Summary
During meiosis in yeast, global splicing efficiency increases and then decreases. Here we provide evidence that splicing improves due to reduced competition for the splicing machinery. The timing of this regulation corresponds to repression and reactivation of ribosomal protein genes (RPGs) during meiosis. In vegetative cells RPG repression by rapamycin treatment also increases splicing efficiency. Down-regulation of the RPG-dedicated transcription factor gene IFH1 genetically suppresses two spliceosome mutations prp11-1 and prp4-1, and globally restores splicing efficiency in prp4-1 cells. We conclude that the splicing apparatus is limiting and pre-mRNAs compete. Splicing efficiency of a pre-mRNA therefore depends not just on its own concentration and affinity for limiting splicing factor(s) but also on those of competing pre-mRNAs. Competition between RNAs for limiting RNA processing factors appears to be a general condition in eukaryotic cells important for function of a variety of post-transcriptional control mechanisms including miRNA repression, polyadenylation and splicing.
Introduction
Pre-mRNA splicing is a fundamental step of eukaryotic gene expression. It can vary in complexity from removal of a single intron to elaborate patterns of alternative splicing that create multiple distinct mRNAs. This complex set of mRNAs diversifies the functionalities of proteins that can be produced from a gene. Alternative splicing patterns arise from differences in key pre-mRNA features such as splice site strength (Roca et al., 2005; Yeo and Burge, 2004), secondary structure (Hiller et al., 2007; Howe and Ares, 1997; Kreahling and Graveley, 2005; Plass et al., 2012; Shepard and Hertel, 2008), or transcription elongation rates (de la Mata et al., 2003; Howe et al., 2003; Kornblihtt, 2005; Roberts et al., 1998), as well as to transacting splicing factors that bind pre-mRNA to differentially enhance or repress spliceosome recruitment (Black, 2003; Nilsen and Graveley, 2010). The regulation of alternative splicing is generally attributed to the changing activities of trans-acting splicing factors that control the likelihood of local spliceosome assembly.
Recent studies have attempted to capture the regulatory networks for individual splicing factors, usually by depleting or overexpressing a specific splicing factor and measuring changes in alternative splicing across the genome. Combining analyses of the global differences in tissue-specific alternative splicing (e. g., Barbosa-Morais et al., 2012; Merkin et al., 2012; Pan et al., 2008; Pan et al., 2004; Sugnet et al., 2006; Wang et al., 2008), tissue-specific splicing factor expression (e. g., Buckanovich et al., 1993; Calarco et al., 2009; Jin et al., 2003; Markovtsov et al., 2000; Underwood et al., 2005; Warzecha et al., 2009), and changes in splicing factor expression and splicing during differentiation (e. g., Boutz et al., 2007; Gabut et al., 2011; Kalsotra et al., 2008) reveals that alternative splicing is deeply integrated into the gene expression programs that define cell identity and state. To understand gene expression, splicing regulatory networks must be connected with transcriptional and post-transcriptional regulatory networks (reviewed in Kalsotra and Cooper, 2011) such as those of miRNAs, so the contribution of splicing regulation to a change in cell identity or state can be understood. A largely ignored aspect of splicing regulation concerns systems-level accounting of substrate concentrations and availability of required factors. Recent reports suggest competition phenomena in splicing (Berg et al., 2012; Du et al., 2010; Kaida et al., 2010; Kanadia et al., 2003; Yin et al., 2012), indicating that splicing may also be regulated by changes in competition for a fixed level of factor activity.
In a previous study of meiosis in Saccharomyces cerevisiae, we identified relationships between two transcriptional regulatory networks and the Mer1 splicing regulatory network, and examined the roles of the four target transcripts controlled by the Mer1 splicing factor (Munding et al., 2010). We also observed a general increase in splicing efficiency during meiosis (see also Juneau et al., 2007) that we could not assign to any particular trans-acting factor. Here we identify the molecular basis for this improvement and provide evidence that the global increase in splicing is due to relief of competition for the splicing apparatus that occurs during the repression of ribosomal protein genes (RPGs) early in meiosis. This phenomenon is not restricted to meiosis since blocking RPG transcription with rapamycin in vegetative cells also improves splicing. Down-regulating transcription of RPGs suppresses temperature sensitive (ts) growth of the prp4-1 and prp11-1 spliceosome mutations, and rescues splicing defects for nearly all intron-containing genes. These results imply that competition for a limiting splicing machinery can be exploited to control splicing of less competitive substrates through transcriptional control of the overall substrate pool.
Results
A global increase in splicing during meiosis
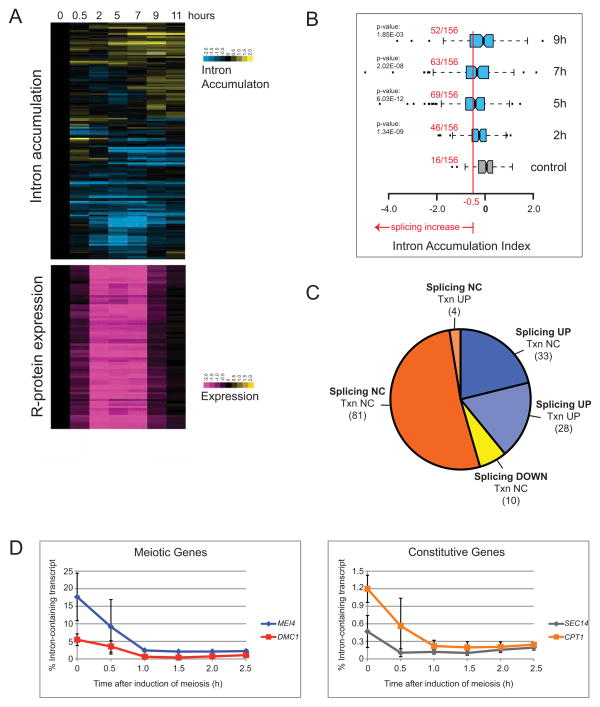
Splicing of numerous meiosis-specific transcripts improves early in meiosis (Juneau et al., 2007; Munding et al., 2010), including four that depend on the meiosis-specific splicing factor Mer1 (Cooper et al., 2000; Davis et al., 2000; Engebrecht et al., 1991; Munding et al., 2010; Nakagawa and Ogawa, 1999). In our previous study, strain SK1 was induced to enter a rapid, synchronous meiosis and RNA was analyzed on splicing-sensitive microarrays (Munding et al., 2010). In addition to meiotic transcripts, we noticed that constitutively expressed transcripts also showed improved splicing. We detect improved splicing by a decrease in Intron Accumulation Index (IAI, a measure of the change in ratios of intron signal to exon 2 signal between two samples, Clark et al., 2002). Measurement of splicing efficiency for genes undergoing transcriptional repression is confounded by the rapid loss of measurable pre-mRNA. For this reason, we examined the 156 intron-containing genes (ICGs) whose expression does not decrease more than 2-fold during mid-meiosis (55% of total ICGs; Fig 1). Splicing improves during mid-meiosis and then declines (Fig 1A, blue indicates reduced IAI, interpreted as improved splicing, data in Table S1).
Figure 1. Splicing improves globally during mid-meiosis.
(A) Top Panel: Changes in splicing during the meiotic time course as represented by Intron Accumulation Indexes. Increased intron accumulation (yellow) represents a decrease in splicing, while decreased intron accumulation (blue) indicates an increase in splicing. See Table S1 for data file. Bottom Panel: Changes in RPG gene expression during the meiotic time course. Purple represents a decrease in gene expression. (B) Distribution of intron accumulation indexes from the microarray data at 2, 5, 7, and 9h meiotic time points relative to the zero time point, and a control distribution from self comparison of replicates (see Experimental Procedures). Red line marks 40% increase in splicing efficiency (IAI < −0.5) used as a threshold for significant splicing change. Numbers in red indicate the fraction of events in each distribution that exceeded the threshold. P-values are derived from a one-tailed t-test comparison of the individual 2, 5, 7, or 9h distributions to the control. (C) Classification of splicing changes at mid-meiotic time points (2, 5, and 7 h) for the 156 events whose expression does not decrease more than 2-fold during mid-meiosis. Bold letters indicate splicing change. “NC” indicates no change. “Txn UP” indicates genes that are transcriptionally induced ≥ 2-fold during mid-meiosis. “Txn NC” indicates genes whose expression changes ≤ 2-fold during mid-meiosis. Numbers in parentheses indicates number of genes in each category. (D) RT-qPCR measurement of percent of intron-containing transcript at the indicated time after induction of meiosis for two meiosis-specific genes (left panel) and two constitutively expressed genes (right panel). Error bars represent ± 1SD. See also Table S1.
To determine a threshold for calling a change in splicing efficiency, we assessed noise in the data by estimating variation in the IAI distribution between replicate samples that should not show splicing changes (see Experimental Procedures, Fig 1B, control distribution, Table S1). We compared the distribution of IAI changes between time zero and the indicated time point for the set of 156 IGCs to this control (background) distribution (Fig 1B). Splicing globally increases in mid-meiosis, peaking at 5 hrs. Of the 156 genes 61 (39%) improve in splicing efficiency by at least 1.4-fold at two of three mid-meiotic time points (2h, 5h, or 7h, Fig 1C). Among the genes whose splicing improves during mid-meiosis, most (48/61) are constitutively expressed without known meiosis-specific functions (Fig 1C). Only a few genes (10/156, 6%) appear to decrease more than 1.4 fold in splicing efficiency more than 1.4 fold, about as expected by chance given the control distribution (Fig 1B, C). We validated improved splicing for four genes by RT-qPCR (Fig 1D). We conclude that splicing for both meiotic and constitutively expressed ICGs globally increases during mid-meiosis. We hypothesize that a splicing regulatory mechanism not specifically restricted to meiotic transcripts is active during mid-meiosis to activate splicing globally.
Splicing is less efficient when ribosomal protein genes are expressed
Meiosis in yeast is triggered in part by nutrient signaling (Mitchell, 1994; Neiman, 2011), which also leads to transcriptional repression of RPGs (Chu et al., 1998; Gasch et al., 2000; Munding et al., 2010; Primig et al., 2000; Warner, 1999). RPGs represent the largest functional class of ICGs in S. cerevisiae (101 of 293 ICGs are RPGs). Given their high expression, RPG pre-mRNAs comprise ~90% of the splicing substrates in a vegetative cell (Ares et al., 1999; Lopez and Seraphin, 1999; Warner, 1999). After their repression early in meiosis, RPGs are induced in late meiosis (Chu et al., 1998; Munding et al., 2010; Primig et al., 2000), even though the starvation conditions continue. We wondered whether the increase in splicing during meiosis might be due to the reduction of RPG pre-mRNAs that normally occupy the spliceosome during vegetative growth. This idea is consistent with the timing of both improved splicing during RPG repression early in meiosis, and loss of efficient splicing during RPG induction at about 9 hours (Fig 1A, B). Based on this, we tested the hypothesis that RPG expression reduces the splicing of other pre-mRNAs.
We first asked whether splicing of meiotic transcripts normally only expressed in the absence of RPG expression, is less efficient during vegetative growth when RPGs are highly expressed. During vegetative growth, meiotic genes are repressed by Ume6 (Mitchell, 1994; Munding et al., 2010; Strich et al., 1994; Williams et al., 2002), thus we evaluated splicing in vegetative ume6Δ cells, where derepressed meiotic genes and RPGs are simultaneously expressed (Fig 2A). Transcripts from SPO22, MEI4, and PCH2 are highly expressed and efficiently spliced during meiosis (Fig 2A, lanes 1, 4, 7), and are not expressed in wild type vegetative cells (Fig 2A, lanes 2, 5, 8). Deletion of UME6 in vegetative cells allows expression and some splicing of SPO22, MEI4, and PCH2 (Fig 2A, lanes 3, 6, 9), however splicing is less efficient in vegetative cells where RPGs are expressed. Quantification confirms that splicing is reduced by 25–45% during vegetative growth as compared to mid-meiosis (Fig 2B).
Figure 2. Splicing of meiotic transcripts is more efficient during meiosis than during vegetative growth.
(A) Expression and splicing of meiotic transcripts SPO22, MEI4, and PCH2 in wild type (+) meiotic (Meio) and vegetative cells (Veg) and in ume6Δ (Δ) vegetative cells. Marker sizes are in base pairs. PCR products representing spliced (S) and unspliced (U) are indicated. (B) Quantification of splicing from at least three biological replicates. Dark gray bar indicates splicing efficiency at t=5h after induction of meiosis; light gray bar indicates splicing efficiency in ume6Δ vegetative cells. Note that ume6Δ also derepresses MER1, which encodes a meiotic splicing fator necessary for SPO22 pre-mRNA splicing (Munding et al., 2010). Error bars represent ± 1SD.
Splicing improves globally when RPGs are repressed
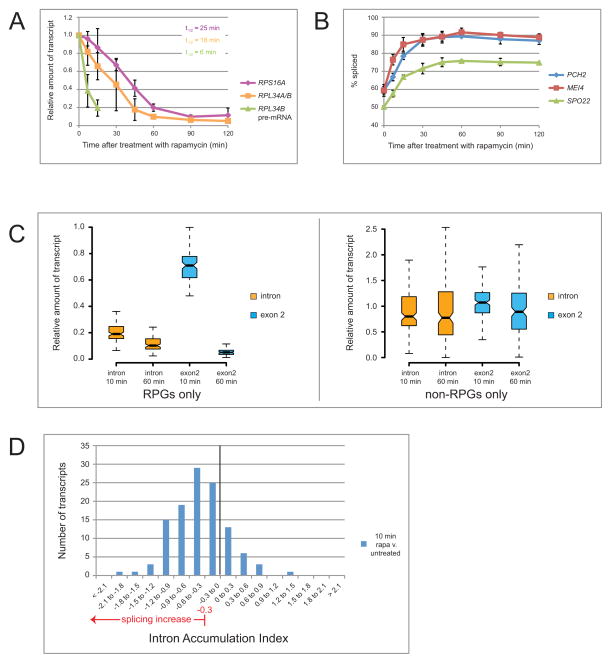
If poor splicing of meiotic transcripts in vegetative ume6Δ cells (Fig 2) is due to RPG expression, then splicing should improve upon repression of RPGs. RPG transcription is promoted by nutrients through the conserved protein kinase TOR (Cardenas et al., 1999; Hardwick et al., 1999; Powers and Walter, 1999). TOR is inactivated by rapamycin (Heitman et al., 1991), leading to rapid RPG repression (Hardwick et al., 1999; Powers and Walter, 1999). We treated vegetative ume6Δ cells with rapamycin (200ng/mL) and monitored RPG pre-mRNA and mRNA levels as well as pre-mRNA and mRNA from non-RPGs. Upon rapamycin addition, steady state levels of RPG pre-mRNA decay immediately with a half-life of <7 minutes (Fig 3A), likely due to the combination of transcription inhibition and rapid splicing. RPG mRNAs decay more slowly than pre-mRNAs, with half-lives similar to those reported by others (Fig 3A, Holstege et al., 1998; Li et al., 1999; Wang et al., 2002). Splicing efficiency of non-RPG pre-mRNAs improves within 7 minutes of rapamycin addition (Fig 3B). This improvement is mediated through TOR because cells lacking Fpr1, a cofactor required for rapamycin binding to TOR (Heitman et al., 1991; Lorenz and Heitman, 1995) do not show improved splicing after rapamycin treatment (Fig S1A).
Figure 3. Splicing increases after treatment with rapamycin.
(A) Quantification of total (exon 2) transcript levels for RPS16A and RPL34A/B and for unspliced RPL34B pre-mRNA by RT-qPCR relative to SEC65, and normalized to t=0 in ume6Δ vegetative cells at indicated times after treatment with rapamycin. Transcript half-lives (t1/2) are indicated in the inset. (B) Quantification of splicing efficiency of meiotic transcripts SPO22, MEI4 and PCH2 by semi-quantitative RT-PCR in ume6Δ vegetative cells at indicated times after treatment with rapamycin. (C) RNA-seq measurement of global expression after rapamycin treatment. Box plot representing change in RPG (n=107 events) (left panel) and non-RPG (n=165 events) (right panel) intron reads vs exon 2 reads after 10 or 60 minutes of treatment with rapamycin, normalized to untreated wild type cells. (D) Global changes in splicing of genes whose expression does not change greater than 2-fold after 10 minutes of rapamycin treatment relative to untreated wild type cells represented by intron accumulation indexes (IAI). Black bar indicates IAI=0 or no change in splicing efficiency. Red arrow indicates splicing changes above the threshold. Error bars represent ± 1SD. See also Fig S1.
Most unspliced pre-mRNAs are decayed by NMD (Burckin et al., 2005; Sayani et al., 2008) after export to the cytoplasm (Kuperwasser et al., 2004). To exclude the possibility that rapamycin mimics improved splicing by increasing NMD, we tested cells lacking the essential NMD factor Upf1 (Leeds et al., 1991). In these cells, the steady state levels of unspliced transcripts are much higher than in wild type (Fig S1B); nonetheless, treatment with rapamycin results in dramatically increased splicing efficiency (Fig S1C).
To explore the transcriptome-wide effect on splicing after RPG repression, we performed RNA sequencing (RNA-seq). We evaluated expression of intron-containing RNA (measured by intronic reads) and total RNA (measured by exon 2 reads) of both RPGs and non-RPGs in cells treated with rapamycin for 10 and 60 minutes (Fig 3C). RPG pre-mRNAs decrease to ~20% of initial levels within 10 minutes of rapamycin treatment, whereas total RPG RNA (mostly mRNA) falls substantially only after 60 minutes (Fig 3C, left panel). In comparison, non-RPG expression remains relatively unchanged (Fig 3C, right panel). We evaluated splicing in cells treated with rapamycin for 10 minutes relative to untreated cells, using a cut-off of 1.25-fold change in splicing (|IAI| ≥ 0.3), a threshold established using a control distribution, see Experimental Procedures, Fig S1D). Of the 116 ICGs whose expression changes less than 2-fold upon rapamycin treatment, 68 improve in splicing efficiency by at least 25% (Fig 3D, Fig S1D). Thus in both vegetative and meiotic cells, RPG expression is associated with inefficient splicing of other transcripts.
Down-regulation of an RPG-dedicated transcription factor suppresses spliceosomal defects
While searching for a way to manipulate RPG expression without rapamycin, we found a report from John Woolford’s lab of extragenic “supersuppressors” that rescued multiple spliceosomal mutations (Maddock et al., 1994). One class of suppressors fell in the SPP42 gene, now also known as FHL1, since shown to encode one of several transcription factors dedicated primarily to RPG transcription (Martin et al., 2004; Rudra et al., 2005; Schawalder et al., 2004; Wade et al., 2004; Zhao et al., 2006). Our hypothesis that pre-mRNAs compete for a limiting splicing apparatus prompted a new interpretation of their suppressor results. If RPG pre-mRNAs compete with essential pre-mRNAs, then competition might be exacerbated in a strain with a compromised spliceosome, for example the ts prp4-1 and prp11-1 strains (Galisson and Legrain, 1993; Hartwell, 1967). Furthermore if ts growth is a consequence of failure to splice growth rate limiting pre-mRNAs, this defect might be suppressed by relieving the competition for the compromised splicing machinery. The ability of spp42-1 to suppress multiple different splicing mutations (Maddock et al., 1994) and its subsequent identification as a dedicated RPG transcription factor suggested it reduced RPG expression and relieved competition.
To test the idea that down-regulation of an RPG-dedicated transcription factor might suppress different ts spliceosome mutations, we constructed strains carrying either the ts prp4-1 or prp11-1 alleles and a glucose-repressible promoter controlling expression of the dedicated RPG transcription factor encoded by IFH1, a protein required by FHL1/SPP42 to promote RPG transcription (Rudra et al., 2005; Schawalder et al., 2004). PRP4 encodes a protein in the U4/U6 snRNP, which enters the spliceosome as part of the U4/U6-U5 trisnRNP, whereas PRP11 encodes a subunit of the U2-associated SF3a complex that establishes U2 snRNP association with the intron branchpoint at an early step (see Will and Luhrmann, 2011 for review). These two proteins contribute to very different steps in the splicing pathway. The prp4-1; GAL-IFH1 and the prp11-1; GAL-IFH1 strains grow similarly to their corresponding IFH1 strains at permissive temperature (26°C) on glucose medium. But at the non-permissive temperature (30°C for prp4-1; IFH1 and 33°C for prp11-1; IFH1), both ts mutations are suppressed by down-regulation of IFH1, as signified by improved growth on glucose-containing media (Fig 4A). Using qPCR, we find that at 26°C on glucose, prp4-1; GAL-IFH1 cells express reduced levels of IFH1 and RPG mRNAs (Fig 4B). These genetic observations suggest a modest decrease in the RPG pre-mRNA pool rescues growth defects of the prp4-1 strain by improving splicing of other essential transcripts.
Figure 4. Splicing defects are suppressed by down-regulation of RPG transcription.
(A) Growth of IFH1 and GAL-IFH1 strains carrying temperature sensitive splicing mutations prp4-1 or prp11-1 on glucose (IFH1 down regulated) at 26°C (permissive temperature) and 30°C (non-permissive temperature for prp4-1) or 33°C (non-permissive for prp11-1). (B) RT-qPCR measurement of IFH1 and RPG expression relative to SEC65 in YPD at 26°C in prp4-1; IFH1, PRP4; GAL-IFH1, and prp4-1; GAL-IFH1 yeast normalized to WT (PRP4; IFH1). (C) Genome-wide changes in splicing of RPG and non-RPG transcripts in prp4-1; GAL-IFH1 cells relative to prp4-1; IFH1 cells. Black bar indicates IAI=0 or no change in splicing efficiency. Red arrow indicates splicing changes above the threshold. (D) RT-qPCR validation of splicing improvement as measured by percent intron-containing transcript for CPT1, HNT1, MOB2, and SEC14 in YPD at 26°C in prp4-1; IFH, PRP4; GAL-IFH1, and prp4-1; GAL-IFH1 yeast normalized to WT. Error bars represent ± 1SD. See also Table S2.
To confirm this we performed RNA-seq and examined the global effect of IFH1 down-regulation on splicing of other transcripts. We compared splicing for genes whose expression does not change more than 2-fold in prp4-1; GAL-IFH1 cells relative to prp4-1; IFH1 cells. Of 225 ICGs, fully 93% improve in splicing by at least 1.25-fold in prp4-1; GAL-IFH1 cells (Fig 4C). This includes most RPG (88/93) as well as non-RPG splicing events (121/132). Validation for four genes by RT-qPCR shows that splicing is restored by down-regulation of IFH1 (Fig 4D). We conclude that subtle down-regulation of a dedicated RPG transcription factor can rescue spliceosomal defects through an unusual suppression mechanism. We infer that by reducing the overall load of RPG pre-mRNAs, the demand on the compromised spliceosome is sufficiently relieved to allow splicing of inefficiently spliced essential transcripts. The RNA-seq data incidentally revealed that the mutant Prp4-1 protein has the substitution F320S in a WD repeat domain (data not shown).
To exclude the possibility that the increase in splicing observed in these three conditions (meiosis, rapamycin treatment, and IFH1 down-regulation) is associated with improved expression of the splicing machinery, we evaluated expression of the five snRNAs and 110 genes encoding splicing proteins in all three treatments (Table S2). Although expression differs across conditions, no global up-regulation of the splicing apparatus is observed under any condition. Furthermore there is no single gene whose expression is correlated with splicing improvement in all conditions (Table S2). Late in meiosis, RPGs are induced and splicing efficiency goes down (Fig 1A and B). In a preliminary attempt to increase competition in vegetative cells, we overexpressed the actin intron from a strong promoter and observed reduced splicing for several weakly competitive pre-mRNAs (data not shown). We conclude that pre-mRNAs compete with each other for a limiting splicing apparatus and that increased splicing is associated with relief of competition by reduced RPG expression.
Pre-mRNA substrates compete at an early step of spliceosome recruitment
Inspection of the splice sites in pre-mRNAs that compete poorly revealed many with canonical splice site and branchpoint sequences, without convincing enrichment for any single feature that might identify a strongly competitive pre-mRNA. To explore whether substrates with suboptimal splicing signals vary in their competitive ability, we used ACT1-CUP1 reporters (Lesser and Guthrie, 1993) containing mutations in the 5′ splice site (5’ss), branchpoint (bp), and 3′ splice site (3’ss, Fig 5A). We tested the effect of rapamycin treatment on reporter splicing in vegetative cells, expecting that a substrate altered in a feature required for competition would show the most improvement in response to RPG repression. Of the seven different mutants tested, only two branchpoint mutants (C256A and A259C) improved in splicing after treatment with rapamycin (Fig 5B). We separately evaluated first and second step splicing and find that rapamycin significantly improves the first step for both C256A and A259C mutant pre-mRNAs (Fig 5C). Other substrates with first step defects, such as the 5’ss mutant U2A, did not significantly improve (Fig 5B). While A259C also shows second step improvement, this effect is likely a consequence of the 2-fold improvement in the first step. The 3’ss mutant U301G (defective in second step catalysis) showed no significant improvement (Fig 5B). Attempts to identify the limiting component by overexpressing individual factors known to act at the branchpoint failed to improve splicing (data not shown). Taken together, these data indicate that competition is likely to involve factors acting with the intron branchpoint to commit the pre-mRNA to splicing.
Figure 5. Competition is imposed at early steps of spliceosome assembly.
(A) ACT1-CUP1 reporter pre-mRNA schematic indicating 5′ splice site, branchpoint, and 3′ splice site mutations used in this study. (B) Quantification of total splicing efficiency as measured by primer extension of wild type and the indicated mutant ACT1-CUP1 reporters before and after (+) treatment for 60min with rapamycin (60′ rapa). Double asterisks indicate p<0.01 in a one-tailed t-test. (C) Quantification of 1st step (dark gray bars) and 2nd step (light gray bars) splicing efficiency as measured by primer extension of WT, C256A, and A259C ACT1-CUP1 reporters before and after (+) treatment for 60′ with rapamycin (60′ rapa). Single asterisk indicates p<0.05 and double asterisks indicate p<0.01 in a one-tailed t-test. Error bars represent ± 1SD
Discussion
These results provide strong evidence that pre-mRNAs compete for the splicing apparatus. For this reason, changes in the composition of the pre-mRNA pool in the nucleus have significant impact on splicing regulation. By manipulating the composition of the pool of competing pre-mRNAs through transcription (Figs 3 and 4) we show that the balance of splicing competition is important for cell function. The ability of competing RNAs to influence splicing by a “trans-competition control” mechanism appears related to a larger group of phenomena described in vertebrate cells in which competition between RNAs for a limiting regulatory factor leads to global changes in gene expression. This mechanism is established for miRNA regulation, whereby repression of an mRNA by a miRNA is affected by the level of other competing RNAs (called “competitive endogenous RNAs,” ceRNAs; Salmena et al., 2011). This process, first described in plants and called “target mimicry” (Franco-Zorrilla et al., 2007), also regulates muscle development (Cesana et al., 2011), and affects cancer progression (Poliseno et al., 2010) in animals. Our results indicate that a parallel mechanism is at work in splicing regulation, whereby pre-mRNAs compete for a limiting splicing machinery, and splicing of many introns is influenced by changes in the composition of the transcript pool. In the case of splicing, the competing RNAs are also substrates, rather than inert decoys.
Evidence that splicing regulation is subject to the composition of a pool of endogenous competing RNAs is not limited to yeast. In models of the human disease myotonic dystrophy, abnormal expression of a CUG repeat expansion RNA acts as a ceRNA for the MBNL1 splicing factor, mimicking a loss of MBNL1 function in splicing (Du et al., 2010; Kanadia et al., 2003; Miller et al., 2000), indicating that pre-mRNAs compete for MBNL1. Similarly sno-lncRNAs have been identified as a kind of ceRNA for pre-mRNAs dependent on the splicing factor RBFOX2 (Yeo et al., 2009; Yin et al., 2012). Under conditions where sno-lncRNAs are depleted (such as in Prader-Willi syndrome, Yin et al., 2012) competition for RBFOX2 is relieved. A third example involves the U1 snRNP, which appears limiting for an activity that influences polyadenylation site selection (Berg et al., 2012; Kaida et al., 2010). When the levels of pre-mRNA increase, the spectrum of polyA sites utilized in the cell changes, creating mRNAs with alternative 3′UTRs, with each pre-mRNA presumably acting as a ceRNA for all the others. Thus understanding post-transcriptional gene regulation requires accounting of changes in the levels of the limiting regulatory factor as well as changes in composition of the larger transcript pool that affect competition for that limiting factor.
What conditions are required for trans-competition control?
Splicing can be regulated by changes in physical levels, specific activity or localization of splicing factors that control the rate-limiting step of splicing in a transcript specific fashion (Black, 2003; Nilsen and Graveley, 2010). Trans-competition control accounts for changes in splicing factor activity observed by altering the effective load of pre-mRNAs that also employ the limiting factor or other RNAs that occupy the factor. Thus splicing regulation may be achieved by either changing the abundance of a limiting factor (or exchanging one limiting factor for another) or by altering the dynamics of competition by changing the composition of the RNA pool (Fig 6A). These systems-level considerations argue that understanding the demand for the splicing machinery and how pre-mRNA competition changes during development will be required to integrate regulatory networks into their gene expression programs. In mammalian systems, induction of gene expression programs can result in large changes in the composition of the transcript pool (Berg et al., 2012), altering competition for the splicing machinery. Under such conditions, the competitive advantage of alternative exons for the splicing machinery may be decreased, resulting in a shift of mRNA isoforms.
Figure 6. Trans-competition control of splicing.
(A) Trans-competition control of alternative splicing. When competitor pre-mRNA levels are low, demand for the limiting factor (LF) is low resulting in efficient inclusion of the weakly competitive cassette exon. When competitor pre-mRNA levels are high, competitor pre-mRNAs titrate increased amounts of the limiting factor, resulting in much less efficient inclusion of the weakly competitive cassette exon. (B) Left Panel: Michaelis-Menten scheme showing two substrates with different affinities (S1 and S2) competing for the same enzyme, E. Formation of products P1 and P2 is determined by the concentration of each substrate and the substrate’s Km when the enzyme is limiting. Right Panel: Splicing scheme of two substrates competing for a limiting splicing machinery (pink circle). In this example, both substrates are present at the same initial concentration, but the orange substrate outcompetes the blue substrate due to its higher affinity (k1 ≫ k2). Note that rates of ES formation will also change between pre-mRNAs of equal affinity when one is at higher concentration. See also Fig S2.
The principles of trans-competition control can be explained using a modification of the general Michaelis-Menten model for competitive inhibition where two different substrates (S1 and S2) compete (Fig 6B). In this case, when the spliceosome is limiting, the amount of mRNA product P1 depends on both the concentration of pre-mRNA S1 ([S1]) and its splicing rate (k1) as well as the concentration ([S2]) and splicing rate (k2) of the competing pre-mRNA substrate (Fig 6B and S2). This simple model shows that splicing regulation can be achieved by altering the competitive status of a target pre-mRNA through modulation of the levels of other RNAs that compete for a limiting factor. In a cell there are thousands of competing introns, each with its own affinity for the spliceosome; as the concentration of any one of them changes, the splicing efficiency of all the others then must change as well. Similar to the queuing theory (Cookson et al., 2011), where degradation of unrelated proteins dependent on a common enzyme become coupled due to competition for the enzyme, change in the demand for the spliceosome couples pre-mRNAs whose splicing is affected after a change to the pool of substrates.
Functional importance of trans-competition control
The striking relationship between RPG expression and the change in splicing efficiency during meiosis suggests a role for trans-competition control in maintaining separation between the meiotic and vegetative gene expression states. Weakly competitive introns reduce the chances that meiotic genes would be expressed during vegetative growth. Repression of RPGs may have become necessary to allow sufficient splicing during meiosis. However, it is not known whether meiosis can proceed in the absence of RPG repression, thus there is no direct evidence that trans-competition control is required for meiosis.
Strong evidence for the functional importance of balanced competition comes from suppression of splicing defects upon down-regulation of RPGs (Fig 4). Rescue of the ts phenotype of prp4-1 and prp11-1 arises from poor splicing of essential pre-mRNAs because they are outcompeted by RPG pre-mRNAs. Restoring the competitive balance decreases the demand on the splicing machinery by reducing the load represented by intron-containing RPGs allows improved splicing of essential non-RPG pre-mRNAs that then increases viability of the prp4-1 and prp11-1 strains.
A number of human diseases are associated with missense mutations in core spliceosome components (reviewed in Padgett, 2012), such as Prp8 and Prp31 (retinitis pigmentosa) and SF3B1 (myelodysplastic syndrome and chronic lymphocytic leukemia). These cases may mirror the subtle loss of splicing capacity observed for the prp4-1 and prp11-1 mutations and alter the competitive landscape for splicing, contributing to disease. Different pre-mRNAs clearly have distinct dependencies on conserved components of the splicing machinery (Burckin et al., 2005; Clark et al., 2002; Park et al., 2004; Pleiss et al., 2007), suggesting transcripts may compete for different limiting factors depending on the context. Thus the key to understanding why certain mutations in conserved splicing factor genes lead to specific diseases may lie in the nature of the composition of the transcript pool in the specific cell type affected, and which pre-mRNA molecules suffer under the altered competitive situation.
Experimental Procedures
Strains and plasmids
Strains are listed in Table S3. GAL-IFH1 strains were constructed (Longtine et al., 1998; Wach et al., 1997) and verified by PCR, so that the GAL1 promoter (marked by the Saccharomyces kluyveri HIS3 gene) was placed upstream of IFH1. Strains carrying the prp4-1 or the prp11-1 mutations were provided by S. Ruby (Ruby et al., 1993). The prp4-1; GAL-IFH1 and the prp11-1; GAL-IFH1 strain were constructed by crossing to the GAL-IFH1 strain. ACT1-CUP1 reporter plasmids (Fig 5) are from (Lesser and Guthrie, 1993).
Media and culture conditions
Standard methods for yeast culture conditions were used (Sherman, 1991). Rapamycin was added cells grown to OD600≈0.5 at 200ng/mL for the indicated time. All yeast strains were grown at 30°C unless otherwise indicated.
RNA isolation
RNA was isolated as described in (Rio et al., 2010). Total meiotic RNA was extracted according to Method 2 to ensure uniform RNA extraction from late spore stages. Total vegetative RNA was prepared from cells grown to OD600=0.5 according to Method 1.
Transcriptome profiling
Microarray data (Munding et al., 2010) is from Gene Expression Omnibus, accession number GSE24686. RNA-Seq data in Fig 3 is from two independent rapamycin time courses. RNA-Seq data in Fig 4 represents one culture from each strain (grown to OD600≈0.5 in YPD at 26°C). Details on methodology and analysis of the microarray and RNA-Seq data are included in Supplemental Information.
RT-PCR and qPCR
Reverse transcribed RNA (cDNA) was amplified using the primers in Table S4. Semi-quantitative RT-PCR was carried out by limiting cycle numbers to 21 and using cDNA derived from 300ng of total RNA. Estimates of splicing efficiency used the Agilent 2100 Bioanalyzer. qPCR was preformed using a master mix (Fermentas). Additional experimental details are included in Supplemental Information.
Primer Extension
At least 3 colonies of BY4741 transformed with each ACT1-CUP1 reporter plasmid were grown to OD=0.5 in SCD medium lacking leucine. 5μg of total RNA was annealed to 0.1ng of PE1 primer (5′-CCTTCATTTTGGAAGTTA-3′) and primer extended as previously described (Perriman and Ares 2007). Extension products were analyzed on a Typhoon imaging system (GE Healthcare). 1st step splicing efficiency was calculated as (M+L)/(M+L+P); 2nd step splicing efficiency was calculated as M/(M+L); total splicing efficiency was calculated as M/(M+L+P) where M is mRNA, L is lariat intermediate, and P is pre-mRNA.
Supplementary Material
Highlights.
Competition between pre-mRNAs for the splicing machinery drives changes in splicing
Down-regulation of an RPG transcription factor rescues ts spliceosomal mutations
Suppression occurs by decreasing competition for the mutant spliceosome
Trans-competition is a general phenomenon of post-transcriptional gene control
Acknowledgments
We would like to thank the UCSC Genomics Core for sequencing, Jon Warner for generosity with suggestions and reagents, and Rhonda Perriman for encouragement and critical reading of the manuscript. Thanks also to Hinrich Boeger, Ted Powers, Grant Hartzog, and Alex Hoffmann for comments and suggestions. This work was primarily supported by GM040478 from the National Institutes of Health to M.A. L.S. and J.P.D. were supported by GM084317. E.M. was partially supported by National Institutes of Health Training Grant T32 GM008646.
Footnotes
Accession numbers
RNA-Seq data has been released through the Gene Expression Omnibus under accession number GSE44219. Additional experimental details are included in Supplemental Information.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ares M, Jr, Grate L, Pauling MH. A handful of intron-containing genes produces the lion’s share of yeast mRNA. RNA. 1999;5:1138–1139. doi: 10.1017/s1355838299991379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, Dreyfuss G. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Posner JB, Darnell RB. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron. 1993;11:657–672. doi: 10.1016/0896-6273(93)90077-5. [DOI] [PubMed] [Google Scholar]
- Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M., Jr Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O’Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Cookson NA, Mather WH, Danino T, Mondragon-Palomino O, Williams RJ, Tsimring LS, Hasty J. Queueing up for enzymatic processing: correlated signaling through coupled degradation. Mol Syst Biol. 2011;7:561. doi: 10.1038/msb.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Egeland DB, Jarnik M, Strich R. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc Natl Acad Sci U S A. 2000;97:14548–14553. doi: 10.1073/pnas.250351297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Grate L, Spingola M, Ares M., Jr Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 2000;28:1700–1706. doi: 10.1093/nar/28.8.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, Ares M., Jr Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht JA, Voelkel-Meiman K, Roeder GS. Meiosis-specific RNA splicing in yeast. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O’Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Galisson F, Legrain P. The biochemical defects of prp4-1 and prp6-1 yeast splicing mutants reveal that the PRP6 protein is required for the accumulation of the [U4/U6.U5] tri-snRNP. Nucleic Acids Res. 1993;21:1555–1562. doi: 10.1093/nar/21.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci U S A. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967;93:1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 2007;3:e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Howe KJ, Ares M., Jr Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc Natl Acad Sci U S A. 1997;94:12467–12472. doi: 10.1073/pnas.94.23.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KJ, Kane CM, Ares M., Jr Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau K, Palm C, Miranda M, Davis RW. High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc Natl Acad Sci U S A. 2007;104:1522–1527. doi: 10.1073/pnas.0610354104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR. Promoter usage and alternative splicing. Curr Opin Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Kreahling JM, Graveley BR. The iStem, a long-range RNA secondary structure element required for efficient exon inclusion in the Drosophila Dscam pre-mRNA. Mol Cell Biol. 2005;25:10251–10260. doi: 10.1128/MCB.25.23.10251-10260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperwasser N, Brogna S, Dower K, Rosbash M. Nonsense-mediated decay does not occur within the yeast nucleus. RNA. 2004;10:1907–1915. doi: 10.1261/rna.7132504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Nierras CR, Warner JR. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol Cell Biol. 1999;19:5393–5404. doi: 10.1128/mcb.19.8.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lopez PJ, Seraphin B. Genomic-scale quantitative analysis of yeast pre-mRNA splicing: implications for splice-site recognition. RNA. 1999;5:1135–1137. doi: 10.1017/s135583829999091x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J Biol Chem. 1995;270:27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- Maddock JR, Weidenhammer EM, Adams CC, Lunz RL, Woolford JL., Jr Extragenic suppressors of Saccharomyces cerevisiae prp4 mutations identify a negative regulator of PRP genes. Genetics. 1994;136:833–847. doi: 10.1093/genetics/136.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AP. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munding EM, Igel AH, Shiue L, Dorighi KM, Trevino LR, Ares M., Jr Integration of a splicing regulatory network within the meiotic gene expression program of Saccharomyces cerevisiae. Genes Dev. 2010;24:2693–2704. doi: 10.1101/gad.1977410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Ogawa H. The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. Embo J. 1999;18:5714–5723. doi: 10.1093/emboj/18.20.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics. 2011;189:737–765. doi: 10.1534/genetics.111.127126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett RA. New connections between splicing and human disease. Trends Genet. 2012;28:147–154. doi: 10.1016/j.tig.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci U S A. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass M, Codony-Servat C, Ferreira PG, Vilardell J, Eyras E. RNA secondary structure mediates alternative 3’ss selection in Saccharomyces cerevisiae. RNA. 2012;18:1103–1115. doi: 10.1261/rna.030767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- Rio DC, Ares M, Jr, Hannon GJ, Nilsen TW. Isolation of Total RNA from Yeast Cell Cultures. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot071456. pdb prot5438. [DOI] [PubMed] [Google Scholar]
- Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CW. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 1998;26:5568–5572. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Sachidanandam R, Krainer AR. Determinants of the inherent strength of human 5′ splice sites. RNA. 2005;11:683–698. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby SW, Chang TH, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- Rudra D, Zhao Y, Warner JR. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 2005;24:533–542. doi: 10.1038/sj.emboj.7600553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol Cell. 2008;31:360–370. doi: 10.1016/j.molcel.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schawalder SB, Kabani M, Howald I, Choudhury U, Werner M, Shore D. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature. 2004;432:1058–1061. doi: 10.1038/nature03200. [DOI] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ. Conserved RNA secondary structures promote alternative splicing. RNA. 2008;14:1463–1469. doi: 10.1261/rna.1069408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Strich R, Surosky RT, Steber C, Dubois E, Messenguy F, Esposito RE. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- Sugnet CW, Srinivasan K, Clark TA, O’Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, Ares M., Jr Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wade JT, Hall DB, Struhl K. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature. 2004;432:1054–1058. doi: 10.1038/nature03175. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci U S A. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Warzecha CC, Shen S, Xing Y, Carstens RP. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009;6:546–562. doi: 10.4161/rna.6.5.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RM, Primig M, Washburn BK, Winzeler EA, Bellis M, Sarrauste de Menthiere C, Davis RW, Esposito RE. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc Natl Acad Sci U S A. 2002;99:13431–13436. doi: 10.1073/pnas.202495299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M, Pfiffner J, Levin JZ, Adiconis X, Gnirke A, Nusbaum C, Thompson DA, Friedman N, Regev A. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol. 2010;11:R87. doi: 10.1186/gb-2010-11-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Zhao Y, McIntosh KB, Rudra D, Schawalder S, Shore D, Warner JR. Fine-structure analysis of ribosomal protein gene transcription. Mol Cell Biol. 2006;26:4853–4862. doi: 10.1128/MCB.02367-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.