Abstract
Q fever is a widespread zoonosis caused by Coxiella burnetii, an obligate intracellular gram-negative bacterium. The investigation of C. burnetii infection in Zambian livestock was carried out using molecular detection techniques. A total of 489 cattle and 53 goat blood samples were collected from Chama, Chongwe, Monze, and Petauke districts in Zambia. Molecular screening by polymerase chain reaction was performed using C. burnetii-species-specific primers. In total, 38 cattle and 4 goat samples were positive. The prevalence of C. burnetii differed among the four sites, with Chama (Eastern province) recording the highest, although Monze (Southern province) did not record any case of the bacteria. This study reports the first genetic detection of C. burnetii in Zambia.
Introduction
Coxiella burnetii, an obligate intracellular gram-negative bacterium, is the causative agent of Q fever in humans and animals. It causes a variety of symptoms such as acute flu-like symptoms, pneumonia, hepatitis, and chronic endocarditis in humans.1,2 It also causes abortion or infertility in animals.1,2 The disease is a ubiquitous zoonosis with worldwide distribution.1 From spring 2007–2011, a Q fever outbreak of unprecedented scale occurred in the Netherlands, involving 4,108 notified human cases including 24 fatal cases.3 The life cycle of C. burnetii is not fully understood, but humans are considered incidental hosts. The reservoir of Q fever is a wide range of domestic and wild animals and arthropods such as ticks4; of note, domestic ruminants including cattle, goats, and sheep are often infected and serve as main sources of human infections.1,5
The sero-prevalence of Q fever in humans has been reported from several African countries,6,7 including Zambia.8,9 In addition, C. burnetii DNA was detected from febrile patients in the malaria-endemic area in Senegal.10 These data may suggest widespread distribution of Q fever in African countries. Discrimination between malaria, other endemic febrile diseases, and Q fever in affected regions including Africa, is important for disease management and control strategies. Therefore, epidemiological surveillance of C. burnetii, and elucidation of the transmission routes are necessary in these areas.
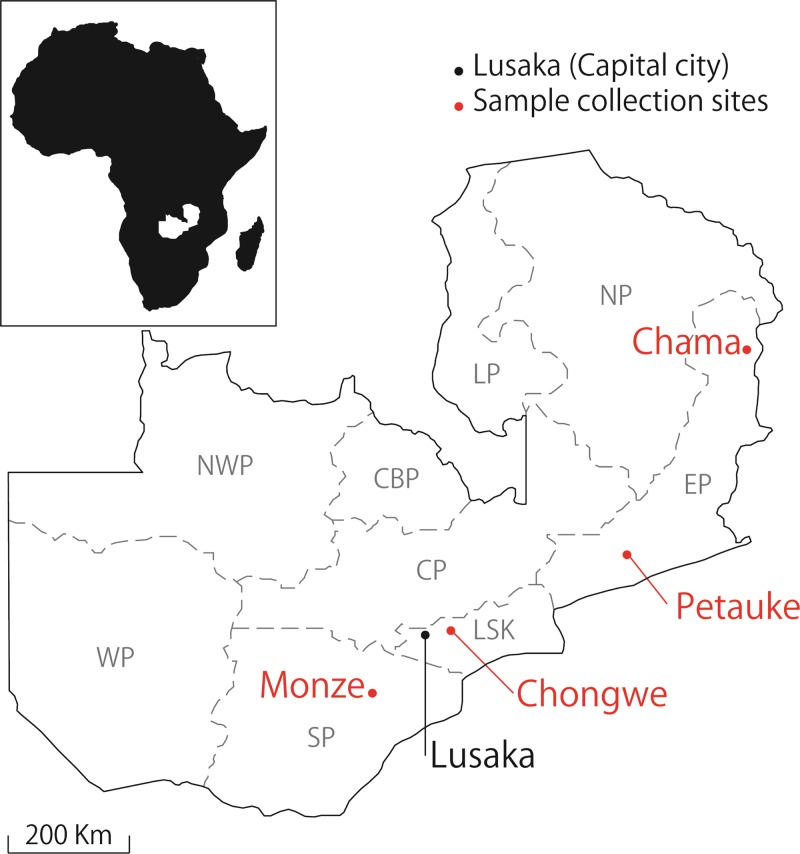
The objective of this study was to investigate the prevalence of C. burnetii in domestic animals in Zambia and to extrapolate the potential infection route to humans. From 2008 to 2010, blood samples were collected from the cattle (Angoni breed) in Chama (N = 295, 11°21′S, 33°16′E), Chongwe (N = 50, 15°33′S, 28°69′E), Monze (N = 80, 16°28′S, 27°49′E), and Petauke (N = 64, 14°24′S, 31°32′E) districts in Zambia (Figure 1). Blood samples were also obtained from goats (Boer breed) in Chama district. In each district, sampling was conducted at 2–7 different sites where the pastured cattle and goats were gathered by the owners. A total of 489 cattle and 53 goat blood samples were collected, from which genomic DNA was extracted using the DNA Isolation Kit for Mammalian Blood (Roche Molecular Biochemical, Boehringer, Germany). The DNA was extracted from 1 mL of EDTA-treated blood and was eluted in a final volume of 200 μL according to the manufacturer's instructions. For the screening of C. burnetii infection, polymerase chain reaction (PCR) was performed with primers designed based on a repetitive transposon-like element of C. burnetii (Trans-PCR).11 The sensitivity and specificity of the assay have been well evaluated.12,13 The PCR reaction was conducted in a final volume of 10 μL, containing 5 μL of KAPA Blood PCR Mix B (Kapa Biosystems, Boston, MA), 0.125 μL of each primer, and 1 μL of template DNA. The PCR conditions were initiated with a 5-min denaturation step at 95°C followed by 40 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min, and final extension step at 72°C for 2 min. The resulting PCR products were electrophoresed on 1% agarose gel, stained with Gel-Red (Biotium, Hayward, CA), and visualized with a UV trans-illuminator.
Figure 1.
Map of Zambia showing its nine provinces: Northern (NP), Eastern (EP), Luapula (LP), Central (CP), Copperbelt (CBP), Lusaka (LSK), Southern (SP), Western (WP), and North-western (NWP) provinces. The Coxiella burnetii samples used in this study were obtained from Monze (SP), Chongwe (LSP), Petauke (EP), and Chama (EP) districts.
Out of 489 cattle samples, Trans-PCR detected 38 (7.8%) C. burnetii-positive samples (Table 1). The prevalence of C. burnetii differed among the four sampling sites, with the highest prevalence observed in Chama (33 of 295, 11.2%), followed by Chongwe (3 of 50, 6.0%), and Petauke (2 of 64, 3.1%). Furthermore, out of the 53 goat samples from Chama, only 4 (7.5%) were positive for C. burnetii (Table 1). On the other hand, all the samples from Monze were negative for C. burnetii.
Table 1.
Prevalence of Coxiella burnetii DNA in Zambian cattle and goats
| Animal | District | Prevalence* |
|---|---|---|
| Goat | Chama | 4/53 (7.5%) |
| Cattle | Chama | 33/295 (11.2%) |
| Chongwe | 3/50 (6.0%) | |
| Monze | 0/80 (0%) | |
| Petauke | 2/64 (3.1%) | |
| Sub-total | 38/489 (7.8%) |
No. of positives/no. of tested samples.
The major infection route of C. burnetii to humans is through the inhalation of aerosol following parturition of an infected animal and the ingestion of contaminated raw milk or milk products.14 This may partially explain why the sero-prevalence of C. burnetii in humans in Zambia was reported to be higher in extensive cattle-breeding areas (Eastern and Western provinces) than less breeding areas (Northern province).9 In our study, the highest prevalence of C. burnetii was obtained in the samples collected from Chama district (Eastern province). Collectively, these data may imply that Eastern province is an endemic area for C. burnetii infection in Zambia. Future studies should be expanded to include the other sampling areas and specimen such as vaginal swab, placenta, and milk, which are likely to contain a higher concentration of C. burnetii than blood samples, and to elucidate the genotypes of C. burnetii circulating in Zambia.6
In conclusion, this is the first report on the genetic detection of C. burnetii in Zambia. People should be aware of the infection of C. burnetii as a cause of non-malarial illness. Further studies should be conducted to assess the potential risk of Q fever in humans.
ACKNOWLEDGMENTS
We thank S. Ando, National Institute of Infectious Diseases, Tokyo, Japan, for providing C. burnetii DNA and K. Hayashida, Research Center for Zoonosis Control, Hokkaido University, for her invaluable suggestions. We also thank the farmers and field veterinary officers in Monze, Chongwe, Petauke, and Chama for their cooperation.
Footnotes
Financial support: This work was financially supported by the program of Funding Research Center for Emerging and Re-emerging Infectious Disease from Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT).
Authors' addresses: Yongjin Qiu and Chihiro Sugimoto, Division of Collaboration and Education, Research Center for Zoonosis Control, Hokkaido University, Kita-ku, Sapporo, Japan, E-mails: yongjin_qiu@czc.hokudai.ac.jp and sugimoto@czc.hokudai.ac.jp. Ryo Nakao, Division of Collaboration and Education, and Division of Bioinformatics, Research Center for Zoonosis Control, Hokkaido University, Sapporo, Japan, E-mail: ryo.nakao@czc.hokudai.ac.jp. Boniface Namangala, Department of Paraclinical studies, School of Veterinary Medicine, University of Zambia, Lusaka, Zambia, E-mail: boniface_1020@yahoo.com.
References
- 1.Marrie TJ. Q fever—a review. Can Vet J. 1990;31:555–563. [PMC free article] [PubMed] [Google Scholar]
- 2.Maurin M, Raoult D. Q fever. Clin Microbiol Rev. 1999;12:518–553. doi: 10.1128/cmr.12.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Loenhout JA, Paget WJ, Vercoulen JH, Wijkmans CJ, Hautvast JL, van der Velden K. Assessing the long-term health impact of Q-fever in The Netherlands: a prospective cohort study started in 2007 on the largest documented Q-fever outbreak to date. BMC Infect Dis. 2012;12:280–285. doi: 10.1186/1471-2334-12-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson HA, Dennis DT, Dasch GA. Q fever. In: Goodman JL, Dennis DT, Sonenshine DE, editors. Tick-Borne Diseases of Humans. Washington, DC: ASM Press; 2005. pp. 328–342. [Google Scholar]
- 5.Woldehiwet Z. Q fever (coxiellosis): epidemiology and pathogenesis. Res Vet Sci. 2004;77:93–100. doi: 10.1016/j.rvsc.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Kelly PJ, Matthewman LA, Mason PR, Raoult D. Q fever in Zimbabwe. A review of the disease and the results of a serosurvey of humans, cattle, goats and dogs. S Afr Med J. 1993;83:21–25. [PubMed] [Google Scholar]
- 7.Dupont HT, Brouqui P, Faugere B, Raoult D. Prevalence of antibodies to Coxiella burnetii, Rickettsia conorii, and Rickettsia typhi in seven African countries. Clin Infect Dis. 1995;21:1126–1133. doi: 10.1093/clinids/21.5.1126. [DOI] [PubMed] [Google Scholar]
- 8.Ghirotti M, Semproni G, De Meneghi D, Mungaba FN, Nannini D, Calzetta G, Paganico G. Sero-prevalences of selected cattle diseases in the Kafue flats of Zambia. Vet Res Commun. 1991;15:25–36. doi: 10.1007/BF00497787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okabayashi T, Hasebe F, Samui KL, Mweene AS, Pandey SG, Yanase T, Muramatsu Y, Ueno H, Morita C. Prevalence of antibodies against spotted fever, murine typhus, and Q fever rickettsiae in humans living in Zambia. Am J Trop Med Hyg. 1999;61:70–72. doi: 10.4269/ajtmh.1999.61.70. [DOI] [PubMed] [Google Scholar]
- 10.Ratmanov P, Bassene H, Fenollar F, Tall A, Sokhna C, Raoult D, Mediannikov O. The correlation of Q fever and Coxiella burnetii DNA in household environments in rural Senegal. Vector Borne Zoonotic Dis. 2013;13:70–72. doi: 10.1089/vbz.2012.1060. [DOI] [PubMed] [Google Scholar]
- 11.Willems H, Thiele D, Frölich-Ritter R, Krauss H. Detection of Coxiella burnetii in cow's milk using the polymerase chain reaction (PCR) Zentralbl Veterinarmed B. 1994;41:580–587. doi: 10.1111/j.1439-0450.1994.tb00267.x. [DOI] [PubMed] [Google Scholar]
- 12.Berri M, Laroucau K, Rodolakis A. The detection of Coxiella burnetii from ovine genital swabs, milk and fecal samples by the use of a single touchdown polymerase chain reaction. Vet Microbiol. 2000;72:285–293. doi: 10.1016/s0378-1135(99)00178-9. [DOI] [PubMed] [Google Scholar]
- 13.Vaidya VM, Malik SV, Kaur S, Kumar S, Barbuddhe SB. Comparison of PCR, immunofluorescence assay, and pathogen isolation for diagnosis of Q fever in humans with spontaneous abortions. J Clin Microbiol. 2008;46:2038–2044. doi: 10.1128/JCM.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fishbein DB, Raoult D. A cluster of Coxiella burnetii infections associated with exposure to vaccinated goats and their unpasteurized dairy products. Am J Trop Med Hyg. 1992;47:35–40. doi: 10.4269/ajtmh.1992.47.35. [DOI] [PubMed] [Google Scholar]