Abstract
MicroRNAs (miRNAs) have been broadly implicated in cancer, but their exact function and mechanism in carcinogenesis remain poorly understood. Elevated miR-17∼92 expression is frequently found in human cancers, mainly due to gene amplification and Myc-mediated transcriptional upregulation. Here we show that B cell-specific miR-17∼92 transgenic mice developed lymphomas with high penetrance and that, conversely, Myc-driven lymphomagenesis stringently requires two intact alleles of miR-17∼92. We experimentally identified miR-17∼92 target genes by PAR-CLIP and validated select target genes in miR-17∼92 transgenic mice. These analyses demonstrate that miR-17∼92 drives lymphomagenesis by suppressing the expression of multiple negative regulators of the PI3K and NFκB pathways and by inhibiting the mitochondrial apoptosis pathway. Accordingly, miR-17∼92-driven lymphoma cells exhibited constitutive activation of the PI3K and NFκB pathways and chemical inhibition of either pathway reduced tumour size and prolonged the survival of lymphoma-bearing mice. These findings establish miR-17∼92 as a powerful cancer driver that coordinates the activation of multiple oncogenic pathways, and demonstrate for the first time that chemical inhibition of miRNA downstream pathways has therapeutic value in treating cancers caused by miRNA dysregulation.
Keywords: lymphoma, miR-17∼92, NFκB, PAR-CLIP, PI3K
Introduction
Studies of human cancer genomes have revealed numerous genetic alterations at the structural and sequence levels. By the time a cancer is diagnosed, it often comprises millions to billions of cells carrying genetic alterations that initiated malignant transformation and many others acquired along the way. Therefore, the contribution of each genetic alteration to cancer varies substantially. Some genetic alterations are strong and causal ‘drivers’ that confer selective growth advantages to cancer cells; others are weaker but important ‘contributors’ to the development of cancer; however, most are incidental ‘passengers’ that have been accumulated by chance during the cancer’s life history (Chin and Gray, 2008). The key challenge in cancer research is to distinguish the drivers and contributors from the passengers. Assessing the contribution of individual alterations to cancer in genetically engineered mice has proven to be the gold standard for establishing their causality in cancer (Frese and Tuveson, 2007), and is essential for the development of cancer therapeutics.
Studies during the past few years have demonstrated that microRNAs (miRNAs), a class of small non-coding RNAs, play important roles in human cancers (Croce, 2009). MiRNAs are endogenously encoded-single stranded RNAs of ∼22 nucleotides (nts) in length that bind to their target mRNAs and regulate their translation and stability (Ambros, 2004; Bartel, 2004; Bushati and Cohen, 2007). Despite the demonstrated importance of miRNAs, determining miRNA targets has been a major obstacle (Thomas et al, 2010). Many bioinformatic algorithms were developed to predict miRNA target genes, mainly based on seed pairing and evolutionary conservation (Rajewsky, 2006). However, these algorithms typically predict hundreds to thousands of target genes for each miRNA, and different algorithms often produce divergent results. Moreover, these algorithms sometimes fail to predict the most biologically important miRNA targets (Thomas et al, 2010). To overcome this problem, two groups recently developed methods to experimentally identify direct miRNA–target mRNA interactions: HITS-CLIP (high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation) and PAR-CLIP (Photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation) (Chi et al, 2009; Hafner et al, 2010). These new methods hold the promise to identify miRNA binding sites in almost any cells and have the potential to revolutionize miRNA research, in the same way that chromatin immunoprecipitation (ChIP) did to transcription factor research, but their utility still awaits validation in animal models with gain- and loss-of-function mutations for individual miRNAs.
Gene expression profiling of human cancer cells has revealed specific miRNA expression signatures in many human cancers (Croce, 2009). However, the development of miRNA-based therapeutics has lagged behind, largely due to the lack of in-depth understanding of function and molecular mechanism of miRNAs in carcinogenesis. We attempt to address this issue through the analysis of miR-17∼92 in the context of lymphomagenesis. MiR-17∼92 is the first miRNA gene implicated in cancer and encodes for six distinct miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92) (Mendell, 2008). These miRNAs fall into four miRNA families (miR-17, miR-18, miR-19, and miR-92 families), with members in each family sharing the same seed sequence. The genomic region encoding human miR-17∼92 is frequently amplified in lymphoma, leukemia, and solid tissue cancers, and the encoded miRNAs are highly expressed in these cancer cells (Mendell, 2008). Previous studies showed that miR-17∼92 overexpression accelerated carcinogenesis initiated by other oncogenic mutations, such as Myc and Notch activation or Rb family deletion (He et al, 2005; Mavrakis et al, 2010; Conkrite et al, 2011). These results established miR-17∼92 as an important contributor to cancer, but it remains unclear whether elevated miR-17∼92 expression per se is sufficient to drive carcinogenesis. The answer to this question will determine whether targeting miR-17∼92 miRNAs or their downstream pathways has any therapeutic value.
In addition to gene amplification, miR-17∼92 expression can be deregulated by other mechanisms. Myc, one of the most common and potent oncogenes (Dang, 2012), activates miR-17∼92 expression by directly binding to its genomic locus (O'Donnell et al, 2005). Myc overexpression is the defining feature of Burkitt lymphoma, a disease state characterized by Myc translocation to the immunoglobulin (Ig) locus (Klapproth and Wirth, 2010). A recent study of Burkitt lymphoma patient biopsies found drastic miR-17∼92 overexpression in all the 28 cases examined (Schmitz et al, 2012), confirming that activation of the Myc→miR-17∼92 axis is a ubiquitous feature of this malignancy. Another study showed that deletion of miR-17∼92 in established Myc-driven lymphoma cell lines slowed down their growth in tissue culture and in immunodeficient hosts, suggesting that miR-17∼92 contributes to the optimal growth of those cancer cell lines (Mu et al, 2009). Established cancer cell lines differ from primary cancers in that the former can survive and proliferate in the absence of their natural tumour microenvironment, probably enabled by additional genetic alterations obtained during the tissue culture process. While the study of cancer cell lines is largely responsible for the early progress in cancer research, recent studies suggested that many of those initial observations need to be re-evaluated in autochthonous tumour models (Frese and Tuveson, 2007). Therefore, it remains unclear how critical miR-17∼92 is in the development of autochthonous lymphomas driven by Myc.
Here we address these issues directly by generating (1) mice with B cell-specific transgenic miR-17∼92 expression, and (2) mice with deletion of the miR-17∼92 gene in a Myc transgenic Burkitt lymphoma model, and then monitoring lymphoma development in the resulting mice over their lifespan. Furthermore, we experimentally identified miR-17∼92 target genes in B cells by PAR-CLIP, validated select target genes in miR-17∼92 transgenic B cells, and explored the possibility of targeting miR-17∼92 downstream pathways to treat miR-17∼92-driven cancers.
Results
B cell-specific miR-17∼92 transgenic mice develop lymphomas
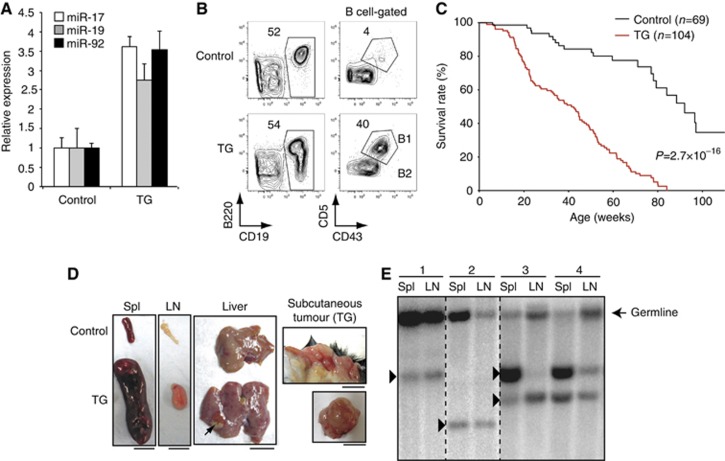
We have previously created a miR-17∼92 transgenic allele (termed miR-17∼92 Tg) by homologous recombination into the Rosa26 locus. The expression of this transgene can be turned on conditionally by Cre recombinase (Xiao et al, 2008). To directly test the role of elevated miR-17∼92 expression in B cell lymphomagenesis, we generated miR-17∼92 Tg/Tg;CD19Cre mice (termed TG mice hereafter) in which the miR-17∼92 transgene is turned on specifically in the B-cell lineage. We detected a 3–4-fold increase in miR-17∼92 expression in TG B cells (Figure 1A). This level of overexpression is relatively modest compared to the drastic increase (5–30-fold) that is routinely seen for miR-17∼92 in human lymphomas (He et al, 2005; Schmitz et al, 2012). Analysis of B-cell development in the bone marrow did not reveal any significant alterations. In the spleen of TG mice, we observed an expansion of CD19+B220lowCD43+CD5+ B1-like cells, which are present mainly in the peritoneal cavity of wild-type mice, as well as a slight increase in the total B-cell number at the age of 2–4 months (Figure 1B and Supplementary Figure S1A).
Figure 1.
Mice with B-cell-specific transgenic miR-17∼92 expression develop lymphoma. (A) miR-17∼92 expression in control and TG B2 cells (TG) were determined by northern blot. miRNA/U6 ratios in control B2 cells was arbitrarily set as 1. (B) Flow cytometry analysis of splenocytes of 2-month-old mice. (C) Kaplan–Meier survival curves of 104 TG and 69 littermate control mice. The P-value was determined by Mantel–Cox log-rank test. (D) Splenomegaly, lymphadenopathy, tumour cell infiltration into liver (arrow), and subcutaneous tumour in TG mice. Spl, spleen; LN, lymph nodes. Scale bars, 1 cm. (E) Representative southern blot analysis of tumour clonality using a JH4 probe. Arrowheads indicate clonal bands corresponding to VDJ or DJ rearrangements. Numbers indicate different lymphoma-bearing TG mice.
We monitored a cohort of 104 TG and 69 littermate control mice for 2 years for lymphoma development. As shown in Figure 1C, all TG mice died during this period, with an average lifespan of 40 weeks. Macroscopic examination of TG mice, sacrificed when they were sick, revealed splenomegaly and lymphadenopathy, in some cases accompanied by tumours in extranodal tissue compartments such as liver and skin (Figure 1D). Clonal expansion of B cells was observed in 24 of 30 sick TG mice analysed (Figure 1E and Supplementary Table S1). We performed histological and immunohistochemical analyses of involved organs from the 24 mice with clonal B cell expansion (Figure 2 and Supplementary Figure S2, Supplementary Table S1). Of these 24, 12 mice developed diffuse large B-cell lymphoma (DLBCL), characterized by sheets of large-to-intermediate-sized, transformed-appearing lymphoid cells with numerous mitotic figures and a moderate-to-high proliferative rate as assessed by Ki-67 staining. Six mice showed lymphoid proliferations characterized by admixed centrocytic (cleaved)- and centroblastic (non-cleaved)-appearing lymphocytes, with a low-to-moderate proliferative rate as assessed by Ki-67 staining, forming a variably nodular pattern; these mice were diagnosed with follicular lymphoma (FL). One mouse showed proliferation characterized by CD138+ plasmablastic-appearing cells with numerous mitoses and a high Ki-67 positivity rate; these features are characteristic of anaplastic plasmacytoma (AP). Two mice had lymphoid proliferations that were categorized as small B-cell lymphoma (SBL). Among the other four mice, one had clonal B cell lymphoid hyperplasia in the peritoneal cavity, one had aggressive B-cell lymphoma similar to Burkitt or high-grade mucosa-associated lymphoid tissue (MALT) lymphoma, and the other two mice had lymphomas whose histologic types were not determined (Figure 2, Supplementary Figure S2, and Supplementary Table S1). Mouse lymphoma cells differ from wild-type B cells in their capacity to transfer disease upon transplantation into healthy mice. To determine whether TG lymphoma cells are able to transfer disease, we transplanted primary lymphoma cells into Rag1−/− (immunodeficient) and wild type-C57BL/6 (immunosufficient) mice. Most TG lymphomas were able to establish secondary lymphomas in Rag1−/− recipients, while lymphoma cells from 6 out of 23 TG mice were able to establish secondary lymphomas in C57BL/6 recipients (Supplementary Table S1). In all transplantable cases, the primary and secondary lymphoma cells exhibited identical immunophenotypes and clonal bands of the IgH locus (Supplementary Figure S1B and C).
Figure 2.
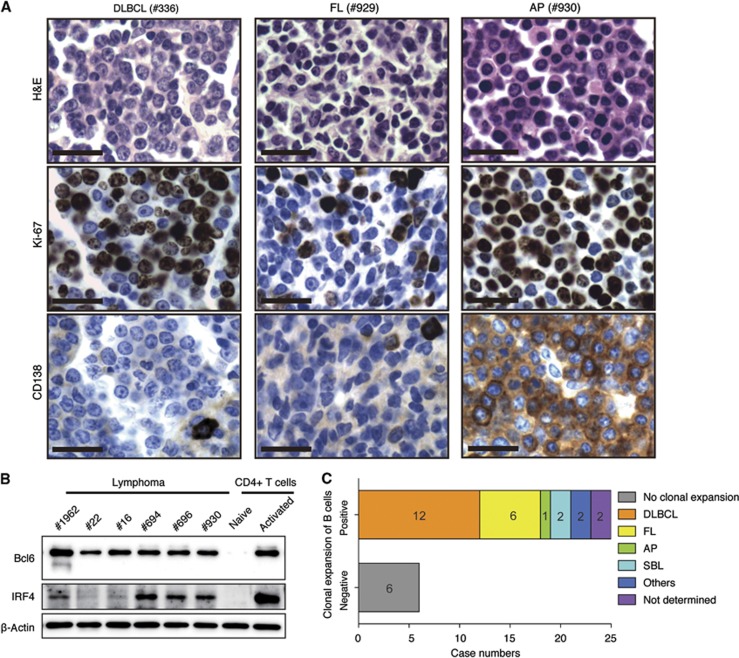
Histological and molecular analyses of lymphomas developed in TG mice. (A) Representative immunohistochemical analyses of tumours in TG mice. DLBCL, diffuse large B cell lymphoma; AP, anaplastic plasmacytoma; FL, follicular lymphoma. Scale bars, 20 μm. (B) Western blot analysis of Bcl6 and IRF4 expression in purified lymphoma cells. Naïve and activated (by anti-CD3/CD28 for 48 h) CD4 T cells were used as negative and positive controls, respectively. (C) Clonality and lymphoma subtypes of TG lymphomas. Note that mouse no. 14 had FL in mesenteric lymph node and DLBCL on small intestine, and was counted in both the FL and DLBCL categories.
Source data for this figure is available on the online supplementary information page.
DLBCL, FL, and AP are thought to originate from germinal centre (GC) or post-GC B cells. For the 18 cases falling into these three categories, we performed immunohistochemical (IHC) analyses of Bcl6 and IRF4 expression, as well as PNA-binding ability (Supplementary Figure S2B and Supplementary Table S1). In addition, we expanded 6 cases of lymphomas (5 DLBCL and 1 AP) by transplanting into Rag1−/− mice and purifying lymphomas cells from the recipient mice, and analysed their expression of Bcl6 and IRF4 protein, as well as V gene mutation status (Figure 2B and Supplementary Table S2). Consistent with their classification as DLBCL, FL, and AP, 13 of these 18 cases are positive in IHC for Bcl6, a signature transcription factor of GC B cells. Among the 5 IHC Bcl6-negative cases, 2 cases (no. 22 and no. 16) are positive in western blot. This discrepancy between IHC and western blot is probably due to the limited sensitivity of Bcl6 IHC (Cattoretti et al, 2005). The expression of IRF4, which denotes B-cell maturation toward plasma cells during late GC B-cell differentiation (De Silva et al, 2012), ranged from negative, dim-positive, to positive (Figure 2B and Supplementary Figure S2B, Supplementary Table S1), suggesting that lymphomas developed in TG mice are heterogeneous in nature, and that some of them originated from late-stage GC B cells or plasmablasts. Only two cases are positive for PNA binding, which is characteristic of GC B cells. The inability of other lymphomas to bind to PNA may be due to constitutive PI3K signalling in these cells (Figure 8D), similar to that reported in a recent study of a new Burkitt lymphoma mouse model that combines constitutive Myc expression and PI3K signalling, in which GC B cell-derived lymphoma cells are also PNA-negative (Sander et al, 2012). Among all the six cases analysed, only one case (no. 1962) contains significant amount of mutations in their V genes (Supplementary Table S2). While most human DLBCL, FL and AP contain somatic mutations in their V genes, our result may reflect the difference between human lymphomas and lymphomas developed in genetically engineered mice, as reported in a recent study of a mouse model of DLBCL lymphomas driven by constitutive NFκB activation and Blimp1 deletion (Calado et al, 2010). Taken together, these results showed that elevated miR-17∼92 expression alone is sufficient to drive lymphomagenesis in mice with a marked preponderance of GC and post-GC B-cell-derived lymphomas.
Myc-driven lymphomagenesis stringently requires two intact alleles of miR-17∼92
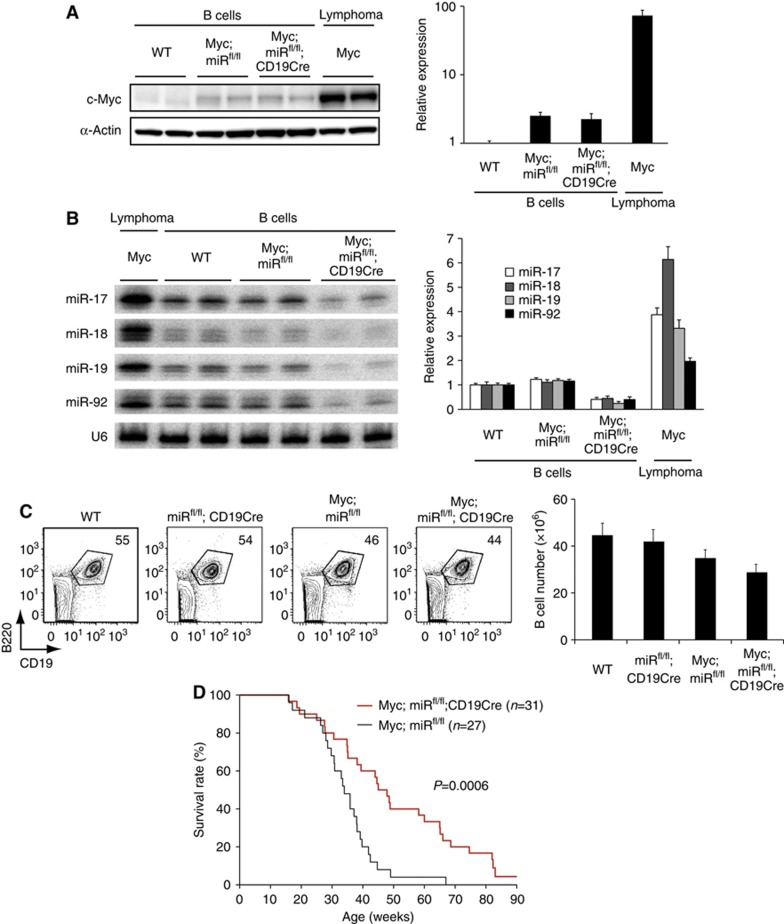
To evaluate the role of miR-17∼92 during the development of autochthonous lymphomas driven by Myc, we generated λ-Myc mice carrying conditional miR-17∼92 knockout alleles (miRfl/fl) under the control of CD19Cre (termed Myc;miRfl/fl; CD19Cre mice hereafter) (Rickert et al, 1997; Kovalchuk et al, 2000; Ventura et al, 2008). λ-Myc mice express a c-Myc transgene under the control of the B cell-specific λ light chain regulatory sequence. These mice develop monoclonal tumours recapitulating features of human Burkitt lymphoma (Kovalchuk et al, 2000). MiR-17∼92 miRNAs were significantly induced at the lymphoma stage, and this correlated with Myc expression (Figure 3A and B). The deletion of miR-17∼92 was confirmed in B cells of young Myc;miRfl/fl;CD19Cre mice (Figure 3B), which exhibited largely normal B-cell development in the bone marrow and spleen (Figure 3C). We monitored a cohort of 31 Myc;miRfl/fl; CD19Cre and 27 Myc;miRfl/fl mice over one and a half years for tumour development and survival (Figure 3D). Lymphoma-free survival of Myc;miRfl/fl;CD19Cre mice was significantly extended.
Figure 3.
Genetic ablation of the miR-17∼92 gene delays Myc-mediated lymphomagenesis. (A, B) The c-Myc protein (A) and miR-17∼92 (B) expression levels in B cells purified from 2–3-month-old mice of indicated genotypes and lymphoma cells purified from λ-Myc mice were determined by western (A) and northern (B) blot analysis, respectively. Left panels show representative blots, and right panels summarize quantification results from three independent experiments (n=5–8 in each group for western blot and n=6∼14 in each group for northern blot). The c-Myc/β-actin and miR/U6 ratios in WT B cells were arbitrarily set as 1. The residual bands in Myc;miRfl/fl;CD19Cre B cells in (B) are likely from cross-hybridization with miRNAs encoded in the two homologous clusters, miR-106a∼363 and miR-106b∼25. (C) Flow cytometry analysis of splenocytes of 2-month-old mice of indicated genotypes (n=5–10 in each group). Splenic B cell (B220+CD19+) numbers are summarized in the right panel. (D) Kaplan–Meier survival curves of 31 Myc;miRfl/fl;CD19Cre and 27 littermate control Myc;miRfl/fl mice. The P-value was determined by Mantel–Cox logrank test.
Source data for this figure is available on the online supplementary information page.
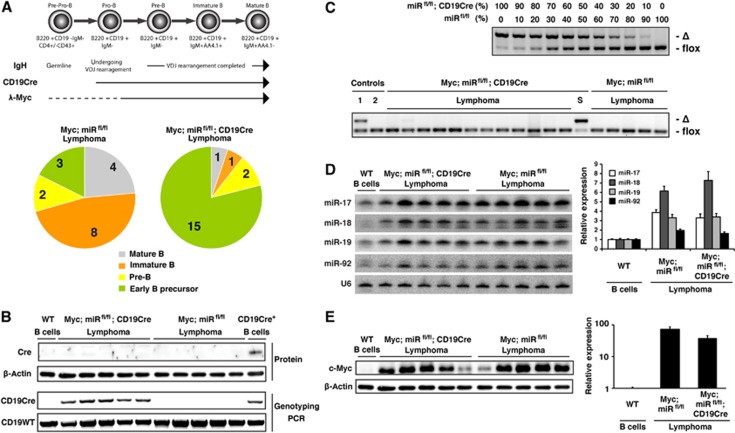
We next performed phenotypic and molecular analyses of those lymphomas to gain insights into mechanisms underlying the delayed lymphoma development in Myc;miRfl/fl; CD19Cre mice. Tumours in the control Myc;miRfl/fl group were predominantly mature and immature B-cell lymphomas, which are typical for λ-Myc mice (Kovalchuk et al, 2000) (Figure 4A and Supplementary Figure S3A). In contrast, the vast majority of Myc;miRfl/fl;CD19Cre lymphomas were IgM− and expressed markers characteristic of early precursor B cells (B220+CD19−CD4+CD43+IgM−) (Welner et al, 2008). These precursor B-cell lymphomas were found in only a small fraction of Myc;miRfl/fl mice (Figure 4A and Supplementary Figure S3A). The drastic shift in lymphoma types was further confirmed by Southern blot analysis of the IgH locus. Mature B and immature B-cell lymphomas in Myc;miRfl/fl mice exhibited clonal bands corresponding to VDJ or DJ rearrangements, whereas the IgH locus of all precursor B cell lymphomas in Myc;miRfl/fl; CD19Cre and Myc;miRfl/fl mice retained the germline configuration (Supplementary Figure S3B).
Figure 4.
Intact miR-17∼92 alleles are required for Myc-driven lymphomagenesis. (A) Distribution of lymphomas in Myc;miRfl/fl and Myc;miRfl/fl;CD19Cre mice based on developmental markers and IgH rearrangement status. Mature B-cell lymphoma, B220+CD19+IgM+AA4.1−; immature B-cell lymphoma, B220+CD19+IgM+AA4.1+; pre-B-cell lymphoma, B220+CD19+IgM−; and early B-lymphocyte precursor-derived lymphoma, B220+CD19−IgM−CD4+. The IgH locus of early B-lymphocyte precursor-derived lymphomas retain germline configuration, whereas other lymphomas display clonal VDJ or DJ rearrangements. Case numbers of each category are indicated. The dash line indicates opportunistic or low Myc expression in early B cell precursors. (B) Western blot analysis of Cre recombinase and Cd19 locus genotyping PCR of lymphoma cells of indicated genotypes. WT and CD19Cre+ B cells were used as controls. Note that the CD19Cre allele was generated by knocking in the Cre recombinase gene into the Cd19 locus. CD19Cre mice used in this study were heterozygous. (C) Genotyping PCR to detect deleted (‘Δ’) and floxed (‘flox’) miR-17∼92 alleles. Upper panel: B cells from miRfl/fl;CD19Cre and miRfl/fl mice were mixed at indicated ratios, and genomic DNA was extracted and used as PCR templates to evaluate the power of the assay. Lower panel: genomic DNA extracted from lymphomas of indicated genotypes were used as PCR templates, in comparison with lymph node samples of young Myc;miRfl/fl;CD19Cre (lane 1) and Myc;miRfl/fl (lane 2) mice, and purified B cells from one 19-month-old lymphoma-free survivor in the Myc;miRfl/fl;CD19Cre group (lane ‘S’). (D, E) Expression levels of miR-17∼92 (D) and c-Myc protein (E) in lymphoma cells of indicated genotypes were determined by northern (D) and western (E) blot analysis, respectively. Left panels show representative blots, and right panels summarize quantification results (n=5–6 in lymphoma groups and n=10 in the WT group for northern blot, and n=5–6 in each group for western blot). The miR/U6 and c-Myc/β-actin ratios in WT B cells were arbitrarily set as 1.
Source data for this figure is available on the online supplementary information page.
The preponderance of CD19-negative precursor B cell lymphomas in Myc;miRfl/fl;CD19Cre mice suggests that those lymphomas lack Cre expression and hence escape CD19Cre-mediated deletion of the floxed miR-17∼92 alleles. Supporting this hypothesis, none of those lymphomas expressed Cre protein (Figure 4B), and they all retained intact miR-17∼92 alleles, a pattern identical to Myc;miRfl/fl lymphomas (Figure 4C, lower panel). In the few cases of CD19-positive pre/immature/mature B-cell lymphomas developed in Myc;miRfl/fl;CD19Cre mice, the complete absence of miR-17∼92 deletion was also observed. It is likely that these tumours were derived from the small fraction of B cells with insufficient amounts of Cre expression (Rickert et al, 1997). Consistent with the presence of intact miR-17∼92 alleles, lymphomas that ultimately developed in Myc;miRfl/fl; CD19Cre mice displayed drastically induced miR-17∼92 miRNAs expression, to the same degree as that in Myc;miRfl/fl lymphomas (Figure 4D). In addition, the c-Myc protein level was comparable between Myc;miRfl/fl;CD19Cre and Myc;miRfl/fl lymphomas (Figure 4E). Thus, the delayed lymphoma development in Myc;miRfl/fl;CD19Cre mice reflects that malignant transformation is restricted to CD19-negative early B-cell precursors, in which the activity of λ-light chain regulatory elements and activation of the Myc transgene might be opportunistic or low, and to a small fraction of CD19-positive B-lineage cells with insufficient Cre expression. Taken together, these results highlight a stringent selection pressure to maintain intact miR-17∼92 alleles in Myc-driven lymphoma cells and emphasize an essential role of miR-17∼92 in Myc-mediated lymphomagenesis.
Identification of miR-17∼92 target genes in B cells by PAR-CLIP
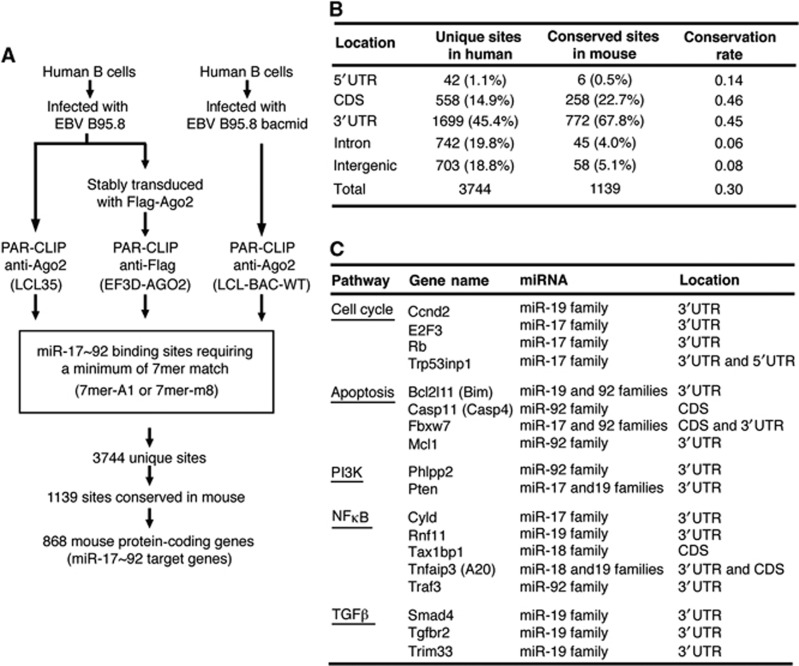
We next explored the molecular pathways through which miR-17∼92 drives lymphomagenesis and evaluated the possibility of targeting these pathways to treat lymphomas harbouring miR-17∼92 overexpression. We employed the recently developed PAR-CLIP method to examine the miR-17∼92 targetome in human B cells immortalized by Epstein-Barr virus (Figure 5A) (Hafner et al, 2010; Skalsky et al, 2012). Data obtained from three Ago2 PAR-CLIP libraries were combined, yielding 3744 unique miR-17∼92 binding sites. The majority (81.2%) of identified miR-17∼92 binding sites were present in protein-coding genes, and the rest (18.8%) were present in intergenic regions. Of the binding sites identified in protein coding genes, 1.1% corresponded to 5′UTRs, 14.9% to coding sequences (CDS), 45.4% to 3′UTRs, and 19.8% to intronic regions (Figure 5B).
Figure 5.
Identification of miR-17∼92 target genes by PAR-CLIP. (A) The scheme of PAR-CLIP analysis to identify miR-17∼92 target genes in B cells. (B) Location and evolutionary conservation of miR-17∼92 binding sites in the human and mouse genomes. (C) Select miR-17∼92 target genes identified by PAR-CLIP and previously implicated in cancer. Gene names in parentheses are commonly known aliases. Targeting miRNAs and locations of their binding sites are indicated. A full list of PAR-CLIP targets can be found in Supplementary Table S3.
Evolutionary conservation of miRNA–mRNA interactions, especially pairing at the seed region, has emerged as a major characteristic of miRNA-mediated gene regulation (Rajewsky, 2006; Bartel, 2009). Since miR-17∼92 family miRNAs are highly conserved in vertebrates (Tanzer and Stadler, 2004), we reasoned that their functionally important binding sites should be conserved between human and mouse. Among the 3744 miR-17∼92 binding sites identified by Ago2 PAR-CLIP, 1139 sites were conserved in mouse (Supplementary Table S3). Strikingly, about half of the CDS and 3′UTR sites were conserved, while the conservation rates for intronic and intergenic sites were only 6 and 8%, respectively, and only 6 of the 42 5′UTR sites were conserved. These findings suggest that miR-17∼92 functions mainly through its binding sites in the CDS and 3′UTR regions of target mRNAs, and are in agreement with previously published PAR-CLIP results from HEK293 cells (Hafner et al, 2010).
The 1139 conserved binding sites corresponded to 868 protein coding genes, including a few miR-17∼92 targets previously validated in various cellular contexts (i.e. Bim, E2F3, Pten, and Tgfbr2) (Sylvestre et al, 2007; Xiao et al, 2008; Dews et al, 2010; Mestdagh et al, 2010; Jiang et al, 2011). Pathway analysis using the Ingenuity software revealed that miR-17∼92 targets are involved in a broad spectrum of biological processes and identified ‘molecular mechanisms of cancer’ as the most enriched pathway (Supplementary Figure S4). From this category, we further narrowed down to a list of 18 target genes most likely implicated in lymphomagenesis based on published literatures, including those involved in cell cycle, apoptosis, nuclear factor kappa B (NFκB) pathway, phosphatidylinositol-3 kinase (PI3K) pathway, and transforming growth factor beta (TGFβ pathway (Figure 5C). We speculate that miR-17∼92 regulation of these genes underlies its role in lymphomagenesis.
Transgenic expression of miR-17∼92 promotes B-cell proliferation and survival
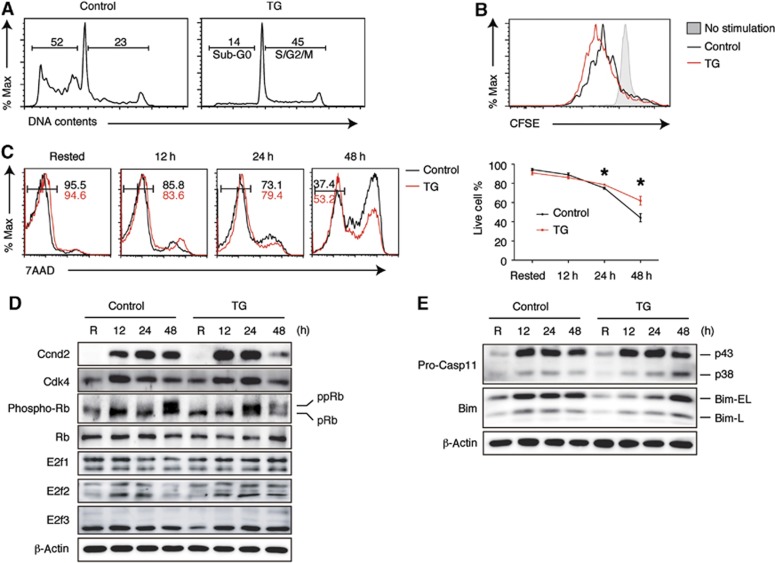
We evaluated the impact of transgenic miR-17∼92 expression on select target genes and pathways identified by PAR-CLIP. For this purpose, we purified naive follicular B cells (also known as B2 cells) from young and disease-free TG and control mice, stimulated them with anti-IgM, and examined the target gene protein levels and target pathway activities. In vitro activation is relevant in this case, because the majority of lymphomas developed in TG mice are DLBCL, FL, and AP, which are thought to originate from previously activated B cells (GC and post-GC B cells) (Shaffer et al, 2012). B2 cells from TG mice exhibited enhanced proliferation and survival upon stimulation when compared to control B2 cells, as demonstrated by an increased percentage of cells in the S/G2/M phases and a decreased percentage of cells in the sub-G0 phase (Figure 6A), accelerated cell division indicated by CFSE dilution (Figure 6B), earlier appearance of phosphorylated Rb (Figure 6D) and increased percentage of live cells in the culture (Figure 6C). Accordingly, PAR-CLIP-identified target genes, such as cell cycle regulator E2F3 and pro-apoptotic protein Bim, showed significantly reduced protein levels in TG B2 cells during early stages of activation (Figure 6D and E). We also examined the protein levels of E2F1 and E2F2, two E2F transcription family members that were previously shown to be miR-17∼92 target genes in various cell culture systems (Sylvestre et al, 2007). Their expression was not significantly affected by transgenic miR-17∼92 expression (Figure 6D). We examined additional target genes of relevance, including Rb and Casp11, as identified by PAR-CLIP, but found no significant downregulation in their protein levels. This suggests the existence of another layer of regulation beyond miRNA–target mRNA interaction (see Discussion). The protein level of another PAR-CLIP-identified target, Cyclin D2 (Ccnd2), was initially increased but decreased at a later time point in TG B2 cells, possibly a consequence of altered proliferation of these cells (Figure 6D). Therefore, transgenic miR-17∼92 expression suppresses the expression of E2F3 and Bim, and this may contribute to the enhanced proliferation and survival of TG B2 cells.
Figure 6.
Enhanced proliferation and survival of miR-17∼92 transgenic B cells. (A, B) B2 cells were stimulated with 10 μg/ml anti-IgM. Cell proliferation was analysed by propidium iodide staining at 48 h (A) and by CFSE dilution at 72 h (B). (C) B2 cells were stimulated with 2 μg/ml anti-IgM and cell death was analysed by 7-AAD staining. Numbers indicate the percentage of 7-AAD-negative cells. Line graph summarizes three independent experiments. Live cell percentage is defined by the percentage of 7-AAD-negative cells. *P<0.05. (D, E) Western blot analysis of miR-17∼92 target genes regulating cell cycle (D) and apoptosis (E) in B2 cells stimulated with 2 μg/ml anti-IgM. R, rested in B cell medium for 3 h without stimulation. pRb, underphosphorylated Rb; ppRB, phosphorylated and hyper-phosphorylated species of Rb. Note that target gene protein level changes at late time points may be affected by cell proliferation.
Source data for this figure is available on the online supplementary information page.
We next examined the TGFβ signalling pathway in TG B2 cells. TGFβ is a potent inhibitor of cell proliferation in the hematopoietic system. Resistance to TGFβ develops during lymphomagenesis, often through decreased expression of TGFβ receptors on the cell surface or repression of TGFβ signalling by oncoproteins (Dong and Blobe, 2006). Our PAR-CLIP analysis identified multiple components of the TGFβ signalling pathway as miR-17∼92 targets (Figure 5C). However, flow cytometry analysis of cell surface Tgfbr2 expression did not reveal any difference between TG and WT B2 cells (Supplementary Figure S5A). Western blot analysis of the protein level of Trim33, a PAR-CLIP-identified miR-17∼92 target and a key mediator of the Smad-independent TGFβ signalling branch (He et al, 2006), and Smad2 phosphorylation, an important step of the Smad-dependent TGFβ signalling branch, did not reveal any significant abnormality in TG B2 cells. Similarly, the protein levels of Smad4, another PAR-CLIP-identified target, are comparable in TG and WT B2 cells (Supplementary Figure S5B). These results suggest that transgenic miR-17∼92 expression does not have a major impact on the TGFβ signalling pathway.
miR-17∼92 enhances the NFκB and PI3K pathway activities by suppressing the expression of multiple negative regulators
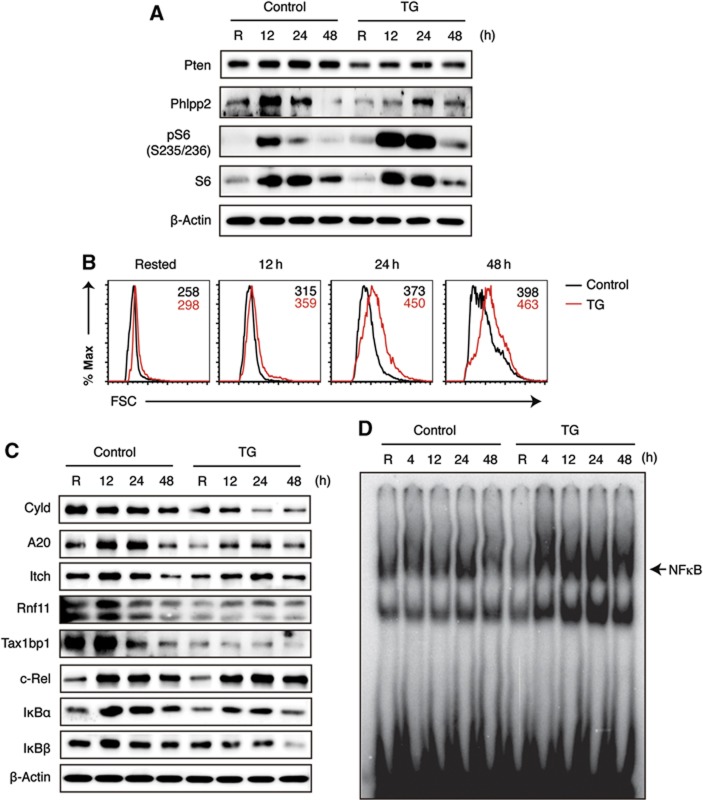
Human lymphomas often activate multiple oncogenic pathways, and genetic alterations of genes in the NFκB and PI3K pathways are among the most frequently found mutations (Shaffer et al, 2012). Interestingly, our PAR-CLIP analysis identified multiple negative regulators of these two pathways as miR-17∼92 target genes. Two miR-17∼92 targets, Pten and Phlpp2, are key inhibitors of the PI3K signalling pathway (Supplementary Figure S6C) (O'Neill et al, 2012; Song et al, 2012), and their expression was significantly repressed by transgenic miR-17∼92 expression (Figure 7A). The phosphorylation of ribosomal protein S6, a direct readout of the PI3K pathway activity, was significantly increased in TG B2 cells. Consistent with previous reports that S6 kinases (S6Ks) and the phosphorylation of its substrate S6 are involved in determining cell size (Ruvinsky and Meyuhas, 2006), TG B2 cells were significantly larger than their WT counterparts (Figure 7B). Therefore, transgenic miR-17∼92 expression enhances the PI3K pathway activity by suppressing the expression of two negative regulators, Pten and Phlpp2.
Figure 7.
Increased PI3K and NFκB pathway activity in miR-17∼92 transgenic B cells. (A) Western blot analysis of miR-17∼92 target genes regulating the PI3K pathway. pS6 indicates the PI3K pathway activity. (B) FACS analysis of cell size. Numbers indicate the geometric mean of forward scatter (FSC). (C) Western blot analysis of miR-17∼92 target genes regulating the NFκB pathway. Note that reduced IκBα and IκBβ protein levels correlate with increased nuclear NFκB activity in TG B2 cells (D). (D) Nuclear NFκB activity was analysed by EMSA. Purified B2 cells were stimulated with 2 μg/ml anti-IgM for indicated amounts of time. R, rested in B cell medium for 3 h without stimulation.
Source data for this figure is available on the online supplementary information page.
NFκB signalling can be dichotomized into the canonical and alternative pathways (Oeckinghaus and Ghosh, 2009). Two deubiquitinases, Cyld and the A20 ubiquitin-editing complex, downregulate the canonical NFκB pathway by removing pathway-activating K63-ubiquitin chains on key signalling molecules (Supplementary Figure S6C) (Harhaj and Dixit, 2011). Remarkably, our PAR-CLIP analysis identified Cyld and three major subunits of the A20 ubiquitin-editing complex (A20, Rnf11, and Tax1bp1) as miR-17∼92 target genes, and their protein levels were all significantly reduced in TG B2 cells (Figure 7C). The fourth major subunit of the A20 ubiquitin-editing complex, Itch, was previously identified as a miR-17∼92 target gene in the PAR-CLIP analysis of HEK293 cell (Hafner et al, 2010). Its expression was also decreased by transgenic miR-17∼92 expression (Figure 7C). The downregulation of these negative regulators led to enhanced BCR-induced activation of the canonical NFκB pathway, as indicated by reduced IκBα and IκBβ protein levels and increased nuclear NFκB activity in TG B2 cells (Figure 7C and D). Our PAR-CLIP analysis also identified Traf3, a negative regulator of the alternative NFκB pathway, as a miR-17∼92 target gene (Figure 5C). The protein level of Traf3 was significantly reduced in TG B2 cells, but this did not lead to an obvious increase in the activity of the alternative NFκB pathway (Supplementary Figure S6A and B). Thus, transgenic miR-17∼92 expression specifically enhances the canonical NFκB pathway activity.
Taken together, our analysis of PAR-CLIP-identified target genes and pathways in TG B2 cells demonstrated that transgenic miR-17∼92 expression promotes cell survival and proliferation by suppressing the expression of multiple inhibitors of the PI3K (Pten and Phlpp2) and NFκB (Cyld, A20, Itch, Rnf11, and Tax1bp1) pathways, the pro-apoptotic protein Bim, and cell cycle regulator E2F3 (Supplementary Figure S6C). The coordinated activation of these oncogenic pathways may eventually drive lymphomagenesis in TG mice.
Chemical inhibition of the PI3K and NFκB pathways prolonged the survival of mice bearing miR-17∼92-driven lymphomas
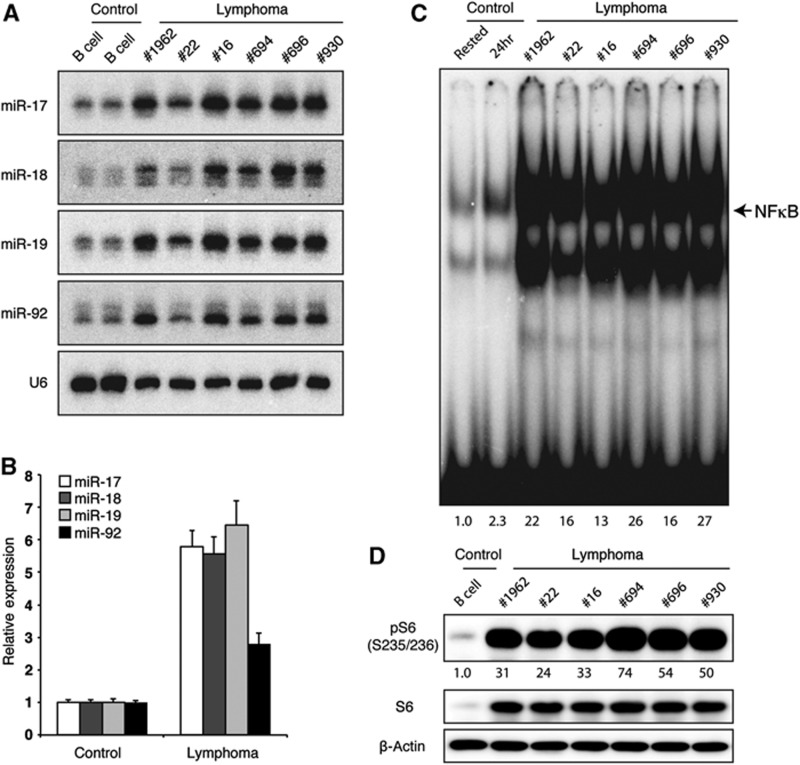
We next analysed the status of NFκB and PI3K pathways in TG lymphomas. For this purpose, we utilized the lymphoma cells purified from Rag1−/−-recipient mice transplanted with primary TG lymphomas. MiR-17∼92 expression in those lymphoma cells is 3–11-fold higher than that in WT B cells, and is similar to those found in human lymphomas (Figure 8A and B) (He et al, 2005; Schmitz et al, 2012). Consistent with our findings that transgenic miR-17∼92 expression promotes the activation of NFκB and PI3K pathways in TG B cells in young mice, all TG lymphomas analysed exhibit high levels of activities of both pathways (Figure 8C and D). These results suggest that miR-17∼92-mediated activation of these two pathways may play important roles in the maintenance of lymphoma cells, in addition to its function in driving lymphomagenesis.
Figure 8.
Constitutive NFκB and PI3K pathway activity in TG lymphoma cells. Primary TG lymphomas were transplanted into Rag1−/− mice and CD19+ lymphoma cells were purified from recipient mice ∼3 weeks post transplantation. (A) miR-17∼92 expression was determined by northern blot. (B) Bar graph summarizes two independent northern blot experiments (n=5 for control B cells and 10 for lymphomas). miR/U6 ratio in freshly purified control B cells was set as 1. (C) Nuclear NFκB activity was determined by EMSA. Purified control B cells rested in B cell medium for 3 h (rested) or stimulated with 2 μg/ml anti-IgM for 24 h (24 h) were used as control. NFκB activity in rested B cells was set as 1. (D) The PI3K pathway activity was determined by western blot analysis of pS6. pS6/β-actin ratio in freshly purified control B cells was set as 1.
Source data for this figure is available on the online supplementary information page.
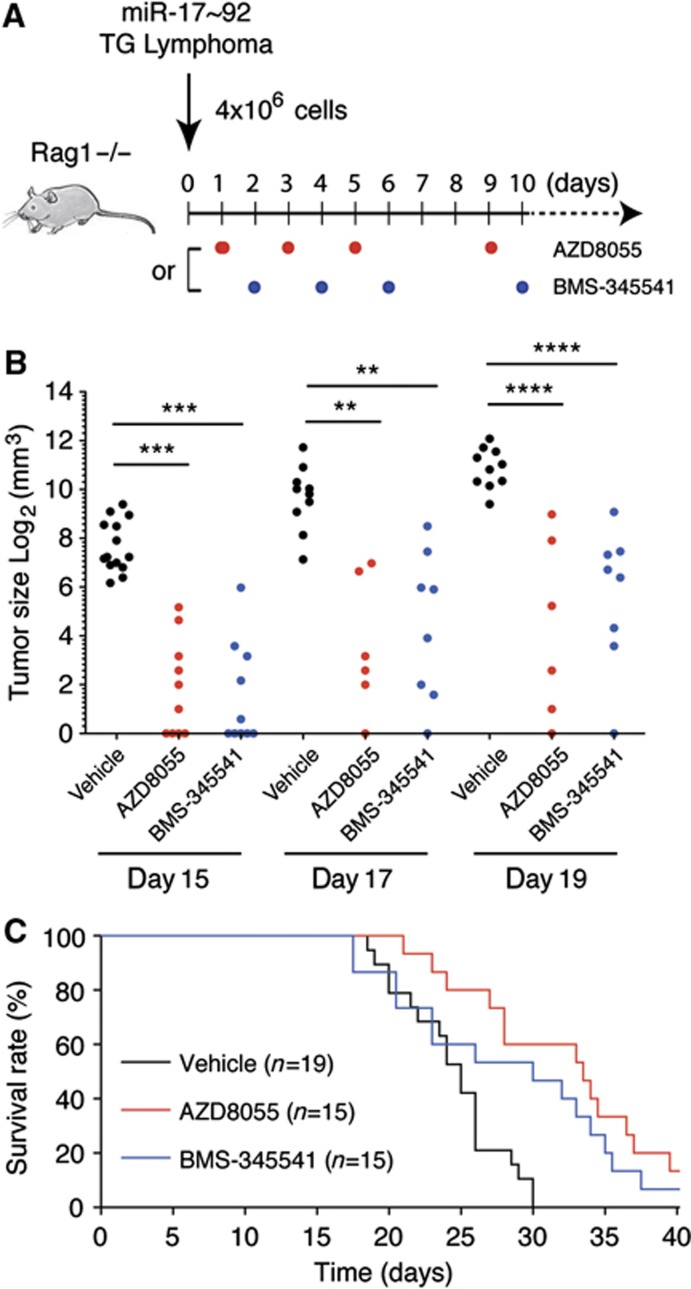
We explored the possibility of treating miR-17∼92-driven lymphomas using chemical inhibitors of the PI3K and NFκB pathways. For this purpose, we transplanted TG lymphoma cells into Rag1−/− mice and treated recipient mice with either PI3K pathway inhibitor AZD8055 (Chresta et al, 2010) or NFκB pathway inhibitor BMS-345541 (Burke et al, 2003). Consistent with previous reports, molecular analysis of lymphoma cells from chemical inhibitor-treated mice showed that AZD8055 inhibited the phosphorylation of Akt and S6 (Supplementary Figure S7A) and reduced lymphoma cell size (Supplementary Figure S7B), while BMS-345541 inhibited IκBα phosphorylation (Supplementary Figure S7C) and nuclear NFκB activity (Supplementary Figure S7D). Strikingly, both inhibitors significantly reduced tumour size and prolonged the survival of recipient mice (Figure 9A–C). Therefore, miR-17∼92-driven lymphomas are addicted to the PI3K and NFκB pathways and targeting these pathways has therapeutic value in treating lymphomas harbouring miR-17∼92 overexpression.
Figure 9.
Chemical inhibition of NFκB and PI3K pathways prolonged the survival of mice bearing miR-17∼92-driven lymphomas. (A) TG lymphoma transplantation and treatment scheme. (B) The size of tumour at injection site on day 15, 17, and 19 post transplantation. Tumour size below the measurable range was given an arbitrary value of 1. Each dot indicates one experimental mouse. **P<0.01; ***P<0.001; ****P<0.0001. (C) Kaplan–Meier survival curve analysis of recipient mice treated with vehicle, AZD8055 (P=0.0002), or BMS-345541 (P=0.0136).
Discussion
The present study shows that elevated miR-17∼92 expression is sufficient to drive lymphomagenesis and, conversely, miR-17∼92 is essential for Myc-mediated lymphoma development. Our molecular analyses revealed that miR-17∼92 drives lymphomagenesis by suppressing the expression of multiple inhibitors of the PI3K and NFκB pathways and by inhibiting the mitochondrial apoptosis pathway, that miR-17∼92-driven lymphoma cells exhibit constitutive activation of the PI3K and NFκB pathways, and that chemical inhibition of these two pathways has therapeutic value in treating miR-17∼92-driven lymphomas.
These findings firmly establish miR-17∼92 as a powerful cancer driver, and therefore, a valid target for cancer therapeutics. This places miR-17∼92 in the pantheon of prominent cancer genes that includes Myc, Ras, Pik3ca (p110α), Pten, and p53 (Schubbert et al, 2007; Liu et al, 2009; Olivier et al, 2010; Dang, 2012). The ubiquitous nature of miR-17∼92 gene amplification and overexpression in human cancers suggests that it plays important roles in other cancer types as well. This warrants a comprehensive investigation of miR-17∼92 in additional cancer models in the future.
Our findings of the essential role of miR-17∼92 in Myc-mediated lymphomagenesis and the promotion of PI3K pathway activation by miR-17∼92 are especially notable in the context of Burkitt lymphoma, which is characterized by Myc overexpression (Klapproth and Wirth, 2010). Recent studies showed that miR-17∼92 was drastically overexpressed and the PI3K pathway was constitutively active in a large number of Burkitt lymphoma patient biopsies (Schmitz et al, 2012), and that combining Myc overexpression and constitutive PI3K pathway activation in germinal centre B cells of the mouse led to the development of lymphomas that are similar to human Burkitt lymphomas (Sander et al, 2012). Considering these two studies together with ours, it becomes highly likely that the Myc →miR-17∼92 Pten/Phlpp2
Pten/Phlpp2 PI3K axis plays a fundamental role in Burkitt lymphomagenesis and may well be the Achilles’ heel of this disease. It will be interesting to investigate in the future whether PI3K pathway inhibitors are effective in treating Myc-driven lymphomas.
PI3K axis plays a fundamental role in Burkitt lymphomagenesis and may well be the Achilles’ heel of this disease. It will be interesting to investigate in the future whether PI3K pathway inhibitors are effective in treating Myc-driven lymphomas.
While bioinfomatic algorithms have predicted thousands of potential target genes for miR-17∼92 and a few of them (Bim, E2F1, E2F2, E2F3, p21, Pten, Rbl2, and Tgfbr2) were experimentally validated in various cellular contexts (O'Donnell et al, 2005; Lu et al, 2007; Sylvestre et al, 2007; Petrocca et al, 2008; Xiao et al, 2008; Dews et al, 2010; Mestdagh et al, 2010; Jiang et al, 2011), it was unclear which ones of the predicted targets are direct targets and whether miR-17∼92 regulates different sets of target genes in different cellular contexts. Our PAR-CLIP analysis identified 868 conserved target genes for miR-17∼92, including a few previously validated targets (Bim, E2F3, Pten, and Tgfbr2). Although the PAR-CLIP experiments were performed in B cells, most identified miR-17∼92 targets are broadly expressed genes, suggesting that miR-17∼92 regulates a common set of target genes in different cell types. Some miR-17∼92 target genes previously validated in other cellular systems (i.e., E2F1, E2F2, p21, and Rbl2) are absent from our PAR-CLIP target list, suggesting that miR-17∼92 regulates additional cell type-specific target genes. The discovery and validation of those target genes awaits CLIP analysis of other cell types, as well as generation and analysis of animal models with miR-17∼92 overexpressed or deleted specifically in the corresponding cell types.
Our validation of PAR-CLIP-identified target genes in miR-17∼92 transgenic B cells led to the elucidation of molecular mechanisms underlying its role in lymphomagenesis. The most striking finding is that miR-17∼92 suppresses the expression of multiple inhibitors of the PI3K and NFκB pathways. The PI3K pathway is perhaps the most commonly activated signalling pathway in human cancers. Activating mutations of Pik3ca (p110α), Pik3cb (p110β), Akt1, Akt2, Akt3, and deletion or loss-of-function mutations of Pten have been found in a broad spectrum of cancers including lymphomas (Liu et al, 2009). Interestingly, deletion of Pten alone in B cells did not cause lymphoma in mice (Miletic et al, 2010). We previously identified Pten as a miR-17∼92 target gene and this was confirmed in our PAR-CLIP experiments (Xiao et al, 2008). While the PI3K pathway activity was drastically increased in activated TG B cells, the reduction in the Pten protein level was relatively mild. This discrepancy was reconciled by the identification of another PI3K pathway inhibitor, Phlpp2, as a miR-17∼92 target gene by PAR-CLIP. Phlpp2 was recently discovered to be the long-sought Akt phosphatase, and its expression is lost or decreased in a variety of cancers (O'Neill et al, 2012). Consistent with a potentially critical role in mediating miR-17∼92 regulation of the PI3K pathway, the Phlpp2 protein level was markedly reduced in TG B cells. Thus, elevated miR-17∼92 expression enhances the PI3K pathway activity by suppressing the expression of two key negative regulators, Pten and Phlpp2.
The most commonly activated signalling pathway in human B-cell lymphomas is the NFκB pathway, which promotes the survival, activation, and proliferation of lymphocytes (Gerondakis and Siebenlist, 2010; Staudt, 2010). Deletion and inactivating mutations of Cyld and A20 are recurrent events in human lymphomas. These, together with genetic alterations of other signalling components, lead to the activation of the canonical NFκB pathway in a large percentage of lymphoma cases (Staudt, 2010). A recent study showed that constitutive activation of the canonical NFκB pathway in B cells, together with genetic ablation of Blimp1, a transcription factor essential for plasma cell differentiation, led to lymphoma development in the mutant mice (Calado et al, 2010). Our study found that miR-17∼92 enhances the canonical NFκB pathway activity by suppressing the expression of two major negative inhibitors of this pathway, Cyld and the A20 ubiquitin-editing complex (Harhaj and Dixit, 2011). The regulation of the latter is remarkable in that miR-17∼92 targets all four major subunits of the A20 complex (A20, Itch, Rnf11, and Tax1bp1). Given the frequent activation of the NFκB pathway in human lymphomas and their demonstrated oncogenic activity in mouse models, miR-17∼92-mediated regulation of this pathway should play an important role in lymphomagenesis.
In addition to the identification of the PI3K and NFκB pathways as two major target pathways of miR-17∼92, our study confirmed the previous findings that miR-17∼92 suppresses the expression of pro-apoptotic protein Bim and cell cycle regulator E2F3. Genetic studies have shown that E2F3 plays a critical role in normal cellular proliferation of mouse embryonic fibroblasts (Humbert et al, 2000; Wu et al, 2001). Further investigation is necessary to determine what roles it may play in lymphocytes and in miR-17∼92-driven lymphomagenesis. Members of the Bcl2 family are major regulators of the mitochondrial pathway of apoptosis in lymphocytes. Bim and Bcl2 associate with and antagonize each other (Rathmell and Thompson, 2002). Bcl2 overexpression by translocation, gene amplification, and transcriptional upregulation is commonly found in lymphomas, especially in follicular lymphomas (Shaffer et al, 2012). Homologous deletion of Bim was also found in mantle cell lymphomas (Tagawa et al, 2005). Accordingly, B cell-specific transgenic expression of Bcl2 led to B-cell hyperplasia and the development of lymphoma in a small fraction of mutant mice at old ages (McDonnell and Korsmeyer, 1991). In addition, both the Bcl2 transgene and heterozygous deletion of Bim drastically accelerated Myc-mediated lymphomagenesis in the Eμ-Myc transgenic mouse model (Strasser et al, 1990; Egle et al, 2004). Therefore, suppression of Bim expression by miR-17∼92 should contribute to its role in lymphomagenesis. Taken together, our PAR-CLIP analysis and target-gene validation in miR-17∼92 transgenic B cells demonstrate that miR-17∼92 coordinates the activation of multiple oncogenic pathways to drive lymphomagenesis (Supplementary Figure S6C).
The functional importance of the PI3K and NFκB pathways was further examined in a lymphoma transplantation model. We showed that chemical inhibitors blocking either of these two pathways significantly controlled tumour growth and prolonged the survival of mice bearing miR-17∼92-driven lymphomas. This demonstrates that miR-17∼92-mediated activation of these two pathways plays important roles in both lymphoma development and maintenance, and targeting these pathways has therapeutic value in treating lymphomas harbouring miR-17∼92 overexpression.
The present study combines target identification by PAR-CLIP and target validation in miRNA mutant animals for the first time, and sheds light on a few basic principles governing miRNA biology and the development of miRNA-based cancer therapeutics. First, our PAR-CLIP results show that the vast majority of evolutionarily conserved miRNA binding sites are found in 3′UTR and CDS of protein-coding genes. Some previous studies found miRNA-binding sites in 5′UTRs and intergenic regions, leading to the speculation that miRNAs can function through those sites (Lytle et al, 2007; Orom et al, 2008). Our results indicate that miRNAs function mainly by regulating the expression of protein-coding genes through their cognate sites in 3′UTR and CDS. This is consistent with the recently published PAR-CLIP analysis of HEK293 cells (Hafner et al, 2010). Second, most PAR-CLIP-identified miR-17∼92 target genes contain only one conserved binding site for miRNAs encoded by this cluster. This observation could have profound implications in interpreting the results from previous studies of miRNA mechanisms of action. Very often those studies employed reporter constructs that contained multiple binding sites for the miRNA of interest. The impact of a miRNA on a reporter mRNA containing multiple closely located binding sites could be very different from that on an endogenous target mRNA with only one binding site, in terms of the relative contribution of translation repression and mRNA degradation, as well as the degree of repression on target protein levels (Huntzinger and Izaurralde, 2011). Third, miRNA binding does not necessarily warrant functional consequence. MiR-17∼92 regulation of Tgfbr2 is a case in point. Several independent studies showed that Tgfbr2 is a direct target of miR-17∼92 and miR-17∼92 suppresses Tgfbr2 expression in various cellular contexts. The PAR-CLIP analysis in this study also identified Tgfbr2 as a miR-17∼92 target gene, with the same binding site as reported before (Dews et al, 2010; Mestdagh et al, 2010; Jiang et al, 2011). However, the surface expression of Tgfbr2, as well as TGFβ-induced Smad2 phosphorylation, was identical between TG and WT B cells. This cellular context dependence suggests that an additional layer of regulation exists and governs the functional consequence of miRNA–target mRNA interactions. RNA-binding proteins may participate in this regulation, as suggested by a few recent studies (van Kouwenhove et al, 2011). This emphasizes the importance of examining miRNA–target gene interactions in desired cells in animal models with gain- or loss-of-function mutations for miRNAs of interest. Fourth, this study confirms that a miRNA targets multiple components of functionally related pathways to generate a significant impact on the biological processes it regulates, as proposed by us and other investigators in previous publications (Krek et al, 2005; Xiao and Rajewsky, 2009). A cluster of miRNAs with different seed sequences may have additional advantages in this context, because they can target several components in each pathway to achieve a synergistic effect. This may explain why a modest increase in miR-17∼92 expression alone is sufficient to drive lymphomagenesis. Finally, this study shows that identification of cancer drivers and elucidation of their molecular mechanism of action are essential for the development of cancer therapeutics. The best-studied modality of miRNA-based therapy to date is to silence or replete cancer-related miRNAs using antisense miRNA inhibitors or miRNA mimics, respectively (van Rooij and Olson, 2012; Nana-Sinkam and Croce, 2013). However, the safety and efficacy of using these oligonucleotide-based agents to treat human cancers remain to be tested. In contrast, small chemical compounds targeting miRNA downstream pathways have been developed and many of them are in clinical trials. Conceptually, chemical compounds targeting functionally important pathways controlled by miRNAs may be effective in treating miRNA-related diseases. A thorough understanding of molecular mechanisms underlying miRNA action would be essential for this approach to work. The present study demonstrates, for the first time to our knowledge, that the combination of experimental target-gene identification by PAR-CLIP and target validation in miRNA mutant animals is a solid way to elucidate molecular pathways controlled by miRNAs, and that chemical inhibition of these pathways has therapeutic value in treating cancers caused by miRNA dysregulation.
Materials and methods
Mice
The generation of miR-17∼92 Tg (Jax stock 008517), miR-17∼92 flox (Jax stock 008458), CD19Cre (Jax stock 006785), and λ-Myc mice was previously reported (Rickert et al, 1997; Kovalchuk et al, 2000; Ventura et al, 2008; Xiao et al, 2008). MiR-17∼92 Tg mice were crossed with CD19Cre mice to generate miR-17∼92 Tg/Tg;CD19Cre (TG) mice. miR-17∼92 flox, CD19Cre, and λ-Myc mice were crossed to generate λ-Myc;miR-17∼92 flox/flox;CD19Cre (Myc;miRfl/fl;CD19Cre) and λ-Myc;miR-17∼92 flox/flox (Myc;miRfl/fl) mice. Cohorts of mice were monitored for disease development for up to 2 years, and euthanized for analysis when they appeared sick. Littermate control mice were euthanized as experimental control and were counted as ‘right-censored’ data in the Kaplan–Meier survival analysis (Stel et al, 2011). All animal experiments were approved by Animal Care and Use Committee of The Scripps Research Institute.
Lymphoma transplantation and chemical inhibitor treatment
Primary TG lymphoma cells (4 × 106) were injected subcutaneously into 2–4-month-old C57BL/6 or Rag1−/− mice. Recipient mice were monitored for up to 6 months for lymphoma development, and were euthanized for analysis when they appeared sick. Primary lymphoma cells from mouse no. 16 were expanded to a large quantity by transplanting into 5 Rag1−/− mice and harvesting ∼3 weeks post transplantation, and were used in the following chemical inhibitor treatment experiments. At day 0, 4 × 106 lymphoma cells were injected subcutaneously into Rag1−/− mice. AZD8055 (Selleckchem, S1555) was formulated in 30% (w/v) Captisol/PBS to produce a 2 mg/ml stock solution and injected i.p. into mice at 15 mg/kg body weight every other day starting on day 1. BMS-345541 (Axon medchem, Axon1731) was dissolved in PBS to produce a 10 mg/ml stock solution and injected i.p into mice at 70 mg/kg body weight every other day starting on day 2. Both chemical inhibitors were injected for three consecutive times, followed by a resting period of 3 days before the treatment is resumed.
Histology
Mouse tissues were fixed in 10% buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E). For diagnosis, serial sections were stained with anti-Ki-67 (Bethyl Laboratories), anti-CD138 (281-2, BD Biosciences), anti-Bcl6 (sc-368, Santa Cruz Biotechnology), anti-IRF4 (sc-56713, Santa Cruz Biotechnology), PNA-bio (B-1075, Vector Labs) using established immunohistological techniques. Staining was performed at San Diego Pathologists Medical Group, Inc. (San Diego, CA), and diagnosis was performed by an experienced subspecialist hematopathologist of human lymphomas according to Bethesda proposals for classification of lymphoid neoplasms in mice (Morse et al, 2002).
PAR-CLIP
Established, human B-cell-derived lymphoblastoid cell lines are infected with EBV B95.8 (EF3D-AGO2, LCL35) or EBV B95.8 Bacmid (LCL-BAC-WT) and were maintained in RPMI 1640 containing 15% FBS. EF3D-AGO2 stably expresses a FLAG-tagged Ago2. PAR-CLIP libraries were generated as described (Hafner et al, 2010; Skalsky et al, 2012). The PAR-CLIP experiments and pathway analysis of miR-17∼92 target genes are described in details in the Supplementary Materials and Methods.
Purification of B cells
Spleen and peripheral lymph nodes were collected from 8-week-old mice with indicated genotypes. B2 cells were purified by depleting cells positive for AA4.1 (CD93), CD43, or CD5 using MACS LD columns (Miltenyi Biotec) according to the manufacturer’s instructions. CD19-positive cells from tumour samples were purified using CD19 enrichment methods with LS columns (Miltenyi Biotec) according to the manufacturer’s instructions.
In vitro B-cell culture and proliferation assay
Single-cell suspension (1 × 106 cells/ml) was cultured in an atmosphere of 7.5% CO2 in B cell media and labelled with CFSE (Invitrogen/Molecular probes, Carlsbad, CA) according to the manufacturer’s instruction and stimulated with 10 μg/ml of anti-IgM antibody (Jackson, 115-006-020). For western blot, EMSA, and 7-AAD staining, purified B2 cells were cultured at 5 × 106 cells/ml and activated with various combinations of the following stimulants: 2 μg/ml anti-IgM, 5 ng/ml TGFβ (R&D 240-B), 100 ng/ml BAFF (R&D, 2106-BF), and 2 μg/ml anti-CD40 (eBioscience, 16-0402).
Statistical analysis
Data were analysed using unpaired two-tailed Student’s t-test (*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001) and Fisher’s exact test. Results are shown as mean with error bars indicating±s.e.m. (standard error of the mean). Kaplan–Meier survival curves were compared using the Log-rank test.
Supplementary Material
Acknowledgments
We thank A Feeney, D Nemazee, A Gonzalez Martin, N Choi, A Baumgart, J Hart, Y Kang, L Liao, G Fu, Y Kuwano, A Ramirez-Borrero, Y Sasaki, M. Schmidt-Supprian, and I Song for advice and technical assistance; TSRI Flow Cytometry Core Facility for their expert support; A Gonzalez Martin, P Sun, D Calado, and J Kefauver for critical reading of the manuscript and discussion. RLS was supported by T32-CA90111. CX is a Pew Scholar in Biomedical Sciences. This study is supported by the PEW Charitable Trusts, Cancer Research Institute, and National Institute of Health (R01 AI067968 to B.R.C., R01 AI087634 and RC1 CA146299 to C.X.).
Author contributions: HO, ML, HYJ, and CX designed research. CX made the initial observation of lymphoma development in mice with B-cell-specific transgenic miR-17∼92 expression in the laboratory of KR before relocating to TSRI. RLS performed PAR-CLIP experiments and and analysed results under the supervision of BRC. MS-G performed bioinformatic analysis of PAR-CLIP data and identified miR-17∼92 binding sites conserved in mouse. KB diagnosed all lymphoma cases. HO, ML, HYJ, JS, SGK, and W-HL performed all other experiments and analysed the data. HYJ, HO, ML, and CX wrote the manuscript with contribution from all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ambros V (2004) The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JR, Pattoli MA, Gregor KR, Brassil PJ, MacMaster JF, McIntyre KW, Yang X, Iotzova VS, Clarke W, Strnad J, Qiu Y, Zusi FC (2003) BMS-345541 is a highly selective inhibitor of I kappa B kinase that binds at an allosteric site of the enzyme and blocks NF-kappa B-dependent transcription in mice. J Biol Chem 278: 1450–1456 [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM (2007) microRNA functions. Annu Rev Cell Dev Biol 23: 175–205 [DOI] [PubMed] [Google Scholar]
- Calado DP, Zhang B, Srinivasan L, Sasaki Y, Seagal J, Unitt C, Rodig S, Kutok J, Tarakhovsky A, Schmidt-Supprian M, Rajewsky K (2010) Constitutive canonical NF-kappaB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer Cell 18: 580–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, Mo T, Murty VV, Dalla-Favera R (2005) Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell 7: 445–455 [DOI] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Gray JW (2008) Translating insights from the cancer genome into clinical practice. Nature 452: 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, Vincent JP, Ellston R, Jones D, Sini P, James D, Howard Z, Dudley P, Hughes G, Smith L, Maguire S, Hummersone M, Malagu K, Menear K, Jenkins R et al. (2010) AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res 70: 288–298 [DOI] [PubMed] [Google Scholar]
- Conkrite K, Sundby M, Mukai S, Thomson JM, Mu D, Hammond SM, MacPherson D (2011) miR-17∼92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev 25: 1734–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM (2009) Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10: 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV (2012) MYC on the path to cancer. Cell 149: 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva NS, Simonetti G, Heise N, Klein U (2012) The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunol Rev 247: 73–92 [DOI] [PubMed] [Google Scholar]
- Dews M, Fox JL, Hultine S, Sundaram P, Wang W, Liu YY, Furth E, Enders GH, El-Deiry W, Schelter JM, Cleary MA, Thomas-Tikhonenko A (2010) The myc-miR-17∼92 axis blunts TGFβ signaling and production of multiple TGFβ-dependent antiangiogenic factors. Cancer Res 70: 8233–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Blobe GC (2006) Role of transforming growth factor-beta in hematologic malignancies. Blood 107: 4589–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egle A, Harris AW, Bouillet P, Cory S (2004) Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci USA 101: 6164–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese KK, Tuveson DA (2007) Maximizing mouse cancer models. Nat Rev Cancer 7: 645–658 [DOI] [PubMed] [Google Scholar]
- Gerondakis S, Siebenlist U (2010) Roles of the NF-kappaB pathway in lymphocyte development and function. Cold Spring Harb Perspect Biol 2: a000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harhaj EW, Dixit VM (2011) Deubiquitinases in the regulation of NF-kappaB signaling. Cell Res 21: 22–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J (2006) Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFβ pathway. Cell 125: 929–941 [DOI] [PubMed] [Google Scholar]
- Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA (2000) E2f3 is critical for normal cellular proliferation. Genes Dev 14: 690–703 [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, Wan Y, He L, Li QJ (2011) Molecular dissection of the miR-17-92 cluster's critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood 118: 5487–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapproth K, Wirth T (2010) Advances in the understanding of MYC-induced lymphomagenesis. Br J Haematol 149: 484–497 [DOI] [PubMed] [Google Scholar]
- Kovalchuk AL, Qi CF, Torrey TA, Taddesse-Heath L, Feigenbaum L, Park SS, Gerbitz A, Klobeck G, Hoertnagel K, Polack A, Bornkamm GW, Janz S, Morse HC 3rd (2000) Burkitt lymphoma in the mouse. J Exp Med 192: 1183–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N (2005) Combinatorial microRNA target predictions. Nat Genet 37: 495–500 [DOI] [PubMed] [Google Scholar]
- Liu P, Cheng H, Roberts TM, Zhao JJ (2009) Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov 8: 627–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL (2007) Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol 310: 442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA (2007) Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3′ UTR. Proc Natl Acad Sci USA 104: 9667–9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, Parker JS, Paddison PJ, Tam W, Ferrando A, Wendel HG (2010) Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol 12: 372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell TJ, Korsmeyer SJ (1991) Progression from lymphoid hyperplasia to high-grade malignant lymphoma in mice transgenic for the t(14; 18). Nature 349: 254–256 [DOI] [PubMed] [Google Scholar]
- Mendell JT (2008) miRiad roles for the miR-17-92 cluster in development and disease. Cell 133: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Bostrom AK, Impens F, Fredlund E, Van Peer G, De Antonellis P, von Stedingk K, Ghesquiere B, Schulte S, Dews M, Thomas-Tikhonenko A, Schulte JH, Zollo M, Schramm A, Gevaert K, Axelson H, Speleman F, Vandesompele J (2010) The miR-17-92 microRNA cluster regulates multiple components of the TGFβ pathway in neuroblastoma. Mol Cell 40: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miletic AV, Anzelon-Mills AN, Mills DM, Omori SA, Pedersen IM, Shin DM, Ravetch JV, Bolland S, Morse HC 3rd, Rickert RC (2010) Coordinate suppression of B cell lymphoma by PTEN and SHIP phosphatases. J Exp Med 207: 2407–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse HC 3rd, Anver MR, Fredrickson TN, Haines DC, Harris AW, Harris NL, Jaffe ES, Kogan SC, MacLennan IC, Pattengale PK, Ward JM (2002) Bethesda proposals for classification of lymphoid neoplasms in mice. Blood 100: 246–258 [DOI] [PubMed] [Google Scholar]
- Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D'Andrea A, Sander C, Ventura A (2009) Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev 23: 2806–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nana-Sinkam SP, Croce CM (2013) Clinical applications for microRNAs in cancer. Clin Pharmacol Ther 93: 98–104 [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- O'Neill AK, Niederst MJ, Newton AC (2012) Suppression of survival signalling pathways by the phosphatase PHLPP. FEBS J 280: 572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeckinghaus A, Ghosh S (2009) The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol 1: a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P (2010) TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2: a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH (2008) MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30: 460–471 [DOI] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A (2008) E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13: 272–286 [DOI] [PubMed] [Google Scholar]
- Rajewsky N (2006) microRNA target predictions in animals. Nat Genet 38(Suppl): S8–S13 [DOI] [PubMed] [Google Scholar]
- Rathmell JC, Thompson CB (2002) Pathways of apoptosis in lymphocyte development, homeostasis, and disease. Cell 109(Suppl): S97–S107 [DOI] [PubMed] [Google Scholar]
- Rickert RC, Roes J, Rajewsky K (1997) B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res 25: 1317–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Meyuhas O (2006) Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci 31: 342–348 [DOI] [PubMed] [Google Scholar]
- Sander S, Calado DP, Srinivasan L, Kochert K, Zhang B, Rosolowski M, Rodig SJ, Holzmann K, Stilgenbauer S, Siebert R, Bullinger L, Rajewsky K (2012) Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell 22: 167–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Shaffer AL, Hodson DJ, Buras E, Liu X, Powell J, Yang Y, Xu W, Zhao H, Kohlhammer H, Rosenwald A, Kluin P, Muller-Hermelink HK, Ott G et al. (2012) Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490: 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G (2007) Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer 7: 295–308 [DOI] [PubMed] [Google Scholar]
- Shaffer AL 3rd, Young RM, Staudt LM (2012) Pathogenesis of human B cell lymphomas. Annu Rev Immunol 30: 565–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA, Tuschl T, Ohler U, Cullen BR (2012) The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog 8: e1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Salmena L, Pandolfi PP (2012) The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol 13: 283–296 [DOI] [PubMed] [Google Scholar]
- Staudt LM (2010) Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol 2: a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stel VS, Dekker FW, Tripepi G, Zoccali C, Jager KJ (2011) Survival analysis I: the Kaplan–Meier method. Nephron Clin Pract 119: c83–c88 [DOI] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Bath ML, Cory S (1990) Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 348: 331–333 [DOI] [PubMed] [Google Scholar]
- Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P (2007) An E2F/miR-20a autoregulatory feedback loop. J Biol Chem 282: 2135–2143 [DOI] [PubMed] [Google Scholar]
- Tagawa H, Karnan S, Suzuki R, Matsuo K, Zhang X, Ota A, Morishima Y, Nakamura S, Seto M (2005) Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene 24: 1348–1358 [DOI] [PubMed] [Google Scholar]
- Tanzer A, Stadler PF (2004) Molecular evolution of a microRNA cluster. J Mol Biol 339: 327–335 [DOI] [PubMed] [Google Scholar]
- Thomas M, Lieberman J, Lal A (2010) Desperately seeking microRNA targets. Nat Struct Mol Biol 17: 1169–1174 [DOI] [PubMed] [Google Scholar]
- van Kouwenhove M, Kedde M, Agami R (2011) MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 11: 644–656 [DOI] [PubMed] [Google Scholar]
- van Rooij E, Olson EN (2012) MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov 11: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T (2008) Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welner RS, Pelayo R, Kincade PW (2008) Evolving views on the genealogy of B cells. Nat Rev Immunol 8: 95–106 [DOI] [PubMed] [Google Scholar]
- Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, Greenberg ME, Orkin S, Nevins JR, Robinson ML, Leone G (2001) The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414: 457–462 [DOI] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K (2009) MicroRNA control in the immune system: basic principles. Cell 136: 26–36 [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K (2008) Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 9: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.