Abstract
Mitochondrial calcium uniporter (MCU) channel is responsible for Ruthenium Red-sensitive mitochondrial calcium uptake. Here, we demonstrate MCU oligomerization by immunoprecipitation and Förster resonance energy transfer (FRET) and characterize a novel protein (MCUb) with two predicted transmembrane domains, 50% sequence similarity and a different expression profile from MCU. Based on computational modelling, MCUb includes critical amino-acid substitutions in the pore region and indeed MCUb does not form a calcium-permeable channel in planar lipid bilayers. In HeLa cells, MCUb is inserted into the oligomer and exerts a dominant-negative effect, reducing the [Ca2+]mt increases evoked by agonist stimulation. Accordingly, in vitro co-expression of MCUb with MCU drastically reduces the probability of observing channel activity in planar lipid bilayer experiments. These data unveil the structural complexity of MCU and demonstrate a novel regulatory mechanism, based on the inclusion of dominant-negative subunits in a multimeric channel, that underlies the fine control of the physiologically and pathologically relevant process of mitochondrial calcium homeostasis.
Keywords: calcium homeostasis, channel, mitochondria, uniporter
Introduction
The notion that energized mitochondria accumulate Ca2+ in the matrix dates half a century ago, even before the chemiosmotic theory postulated the generation by the mitochondrial respiratory chain of an electrochemical gradient, negative inside, that provides the thermodynamic basis for cation accumulation into the matrix (Deluca and Engstrom, 1961). In the following years, the fundamental transport mechanisms were characterized (for a review, see Berridge et al, 2003 and Carafoli, 2010). Ca2+ uptake was shown to occur through a low-affinity electrogenic mechanism (hence the name mitochondrial calcium uniporter, MCU), inhibited by Ruthenium Red and lanthanides and was likely to be a channel (as directly demonstrated in 2004 by patch-clamp measurements; Kirichok et al, 2004).
In the last two decades, the direct measurement of [Ca2+] in the matrix ([Ca2+]mt) with recombinant targeted indicators (Rizzuto et al, 1992) demonstrated that mitochondria, upon cell stimulation, rapidly accumulate Ca2+ up to concentrations >100 μM (Montero et al, 2000). The apparent discrepancy with the low affinity of MCU was solved by the demonstration that mitochondria are located in close proximity to the Ca2+ source and exposed to microdomains of high [Ca2+] (Rizzuto et al, 1993, 1998; Csordas et al, 1999, 2010; Giacomello et al, 2010). As to the role of mitochondrial Ca2+ homeostasis, it became soon clear that the [Ca2+]mt changes modulate key cellular processes, such as aerobic metabolism (through Ca2+-sensitive dehydrogenases; McCormack et al, 1990; Denton, 2009) and the release of pro-apoptotic factors (Szalai et al, 1999 Pinton et al, 2001; Scorrano et al, 2003). On the cytosolic side, mitochondria act as large-capacity Ca2+ buffers that are responsible for compartmentalization of Ca2+ increases (Tinel et al, 1999) as well as local modulation of the activity of channels and enzymes (Boitier et al, 1999; Hajnoczky et al, 1999; Gilabert and Parekh, 2000; Hoth et al, 2000). These observations restored a great interest in mitochondria in the signalling field that was however frustrated by the lack of molecular insight into the process.
The past 2 years have witnessed the molecular unveiling of mitochondrial Ca2+ homeostasis. In 2010, the main efflux pathway (the NCX) (Palty et al, 2010), as well as a protein (named MICU1), that although not being itself a channel, appeared necessary for mitochondrial Ca2+ uptake (Perocchi et al, 2010) were identified. In 2011, a protein (named MCU) was identified (Baughman et al, 2011; De Stefani et al, 2011) and was shown, in intact cells and in reconstitution experiments, to be necessary and sufficient for channel activity with electrophysiological properties and inhibitor sensitivity of the MCU (De Stefani et al, 2011). Thus, the long awaited molecules corresponding to the uniporter and the exchangers characterized in the sixties and seventies were set in place, but fundamental issues remained to be solved, that in the case of MCU appeared truly critical. How can MCU, that possesses only two predicted transmembrane domains, form a functional channel? Genomic analysis identified a gene closely related to MCU: is this gene functional? And does the encoded protein play a role in MCU function? In this contribution, we addressed all these issues, with the aim of obtaining a comprehensive understanding of this novel channel that has no similarity to all other known calcium channels. Overall, the data show that the physiological activity of MCU is compatible with the interaction of functional and inactive pore-forming subunits, a unique regulatory mechanism allowing a great plasticity in the control of the fundamental process of mitochondrial Ca2+ uptake.
Results
The MCU isogene
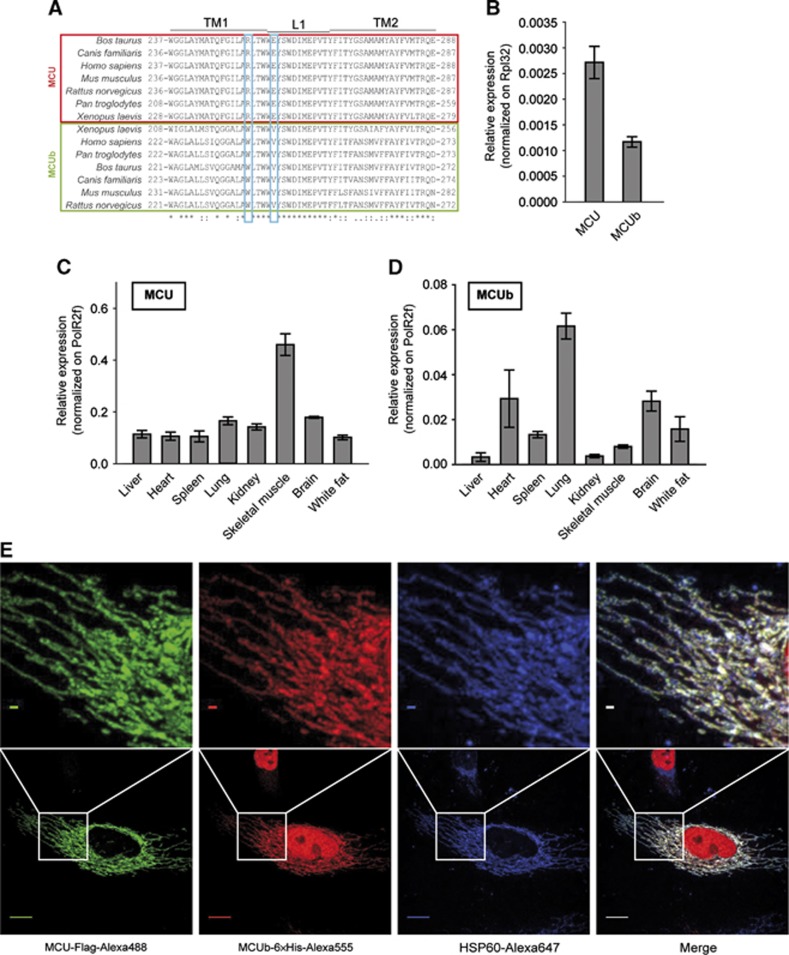
Sequence analysis of MCU (i.e., the gene originally named Ccdc109a) identified a related gene (Ccdc109b, NCBI GeneID 66815) located on Mus musculus chromosome 3 (chromosome 4 for the Homo sapiens orthologue). The gene is present in vertebrates but absent in other organisms in which MCU is present (e.g., plants, kinetoplastids, Nematoda, and Arthropoda). The encoded protein (∼330 amino acids long) is highly conserved among all species and shares a 50% similarity with MCU. It has two predicted transmembrane domains similar in sequence to MCU, although some conserved differences in the primary sequence are present (Figure 1A). RT–PCR analysis of HeLa cells and of a panel of mouse tissues reveals that CCDC109B has a lower expression level and a different expression profile from MCU (Figure 1B–D). Indeed, the mRNA encoded by the CCDC109B gene (hereafter named MCUb) is expressed at a ratio with MCU (MCU/MCUb) that, based on the RT–PCR data, varies from ∼3:1 (e.g., heart or lung) to >40:1 (skeletal muscle). We thus cloned and expressed the protein in HeLa cells. Immunofluorescence of transfected cells shows a complete overlap with MCU and the mitochondrial marker HSP60 (Figure 1E). However, the lack of any structural data about the native structure of the channel seriously limits all hypotheses on ion permeation through the channel. To circumvent this problem, we developed an in silico comparative model of the pore domain of the MCU.
Figure 1.
The MCU isogene. (A) Multiple alignment of the TM1, L1, and TM2 regions of MCU (red) and MCUb (green) in seven different species. Blue boxes show the two critical conserved substitutions. (B–D) Quantitative real-time PCR analysis of HeLa cells and mouse tissues of MCU and MCUb. (B) MCU and MCUb relative expression in HeLa cells. (C) MCU and (D) MCUb relative expression in the indicated mouse tissues as described in Materials and methods. All values are normalized to the indicated housekeeping genes. (E) Immunolocalization of MCUb. HeLa cells were transfected with MCUb-6 × His and MCU-Flag. After 24 h, the cells were fixed and immunocytochemistry was performed with α-Flag, α-6 × His, and α-HSP60 antibodies followed by incubation with Alexa488-, Alexa555-, and Alexa647-conjugated secondary antibodies as described in Materials and methods. Confocal images were taken (scale bar: 10 μm), and a region is expanded to appreciate co-localization (scale bar: 1 μm).
Predicted quaternary structure of the MCU
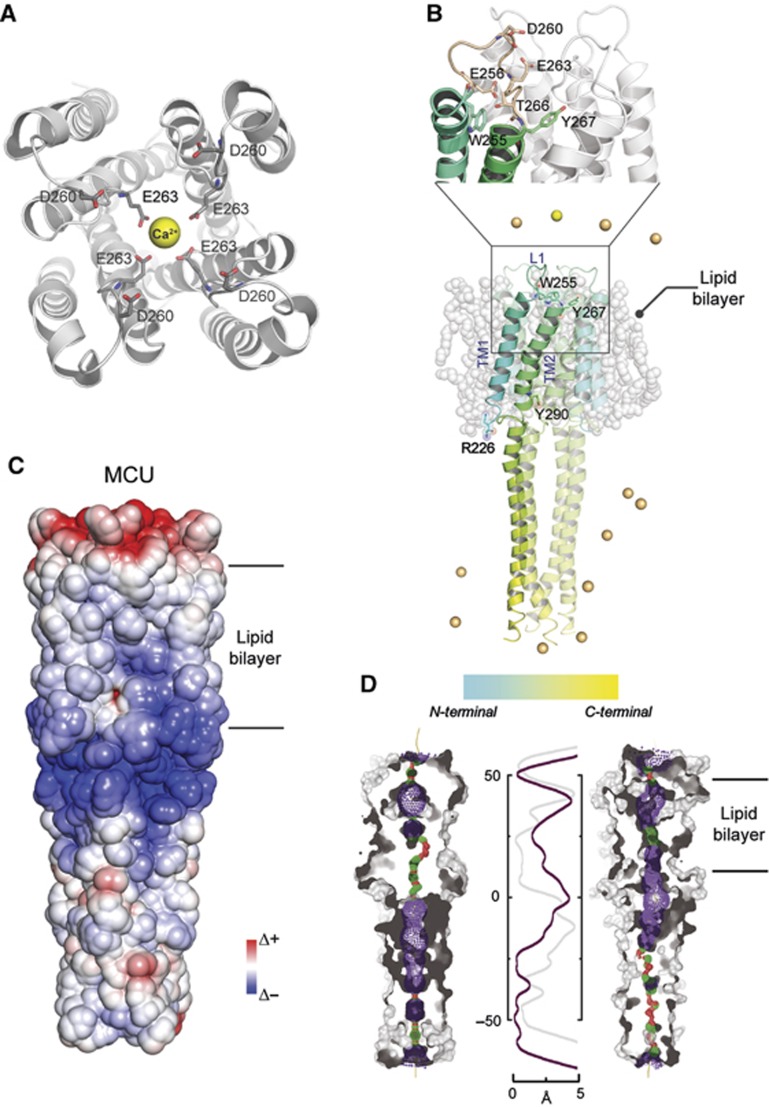
The combination of structural bioinformatics techniques and molecular dynamics (MD) simulations provides hypothesis of ion channel topologies for which the three-dimensional structure is yet unknown and of their behaviour in a lipid bilayer environment, in particular regarding the mechanism of ion permeation. We thus developed a comparative model of the pore domain of the MCU, and used it for membrane MD simulations on a nanosecond scale, as described in Materials and methods. Briefly, a multiple-template approach has been used to identify the possible structural organization. Four-fold rotational symmetry was imposed to the oligomer construction, as suggested by most of the available crystallographic data. A refinement procedure to optimize the quaternary assembly was carried out evaluating the best surface complimentary among each subunit using a protein–protein docking approach. The three-dimensional averaged structure obtained from the last 5 ns of MD simulation of membrane-embedded MCU model and the starting conformation of the channel protein have been used to investigate the effect of a membrane-like environment on modulating tetramer packing and its effect on the circumscribed aqueous pore topology evolution. The sequence identity between MCU and all crystallized ion channels is rather low, so their initial alignment was adjusted to maximize overlap between the predicted locations of the TM helices in MCU and their locations in the X-ray structure of different templates.
The final hypothetical model of the MCU pore domain linked to its C-terminus (residues 224–334) includes four identical subunits (Figure 2A), composed of two helical membrane spanning domains, connected by a short loop containing a DIME motif (Figure 2B). In particular, the region between R226 and W255 constitutes the first membrane spanning domain (TM1), whereas residues from Y267 to Y290 are part of the second helical segment (TM2), which protrudes outside the membrane region forming a long water exposed helical tail, as shown in Figure 2B. Finally, the region between E256 and T266 constitutes the water exposed loop (L1) where the DIME motif is located. This region includes a few negatively charged amino acids (such as D260 and E263) that have been shown to play an essential role in MCU-mediated mitochondrial Ca2+ uptake (Figure 2C) (Baughman et al, 2011; De Stefani et al, 2011). The three-dimensional averaged structure obtained from the last 5 ns of MD simulation of a membrane-embedded MCU model reveals the presence of a narrow selectivity filter constituted by the conserved acidic residues cited above which are able to locate a single Ca2+ above the channel pore region (Figure 2D). This feature is connected to a progressively widening chamber (3–5 Å in diameter), which extends underneath the channel mouth, through all the MCU membrane spanning domains. Surprisingly, the C-terminal tails, which are TM2 extensions directed through the solvent, define a pore of ∼1 Å wide in diameter suggesting that our MCU model is likely to stably adopt a closed state conformation, at least in the nanoseconds time scale. Such a pore topology is common among Ca2+ channels (Corry et al, 2001).
Figure 2.
Predicted quaternary structure of MCU. (A) Top view of the pore region of the predicted MCU tetramer. (B) Representation of the MCU model inserted into a POPC lipid bilayer. Indicated amino acids locate the boundaries among TM1, L1, and TM2. Zoomed region: E263 and D260 side chains face the pore region of the channel. E256 and T266 interaction is critical for loop conformation and dynamics. W255 and Y267 locate the upper boundaries of TM1 and TM2, respectively. N and C-terminal portion of the MCU monomers is highlighted according to the reported colour gradient bar. Chlorine and calcium ions are depicted as green and yellow spheres, respectively. (C) Electrostatic properties surface distribution of MCU. (D) Comparison of the pore width before (left) and after (right) insertion and equilibration into a lipid bilayer. The central panel shows the calculated width along the pore (before, grey trace; after, purple trace). Predicted MCU pore surface is depicted using red, green, and violet marks. Red: pore radius (R) is below 0.6 Å, green: 0.6 Å<R<1.15 Å and blue marks place where R is above 1.15 Å.
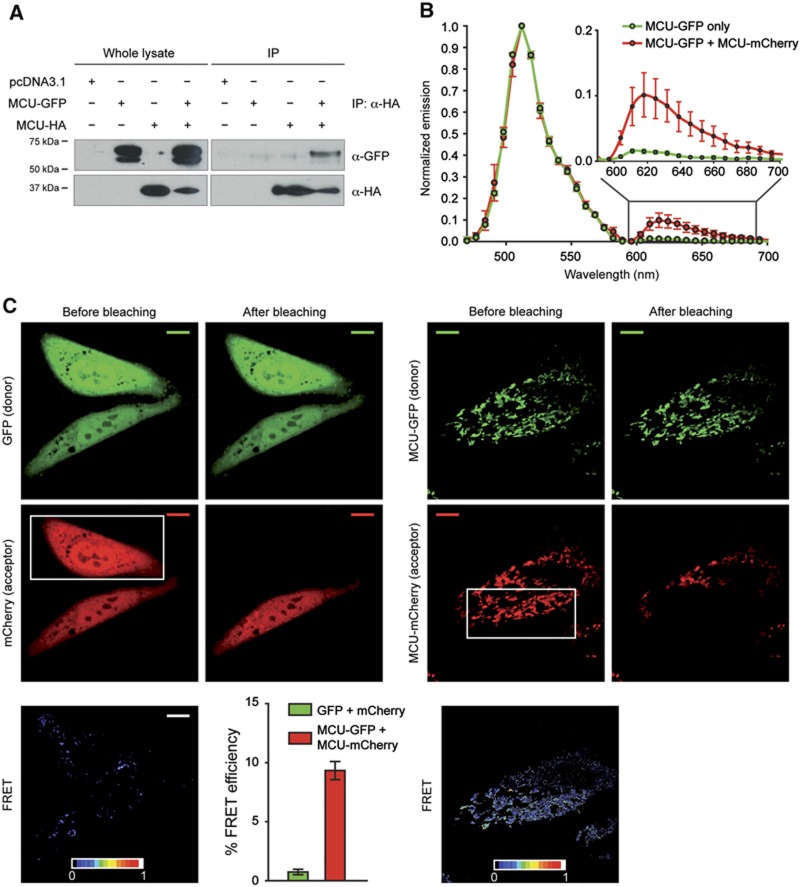
We then looked for experimental confirmation of the proposed oligomeric structure. Three approaches were followed. In the first, two different tags were added to MCU (MCU-GFP and MCU-HA) and the modified MCU constructs were recombinantly expressed in HeLa cells. Thirty-six hours after transfection, the cells were lysed and immunoprecipitation was carried out with the α-HA antibody. When MCU-GFP and MCU-HA were co-expressed, the α-HA antibody immunoprecipitated MCU-GFP (Figure 3A), thus revealing the interaction in situ of MCU monomers in a higher order complex. Then, we looked for confirmation of this result in a Förster resonance energy transfer (FRET) experiment in living cells (Figure 3B). For this purpose, an MCU-GFP (donor) and an MCU-mCherry (acceptor) chimera were generated and compared with two non-interacting fluorophores (GFP and mCherry). These two chimeras proved to be properly folded and functional (Supplementary Figure S1A). FRET was evaluated by emission spectrum analysis and acceptor photobleaching. In the first set of experiments, HeLa cells were transfected with the MCU-GFP or MCU-mCherry expression plasmids. Fluorescence emission spectra were recorded in the 470–700 nm range by exciting the donor (GFP) with the 458-nm laser line, in order to minimize (<1%) the cross-excitation of the acceptor (mCherry), while donor-only transfected cells show the expected spectrum of the GFP (Figure 3B). In contrast, cells cotransfected with both MCU-GFP and MCU-mCherry clearly show a secondary emission peak at 615 nm, due to the energy transfer between donor and acceptor molecules (Figure 3B). We confirmed the occurrence of FRET by monitoring fluorescence acceptor photobleaching in MCU-GFP and MCU-mCherry expressing HeLa cells. mCherry was bleached in a defined region with a 592-nm high power laser, and the changes in MCU-GFP (excitation at 488 nm) and MCU-mCherry (excitation at 543 nm) fluorescence were measured. When FRET occurs, acceptor photobleaching leads to the de-quenching and the consequent increase in donor fluorescence. Thus, FRET was calculated as the normalized increase in donor fluorescence after acceptor bleaching. Figure 3C shows a representative experiment: when MCU-GFP and MCU-mCherry were co-expressed, a significant FRET occurred (9.333±3.256%), whereas minimal FRET was detected when GFP and mCherry were not fused to MCU (0.763±0.980%). The calculated efficiency is in line with other reports using the same FRET pair (van der Krogt et al, 2008; Goh et al, 2011) and does not correlate with the expression level of the fluorescent proteins. Finally, we loaded in a native gel and immunoblotted in vitro translated MCU (De Stefani et al, 2011), detecting a band at the expected molecular weight of the monomer (40 kDa) and a higher band at 170 kDa (reactive with 6 × His antibody), which is compatible with a tetramer (Figure 4C, left panel), indicating that MCU monomers oligomerize both in vitro and in vivo in higher order complexes, and thus support the tetrameric model of the computational analysis.
Figure 3.
MCU forms oligomers in vitro and in vivo. (A) Co-immunoprecipitation experiments. HeLa cells were transfected with the indicated constructs. HA-tagged MCU was immunoprecipitated from cell extracts with a specific α-HA antibody. The precipitated proteins were immunoblotted with α-HA and α-GFP antibodies. (B) Emission spectra analysis of HeLa cells transfected with MCU-GFP or MCU-GFP and MCU-mCherry and analysed after 24 h. (C) FRET analysis. HeLa cells were transfected with GFP and mCherry or MCU-GFP and MCU-mCherry and analysed after 24 h. Images of donor and acceptor were taken before and after photobleaching the indicated region (white box). FRET was calculated as detailed in Materials and methods. Histogram bar diagram shows FRET efficiency of the indicated donor and acceptor pairs. Descriptive statistics can be found in Supplementary Table S1.
Figure 4.
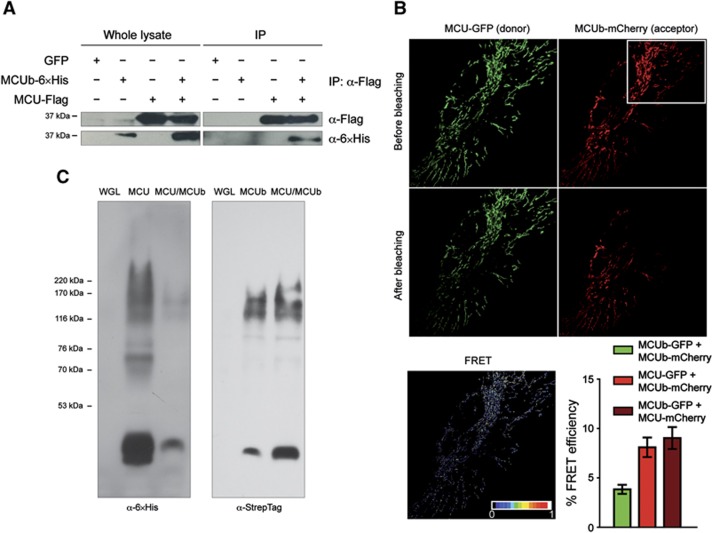
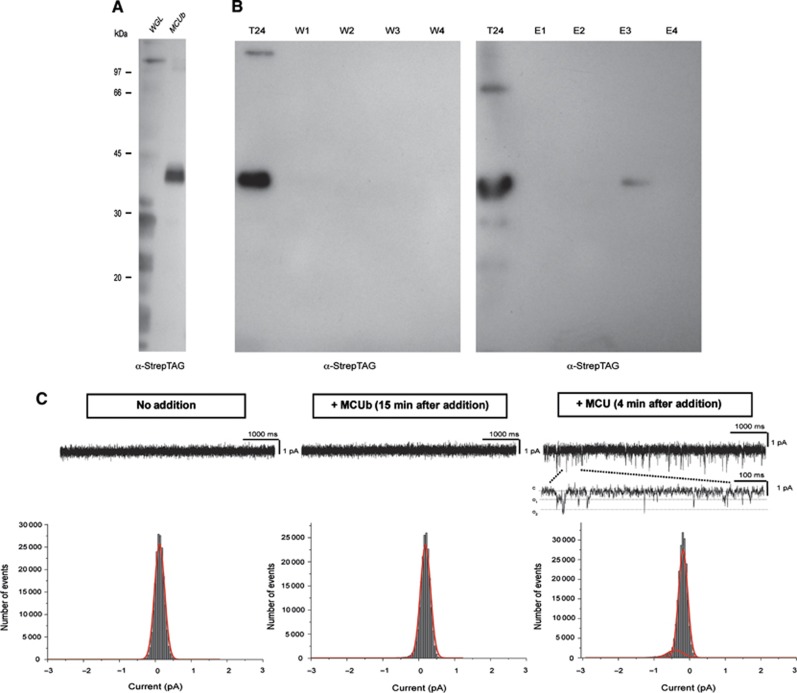
MCU and MCUb form hetero-oligomers. (A) Co-immunoprecipitation experiments. HeLa cells were infected with the indicated adenoviruses. Flag-tagged MCU was immunoprecipitated from cell extracts with a specific α-Flag antibody. The co-immunoprecipitated proteins were immunoblotted with α-Flag and α-6 × His antibodies. (B) FRET analysis. HeLa cells were transfected with MCU-GFP and MCUb-mCherry and analysed after 24 h. Images of donor and acceptor were taken before and after photobleaching of the indicated region (white box). FRET was calculated as detailed in Materials and methods. Histogram bar diagram shows FRET efficiency of the indicated donor and acceptor pairs. Descriptive statistics can be found in Supplementary Table S1. (C) In vitro expression. wheat germ lysate expressing MCU-6 × His or MCUb-StrepTag alone and co-expressing MCU-6 × His/MCUb-StrepTag (2:2 ratio) was loaded on a native polyacrylamide gel without denaturing the samples. Blots were developed with anti-6 × His and anti-StrepTag antibodies.
MCU and MCUb form hetero-oligomers
In view of the proposed oligomeric structure of MCU and given the predicted structural similarity with MCUb, we investigated whether the two proteins interact within the MCU oligomer with the same approach employed in Figure 3. At first, MCU-Flag and MCUb-6 × His were expressed in HeLa cells, and the α-Flag antibody immunoprecipitated also MCUb-6 × His, thus revealing the in situ interaction of MCU and MCUb (Figure 4A). We then carried out FRET analysis of the interaction, by generating and imaging different combinations of GFP- and mCherry-tagged MCU and MCUb proteins. As for MCU, also MCUb chimeras proved to be functional as they affected mitochondrial calcium uptake in intact cells (Supplementary Figure S1B). FRET was evaluated by acceptor photobleaching as in Figure 3. Representative fluorescence images of the MCU-GFP (donor) and MCUb-mCherry (acceptor) pair are shown in Figure 4B. A significant FRET was observed (8.090±3.700%), with an efficiency very similar to that generated by the MCU self-oligomerization. Similar results were obtained by switching donor and acceptor (with MCUb-GFP as a donor and MCU-mCherry as an acceptor, FRET efficiency is 9.029±4.151%). Importantly, a detectable, but lower FRET was measured also between MCUb monomers, using an MCUb-GFP and MCUb-mCherry pair (3.831±1.660%), thus indicating that MCUb also can self-oligomerize (Figure 4B). This experiment was repeated in cells silenced for MCU in order to exclude the indirect interaction due to the endogenous MCU (Supplementary Figure S2). Finally, wheat germ lysate expressing MCU or MCUb alone and co-expressing MCU/MCUb was loaded on a native polyacrylamide gel without denaturing the samples, clearly showing MCU and MCUb monomers at the expected molecular weight (40 kDa) and a higher band (170 kDa) compatible with a tetramer and reactive with both anti-6 × His (for MCU) and anti-StrepTag (for MCUb) antibodies (Figure 4C).
MCUb acts as an endogenous dominant-negative MCU subunit
We then investigated the function of MCUb. First, we obtained indication of an altered ion permeation pathway from molecular modelling. From a structural point of view, crucial differences between MCU and MCUb are located in the ‘DIME motif’ such as the replacement with a valine of one of the three conserved negatively charged residues of the N-terminal portion of the loop region (V251, corresponding to E256 in MCU as also depicted in Figure 1A, blue boxes). This crucial E256V substitution in MCUb might have an important impact on the kinetic of Ca2+ permeation as expected by the comparison of the surface electrostatic potentials between MCU (more negatively charged) and MCUb (less negatively charged).
Based on the computational model, MCUb was thus expected to be poorly permeable to Ca2+. We tested this prediction by two different approaches: (i) in vitro analysis of MCUb channel properties and (ii) in situ investigation of the role of MCUb in mitochondrial Ca2+ handling. In the first case, the MCUb protein was either produced in vitro or expressed in E. coli (Figure 5A and B), purified and inserted in planar lipid bilayers, then electrophysiological recordings were carried out (Figure 5C). Under our recording conditions, in 100 mM calcium-gluconate, no channel activity was detected upon addition of purified MCUb into the cis chamber, whereas the subsequent addition of MCU to the same membrane gave rise to channel activity with a conductance of 7 pS, typical of MCU (Figure 5C). The lack of channel activity might have been due to misfolding of MCUb. To prove that this was not the case, we recorded the activity of the same protein preparation in a sodium-gluconate low divalent solution (10 pM calculated [Ca2+]), given the known characteristic of calcium channels (Hess and Tsien, 1984; Lepple-Wienhues and Cahalan, 1996; Talavera and Nilius, 2006) and of MCU (Kirichok et al, 2004) to allow the passage of Na+ upon removal of Ca2+ (Supplementary Figure S3). Indeed, an Na+ current was observed indicating that MCUb gives rise to a functional channel, albeit incapable of significant Ca2+ permeation.
Figure 5.
MCUb has no channel activity in planar lipid bilayer. (A) In vitro expression of MCUb. Empty wheat germ lysate (WGL) and WGL after expression of MCUb-StrepTag were loaded on SDS–PAGE and blotted with α-StrepTag antibody. (B) Induction and purification of MCUb in E. coli. Bacteria were harvested after induction (T24) to check for the expression of the protein. Solubilized membranous fraction was passed through Strep-Tactin column; after washing (W1–W4), protein was eluted with 2.5 mM desthiobiotin (E1–E4). All samples were blotted and developed with α-StrepTag antibody. In all, 30 μl of eluted fractions/lane was loaded. (C) Electrophysiological recordings: in vitro expressed MCUb was added to the cis side (middle panel) and current was recorded for at least 10 min (n=5) without observing channel activity in 100 mM calcium-gluconate solution. Amplitude histograms, obtained from analysis of 50 s current traces recorded at −80 mV Vcis before (left panel) and 15 min after addition of MCUb (middle panel). Following addition of excess MCU (not incorporated into liposome) to the same experiment (right panel), spiky channel activity with a conductance of 7 pS has appeared (n=3). In the lower current trace, representative channel activity is shown in an extended time scale. The open probability of MCU was compatible with that previously reported for the channel recorded in the same condition (De Stefani et al, 2011). Lack of channel activity for MCUb in calcium-gluconate was also observed using the protein incorporated into liposomes (n=4).
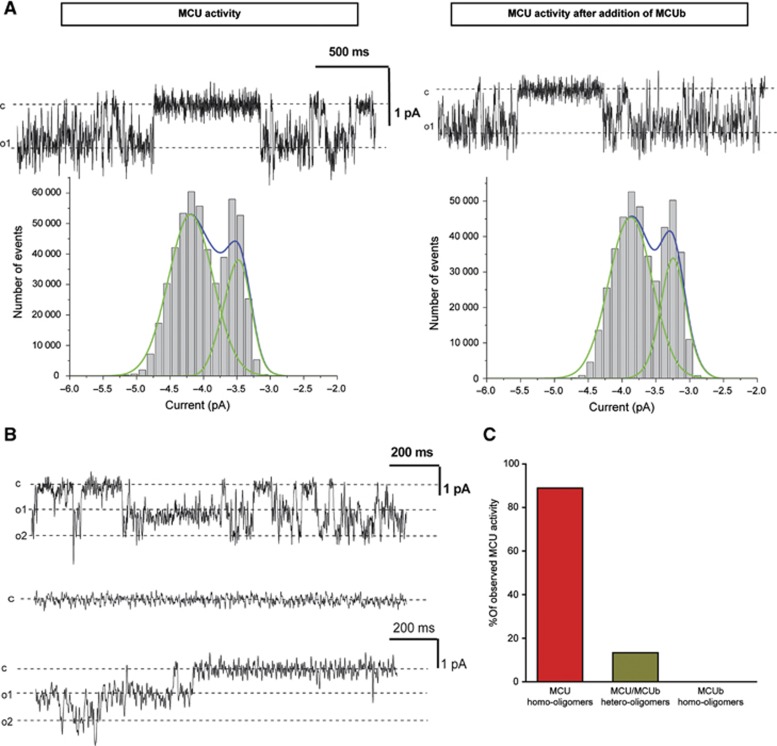
The lack of MCUb channel activity in calcium is compatible with MCUb being a dominant-negative form of MCU, similarly to the silent mutant subunits observed for various ion channels (Lafreniere et al, 2010; Jeanguenin et al, 2011). Addition of MCUb to active homomeric MCU, already incorporated into the bilayer, did not change either conductance or open probability and kinetic behaviour of the MCU channel (as expected, given that subunit switch is unlikely to occur in lipid bilayer experiments) (Figure 6A). Therefore, we co-expressed in vitro MCU and MCUb using ratios of plasmid DNA yielding different protein expression levels (Supplementary Figure S4A), selecting a plasmid ratio (MCUb:MCU=2:2 or 3:1) that gave near equimolar or 2:1 amounts of the two proteins. MCU-only, MCUb-only or the co-expressed proteins were incorporated into liposomes (Supplementary Figure S4B) and their activity was assessed in electrophysiological experiments (Figure 6B). When the two proteins were co-expressed, the number of experiments in which we observed MCU activity in calcium (due to the presence of homomeric MCU, statistically expected to be present in the co-expressed preparation) became drastically reduced to 13% compared to MCU alone (89%) under the same recording conditions (Figure 6C). In 13% of the experiments, we observed activity with the same conductance of the MCU homomer (7 pS). These data thus indicate that MCUb subunits, when forming heteromers with MCU alter calcium permeation across the heteromeric channel, thus acting as a bona fide dominant-negative subunit.
Figure 6.
MCU activity in the presence of MCUb. (A) Addition of excess MCUb during the same experiment does not alter the electrophysiological properties of MCU activity. Current traces recorded at −100 mV before and 6 min after addition are shown. Conductance values are 6.7 and 6.4 pS, respectively. Mean open time constants (280 ms for MCU and 360 ms after addition of MCUb) were similar. Below: respective amplitude histograms are shown. The open probability was 0.498 before and 0.513 after addition of MCUb. Similar results were obtained in two other experiments. (B) Activities observed with homomeric MCU (upper trace, representative of 8 experiments) or heteromeric MCU/MCUb (representative of 13 experiments) in liposome recorded at −140 mV are shown (middle trace). Lower current trace: in 2 cases out of 15 we recorded the activity shown using the heteromer preparation (3:1 ratio), which displayed the same characteristics as homomeric MCU. (C) Histogram showing the percentage of experiments in which activity was observed with the different preparations studied under the same conditions (MCU in 8 out of 9 cases (89%); MCU/MCUb co-expressed in 2 out of 15 cases (13%); MCUb in 0 out of 4 cases (0%).
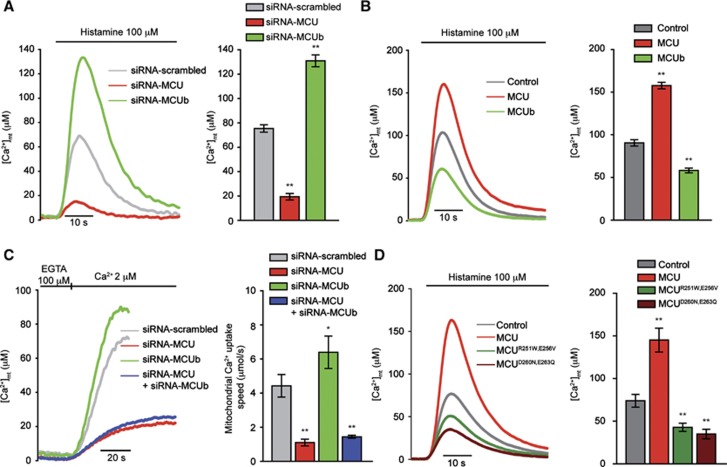
Then, we proceeded to Ca2+ measurements in intact cells in which MCUb was either silenced or overexpressed. In the first case, siRNAs were synthesized and tested by quantitative real-time PCR and western blot (Supplementary Figure S5A and B), and then mitochondrial Ca2+ measurements were carried out. MCUb silencing caused a significant increase in the histamine-induced [Ca2+]mt peak, that is, the opposite effect of MCU silencing (Figure 7A; Supplementary Figure S6). MCUb overexpression not only did not increase the [Ca2+]mt rise evoked by 100 μM histamine (as expected for an inactive protein), but rather markedly reduced it, indicating that it interferes with the function of the endogenous protein (Figure 7B). The same effect was observed in other cell lines (Human embryonic kidney 293 cells (HEK 293) and neonatal mouse cardiac fibroblasts) (Supplementary Figure S7A and B). Experiments were then carried out to exclude that the observed effect was secondary to changes in MCU protein expression, cytosolic Ca2+ transients or in the driving force for mitochondrial Ca2+ accumulation. HeLa cells overexpressing MCUb show no changes in MCU protein expression (Supplementary Figure S8) and cytosolic Ca2+ transients were unaffected by MCUb silencing and overexpression (Supplementary Figure S9A). As for the membrane potential, both MCUb silencing and overexpression did not alter mitochondrial TMRM loading (Supplementary Figure S9B). This modulation of mitochondrial Ca2+ uptake by MCUb was confirmed by experiments carried out in permeabilized cells, in which mitochondrial Ca2+ accumulation was initiated by switching the perfusion medium from a Ca2+-free intracellular buffer (IB) to IB supplemented with an EGTA-buffered [Ca2+] of 2 μM. Also in this experiment, MCUb silencing increased the uptake rate, whereas MCU silencing markedly reduced it; in the latter case, MCUb silencing did not rescue Ca2+ uptake (Figure 7C).
Figure 7.
MCUb acts as a dominant negative on MCU. (A) [Ca2+]mt measurements in control, MCU- and MCUb-silenced intact HeLa cells challenged with 100 μM histamine. (B) [Ca2+]mt measurements in intact HeLa cells overexpressing MCU or MCUb. (C) [Ca2+]mt measurements in control, MCU-, MCUb- and MCU/MCUb-silenced permeabilized HeLa cells perfused with 2 μM buffered [Ca2+]. (D) [Ca2+]mt measurements in intact HeLa cells overexpressing MCU, MCUR251W,E256V, and MCUD260N,E263Q. Descriptive statistics can be found in Supplementary Table S1. *P<0.05, **P<0.001.
Finally, we investigated whether introducing in MCU the critical amino-acid residues of MCUb could affect mitochondrial Ca2+ uptake. Specifically, the Arg 251 and the Glu 256 residues were mutated in Trp and Val, respectively (Figure 1A). The mutant protein was then expressed in HeLa cells, and mitochondrial Ca2+ transients evoked by histamine stimulation were assessed. The results (Figure 7D) show that in MCUR251W,E256V expressing cells, the [Ca2+]mt rises evoked by 100 μM histamine were markedly reduced. This effect is qualitatively and quantitatively very similar to the MCUD260N,E263Q mutant that we previously characterized (Figure 7D) and to MCUb.
Discussion
The recent identification of MCU has opened the way to the molecular elucidation and experimental manipulation of a key event in calcium signalling, but also raised new intriguing biological questions. Structure/function relationship cannot be easily inferred, due to the lack of experimental structural information and poor homology with any known Ca2+ channels. To circumvent this problem, we pursued a computational approach that allowed us to model the pore-forming domain of MCU and to test its predicted stability in an artificial membrane environment. Oligomerization appeared the only possible conformation allowing ion permeation and, although other stoichiometries were in principle thermodynamically possible, a tetramer was by far the most likely quaternary structure. The computational model of an oligomer was confirmed by three independent approaches, co-immunoprecipitation of tagged MCU monomers, FRET analyses, and immunoblot of in vitro expressed MCU. In the predicted quaternary structure eight helices line the putative pore region, in good agreement with other cation channels, and the clustering of charged residues in proximity of the pore of the channel generates a negative electrostatic potential favouring the flux of a cation.
The concept of MCU oligomerization opens the possibility that, together with MCU, other related proteins could be components of the channel complex. We were thus intrigued by the presence of a gene closely related to that encoding MCU. The encoded protein has a high similarity, and also includes two highly conserved putative transmembrane domains, separated by a short loop region including negatively charged residues. However, amino-acid sequence differences in the protein domain predicted to be critical for ion permeation suggested that this protein has a lower efficiency in calcium conduction. The data in reconstituted lipid bilayers showed that indeed MCUb in 100 mM Ca2+ has no channel activity at all and upon co-expression with MCU, it abolishes the calcium channel activity of the latter protein. In intact cells, MCUb overexpression reduces the mitochondrial Ca2+ responses of the cell, indicating that it interferes with the function of the endogenous channel. In support of this notion, MCUb was shown, by immunoprecipitation and FRET analyses, to directly interact with MCU thus suggesting that it may get incorporated into the channel oligomer and act within the channel as an endogenous dominant-negative subunit. In the case of some potassium channels, it is known that dominant-negative and wild-type subunits can assemble, thus forming tetrameric structures and introducing large positively charged residues into the pore that alter the positioning of carbonyl oxygens and disrupt the permeation pathway. This effect underlies several disease states, including one associated with two-pore potassium channels (Barel et al, 2008). Another example is the plant silent shaker-subunit AtKC1 which co-assembles with KAT1 and AKT2, largely inhibiting their normal activity (Jeanguenin et al, 2011). As indicated by our data, the MCUb isoform gets inserted into the channel oligomer and alters its ion permeation properties. The fact that mutation of Glu residue in the pore alters calcium permeation in MCUb is in agreement with findings describing single glutamate mutations in the pore of L-type calcium channel leading to large reduction/loss in barium permeability (e.g., Tang et al, 1993; Yang et al, 1993). In agreement with the examples of these Ca2+ channels, these mutations preserved the permeability to sodium in low-divalent solution. Thus, although the possibility of other mechanisms cannot be excluded (e.g., a change in gating properties), the marked effect of MCUb on ion permeation strongly argues in favour of a direct regulation of the pore region.
In the context of MCUb acting as a dominant-negative subunit, the expression data of the two isoforms appear very interesting. Indeed, publicly available gene expression data (NCBI GEO database) suggest a more restricted expression pattern for MCUb, with high levels in haematopoietic and immune organs. In our analyses of a broad panel of mouse tissues, we detected in general a low level of expression that however did not correlate with that of MCU. Thus, at least based on the mRNA data, the MCU/MCUb ratio appears to widely differ in the various tissues, possibly providing a molecular mechanism for tuning the efficiency of mitochondrial Ca2+ uptake. In this respect, it is interesting to note that recent recording of the mitochondrial uniporter from mitoplasts isolated from different tissues highlighted an ∼20-fold higher current density in mitochondria of skeletal muscle with respect to heart (Fieni et al, 2012). We found an ∼4-fold higher expression of MCU and a 3-fold lower expression of MCUb in skeletal muscle compared to heart, suggesting that MCU/MCUb ratios might indeed contribute to different tissue-dependent expression of active channels. Further analyses of the regulation of the two genes are likely to reveal conditions, during development, physiological stimulations, or pathological challenges, in which the MCU/MCUb ratio is changed and thus mitochondrial sensitivity to Ca2+-mediated stimuli is modified.
Overall, the data reveal an unsuspected complexity and plasticity of the mitochondrial Ca2+ uptake machinery. Indeed, MCU is confirmed to be necessary and sufficient for channel activity (De Stefani et al, 2011). Uniporter activity requires the formation of a functional oligomer that allows an intrinsic regulatory mechanism: the inclusion of an endogenous dominant-negative subunit within the complex. It can thus be envisaged that, besides other regulatory mechanisms that indirectly affect mitochondrial Ca2+ accumulation (e.g., ER Ca2+ loading, mitochondrial location in proximity of intracellular stores or plasma membrane, organelle fusion/fission), the intrinsic properties of the channel set the mitochondrial responsiveness to Ca2+-mediated signals in a defined cell type. This appears to be crucial, given the highly pleiotropic functions of mitochondrial Ca2+ homeostasis. Moreover, this complexity is currently growing: novel MCU interactors, MCUR1 (Mallilankaraman et al, 2012a), and MICU2/3 (Plovanich et al, 2013), have been recently identified, and the role of the other known uniporter modulator, MICU1 has been recently revisited (Mallilankaraman et al, 2012b). The deciphering of this signalling complex will be an experimental challenge in the next years, as a detailed understanding of the functional interaction and of the individual regulatory mechanisms of the four known molecular components (MCU, MCUb, MICU1, MICU2, MICU3, and MCUR1) will be required for understanding how cell-specific signalling signatures are determined and dynamically changed. At the same time, this complexity, with defined functions for each component, identify novel, specific molecular targets that can be exploited for developing novel drugs acting on the mitochondrial signalling checkpoint. Given the key role of mitochondria in the pathogenesis of a variety of human disorders (ranging from neuromuscular disorders to metabolic diseases and cancer) (Duchen and Szabadkai, 2010), this opportunity is an exciting option for the next future.
Materials and methods
Adenovirus production
The adenovirus expressing MCU-Flag, MCUR251W,E256V-Flag, MCUD260N,E263Q-Flag, and MCUb-6 × His were created using the AdEasy strategy (Stratagene). Mouse MCU and MCUb cDNAs were cloned in the pAdTrack-CMV vector (Stratagene) using the following primers:
- For the cloning of MCU-Flag, MCUR251W,E256V-Flag, and MCUD260N,E263Q-Flag:
fw: 5′-GGTACCGCCACCATGGCGGCCGCCGCAGGTAG-3′
rv: 5′-CTCGAGTCACTTATCGTCGTCATCCTTGTAATCTTCCTTTTCTCCGATCTGTC-3′
The PCR fragment was cloned into KpnI and XhoI sites in pAdTrack-CMV.
- For the cloning of MCUb-6 × His:
fw: 5′-GGTACCGCCACCATGCCAGGAGCTCTGTCCGG-3′
rv: 5′-CTCGAGCTAGTGATGGTGATGGTGATGGTTCTTCTCGCTGGCTT-3′
The PCR fragment was cloned into KpnI and XhoI sites in pAdTrack-CMV.
Subsequent steps were performed according to the manufacturer’s instructions (Stratagene). Adenoviral vectors contain two distinct promoters that independently drive the expression of the gene of interest and of GFP. Therefore, mock plasmid expresses only GFP protein.
Contructs and siRNAs
Mouse MCUb (NM_025779) was amplified from mouse skeletal muscle cDNA by PCR using the following primers:
- For the cloning in pEGFP-N1:
fw: 5′-CTCGAGATGCCAGGAGCTCTGTCCGG-3′
rv: 5′-GGTACCCGGTTCTTCTCGCTGGCTTCCT-3′
The PCR fragment was cloned into XhoI and KpnI sites in pEGFP-N1 (Clontech).
- For the cloning of MCUb-6 × His in pcDNA3.1:
fw: 5′-CCGGTACCGCCACCATGCCAGGAGCTCTGTCCGG-3′
rv: 5′-CTCGAGCTAGTGATGGTGATGGTGATGGTTCTTCTCGCTGGCTT-3′
The PCR fragment was cloned into KpnI and XhoI sites in pcDNA3.1 (Invitrogen).
- For the cloning of MCU and MCUb in pmCherry-N1, the two cDNAs were subcloned from pEGFP-N1-MCU (De Stefani et al, 2011) and pEGFP-N1-MCUb to pmCherry-N1 (Clontech).
The generation of the pcDNA3.1-MCUD260N,E263Q-Flag was performed by mutagenesis PCR using the wild-type pcDNA3.1-MCU-Flag as a template and the mutagenesis primer:
5′-CTGGTGGGAGTACTCGTGGAACATCATGCAACCCGTCACCTACTTCATCAC-3′
The generation of the pcDNA3.1-MCUR251W,E256V-Flag was performed by mutagenesis PCR using the wild-type pcDNA3.1-MCU-Flag as a template and the mutagenesis primer:
5′-CCAGTTTGGCATTCTGGCCTGGCTCACCTGGTGGGTGTACTCGTGGGACATCATGG-3′
- For the cloning of Strep-MCUb in pET-28A(+) and pIVEX 1.3 WG:
fw: 5′-CCATGGTTTGGTCCCACCCCCAGTTCGAGAAGCCAGGAGCTCTGTCCGGCAG-3′
rv: 5′-CTCGAGCTAGTTCTTCTCGCTGGCTT-3′
The PCR fragment was cloned into NcoI and XhoI sites in pET-28A(+) (Novagen) and pIVEX 1.3 WG (Roche).
- For the cloning of MCU-Flag-thrombin-HA in pcDNA3.1:
fw: 5′-GGTACCGCCACCATGGCGGCCGCCGCAGGTAG-3′
rv: 5′-GAATTCTCACAGGGAAGCGTAGTCAGGCACATCGTAGGGGTAGCTGCCGCGCGGCACCAGCTTATCGTCGTCATCCTTGT-3′
The PCR was performed amplifying MCU-Flag cloned in pcDNA3.1 (De Stefani et al, 2011) fragment was cloned into KpnI and EcoRI sites in pcDNA3.1 (Invitrogen).
- To silence MCUb specific siRNA was designed:
siRNA-MCUb#1: nucleotides 130–148 of the corresponding mRNA
5′-AUACUACCAGUCACACCAUtt-3′
3′-ttUAUGAUGGUCAGUGUGGUA-5′
siRNA-MCUb#2: nucleotides 848–866 of the corresponding mRNA
5′-UUUCUUCAGUUCUUCCACAtt-3′
3′-ttAAAGAAGUCAAGAAGGUGU-5′
The non-targeting siRNA (scrambled) is the following:
5′-GCCUAAGAACGACAAAUCAtt-3′
3′-ttCGGAUUCUUGCUGUUUAGU-5′
siRNA against 3′-UTR of MCU mRNA and the relative control (IBONI Negative Control-N3, cat no. K-00301-0001-N3) were designed by Riboxx. For siRNA-MCU, the following sequences were used:
- Guide sequence: 5′-AUCAUCCUUUCCAUCCUGCCCCC-3′
- Passenger sequence: 5′-GGGGGCAGGAUGGAAAGGAUGAU-3′.
Materials, cell culture, transfection, and adenoviral infection
All chemicals were purchased from Sigma-Aldrich, unless specified. α-Flag (WB, 1:1000; IF, 1:100), α-HA (WB, 1:1000; IF, 1:100), and α-MCU (WB, 1:1000) were purchased from Sigma-Aldrich. α-6 × His (WB, 1:1000; IF, 1:100) was purchased from AbCam. α-HSP60 (IF, 1:100) and α-GFP (WB: 1:1000) were purchased from Santa Cruz, α-StrepTag (WB, 1:1000) was purchased from IBA. Secondary, HRP-conjugated antibodies (WB, 1:5000) were purchased from Bio-Rad.
HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen), supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and transfected with a standard calcium-phosphate procedure or infected with the different adenoviruses at an MOI (multiplicity of infection) of 50 pfu/cell. The infection efficiency was typically greater than 90%. For aequorin measurements, the cells were seeded 24 h before transfection or infection onto 13 mm glass coverslips and allowed to grow to 50% confluence before transfection or to 80% before infection. For morphologic analyses, cells were seeded 24 h before transfection onto 24 mm glass coverslips and allowed to grow to 50% confluence before transfection, unless otherwise specified. For HEK 293 cells were cultured and transfected as HeLa cells. Neonatal mouse cardiac fibroblasts were prepared and cultured as previously described (Terrin et al, 2012) and infected with the different adenoviruses at an MOI of 50 pfu/cell. The infection efficiency was typically greater than 90%.
RNA extraction, reverse transcription, and quantitative real-time PCR
For the expression analysis of MCU and MCUb in mouse tissues, adult male C57BL/6 mice (28–30 g) and HeLa cells were used. Skeletal muscles (a pool of tibialis anterior, gastrocnemius, soleus, and extensor digitorum longus), heart, brain, spleen, lung, liver, kidney, and white fat were excised from four age-matched animals. At least three samples were prepared for each tissue and for HeLa cells. Total RNA was extracted from 15–60 mg of frozen tissues or 1 × 106 HeLa cells using the SV Total RNA Isolation Kit (Promega) following the manufacturer’s instructions. The RNA was quantified with an Eppendorf Bio photometer Plus. From an equal amount of total RNA of each sample, complementary DNA was generated with a cDNA synthesis kit (SuperScript II, Invitrogen) and analysed by real-time PCR using the SYBR green chemistry (Bio-Rad). The primers were designed and analysed with Primer3 (Rozen and Skaletsky, 2000). Identity of the amplicons was confirmed by their dissociation profiles and gel analysis. Real-time PCR standard curves were constructed by using serial dilution of cDNAs of the analysed samples, using at least four dilution points and the efficiency of all primer sets was between 95 and 105%. The housekeeping genes polymerase (RNA) II (DNA directed) polypeptide F (Pol2RF) and ribosomal protein 32 (Rpl32) were used as an internal control for cDNA quantification and normalization of the amplified products. Real-time PCR primer sequences were as follows:
To amplify MCU and MCUb cDNAs in mouse tissues:
PolR2f-fw: 5′-CGACGACTTTGATGACGTTG-3′
PolR2f-rv: 5′-GCTCACCAGATGGGAGAATC-3′
These primers amplify a fragment of 101 base pairs.
MCU-fw: 5′-AAAGGAGCCAAAAAGTCACG-3′
MCU-rv: 5′-AACGGCGTGAGTTACAAACA-3′
These primers amplify a fragment of 200 base pairs.
MCUb-fw: 5′-AGTTACCTTCTTCCTGTCGTTTGCG-3′
MCUb-rv: 5′-CAGGGATTCTGTAGCCTCAGCAAGG-3′
These primers amplify a fragment of 198 base pairs.
To amplify MCU and MCUb in HeLa cells:
Rpl32-fw: 5′-CATCTCCTTCTCGGCATCA-3′
Rpl32-rv: 5′-CTGGGTTTCCGCCAGTTAC-3′
These primers amplify a fragment of 133 base pairs.
MCU-fw: 5′-GCAGAATTTGGGAGCTGTTT-3′
MCU-rv: 5′-GTCAATTCCCCGATCCTCTT-3′
These primers amplify a fragment of 195 base pairs.
MCUb-fw: 5′-GGCCTTCCCTTGGTAACACT-3′
MCUb-rv: 5′-GTTGCCATCTGCTGTGAAGA-3′
These primers amplify a fragment of 155 base pairs.
Co-immunoprecipitation
For the interaction between MCU-GFP and MCU-HA, HeLa cells were transfected with a calcium-phosphate procedure with pcDNA3.1, pEFGP-N1-MCU, pcDNA3.1-MCU-Flag-thrombin-HA and pEFGP-N1-MCU together with pcDNA3.1-MCU-Flag-thrombin-HA. After 48 h of expression, cells were lysed in an appropriate volume of lysis buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris–HCl pH 7.4, 1 mM EGTA-Tris pH 7.4 and Complete EDTA-free protease inhibitor mixture; Roche Applied Science). In all, 1 mg of proteins from the different conditions was incubated with monoclonal α-HA-Agarose antibody (Sigma), and the co-immunoprecipitation was performed following the manufacturer’s instructions. The whole lysate and the immunoprecipitated samples were separated by SDS–PAGE, transferred onto Hybond-C Extra membrane (Amersham) and stained with Ponceau S solution. MCU-GFP was visualized with polyclonal α-GFP antibody (Santa Cruz Biotechnology) and MCU-Flag-thrombin-HA with polyclonal α-HA antibody (Sigma).
For the interaction between MCUb and MCU, HeLa cells at 80% confluence were infected with GFP, MCUb, MCU, or MCUb together with MCU. The cells were collected 2 days after the infection, and total proteins were extracted in an appropriate volume of lysis buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris–HCl pH 7.4, 1 mM EGTA-Tris pH 7.4 and Complete EDTA-free protease inhibitor mixture; Roche Applied Science). In all, 1 mg of proteins from the different conditions was incubated with monoclonal α-Flag-Agarose antibody (Sigma), and the co-immunoprecipitation was performed following the manufacturer’s instructions. The whole lysate and the immunoprecipitated samples were separated by SDS–PAGE, transferred onto Hybond-C Extra membrane (Amersham) and stained with Ponceau S solution. MCUb was visualized with polyclonal α-6 × His antibody (Abcam) and MCU-Flag with polyclonal α-Flag antibody (Sigma).
Protein expression and purification
Escherichia coli. DE3 competent cells (Stratagene) were transformed with pET28A(+)-MCUb and with pET28A(+)-MCU (De Stefani et al, 2011). Induction was performed (OD600 0.4) with 0.35 mM IPTG for 24 h.
Purification of MCUb. Bacteria were disrupted by using a French press (Constant Cell Disruption System, at 2400 Bar) in buffer W (100 mM Tris/HCl pH 8, 150 mM NaCl, 1 mM EDTA+protease inhibitor cocktail); E. coli total lysate was solubilized with 2.5% Nonidet P-40 (ICN Biomedicals). The non-solubilized material was removed by centrifugation (10 min at 12 000 g) and the supernatant was loaded on a Strep-Tactin affinity column. Column was washed with buffer W and the protein was eluted with 2.5 mM desthiobiotin. In all, 0.025% Nonidet P-40 was added in all buffers used for the purification.
Purification of MCU was performed as described in De Stefani et al (2011).
In vitro. In vitro expression of pIVEX 1.3 WG-MCUb and pIVEX 1.4 WG-MCU was performed as previously described (De Stefani et al, 2011). For co-expression experiment, different ratios of plamidic DNA were used to achieve different expression levels. After expression, the MCUb and MCU reaction mix was solubilized with 2% Triton X-100 under shaking.
Reconstitution into proteoliposomes. In vitro expressed proteins were incorporated into liposomes.
Purified soybean azolectin was used to produce liposomes at 2 mg/ml in 10 mM HEPES, 10 mM CaCl2 pH 7.3. After solubilization with 2% Triton X-100, the reaction mix (expressing MCU and/or MCUb) was incubated with liposomes for 15′ at RT. Liposomes were pelleted and suspended in the same volume and subjected to alkaline extraction by adding 1/10 V of 2 M NaHCO3.
Gel electrophoresis. If not specified otherwise, SDS–PAGE was performed using 6 M urea and standard protocols. In all, 30 μl of eluted fractions and 1 μl of the reaction mix/lane were loaded. Native polyacrylamide gel electrophoresis was performed by loading the protein samples without denaturation following standard protocol. A sample buffer containing only glycerol was used to load the samples.
Electrophysiology
Planar lipid bilayer experiments were performed as described in De Stefani et al (2011). Briefly, bilayers of ∼150–200 pF capacity were prepared using purified soybean azolectin. The standard experimental medium was 100 mM calcium-gluconate, 10 mM Hepes/pH 7.2. Gluconate was used as an anion to exclude anion channel activity. As low divalent solution, 100 mM Na-gluconate, 10 mM Hepes/pH 7.2, and 5 mM EDTA were used. For calculation of the free calcium concentration, WebMaxC v2.2 program was used. Control experiments with empty membrane or with detergents used for the purification showed no activity in any of the above solutions. All voltages reported are those of the cis chamber, zero being assigned by convention to the trans (grounded) side. Currents are considered as positive when carried by cations flowing from the cis to the trans compartment. Data were acquired at 100 μs/point, filtered at 200 or 300 Hz and analysed offline using the pClamp program set (Axon Instruments, Union City, CA, USA). Histograms were fitted using the Origin7.5 program set. Leak was not subtracted.
Aequorin measurements
HeLa cells grown on 13 mm round glass coverslips at 50% confluence were infected with indicated adenoviruses or transfected with the cytosolic (cytAEQ) or the low-affinity mitochondrial (mtAEQmut, referred in the text as mtAEQ) probe (as previously described; Pinton et al, 2007) together with the indicated siRNA or plasmid. AdGFP and pcDNA3.1 were used as controls for adenoviral transduction and transfection, respectively, unless otherwise indicated. The coverslip with the cells was incubated with 5 μM coelenterazine for 1–2 h in KRB (Krebs-Ringer modified buffer: 125 mM NaCl, 5 mM KCl, 1 mM Na3PO4, 1 mM MgSO4, 5.5 mM glucose, 20 mM HEPES, pH 7.4, 37°C) supplemented with 1 mM CaCl2, and then transferred to the perfusion chamber. All aequorin measurements were carried out in KRB. Agonists and other drugs were added to the same medium, as specified in the text. The experiments were terminated by lysing the cells with 100 μM digitonin in a hypotonic Ca2+-rich solution (10 mM CaCl2 in H2O), thus discharging the remaining aequorin pool. The light signal was collected and calibrated into [Ca2+] values by an algorithm based on the Ca2+ response curve of aequorin at physiological conditions of pH, [Mg2+] and ionic strength, as previously described (Pinton et al, 2007). Representative traces are shown in the figures whereas the full data set is included in Supplementary Table S1. In the experiments with permeabilized cells, a buffer mimicking the cytosolic ionic composition (IB) was employed: 130 mM KCl, 10 mM NaCl, 2 mM K2HPO4, 5 mM succinic acid, 5 mM malic acid, 1 mM MgCl, 20 mM HEPES, 1 mM pyruvate, 0.5 mM ATP and 0.1 mM ADP (pH 7 at 37°C). IB was supplemented with either 100 μM EGTA (IB/EGTA) or a 2-mM EGTA and 2 mM HEEDTA-buffered [Ca2+] of 1 or 2 μM (IB/Ca2+), calculated with Chelator software (Schoenmakers et al, 1992). HeLa cells were permeabilized by a 1-min perfusion with 50 μM digitonin (added to IB/EGTA) during luminescence measurements. Mitochondrial Ca2+ uptake speed was calculated as the first derivative by using the SLOPE excel function and smoothed for three time points. The higher value reached during Ca2+ addition represents the maximal Ca2+ uptake speed. All of the results are expressed as means±s.e.m., and Student’s t-test was used for the statistic. All the materials were from Sigma-Aldrich unless specified.
Measurements of mitochondrial Δ Ψ
Mitochondrial ΔΨ was measured by loading cells with 20 nM tetramethyl rhodamine methyl ester (TMRM) for 30 min at 37°C. Images were taken on an inverted microscope (Zeiss Axiovert 200) equipped with a PlanFluar × 40/1.3 NA objective, a Photometrics Evolve Delta EMCCD, and a 75-W Xenon arc lamp coupled to a monochromator (PTI Deltaram V). The system was assembled by Crisel Instruments (Rome, Italy). TMRM excitation was performed at 560 nm and emission was collected through a 590–650 nm bandpass filter. Images were taken every 10 s with a fixed 200 ms exposure time. In all, 10 μM CCCP (carbonyl cyanide p-triclooromethoxyphenylhydrazone), an uncoupler of oxidative phosphorylation, was added after 12 acquisitions to completely collapse the electrical gradient established by the respiratory chain (ΔΨ). After background correction, the fluorescence value after FCCP addition (i.e., TMRM fluorescence not due to membrane potential) was subtracted for each cell. Data are presented as the corrected average fluorescence of the first five frames.
Immunofluorescence
HeLa cells were grown on 13 mm coverslips and transfected with MCU-Flag and MCUb-6 × His encoding plasmids when 50% confluent. After 24 h, cells were washed with PBS, fixed in 4% formaldehyde for 10 min and quenched with 50 mM NH4Cl in PBS. Cells were permeabilized for 10 min with 0.1% Triton X-100 in PBS and blocked in PBS containing 2% BSA for 1 h. Cells were then incubated with primary antibodies (α-HSP60, α-Flag, and α-6 × His) for 3 h at room temperature and washed three times with 0.1% Triton X-100 in PBS. The appropriate isotype matched, AlexaFluor conjugated secondary antibodies (Invitrogen) were used and the coverslips were mounted with ProLong Gold Antifade reagent (Invitrogen). Images were taken on a Leica TCS-SP5-II equipped with a × 100, 1.4 NA Plan Apochromat objective. AlexaFluor488 and Alexa-Fluor647 were excited simultaneously by 488 and 633 nm laser lines and images were collected in the 495–535 nm and 645–720 nm ranges. AlexaFluor555 was excited with the 543-nm laser line and the signal was collected in the 555–600 nm range. Pixel size was set to 75 nm. For each cell, a z-stack of the whole cell was taken, with a step size of 130 nm. Images are presented as maximum projections of the whole stack.
Förster resonance energy transfer (FRET)
HeLa cells were grown on 24 mm coverslips and transfected with the indicated combination of GFP, MCU-GFP, MCUb-GFP, mCherry, MCU-mCherry, or MCUb-mCherry encoding plasmids when 50% confluent. GFP and mCherry were excited at 488 and 543 nm, and their signals were collected in the 495–535 and 598–670 nm range, respectively. A specific region of the specimen was bleached with a 1.5-W 592 nm laser (4 passes at 30% power). Donor and acceptor images were collected before and after bleaching, and FRET efficiency was calculated from the background subtracted images with the formula:
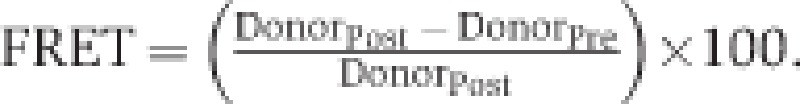 .where DonorPre and DonorPost are the mean fluorescence intensities in the selected region before and after bleaching. As an internal control, no significant FRET was observed outside the bleached region. No bleaching of the donor was observed with these experimental settings. Pinhole was set to 1 airy unit and the pixel size was adjusted to 150 nm. Representative images were generated using the AccPbFRET plugin (Roszik et al, 2008) for ImageJ. For the analysis of emission spectra, 458 nm Argon laser line was used to excite the donor (in order to minimize cross-excitation of the mCherry). Emission slit was set to a 10-nm window and progressively shifted in the 470–700 nm range in 34 subsequent steps. Pinhole was set to 3 airy units in order to maximize the signal and the pixel size was set to 300 nm. Collected images were then background corrected and fluorescence was quantified in a defined ROI. Absolute florescence intensities were then normalized to the maximum value. With these experimental settings, no signal could be detected from acceptor only (MCU-mCherry) transfected cells in the whole range: however, excitation of these cells with the 543 laser line generated the expected emission spectrum of the mCherry. All images were taken on a Leica TCS STED CW system equipped with a × 100/1.4 NA Plan Apochromat objective.
.where DonorPre and DonorPost are the mean fluorescence intensities in the selected region before and after bleaching. As an internal control, no significant FRET was observed outside the bleached region. No bleaching of the donor was observed with these experimental settings. Pinhole was set to 1 airy unit and the pixel size was adjusted to 150 nm. Representative images were generated using the AccPbFRET plugin (Roszik et al, 2008) for ImageJ. For the analysis of emission spectra, 458 nm Argon laser line was used to excite the donor (in order to minimize cross-excitation of the mCherry). Emission slit was set to a 10-nm window and progressively shifted in the 470–700 nm range in 34 subsequent steps. Pinhole was set to 3 airy units in order to maximize the signal and the pixel size was set to 300 nm. Collected images were then background corrected and fluorescence was quantified in a defined ROI. Absolute florescence intensities were then normalized to the maximum value. With these experimental settings, no signal could be detected from acceptor only (MCU-mCherry) transfected cells in the whole range: however, excitation of these cells with the 543 laser line generated the expected emission spectrum of the mCherry. All images were taken on a Leica TCS STED CW system equipped with a × 100/1.4 NA Plan Apochromat objective.
Sequence alignment
Sequence alignments were performed using Molecular Operating Environment (MOE, ver.2010.10) aided by various other web-based alignment tools. Prediction of the location of the TM helices in MCU has been carried out using TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/).
Comparative modelling
MCU comparative models were generated using MOE suite using a multi-template approach. In particular, ‘3EAM’ PDB structure has been used to model the region from 224 to 291 whereas ‘3EFF.K’ PDB structure has been use to model the region from 294 to 334. An ensemble of 25 model structures was generated. These were ranked by analysis of their stereochemistry, using the ‘Protein Geometry’ tool implemented by MOE. From the ensemble, a single structure was selected for further analysis and as a starting structure for the assembly of the tetrameric multimer of MCU and for MD simulations. The assembly of the modelled MCU subunits was performed using the symmetric multimer docking program M-ZDOCK (Pierce et al, 2005). Channel forming proposed solutions were analysed and the best scoring ones, according to energetic criteria, have been subjected to a simulated annealing minimization using Yasara Structure Suite (v. 11.4.18) and used as further specified. Convergence was considered reached as soon as the energy of the entire system improved by <0.05 kJ/mol per atom during 200 steps of minimization. Homology Model of the MCUb isoform was built with Yasara Structure Suite (v. 11.4.18) using the geometrical information of MCU, as template structure. Side chains of the entire model were optimized using the SCWALL method (Canutescu et al, 2003) with YASARA2 force field (Krieger et al, 2009). All modelled parts, excluding backbone atoms, were subjected to a combined steepest descent and simulated annealing minimization. Then, a full unrestrained simulated annealing minimization was run for the entire model. Convergence was considered reached as soon as the energy of the entire system improved by <0.05 kJ/mol per atom during 200 steps of minimization.
Membrane MD
The solvent exposed area of the channel has been solvated using the program Solvate 1.0 (http://www.mpibpc.mpg.de/home/grubmueller/downloads/solvate/index.html) then the protein has been embedded in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid bilayer (63 × 63 Å wide) and oriented through the membrane using an in-house tcl script, using Visual Molecular Dynamics. Bumping lipids (distance<0.45 Å from protein atoms) and eventual water located in the hydrophobic protein-membrane interface (distance<3 Å from lipids molecules) were removed upon insertion of the protein, then the entire complex was solvated using atomistic TIP3P water (Jorgensen et al, 1983) and electrically neutralized with a total ionic concentration (Ca2+ and Cl− ions) of 0.1 M. Final system was composed by ∼48 000 atoms including solvation water molecules and ions. MD simulations were carried out using ACEMD program (Harvey et al, 2009) on a local cluster. Protein atom positions were restrained by a 1-kcal/mol/Å2 spring constant and conjugate-gradient minimization was performed for 500 steps. The magnitude of the restraining spring constant was kept at 1 kcal/mol/Å2 for the first 5 ns, then scaled to 0.1 kcal/mol/Å2 till 10 ns. A randomly chosen calcium ion was placed 10 Å above the channel centre and its position has been restrained, until lipid atoms surrounding the channel reached equilibrium, using a harmonic potential with a spring constant value of 1 kcal/mol/Å2 and then set free to move along the Z axis. The temperature was maintained at 298 K using a Langevin thermostat with a low damping constant of 1 ps, and the pressure was maintained at 1 atm using a Berendensen barostat. Atoms composing the entire system were finally unrestrained in an NVT ensemble using a low damping constant of 0.1 ps. The long-range Coulomb interaction was handled using the particle mesh Ewald summation method (PME) (Essmann et al, 1995) with grid size rounded to the approximate integer value of cell wall dimensions.
A non-bonded cutoff distance of 9 Å was used with a switching distance of 7.5 Å. SHAKE algorithm has been used on all atoms covalently bonded to a hydrogen atom with an integration time step of 2 fs. All MD simulations were performed using Charmm27 (MacKerell et al, 1998) Force Field. Molecular graphics has been created using the Linux distributed version of Pymol (v. 1.4.1) (http://www.pymol.org/) and PovRay (http://www.povray.org/). Trajectory manipulations have been carried out using Wordom (v. 0.22-rc2) (Seeber et al, 2007). Hole program (v. 2.2.002) has been used to evaluate channel pore topologies (Smart et al, 1993) and APBS program (v. 1.2.1) has been used to evaluate electrostatic properties of the built models (Baker et al, 2001).
Statistical analysis of data
Statistical data are presented as mean±s.e.m. unless specified, significance was calculated by Student’s t-test (*P<0.05, **P<0.001), and correlation analysis was performed with the SigmaPlot 11.0 software (Systat Software Inc.).
Supplementary Material
Acknowledgments
We are grateful to Tullio Pozzan, Richard Wagner and Mario Zoratti for helpful discussion, and Angela Paggio for carrying out some of the experiments. This research was supported by grants from the Italian Ministries of Health (Ricerca Finalizzata) and of Education, University and Research (PRIN, FIRB), the European Union (ERC mitoCalcium, no. 294777 and FP7 ‘MyoAGE’, no. 223576), NIA (2P01AG025532–06A1), Cariparo and Cariplo Foundations (Padua), the Italian Association for Cancer Research (AIRC) and Telethon-Italy (GPP1005A, GGP11082).
Author contributions: AR performed molecular biology and biochemical analysis. DDS performed Ca2+ protein immunolocalization and FRET analysis. DDS and AR contributed equally to the study. DS and SM performed computational structural biology studies. ET expressed and purified the protein in heterologous systems; GM performed Ca2+ measurements. AP performed gene expression analysis. IS designed, performed, and analysed the electrophysiology experiments; VC performed the electrophysiology experiments; RR discussed the results and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barel O, Shalev SA, Ofir R, Cohen A, Zlotogora J, Shorer Z, Mazor G, Finer G, Khateeb S, Zilberberg N, Birk OS (2008) Maternally inherited Birk Barel mental retardation dysmorphism syndrome caused by a mutation in the genomically imprinted potassium channel KCNK9. Am J Hum Genet 83: 193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, Koteliansky V, Mootha VK (2011) Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ, Bootman MD, Roderick HL (2003) Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol 4: 517–529 [DOI] [PubMed] [Google Scholar]
- Boitier E, Rea R, Duchen MR (1999) Mitochondria exert a negative feedback on the propagation of intracellular Ca2+ waves in rat cortical astrocytes. J Cell Biol 145: 795–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canutescu AA, Shelenkov AA, Dunbrack RL Jr (2003) A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci 12: 2001–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafoli E (2010) The fateful encounter of mitochondria with calcium: how did it happen? Biochim Biophys Acta 1797: 595–606 [DOI] [PubMed] [Google Scholar]
- Corry B, Allen TW, Kuyucak S, Chung SH (2001) Mechanisms of permeation and selectivity in calcium channels. Biophys J 80: 195–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Thomas AP, Hajnoczky G (1999) Quasi-synaptic calcium signal transmission between endoplasmic reticulum and mitochondria. EMBO J 18: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Varnai P, Golenar T, Roy S, Purkins G, Schneider TG, Balla T, Hajnoczky G (2010) Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol Cell 39: 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R (2011) A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca HF, Engstrom GW (1961) Calcium uptake by rat kidney mitochondria. Proc Natl Acad Sci USA 47: 1744–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton RM (2009) Regulation of mitochondrial dehydrogenases by calcium ions. Biochim Biophys Acta 1787: 1309–1316 [DOI] [PubMed] [Google Scholar]
- Duchen MR, Szabadkai G (2010) Roles of mitochondria in human disease. Essays Biochem 47: 115–137 [DOI] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103: 8577–8593 [Google Scholar]
- Fieni F, Lee SB, Jan YN, Kirichok Y (2012) Activity of the mitochondrial calcium uniporter varies greatly between tissues. Nat Commun 3: 1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomello M, Drago I, Bortolozzi M, Scorzeto M, Gianelle A, Pizzo P, Pozzan T (2010) Ca2+ hot spots on the mitochondrial surface are generated by Ca2+ mobilization from stores, but not by activation of store-operated Ca2+ channels. Mol Cell 38: 280–290 [DOI] [PubMed] [Google Scholar]
- Gilabert JA, Parekh AB (2000) Respiring mitochondria determine the pattern of activation and inactivation of the store-operated Ca(2+) current I(CRAC). EMBO J 19: 6401–6407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WI, Lim KB, Sudhaharan T, Sem KP, Bu W, Chou AM, Ahmed S (2011) mDia1 and WAVE2 interact directly with IRSp53 in filopodia and are involved in filopodium formation. J Biol Chem 287: 4702–4714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnoczky G, Hager R, Thomas AP (1999) Mitochondria suppress local feedback activation of inositol 1,4, 5-trisphosphate receptors by Ca2+. J Biol Chem 274: 14157–14162 [DOI] [PubMed] [Google Scholar]
- Harvey MJ, Giupponi G, De FG (2009) ACEMD: accelerating biomolecular dynamics in the microsecond time scale. J Chem Theory Comput 5: 1632–1639 [DOI] [PubMed] [Google Scholar]
- Hess P, Tsien RW (1984) Mechanism of ion permeation through calcium channels. Nature 309: 453–456 [DOI] [PubMed] [Google Scholar]
- Hoth M, Button DC, Lewis RS (2000) Mitochondrial control of calcium-channel gating: a mechanism for sustained signaling and transcriptional activation in T lymphocytes. Proc Natl Acad Sci USA 97: 10607–10612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanguenin L, Alcon C, Duby G, Boeglin M, Cherel I, Gaillard I, Zimmermann S, Sentenac H, Very AA (2011) AtKC1 is a general modulator of Arabidopsis inward Shaker channel activity. Plant J 67: 570–582 [DOI] [PubMed] [Google Scholar]
- Jorgensen W, Chandrasekhar J, Madura J, Impey R, Klein M (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79: 926–935 [Google Scholar]
- Kirichok Y, Krapivinsky G, Clapham DE (2004) The mitochondrial calcium uniporter is a highly selective ion channel. Nature 427: 360–364 [DOI] [PubMed] [Google Scholar]
- Krieger E, Joo K, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77: Suppl 9114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafreniere RG, Cader MZ, Poulin JF, Andres-Enguix I, Simoneau M, Gupta N, Boisvert K, Lafreniere F, McLaughlan S, Dube MP, Marcinkiewicz MM, Ramagopalan S, Ansorge O, Brais B, Sequeiros J, Pereira-Monteiro JM, Griffiths LR, Tucker SJ, Ebers G, Rouleau GA (2010) A dominant-negative mutation in the TRESK potassium channel is linked to familial migraine with aura. Nat Med 16: 1157–1160 [DOI] [PubMed] [Google Scholar]
- Lepple-Wienhues A, Cahalan MD (1996) Conductance and permeation of monovalent cations through depletion-activated Ca2+ channels (ICRAC) in Jurkat T cells. Biophys J 71: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell AD Jr, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE III et al. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102: 3586–3616 [DOI] [PubMed] [Google Scholar]
- Mallilankaraman K, Cardenas C, Doonan PJ, Chandramoorthy HC, Irrinki KM, Golenar T, Csordas G, Madireddi P, Yang J, Muller M, Miller R, Kolesar JE, Molgo J, Kaufman B, Hajnoczky G, Foskett JK, Madesh M (2012a) MCUR1 is an essential component of mitochondrial Ca(2+) uptake that regulates cellular metabolism. Nat Cell Biol 14: 1336–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K, Doonan P, Cardenas C, Chandramoorthy HC, Muller M, Miller R, Hoffman NE, Gandhirajan RK, Molgo J, Birnbaum MJ, Rothberg BS, Mak DO, Foskett JK, Madesh M (2012b) MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca(2+) uptake that regulates cell survival. Cell 151: 630–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack JG, Halestrap AP, Denton RM (1990) Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol Rev 70: 391–425 [DOI] [PubMed] [Google Scholar]
- Montero M, Alonso MT, Carnicero E, Cuchillo-Ibanez I, Albillos A, Garcia AG, Garcia-Sancho J, Alvarez J (2000) Chromaffin-cell stimulation triggers fast millimolar mitochondrial Ca2+ transients that modulate secretion. Nat Cell Biol 2: 57–61 [DOI] [PubMed] [Google Scholar]
- Palty R, Silverman WF, Hershfinkel M, Caporale T, Sensi SL, Parnis J, Nolte C, Fishman D, Shoshan-Barmatz V, Herrmann S, Khananshvili D, Sekler I (2010) NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc Natl Acad Sci USA 107: 436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, Mootha VK (2010) MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature 467: 291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce B, Tong W, Weng Z (2005) M-ZDOCK: a grid-based approach for Cn symmetric multimer docking. Bioinformatics 21: 1472–1478 [DOI] [PubMed] [Google Scholar]
- Pinton P, Ferrari D, Rapizzi E, Di Virgilio F, Pozzan T, Rizzuto R (2001) The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: significance for the molecular mechanism of Bcl-2 action. EMBO J 20: 2690–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinton P, Rimessi A, Romagnoli A, Prandini A, Rizzuto R (2007) Biosensors for the detection of calcium and pH. Methods Cell Biol 80: 297–325 [DOI] [PubMed] [Google Scholar]
- Plovanich M, Bogorad RL, Sancak Y, Kamer KJ, Strittmatter L, Li AA, Girgis HS, Kuchimanchi S, De Groot J, Speciner L, Taneja N, Oshea J, Koteliansky V, Mootha VK (2013) MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS One 8: e55785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, Murgia M, Pozzan T (1993) Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science 262: 744–747 [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T (1998) Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280: 1763–1766 [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Simpson AW, Brini M, Pozzan T (1992) Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature 358: 325–327 [DOI] [PubMed] [Google Scholar]
- Roszik J, Szollosi J, Vereb G (2008) AccPbFRET: an ImageJ plugin for semi-automatic, fully corrected analysis of acceptor photobleaching FRET images. BMC Bioinformatics 9: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Schoenmakers TJ, Visser GJ, Flik G, Theuvenet AP (1992) CHELATOR: an improved method for computing metal ion concentrations in physiological solutions. Biotechniques 12: 876–879 [PubMed] [Google Scholar]
- Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, Korsmeyer SJ (2003) BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science 300: 135–139 [DOI] [PubMed] [Google Scholar]
- Seeber M, Cecchini M, Rao F, Settanni G, Caflisch A (2007) Wordom: a program for efficient analysis of molecular dynamics simulations. Bioinformatics 23: 2625–2627 [DOI] [PubMed] [Google Scholar]
- Smart OS, Goodfellow JM, Wallace BA (1993) The pore dimensions of gramicidin A. Biophys J 65: 2455–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalai G, Krishnamurthy R, Hajnoczky G (1999) Apoptosis driven by IP(3)-linked mitochondrial calcium signals. EMBO J 18: 6349–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera K, Nilius B (2006) Biophysics and structure-function relationship of T-type Ca2+ channels. Cell Calcium 40: 97–114 [DOI] [PubMed] [Google Scholar]
- Tang S, Mikala G, Bahinski A, Yatani A, Varadi G, Schwartz A (1993) Molecular localization of ion selectivity sites within the pore of a human L-type cardiac calcium channel. J Biol Chem 268: 13026–13029 [PubMed] [Google Scholar]
- Terrin A, Monterisi S, Stangherlin A, Zoccarato A, Koschinski A, Surdo NC, Mongillo M, Sawa A, Jordanides NE, Mountford JC, Zaccolo M (2012) PKA and PDE4D3 anchoring to AKAP9 provides distinct regulation of cAMP signals at the centrosome. J Cell Biol 198: 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel H, Cancela JM, Mogami H, Gerasimenko JV, Gerasimenko OV, Tepikin AV, Petersen OH (1999) Active mitochondria surrounding the pancreatic acinar granule region prevent spreading of inositol trisphosphate-evoked local cytosolic Ca(2+) signals. EMBO J 18: 4999–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krogt GN, Ogink J, Ponsioen B, Jalink K (2008) A comparison of donor-acceptor pairs for genetically encoded FRET sensors: application to the Epac cAMP sensor as an example. PLoS One 3: e1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Ellinor PT, Sather WA, Zhang JF, Tsien RW (1993) Molecular determinants of Ca2+ selectivity and ion permeation in L-type Ca2+ channels. Nature 366: 158–161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.