Abstract
The presence of Bartonella species in Xenopsylla cheopis fleas collected from Rattus spp. (R. exulans, R. norvegicus, and R. rattus) in Khon Kaen Province, Thailand was investigated. One hundred ninety-three fleas obtained from 62 rats, were screened by polymerase chain reaction using primers specific for the 16S–23S intergenic spacer region, and the presence of Bartonella DNA was confirmed by using the citrate synthase gene. Bartonella DNA was detected in 59.1% (114 of 193) of fleas examined. Sequencing demonstrated the presence of Bartonella spp. similar to B. elizabethae, B. rattimassiliensis, B. rochalimae, and B. tribocorum in the samples tested with a cutoff for sequence similarity ≥ 96% and 4 clustered together with the closest match with B. grahamii (95.5% identity). If X. cheopis proves to be a competent vector of these species, our results suggest that humans and animals residing in this area may be at risk for infection by several zoonotic Bartonella species.
Bartonella species are small, pleomorphic, gram-negative bacteria that infect a variety of mammalian hosts, including cats, dogs, rodents, ruminants, and humans. Clinical symptoms associated with Bartonella range from mild, influenza-like symptoms to more severe manifestations such as endocarditis, myocarditis, uveitis, bacillary angiomatosis, and peliosis hepatis.1 Approximately half of the 20 Bartonella species or subspecies identified to date are known or suspected human pathogens,2 and most are believed to be transmitted by arthropod vectors (fleas, lice, sandflies, and ticks).3
Xenopsylla cheopis, the Oriental rat flea, is a suspected vector of several Bartonella species (B. tribocorum, B. elizabethae, B. queenslandensis, B. rochalimae, and novel Bartonella genotypes), and Bartonella DNA has been detected in these fleas from various locations worldwide.3–8 Although generally found on rodents, X. cheopis have been found to parasitize humans and are known vectors of the zoonotic agents Yersinia pestis (plague) and Rickettsia typhi (murine typhus).9
Numerous surveys have been performed to identify the presence of Bartonella species affecting humans and domestic and peri-domestic animals in Thailand.10–17 Bartonella henselae, (the agent of cat scratch disease),14 B. tamiae,10 B. elizabethae, B. rattimassiliensis, and B. tribocorum have been isolated from febrile patients,15 B. henselae and B. clarridgeiae have been reported in cats,11 and B. clarridgeiae, B. vinsonii subsp. arupensis, B. elizabethae, B. grahamii, B. quintana, B. taylorii, and novel Bartonella genotypes have been found in dogs.11,16 In rodent species, B. grahamii, B. elizabethae, Candidatus Bartonella thailandensis, B. coopersplainensis, B. phoceensis, B. rattimassiliensis, B. tribocorum, and novel Bartonella genotypes have been detected by culture and polymerase chain reaction (PCR) analysis.12,13,17 However, little information has been obtained to identify potential arthropod vectors of Bartonella species in Thailand. Bartonella henselae, B. clarridgeiae and B. koehlerae were detected in Ctenocephalides felis fleas removed from cats18–20 and B. henselae was identified in two C. canis19 also collected from cats. Furthermore, a Bartonella sp., similar to B. grahamii, was found in a rodent flea, Nosopsyllus fasciatus, obtained from Rattus surifer.18 Bartonella tamiae DNA has also been found in chigger mites (genera Leptotrombidium, Schoengastia, and Blankarrtia) and in a tick (genus Haemaphysalis) collected from rodents in Thailand, suggesting a potential role for these arthropods in the transmission of B. tamiae.21
The aim of the current study was to investigate the prevalence of Bartonella species in rodent-associated fleas collected in Khon Kaen Province, Thailand, and to determine what potential role, if any, these fleas may play in the transmission of Bartonella species to individuals residing in this area.
For this study, 62 rats (10 R. norvegicus, 9 R. rattus, and 43 R. exulans) were trapped in and around homes in 4 villages, 1 market, and on farm land (a pig farm and 2 rice fields) in Khon Kaen Province, Thailand during May–June 2011 (Table 1). Fleas were collected from rats and placed in tubes containing isopropanol. Samples were shipped to Bartonella Laboratory at the Centers for Disease Control and Prevention (Fort Collins, CO) on dry ice and stored at −20°C until further analysis. All fleas were subsequently identified as X. cheopis by using a taxonomic key.22 Work involving rodents was conducted as outlined in our approved animal use protocol (#11-003), under the supervision of the Institutional Animal Care and Use Committee of the Division of Vector Borne Diseases.
Table 1.
Number of rats trapped per site by species and total number of fleas examined per site, northeastern Thailand
| Site designation | No. Rattus exulans/site | No. R. norvegicus/site | No. R. rattus/site | No. fleas examined/site* |
|---|---|---|---|---|
| Village 1 | 7 | 8 | 2 | 60 |
| Village 2 | 1 | 1 | 0 | 6 |
| Village 3 | 15 | 0 | 1 | 46 |
| Village 4 | 17 | 0 | 0 | 60 |
| Neighborhood market | 1 | 0 | 1 | 5 |
| Farmland (pig farm and rice fields) | 2 | 1 | 5 | 16 |
| Total | 43 | 10 | 9 | 193 |
Total number of fleas per rat was not determined. No more than five fleas/rat were screened for Bartonella DNA.
Individual fleas were triturated by using a bead beater protocol,23 and DNA was extracted by using a Qiagen QIAamp tissue kit (QIAGEN, Valencia, CA) according to the manufacturer's instruction. DNA was extracted from 1–5 fleas/rat (depending upon the number of fleas collected: in most cases, > 5 fleas per rat were recovered); a total of 193 fleas were examined. Fleas were initially screened by conventional PCR using primers specific for the 16S–23S intergenic spacer region (ITS),24 and the presence of Bartonella DNA was confirmed by using citrate synthase gene (gltA)–specific primers.8 Bartonella doshiae DNA was used as a positive control, and nuclease-free water was used as a negative control.
GltA amplicons were purified by using the QIAquick PCR purification kit (QIAGEN) and sequenced by using a Model 3130 genetic analyzer (Applied Biosystems, Foster City, CA). DNA sequences were analyzed by using the Lasergene version 8 sequence analysis software (DNASTAR, Madison, WI). All gltA sequences for this study were shortened to ≈379 basepairs to enable further phylogenetic analysis. Sequences obtained in this study were considered similar to validated Bartonella spp. if similarity over the 379-base-pairs gltA fragment was ≥ 96%.25 The Clustal W program in Megalign (Lasergene) was used to compare sequences obtained from this study to Bartonella sequences available in GenBank. The neighbor-joining (NJ) method by Kimura's two-parameter distance method and bootstrap calculation was carried out with 1,000 resamplings. GltA sequences were submitted to GenBank (accession numbers JX123018–JX123023).
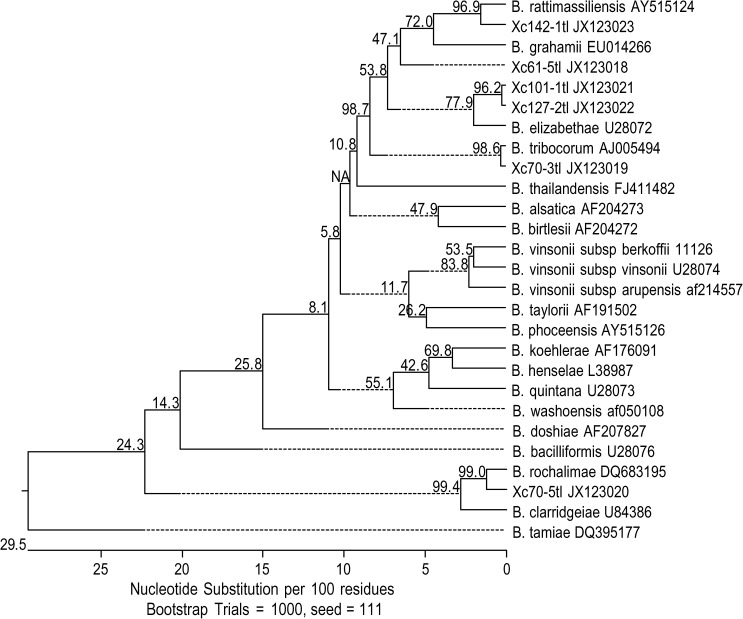
Of the 193 X. cheopis fleas examined, 59.1% (114) were positive for Bartonella DNA by using ITS and gltA primers (113 fleas ITS positive and 107 fleas gltA positive). A total of 80 gltA amplicons were sequenced. Six genotypes, with at least one nucleotide difference, were found and sequence similarity between genotypes ranged between 87.6% and 99.5% (Table 2). These six genotypes were clustered around B. elizabethae (U28072) (genotypes 1 and 2 with sequence similarity of 96.2%, GenBank accession nos. JX123021 and JX123022), B. grahamii (EU014266) (genotypes 3 with sequence similarity of 95.5%, GenBank accession no. JX123018), B. rattimassiliensis 15908T (AY515124) (genotype 4 with sequence similarity of 96.6%, GenBank accession no. JX123023), B. rochalimae BMGH (DQ683195) (genotype 5 with sequence similarity of 98.8%, GenBank accession no. JX123020), or B. tribocorum IBS506T (AJ005494) (genotypes 6 with sequence similarity of 99.7%, GenBank accession no. JX123019) (Figure 1 ).
Table 2.
Bartonella citrate synthase A genotypes detected in Xenopsylla cheopis, number of sequences of each genotype, and flea rodent host, northeastern Thailand*
| GenBank accession no. | Bartonella genotype | No. sequences/genotype | Flea rodent host* |
|---|---|---|---|
| JX123018 | Xc61-5tl | 4 | RE (1), RN (1), RR (1) |
| JX123019 | Xc70-3tl | 14 | RE (5), RN (4), RR (1) |
| JX123020 | Xc70-5tl | 24 | RE (5), RN (8), RR (2) |
| JX123021 | Xc101-1tl | 1 | RN (1) |
| JX123022 | Xc127-2tl | 36 | RE (12), RN (5) |
| JX123023 | Xc142-1tl | 1 | RR (1) |
RE = Rattus exulans; RN = Rattus norvegicus; RR = Rattus rattus.
Figure 1.
Tree topology displaying similarity of Bartonella DNA detected in Xenopsylla cheopis with known Bartonella sequences based upon partial citrate synthase gene (gltA) sequences, northeastern Thailand. GltA sequences obtained from fleas are represented by GenBank Accession nos. JX123018–JX123023.
The B. elizabethae group (genotypes 1 and 2), detected in fleas recovered from 18 rats (12 R. exulans and 6 R. norvegicus), contained 36 identical sequences and a distinct sequence, respectively. This group was also similar to a Bartonella sp. detected in R. norvegicus from Beijing, China (EF213769) and Praomys delectorum from Tanzania (FJ851115) with 98.9–99.5% and 99.2% sequence similarity, respectively. Genotype 3, most closely related to B. grahamii with 95.5% similarity and a Bartonella sp. detected in stray animals from Taiwan (GU056195) with 99.2% similarity, contained 4 identical sequences and was detected in fleas collected from 3 rats (1 R. exulans, 1 R. norvegicus, and 1 R. rattus). The B. rattimassilienis sequence (genotype 4) was detected in a flea collected from a R. rattus and was also 98.9% similar to a bartonellae isolated from the blood of a R. argentiventer from Thailand (FJ655402). The B. rochalimae group (genotype 5) contained 24 identical sequences found in fleas removed from 15 rats (5 R. exulans, 8 R. norvegicus, and 2 R. rattus). This genogroup was also 100% identical to Bartonella sp. 1-1C detected in a R. norvegicus from Taiwan (FN545495). The B. tribocorum group (genotype 6) contained 14 identical sequences and was detected in fleas recovered from 10 rats (5 R. exulans, 4 R. norvegicus, and 1 R. rattus). This group was also 99.5–99.9% similar to a Bartonella sp. detected in rodents from Nepal (GU143516) and Yunnan, China (FJ589051).
Humans and animals residing in this area commonly come into contact with rodents and are potentially at risk for infection with rodent-borne diseases. A large percentage of rodents in this study were trapped either in or around homes or in food storage areas, increasing the likelihood of disease transmission. In a separate survey, Kosoy and others15 screened the blood of 261 patients to identify what role Bartonella species play in acute febrile illness in Thailand; Bartonella spp. were detected in 7.7% (20) of these samples. Sequencing demonstrated the presence of rodent-borne Bartonella species in half of these samples, specifically B. rattimassiliensis, B. vinsonii subsp. arupensis, B. vinsonii subsp. vinsonii, B. tribocorum, and B. elizabethae, and 71% of patients reported exposure to rats during the two weeks before the onset of illness.15 An additional study was conducted in rural Thailand to screen febrile and non-febrile patients who came to local hospitals for Bartonella-specific antibodies.26 Of the 521 serum samples screened, 9.8% (51) were seropositive for B. elizabethae and 3.6% (19) for B. vinsonii subsp. vinsonii. Interestingly, 18 patients were seroreactive against B. elizabethae and B. vinsonii subsp. vinsonii, 1 patient was seroreactive against B. elizabethae, B. henselae, and B. quintana, 4 patients were seroreactive against B. elizabethae, B. vinsonii subsp. vinsonii, and B. quintana, and 6 patients harbored antibodies against B. elizabethae, B. vinsonii subsp. vinsonii, B. henselae, and B. quintana.26 These results further strengthen the supposition that contact with rodents is quite common in Thailand and rodents might serve as reservoirs for human Bartonella infections.
Almost 60% of fleas examined in this study harbored Bartonella DNA. Parola and others18 found a much lower Bartonella prevalence in rodent fleas collected along the Thailand–Myanmar border. In this study, 10 X. cheopis and 26 N. fasciatus were tested and 1 flea (2.8% positivity), a N. fasciatus collected from a R. surifer, contained a species closely related to B. grahamii.18 The results from our study demonstrate that a large percentage of X. cheopis from northeastern Thailand harbor Bartonella species, including known zoonotic pathogens. What role, if any, X. cheopis plays in the transmission of Bartonella species remains unclear. Currently, studies are being performed in our laboratory to determine if X. cheopis are competent vectors of Bartonella species.
ACKNOWLEDGMENTS
We thank John Monteneiri for assistance with initial flea identifications and the staff and faculty members of Khon Kaen University College of Veterinary and Dr. Ratree Takhampunya (Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand) for support during field collections.
Disclaimer: The mention of trade names or commercial products does not constitute endorsement or recommendation for use by the Department of Defense. The opinions and assertions are those of the authors and are not to be construed as official or reflecting the views of the Department of Defense.
Footnotes
Financial support: This study was supported by an American Society of Tropical Medicine and Hygiene Robert E. Shope International Fellowship to Sarah A. Billeter.
Authors' addresses: Sarah A. Billeter, Leah Colton, and Michael Y. Kosoy, Division of Vector Borne Diseases, Centers for Disease Control and Prevention, Fort Collins, CO, E-mails: ina8@cdc.gov, ant6@cdc.gov, and mck3@cdc.gov. Somboon Sangmaneedet and Fanan Suksawat, Faculty of Veterinary Medicine, Khon Kaen University, Amphur Mueng, Khon Kaen, Thailand, E-mails: sombn_sa@kku.ac.th and jsvetmed@yahoo.com. Brian P. Evans, Department of Entomology, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand, E-mail: brian.patrick.evans@us.army.mil.
References
- 1.Chomel BB, Boulouis HJ, Maruyama S, Breitschwerdt EB. Bartonella spp. in pets and effect on human health. Emerg Infect Dis. 2006;12:389–394. doi: 10.3201/eid1203.050931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vayssier-Taussat M, Le Rhun D, Bonnet S, Cotté V. Insights in Bartonella host specificity. Ann N Y Acad Sci. 2009;1166:127–132. doi: 10.1111/j.1749-6632.2009.04531.x. [DOI] [PubMed] [Google Scholar]
- 3.Billeter SA, Levy MG, Chomel BB, Breitschwerdt EB. Vector transmission of Bartonella species with emphasis on the potential for tick transmission. Med Vet Entomol. 2008;22:1–15. doi: 10.1111/j.1365-2915.2008.00713.x. [DOI] [PubMed] [Google Scholar]
- 4.Loftis AD, Reeves WK, Szumlas DE, Abbassy MM, Helmy IM, Moriarity JR, Dasch GA. Surveillance of Egyptian fleas for agents of public health significance: Anaplasma, Bartonella, Coxiella, Ehrlichia, Rickettsia, and Yersinia pestis. Am J Trop Med Hyg. 2006;75:41–48. [PubMed] [Google Scholar]
- 5.Li DM, Liu QY, Yu DZ, Zhang JZ, Gong ZD, Song XP. Phylogenetic analysis of Bartonella detected in rodent fleas in Yunnan, China. J Wildl Dis. 2007;43:609–617. doi: 10.7589/0090-3558-43.4.609. [DOI] [PubMed] [Google Scholar]
- 6.Reeves WK, Rogers TE, Durden LA, Dasch GA. Association of Bartonella with the fleas (Siphonaptera) of rodents and bats using molecular techniques. J Vector Ecol. 2007;32:118–122. doi: 10.3376/1081-1710(2007)32[118:aobwtf]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 7.Tsai YL, Chuang ST, Chang CC, Kass PH, Chomel BB. Bartonella species in small mammals and their ectoparasites in Taiwan. Am J Trop Med Hyg. 2010;83:917–923. doi: 10.4269/ajtmh.2010.10-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billeter SA, Gundi VA, Rood MP, Kosoy MY. Molecular detection and identification of Bartonella species in Xenopsylla cheopis fleas (Siphonaptera: Pulicidae) collected from Rattus norvegicus rats in Los Angeles, California. Appl Environ Microbiol. 2011;77:7850–7852. doi: 10.1128/AEM.06012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brouqui P, Raoult D. Arthropod-borne diseases in homeless. Ann N Y Acad Sci. 2006;1078:223–235. doi: 10.1196/annals.1374.041. [DOI] [PubMed] [Google Scholar]
- 10.Kosoy M, Morway C, Sheff KW, Bai Y, Colborn J, Chalcraft L, Dowell SF, Peruski LF, Maloney SA, Baggett H, Sutthirattana S, Sidhirat A, Maruyama S, Kabeya H, Chomel BB, Kasten R, Popov V, Robinson J, Kruglov A, Petersen LR. Bartonella tamiae sp. nov., a newly recognized pathogen isolated from three human patients from Thailand. J Clin Microbiol. 2008;46:772–775. doi: 10.1128/JCM.02120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue K, Maruyama S, Kabeya H, Kawanami K, Yanai K, Jitchum S, Jittaparapong S. Prevalence of Bartonella infection in cats and dogs in a metropolitan area, Thailand. Epidemiol Infect. 2009;137:1568–1573. doi: 10.1017/S095026880900257X. [DOI] [PubMed] [Google Scholar]
- 12.Saisongkorh W, Wootta W, Sawanpanyalert P, Raoult D, Rolain JM. “Candidatus Bartonella thailandensis”: a new genotype of Bartonella identified from rodents. Vet Microbiol. 2009;139:197–201. doi: 10.1016/j.vetmic.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 13.Bai Y, Kosoy MY, Lerdthusnee K, Peruski LF, Richardson JH. Prevalence and genetic heterogeneity of Bartonella strains cultured from rodents from 17 provinces in Thailand. Am J Trop Med Hyg. 2009;81:811–816. doi: 10.4269/ajtmh.2009.09-0294. [DOI] [PubMed] [Google Scholar]
- 14.Paitoonpong L, Chitsomkasem A, Chantrakooptungool S, Kanjanahareutai S, Tribuddharat C, Srifeungfung S. Bartonella henselae: first reported isolate in a human in Thailand. Southeast Asian J Trop Med Public Health. 2008;39:123–129. [PubMed] [Google Scholar]
- 15.Kosoy M, Bai Y, Sheff K, Morway C, Baggett H, Maloney SA, Boonmar S, Bhengsri S, Dowell SF, Sitdhirasdr A, Lerdthusnee K, Richardson J, Peruski LF. Identification of Bartonella infections in febrile patients from Thailand and their potential animal reservoirs. Am J Trop Med Hyg. 2010;82:1140–1145. doi: 10.4269/ajtmh.2010.09-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai Y, Kosoy MY, Boonmar S, Sawatwong P, Sangmaneedet S, Peruski LF. Enrichment culture and molecular identification of diverse Bartonella species in stray dogs. Vet Microbiol. 2010;146:314–319. doi: 10.1016/j.vetmic.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 17.Castle KT, Kosoy M, Lerdthusnee K, Phelan L, Bai Y, Gage KL, Leepitakrat W, Monkanna T, Khlaimanee N, Chandranoi K, Jones JW, Coleman RE. Prevalence and diversity of Bartonella in rodents of northern Thailand: a comparison with Bartonella in rodents from southern China. Am J Trop Med Hyg. 2004;70:429–433. [PubMed] [Google Scholar]
- 18.Parola P, Sanogo OY, Lerdthusnee K, Zeaiter Z, Chauvancy G, Gonzalez JP, Miller RS, Telford SR III, Wongsrichanalai C, Raoult D. Identification of Rickettsia spp. and Bartonella spp. in fleas from the Thai–Myanmar border. Ann N Y Acad Sci. 2003;990:173–181. doi: 10.1111/j.1749-6632.2003.tb07359.x. [DOI] [PubMed] [Google Scholar]
- 19.Foongladda S, Inthawong D, Kositanont U, Gaywee J. Rickettsia, Ehrlichia, Anaplasma, and Bartonella in ticks and fleas from dogs and cats in Bangkok. Vector Borne Zoonotic Dis. 2011;11:1335–1341. doi: 10.1089/vbz.2010.0174. [DOI] [PubMed] [Google Scholar]
- 20.Assarasakorn S, Veir JK, Hawley JR, Brewer MM, Morris AK, Hill AE, Lappin MR. Prevalence of Bartonella species, hemoplasmas, and Rickettsia felis DNA in blood and fleas of cats in Bangkok, Thailand. Res Vet Sci. 2012;93:1213–1216. doi: 10.1016/j.rvsc.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Kabeya H, Colborn JM, Bai Y, Lerdthusnee K, Richardson JH, Maruyama S, Kosoy MY. Detection of Bartonella tamiae DNA in ectoparasites from rodents in Thailand and their sequence similarity with bacterial cultures from Thai patients. Vector Borne Zoonotic Dis. 2010;10:429–434. doi: 10.1089/vbz.2009.0124. [DOI] [PubMed] [Google Scholar]
- 22.Furman DP, Catts EP. Manual of Medical Entomology. Fourth Edition. New York: Cambridge University Press; 1982. pp. 138–157. [Google Scholar]
- 23.Halos L, Jamal T, Vial L, Maillard R, Suau A, Le Menach A, Boulouis HJ, Vayssier-Taussat M. Determination of an efficient and reliable method for DNA extraction from ticks. Vet Res. 2004;35:709–713. doi: 10.1051/vetres:2004038. [DOI] [PubMed] [Google Scholar]
- 24.Billeter SA, Miller MK, Breitschwerdt EB, Levy MG. Detection of two Bartonella tamiae-like sequences in Amblyomma americanum (Acari: Ixodidae) using 16S-23S intergenic spacer region-specific primers. J Med Entomol. 2008;45:176–179. doi: 10.1603/0022-2585(2008)45[176:dotbts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 25.La Scola B, Zeaiter Z, Khamis A, Raoult D. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 2003;11:318–321. doi: 10.1016/s0966-842x(03)00143-4. [DOI] [PubMed] [Google Scholar]
- 26.Bhengsri S, Bagget HC, Peruski LF, Morway C, Bai Y, Fisk TL, Sitdhirasdr A, Maloney SA, Dowell SF, Kosoy M. Bartonella seroprevalence in rural Thailand. Southeast Asian J Trop Med Public Health. 2011;42:687–692. [PubMed] [Google Scholar]