological problems, the evolution of Rosalind.info may permit the computationally naïve researcher to acquire even simple skills in file manipulation and query.
Going forward, the CBW will need to continue to evolve its bioinformatics continuing education programs if it wants to remain relevant in the world of open online learning. CBW’s current strategy to blend face-to-face instruction with online learning works for current bioinformatics continuing education needs in Canada. Participant feedback indicates that interaction with faculty and the face-to-face discussions of informatic analysis approaches is the most highly valued component of the current strategy; feedback also indicates that online support tools used to prepare for and execute a workshop are also well liked. This is likely a product of the current workshop focus on specific data-centric topics where the tools and analysis processes are presented together as a workflow, and the absence of such comprehensive training in currently available online materials. The CBW, however, as with other bioinformatics continuing education programs, will need to stay aware of new developments in the online learning space in bioinformatics and continuously update its programming accordingly, as from experience, needs will change as the learning landscape changes. What exactly such future programs will look like though, remains an exciting predictive problem in a rapidly changing landscape.
Key points.
Computers and the internet have shifted access to information and mechanisms of communication
Initial bioinformatics continuing education programs followed the traditional didactic model with exercises
Online bioinformatics resources shifted bioinformatics training needs
Current bioinformatics continuing education programs at bioinformatics.ca mix face-to-face and online learning
Online resources and learning technologies will continue to impact bioinformatics continuing education programs
Acknowledgements
Bioinformatics.ca and the CBWs would like to acknowledge the dedicated faculty (bioinformatics.ca/workshops/faculty) who develop the learning materials for the CBW workshops, and who freely contribute these resources under our Creative Commons Attribution-Share Alike 2.5 license.
Biographies
Michelle D. Brazas is a specialist of knowledge and research translation at the Ontario Institute for Cancer Research, where she manages bioinformatics.ca, and develops the bioinformatics continuing education programs offered by the Canadian Bioinformatics Workshop series.
Francis Ouellette is Associate Director of Informatics and Bio-computing at the Ontario Institute for Cancer Research and Associate Professor in the department of Cell and Systems Biology at the University of Toronto. He is the scientific director and one of the founders of bioinformatics.ca and the Canadian Bioinformatics Workshop series. @bffo on twitter.
FUNDING
This work was supported by the Ontario Institute for Cancer Research; the Ontario Provincial Government; the Canadian Institutes of Health Research; Genome Canada; the Ontario Genomics Institute; The Centre for Applied Genomics and in-kind contributions from Amazon Web Services.
References
- 1.OED. Oxford English Dictionary June 2006 Update. 2006. http://public.oed.com/the-oed-today/recent-updates-to-the-oed/previous-updates/june-2006-update/ (21 January 2013, date last accessed) [Google Scholar]
- 2.Merriam-Webster. A Sample of New Dictionary Words for 2012. http://www.merriam-webster.com/info/newwords12.htm (21 January 2013, date last accessed) [Google Scholar]
- 3.Google. Year-end Look at Google Search Trends. 2005. http://www.google.ca/corporate/timeline/ - 2005-year-end-look-at-google-search-trends (21 January 2013, date last accessed) [Google Scholar]
- 4.Bosman J. After 244 Years, Encyclopaedia Britannica Stops the Presses. Media Decoder 2012. http://mediadecoder.blogs.nytimes.com/2012/03/13/after-244-years-encyclopaedia-britannica-stops-the-presses/ (21 January 2013, date last accessed) [Google Scholar]
- 5.Bergmann J, Sams A. How the Flipped Classsroom is Radically Transforming Learning. The Daily Riff, 2012. pp. 1–3. [Google Scholar]
- 6.McKinney D, Dyck JL, Luber ES. iTunes University and the classroom: can podcasts replace Professors? Comput Educ. 2009;52:617–23. [Google Scholar]
- 7.Martin S, Diaz G, Sancristobal E, et al. New technology trends in education: seven years of forecasts and convergence. Comput Educ. 2011;57:1893–906. [Google Scholar]
- 8.Counsell D. A review of bioinformatics education in the UK. Brief Bioinform. 2003;4:7–21. doi: 10.1093/bib/4.1.7. [DOI] [PubMed] [Google Scholar]
- 9.Network CGD. White Paper: Bioinformatics Curriculum Recommendations for Undergraduate, Graduate and Professional programs. http://www.biotalent.ca/sites/biotalent/files/PDF/BioinformaticsWhitePaper.pdf (21 January 2013, date last accessed) [Google Scholar]
- 10.Benson DA, Cavanaugh M, Clark K, et al. GenBank. Nucleic Acids Res. 2013;41(Database issue):D36–42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rose PW, Bi C, Bluhm WF, et al. The RCSB Protein Data Bank: new resources for research and education. Nucleic Acids Res. 2013;41(Database issue):D475–82. doi: 10.1093/nar/gks1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stonebank M. UNIX Tutorial for Beginners. http://web.archive.org/liveweb/http://www.ee.surrey.ac.uk/Teaching/Unix/ (21 January 2013, date last accessed) [Google Scholar]
- 13.web.archive.org. Perl Tutorial: Start. http://web.archive.org/web/20000619112311/http://agora.leeds.ac.uk/Perl/start.html (21 January 2013, date last accessed) [Google Scholar]
- 14.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Google. Original Prototype. 1998. http://www.google.ca/corporate/timeline/ - homepage-original-prototype (21 January 2013, date last accessed) [Google Scholar]
- 16.Google. Homepage Through the Years. 2005. http://www.google.ca/corporate/timeline/ - homepage-hurricane-katrina-relief-effort (21 January 2013, date last accessed) [Google Scholar]
- 17.Madden T. Chapter 16. The BLAST Sequence Analysis Tool. The NCBI Handbook; 2002. http://www.ncbi.nlm.nih.gov/books/NBK21101/ [Google Scholar]
- 18.Wolfsberg TG, Wetterstrand KA, Guyer MS, et al. A user's guide to the human genome. Nat Genet. 2002;32(Suppl):1–79. doi: 10.1038/ng961. [DOI] [PubMed] [Google Scholar]
- 19.Brazas MD, Yim D, Yeung W, et al. A decade of Web Server updates at the Bioinformatics Links Directory: 2003-2012. Nucleic Acids Res. 2012;40(Web Server issue):W3–12. doi: 10.1093/nar/gks632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Searls DB. An online Bioinformatics curriculum. PLoS Comput Biol. 2012;8:e1002632. doi: 10.1371/journal.pcbi.1002632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.YouTube. Timeline. 2005. http://www.youtube.com/t/press_timeline (21 January 2013, date last accessed) [Google Scholar]
- 22.NCBI-NLM. NCBI YouTube Channel. 2010. http://www.youtube.com/user/NCBINLM (21 January 2013, date last accessed) [Google Scholar]
- 23.Translational Bioinformatics. 2012. http://www.ploscollections.org/article/browse/issue/info%3Adoi%2F10.1371%2Fissue.pcol.v03.i11 (18 February 2013, date last cited) [Google Scholar]
- 24.Bioinformatics. WikiBooks; http://en.wikibooks.org/wiki/Bioinformatics (18 February 2013, date last cited) [Google Scholar]
- 25.Bader G. Module 4: Network Visualization. 2012. http://bioinformatics.ca//files/public/Pathways_2012_Module4.pdf (21 January 2013, date last accessed) [Google Scholar]
- 26.Li JW, Schmieder R, Ward RM, et al. SEQanswers: an open access community for collaboratively decoding genomes. Bioinformatics. 2012;28:1272–3. doi: 10.1093/bioinformatics/bts128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parnell LD, Lindenbaum P, Shameer K, et al. BioStar: an online question & answer resource for the bioinformatics community. PLoS Comput. Biol. 2011;7:e1002216. doi: 10.1371/journal.pcbi.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]