Abstract
This paper will describe the biosynthesis of the thiamin thiazole in Bacillus subtilis and Saccharomyces cerevisiae. The two pathways are quite different: in B. subtilis, the thiazole is formed by an oxidative condensation of glycine, deoxy-D-xylulose- 5-phosphate and a protein thiocarboxylate, while in S. cerevisiae the thiazole is assembled from glycine, NAD and Cys205 of the thiazole synthase.
Keywords: Thiamin biosynthesis, thiazole, suicide enzyme, THI4, ThiG
The major thiamin biosynthetic pathway in bacteria is outlined in Figure 1.(1, 2) In this pathway, glycine 3 undergoes an oxidative condensation with deoxy-D-xylulose- 5-phosphate 5 and ThiS-thiocarboxylate 6, to give the thiazole tautomer 7, which then aromatizes to form carboxythiazole 8.(3, 4) The thiamin pyrimidine 15 is formed by a remarkable rearrangement of AIR 14, an intermediate on the purine pathway.(5) Coupling the thiazole and the pyrimidine, with concomitant decarboxylation, yields thiamin phosphate 2.(6, 7) A final phosphorylation gives thiamin pyrophosphate 1, the biochemically active form of the cofactor.(8)
Figure 1.
The bacterial thiamin biosynthetic pathway.
Our understanding of this biosynthetic pathway is now at an advanced stage. All the biosynthetic genes have been identified and cloned, all of the enzymes have been overexpressed, reconstituted and structurally characterized and mechanisms for all of the biosynthetic reactions, except for the pyrimidine synthase (ThiC) are reasonably clear (1, 5). The entire biosynthetic pathway has been fully reconstituted using pure enzymes. In this lecture, I will describe the biosynthesis of the thiamin thiazole in B. subtilis and compare this pathway with the very different thiazole biosynthesis recently elucidated in S. cerevisiae.
Thiamin thiazole biosynthesis in B. subtilis
Each of the steps involved in the assembly of the thiamin thiazole in bacteria will be described in the following sections.
Glycine Oxidation
The ThiO gene product encodes a flavin-dependent glycine oxidase that catalyzes the oxidation of glycine 3 to the glycine imine 4 (9). In the absence of the other thiazole biosynthetic enzymes, the glycine imine is hydrolyzed to glyoxal.
The structure of this enzyme, with N-acetyl glycine bound at the active site, has been determined (PDB code = 1NG3). This structure and studies with substrate analogs are consistent with a hydride transfer mechanism for glycine oxidation (9).
In anaerobes, the glycine imine is formed from tyrosine in a reaction catalyzed by ThiH, a radical SAM enzyme (10–12).
ThiS-thiocarboxylate Formation
The chemistry involved in the formation of ThiS-thiocarboxylate is outlined in Figure 1. Activation of ThiS-COOH 9, at its carboxy terminus, by adenylation, gives 10 which then acylates the IscS persulfide to give 13. Reduction of 13, by DTT in the reconstitution reaction mixture, gives the ThiS thiocarboxylate 6 (13, 14). The biochemical reduction of 13 is not yet understood. In some bacteria, an additional protein (ThiI) mediates the sulfur transfer to 10.(15) ThiS-thiocarboxylate 6 can also be efficiently synthesized by treating intein-activated ThiS-COOH with ammonium sulfide (16).
The structures of the ThiF/ThiS complex and the ThiF/ATP complex have been determined (17, 18) (PDB codes = 1ZUD and 1ZFN). The IscS protein probably does not form a specific complex with ThiS/ThiF because all four IscS paralogs in B. subtilis are competent persulfide donors. (14, 19)
Protein thiocarboxylates as sulfide carriers in other biosynthetic pathways
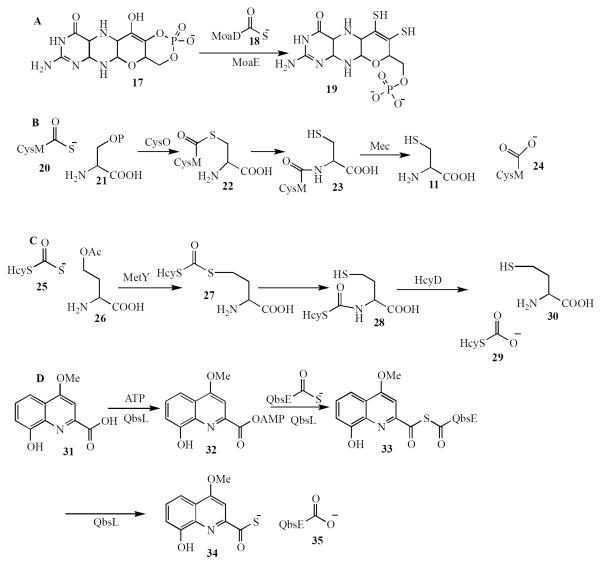
Protein thiocarboxylates have now been found to play a role as sulfide carriers in several other biosynthetic pathways and sequence analysis suggests that this strategy may be quite general (Figure 2).
Figure 2.
Four additional examples of protein-thiocarboxylate-dependent biosynthetic pathways. A) Molybdopterin biosynthesis in bacteria, B) Cysteine biosynthesis in Mycobacterium tuberculosis, C) Homocysteine biosynthesis in Wolinella succinogenes, D) Thioquinolobactin biosynthesis in Pseudomonas fluorescens.
In molybdopterin biosynthesis, MoaE catalyzes the transfer of sulfide from MoaD-thiocarboxylate to give 19 (Figure 2A)(20, 21). A protein thiocarboxylate dependent cysteine biosynthetic pathway has been found in M. tuberculosis (Figure 2B). In this pathway, CysM thiocarboxylate reacts with phosphoserine 21 in a PLP-mediated reaction to form thioester 22. This then undergoes an N/S acyl shift to give 23 followed by release of cysteine in a hydrolysis reaction catalyzed by the Mec protease (22–26). A closely related pathway for the biosynthesis of homocysteine was discovered in Wolinella succinogenes (Figure 2C). In this pathway HcyS-thiocarboxylate 25 adds to O-acetyl homoserine 26 to give thioester 27. An N/S acyl shift to give 28 followed by HcyD-catalyzed amide hydrolysis generates homocysteine 30 (27, 28). A fourth example is found in the biosynthesis of the siderophore thioquinolobactin 34 (Figure 2D). In this pathway, QbsE thiocarboxylate forms a mixed thioanhydride 33 with quinolobactin 31. Hydrolysis of 33 generates the siderophore 34.(29, 30) A reagent for the sensitive detection of protein thiocarboxylates in a proteome, that uses a click reaction between the protein thiocarboxylate and a fluorophore-tagged sulfonyl azide, has been described. (31)
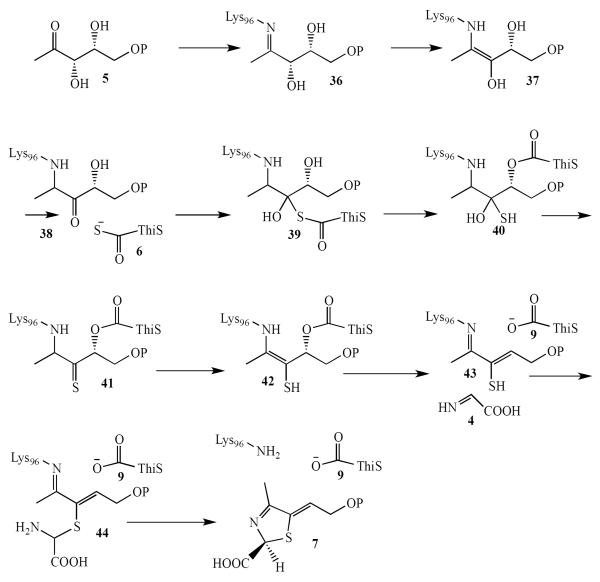
Formation of the thiazole tautomer 7
The bacterial thiazole synthase catalyzes the condensation of DXP (5), ThiS-COSH 6 and the glycine imine 4 to form the thiazole tautomer 7 (Figure 1)(3). A mechanistic proposal for this reaction is outlined in Figure 3. In this mechanism, DXP 5 forms an imine with Lysine 96 of the thiazole synthase. Tautomerization to 38 followed by thiocarboxylate addition gives 39. An O/S acyl shift followed by loss of water generates thioketone 41. Tautomerization of 41, followed by loss of ThiS-COOH generates 43. Addition of the glycine imine 4 followed by transimination gives the thiazole tautomer 7.
Figure 3.
Mechanistic proposal for the formation of the thiazole tautomer 7.
In support of this mechanism, enzyme-catalyzed exchange of the DXP carbonyl oxygen has been observed and the DXP/K96 imine has been trapped by borohydride reduction and characterized by MS analysis. Intermediate 37 is supported by the observation of enzyme-catalyzed exchange of the C3 proton of DXP. The unanticipated O/S acyl shift to give 40 is supported by the observation of oxygen incorporation from DXP and not the buffer into the nascent ThiS-COOH. Thioenol 42 has also been trapped and characterized by MS analysis and the final product 7 has been fully characterized by spectroscopic analysis (3, 14).
The structure of the ThiG/ThiS complex, with phosphate bound at the active site, has been determined (PDB code = 1TYG). In this structure the phosphate and Lys96 define the DXP binding site, which suggests that Glu98 and Asp182 are also likely to play a role in the catalysis of thiazole formation (32).
Thiazole tautomerase
The thiazole tautomer 7 is surprisingly stable and the aromatization reaction to produce the thiazole 8 requires enzymatic catalysis. In B. subtilis, the TenI protein has recently been identified as the thiazole tautomerase.
The structure of the enzyme product 8 complex has been determined (PDB code = 3QH2). A model of the enzyme substrate complex generated from this structure suggests that His122 mediates the deprotonation at C2 and that the substrate phosphate group functions as the proton donor for the exocyclic double bond protonation (4). TenI shows high sequence similarity to thiamin phosphate synthase and the two enzymes are frequently incorrectly assigned in genome annotation.
Thiamin thiazole biosynthesis in S. cerevisiae
The thiamin biosynthetic pathway in S. cerevisiae is outlined in Figure 4.(33) The biosynthesis of the thiazole and the pyrimidine heterocycles (5 &10) occurs by very different chemistry from that used for the bacterial biosynthesis. Labeling studies have demonstrated that the thiazole is formed from an unidentified C5 carbohydrate, glycine 3 and cysteine 11(34–36) and that the pyrimidine 10 is formed from histidine 48 and PLP 49 (37–39). Thiamin biosynthesis in yeast requires fewer enzymes than the bacterial pathway. The biosynthesis of the thiazole requires only one protein (THI4p) in contrast to the bacterial pathway, which requires six (ThiOFSG, IscS and TenI).
Figure 4.
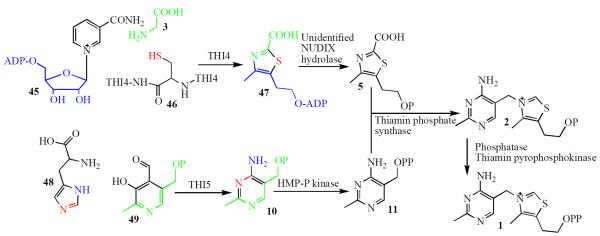
Thiamin pyrophosphate biosynthesis in S. cerevisiae.
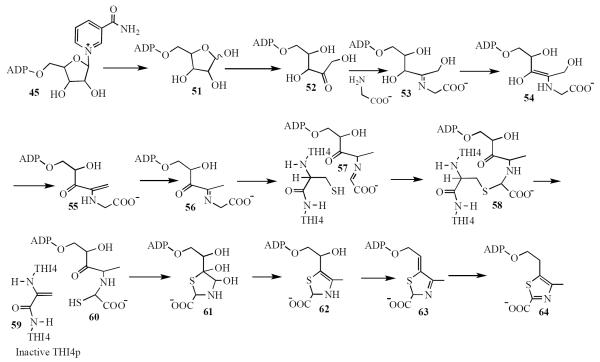
All attempts to reconstitute the THI4p-catalyzed reaction, using a variety of C5 carbohydrates, initially failed. However, a breakthrough was achieved by the detection of three metabolites (56, 63 and 64 in Figure 5) released from the protein by heat denaturation (40, 41).
Figure 5.
Mechanistic proposal for the THI4 mediated formation of ADP-thiazole 64.
The identification of product 64 demonstrated that complete thiazole biosynthesis could be achieved using THI4p expressed in E. coli. In addition, this structure demonstrated that the thiazole was adenylated, suggesting that NAD 45, and not a simple pentose, might be the donor of the C5 carbohydrate. Initial attempts to detect Thi4p-catalyzed modification of NAD failed. However, after the structure of THI4p was determined (PDB code = 3FPZ)(42) it was possible to prepare an active site mutant (C204A) that was free of the tightly bound metabolites 56, 63 and 64 (42). This form of the enzyme catalyzed the conversion of NAD 45 and glycine 3 to 56 via intermediates 51 and 52 and confirmed NAD at the C5 carbohydrate donor. (43)
The discovery that metabolite-free THI4p could be isolated when the E. coli overexpression strain was grown at low iron concentrations provided a source of native enzyme with an unoccupied active site. Treatment of NAD and glycine with this form of the enzyme generated intermediate 56. Addition of Fe(III) to this reaction mixture resulted in the transfer of sulfide from Cys205 of THI4p to generate 63 and 64. MS analysis of the protein in this reaction mixture confirmed Cys205 as the sulfide donor.(44) These observations led to the mechanistic proposal outlined in Figure 5.
In this proposal hydrolysis of the N-glycosyl bond of NAD 45 gives 51. Ring opening, tautomerization and imine formation give 53. Tautomerization, loss of water and a second tautomerization generates compound 56, the most labile of the three intermediates released in the heat denaturation experiment. Tautomerization to 57 followed by sulfide transfer from Cys205 of the THI4 protein gives 60. Cyclization and two dehydrations gives the thiazole tautomer 63, the second of the heat released metabolites. A final tautomerization completes the thiazole formation. Our mechanism suggests that the THI4 protein may be a single turnover enzyme. This was confirmed by demonstrating a 1:1 ratio of THI4p to thiamin produced.
In conclusion, we have explored here the mechanistic biochemistry of thiamin thiazole biosynthesis in B. subtilis as a representative prokaryote and in S. cerevisiae as a representative eukaryote. The biosynthetic routes are quite different between the two systems and the reasons for these differences are not yet known. The mechanism of thiazole biosynthesis in bacteria is at an advanced stage, while our understanding of the mechanism of thiazole biosynthesis in yeast is still growing with many unanswered questions remaining. We have not yet identified most of the residues involved in catalyzing the conversion of 45 to 64. We also do not yet understand the role of iron in the sulfur transfer or the physiological role of inactive THI4p.
Acknowledgements
The research described in this lecture was a collaborative effort between the Begley, Ealick and McLafferty groups. We would like to thank the capable graduate students and postdoctoral associates who carried out all of the experimental work. Begley Group: Dinuka Abeydeera, Alison Backstrom, Kristin Burns, Abhiskek Chatterjee, Pieter Dorrestein, Amrita Hazra, Amy Godert, Neil Kelleher, Cynthia Kinsland, Kalyan Krishnamoorthy, Rung-Yi Lai, Sean O'Leary, Joo-Heon Park and Sean Taylor. Ealick Group: Jessica Chiu, Ying Han, Chris Jurgenson, Christopher Lehmann, Ethan Settembre, Tim Tran and Yang Zhang. McLafferty Group: Sabine Baumgart, Ying Ge, Mi Jin, Neil Kelleher and Huili Zhai. Their individual accomplishments are listed in the references. The thiamin project was supported by the Robert A. Welch Foundation (A-0034) and NIH Grants DK44083 to T.P.B. DK67081 to S.E.E. and GM16609 to F.W.M.
References
- 1.Jurgenson CT, Begley TP, Ealick SE. The structural and biochemical foundations of thiamin biosynthesis. Annual review of biochemistry. 2009;78:569–603. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begley TP, Chatterjee A, Hanes JW, Hazra A, Ealick SE. Cofactor biosynthesis--still yielding fascinating new biological chemistry. Current Opinion in Chemical Biology. 2008;12:118–125. doi: 10.1016/j.cbpa.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazra A, Chatterjee A, Begley TP. Biosynthesis of the Thiamin Thiazole in Bacillus subtilis: Identification of the Product of the Thiazole Synthase-Catalyzed Reaction. Journal of the American Chemical Society. 2009;131:3225–3229. doi: 10.1021/ja806752h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazra AB, Han Y, Chatterjee A, Zhang Y, Lai R-Y, Ealick SE, Begley TP. A missing enzyme in thiamin thiazole biosynthesis: Identification of TenI as a thiazole tautomerase. Journal of the American Chemical Society. 2011;133:9311–9319. doi: 10.1021/ja1110514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee A, Hazra AB, Abdelwahed S, Hilmey DG, Begley TP. A “Radical Dance” in Thiamin Biosynthesis: mechanistic Analysis of the Bacterial Hydroxymethylpyrimidine Phosphate Synthase. Angewandte Chemie, International Edition. 2010;49:8653–8656. doi: 10.1002/anie.201003419. S8653/8651-S8653/8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peapus DH, Chiu H-J, Campobasso N, Reddick JJ, Begley TP, Ealick SE. Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase. Biochemistry. 2001;40:10103–10114. doi: 10.1021/bi0104726. [DOI] [PubMed] [Google Scholar]
- 7.Hanes JW, Ealick SE, Begley TP. Thiamin Phosphate Synthase: The Rate of Pyrimidine Carbocation Formation. Journal of the American Chemical Society. 2007;129:4860–4861. doi: 10.1021/ja0679634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCulloch KM, Kinsland C, Begley TP, Ealick SE. Structural Studies of Thiamin Monophosphate Kinase in Complex with Substrates and Products. Biochemistry. 2008;47:3810–3821. doi: 10.1021/bi800041h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Settembre EC, Dorrestein PC, Park J-H, Augustine AM, Begley TP, Ealick SE. Structural and Mechanistic Studies on ThiO, a Glycine Oxidase Essential for Thiamin Biosynthesis in Bacillus subtilis. Biochemistry. 2003;42:2971–2981. doi: 10.1021/bi026916v. [DOI] [PubMed] [Google Scholar]
- 10.Challand MR, Martins FT, Roach PL. Catalytic Activity of the Anaerobic Tyrosine Lyase Required for Thiamine Biosynthesis in Escherichia coli. Journal of Biological Chemistry. 2010;285:5240–5248. doi: 10.1074/jbc.M109.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kriek M, Martins F, Challand MR, Croft A, Roach PL. Thiamine biosynthesis in Escherichia coli: identification of the intermediate and by-product derived from tyrosine. Angewandte Chemie, International Edition. 2007;46:9223–9226. doi: 10.1002/anie.200702554. [DOI] [PubMed] [Google Scholar]
- 12.Kriek M, Martins F, Leonardi R, Fairhurst SA, Lowe DJ, Roach PL. Thiazole Synthase from Escherichia coli: An investigation of the substates and purified proteins required for activity in vitro. Journal of Biological Chemistry. 2007;282:17413–17423. doi: 10.1074/jbc.M700782200. [DOI] [PubMed] [Google Scholar]
- 13.Taylor SV, Kelleher NL, Kinsland C, Chiu H-J, Costello CA, Backstrom AD, McLafferty FW, Begley TP. Thiamin biosynthesis in Escherichia coli. Identification of the thiocarboxylate as the immediate sulfur donor in the thiazole formation. Journal of Biological Chemistry. 1998;273:16555–16560. doi: 10.1074/jbc.273.26.16555. [DOI] [PubMed] [Google Scholar]
- 14.Dorrestein PC, Zhai H, McLafferty FW, Begley TP. The biosynthesis of the thiazole phosphate moiety of thiamin: the sulfur transfer mediated by the sulfur carrier protein ThiS. Chemistry & Biology. 2004;11:1373–1381. doi: 10.1016/j.chembiol.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Gomez NC, Palmer LD, Vivas E, Roach PL, Downs DM. The rhodanese domain of ThiI is both necessary and sufficient for synthesis of the thiazole moiety of thiamine in Salmonella enterica. Journal of Bacteriology. 2011;193:4582–4587. doi: 10.1128/JB.05325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinsland C, Taylor SV, Kelleher NL, McLafferty FW, Begley TP. Overexpression of recombinant proteins with a C-terminal thiocarboxylate: implications for protein semisynthesis and thiamin biosynthesis. Protein Science. 1998;7:1839–1842. doi: 10.1002/pro.5560070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann C, Begley TP, Ealick SE. Structure of the Escherichia coli ThiS-ThiF Complex, a Key Component of the Sulfur Transfer System in Thiamin Biosynthesis. Biochemistry. 2006;45:11–19. doi: 10.1021/bi051502y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duda DM, Walden H, Sfondouris J, Schulman BA. Structural analysis of Escherichia coli ThiF. Journal of Molecular Biology. 2005;349:774–786. doi: 10.1016/j.jmb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Park J-H, Dorrestein PC, Zhai H, Kinsland C, McLafferty FW, Begley TP. Biosynthesis of the Thiazole Moiety of Thiamin Pyrophosphate (Vitamin B1) Biochemistry. 2003;42:12430–12438. doi: 10.1021/bi034902z. [DOI] [PubMed] [Google Scholar]
- 20.Wuebbens MM, Rajagopalan KV. Mechanistic and Mutational Studies of Escherichia coli Molybdopterin Synthase Clarify the Final Step of Molybdopterin Biosynthesis. Journal of Biological Chemistry. 2003;278:14523–14532. doi: 10.1074/jbc.M300453200. [DOI] [PubMed] [Google Scholar]
- 21.Rudolph MJ, Wuebbens MM, Turque O, Rajagopalan KV, Schindelin H. Structural studies of molybdopterin synthase provide insights into its catalytic mechanism. Journal of Biological Chemistry. 2003;278:14514–14522. doi: 10.1074/jbc.M300449200. [DOI] [PubMed] [Google Scholar]
- 22.Burns KE, Baumgart S, Dorrestein PC, Zhai H, McLafferty FW, Begley TP. Reconstitution of a new cysteine biosynthetic pathway in Mycobacterium tuberculosis. Journal of the American Chemical Society. 2005;127:11602–11603. doi: 10.1021/ja053476x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Leary SE, Jurgenson CT, Ealick SE, Begley TP. O-Phospho-L-serine and the Thiocarboxylated Sulfur Carrier Protein CysO-COSH Are Substrates for CysM, a Cysteine Synthase from Mycobacterium tuberculosis. Biochemistry. 2008;47:11606–11615. doi: 10.1021/bi8013664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnell R, Schneider G. Structural enzymology of sulphur metabolism in Mycobacterium tuberculosis. Biochemical and Biophysical Research Communications. 2010;396:33–38. doi: 10.1016/j.bbrc.2010.02.118. [DOI] [PubMed] [Google Scholar]
- 25.Jurgenson CT, Burns KE, Begley TP, Ealick SE. Crystal Structure of a Sulfur Carrier Protein Complex Found in the Cysteine Biosynthetic Pathway of Mycobacterium tuberculosis. Biochemistry. 2008;47:10354–10364. doi: 10.1021/bi800915j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aagren D, Schnell R, Oehlmann W, Singh M, Schneider G. Cysteine Synthase (CysM) of Mycobacterium tuberculosis Is an O-Phosphoserine Sulfhydrylase: evidence for an alternative cysteine biosynthesis pathway in mycobacteria. Journal of Biological Chemistry. 2008;283:31567–31574. doi: 10.1074/jbc.M804877200. [DOI] [PubMed] [Google Scholar]
- 27.Krishnamoorthy K, Begley TP. Protein Thiocarboxylate-Dependent Methionine Biosynthesis in Wolinella succinogenes. Journal of the American Chemical Society. 2011;133:379–386. doi: 10.1021/ja107424t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran TH, Krishnamoorthy K, Begley TP, Ealick SE. A novel mechanism of sulfur transfer catalyzed by O-acetylhomoserine sulfhydrylase in the methionine-biosynthetic pathway of Wolinella succinogenes. Acta Crystallographica, Section D: Biological Crystallography. 2011;D67:831–838. doi: 10.1107/S0907444911028010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godert AM, Jin M, McLafferty FW, Begley TP. Biosynthesis of the thioquinolobactin siderophore: An interesting variation on sulfur transfer. Journal of Bacteriology. 2007;189:2941–2944. doi: 10.1128/JB.01200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthijs S, Baysse C, Koedam N, Tehrani Kourosh A, Verheyden L, Budzikiewicz H, Schafer M, Hoorelbeke B, Meyer J-M, De Greve H, Cornelis P. The Pseudomonas siderophore quinolobactin is synthesized from xanthurenic acid, an intermediate of the kynurenine pathway. Molecular microbiology. 2004;52:371–384. doi: 10.1111/j.1365-2958.2004.03999.x. [DOI] [PubMed] [Google Scholar]
- 31.Krishnamoorthy K, Begley TP. Reagent for the Detection of Protein Thiocarboxylates in the Bacterial Proteome: Lissamine Rhodamine B Sulfonyl Azide. Journal of the American Chemical Society. 2010;132:11608–11612. doi: 10.1021/ja1034107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Settembre EC, Dorrestein PC, Zhai H, Chatterjee A, McLafferty FW, Begley TP, Ealick SE. Thiamin biosynthesis in Bacillus subtilis: Structure of the thiazole synthase/sulfur carrier protein complex. Biochemistry. 2004;43:11647–11657. doi: 10.1021/bi0488911. [DOI] [PubMed] [Google Scholar]
- 33.Nosaka K. Recent progress in understanding thiamin biosynthesis and its genetic regulation in Saccharomyces cerevisiae. Applied Microbiology and Biotechnology. 2006;72:30–40. doi: 10.1007/s00253-006-0464-9. [DOI] [PubMed] [Google Scholar]
- 34.White RL, Spenser ID. Thiamin biosynthesis in Saccharomyces cerevisiae. Origin of carbon-2 of the thiazole moiety. Biochemical Journal. 1979;179:315–325. doi: 10.1042/bj1790315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White RL, Spenser ID. Biosynthesis of vitamin B1 in yeast. Origin of the thiazole unit. Journal of the American Chemical Society. 1979;101:5102–5104. [Google Scholar]
- 36.White RL, Spenser ID. Thiamin biosynthesis in yeast. Origin of the five-carbon unit of the thiazole moiety. Journal of the American Chemical Society. 1982;104:4934–4943. [Google Scholar]
- 37.Ishida S, Tazuya-Murayama K, Kijima Y, Yamada K. The direct precursor of the pyrimidine moiety of thiamin is not urocanic acid but histidine in Saccharomyces cerevisiae. Journal of Nutritional Science and Vitaminology. 2008;54:7–10. doi: 10.3177/jnsv.54.7. [DOI] [PubMed] [Google Scholar]
- 38.Tazuya K, Azumi C, Yamada K, Kumaoka H. Pyrimidine moiety of thiamin is biosynthesized from pyridoxine and histidine in Saccharomyces cerevisiae. Biochemistry and Molecular Biology International. 1995;36:883–888. [PubMed] [Google Scholar]
- 39.Himmeldirk K, Sayer BG, Spenser ID. Comparative Biogenetic Anatomy of Vitamin B1: A 13C NMR Investigation of the Biosynthesis of Thiamin in Escherichia coli and in Saccharomyces cerevisiae. Journal of the American Chemical Society. 1998;120:3581–3589. [Google Scholar]
- 40.Chatterjee A, Jurgenson CT, Schroeder FC, Ealick SE, Begley TP. Thiamin Biosynthesis in Eukaryotes: Characterization of the Enzyme-Bound Product of Thiazole Synthase from Saccharomyces cerevisiae and Its Implications in Thiazole Biosynthesis. Journal of the American Chemical Society. 2006;128:7158–7159. doi: 10.1021/ja061413o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chatterjee A, Schroeder FC, Jurgenson CT, Ealick SE, Begley TP. Biosynthesis of the Thiamin-Thiazole in Eukaryotes: Identification of a Thiazole Tautomer Intermediate. Journal of the American Chemical Society. 2008;130:11394–11398. doi: 10.1021/ja802140a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jurgenson CT, Chatterjee A, Begley TP, Ealick SE. Structural Insights into the Function of the Thiamin Biosynthetic Enzyme Thi4 from Saccharomyces cerevisiae. Biochemistry. 2006;45:11061–11070. doi: 10.1021/bi061025z. [DOI] [PubMed] [Google Scholar]
- 43.Chatterjee A, Jurgenson CT, Schroeder FC, Ealick SE, Begley TP. Biosynthesis of Thiamin Thiazole in Eukaryotes: Conversion of NAD to an Advanced Intermediate. Journal of the American Chemical Society. 2007;129:2914–2922. doi: 10.1021/ja067606t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee A, Abeydeera ND, Bale S, Pai P-J, Dorrestein Pieter C, Russell David H, Ealick Steven E, Begley Tadhg P. Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature. 2011;478:542–546. doi: 10.1038/nature10503. [DOI] [PMC free article] [PubMed] [Google Scholar]