Abstract
The presence of aggregates of abnormally expanded polyglutamine (polyQ)-containing proteins are a pathological hallmark of a number of neurodegenerative diseases including Huntington’s disease (HD) and spinocerebellar ataxia-3 (SCA3). Previous studies in cellular, Drosophila, and mouse models of HD and SCA have shown that neurodegeneration can be prevented by manipulations that inhibit polyQ aggregation. We have shown that the UL97 kinase of the human cytomegalovirus (HCMV) prevents aggregation of the pp71 and pp65 viral tegument proteins. To explore whether UL97 may act as a general antiaggregation factor, we examined whether UL97 prevents aggregation of cellular non-polyQ and polyQ proteins. We report that UL97 prevents the deposition of aggregates of two non-polyQ proteins: a protein chimera (GFP170*) composed of the green fluorescent protein and a fragment of the Golgi Complex protein (GCP-170) and a chimera composed of the red fluorescent protein (RFP) fused to the Werner syndrome protein (WRN), a RecQ helicase and exonuclease involved in DNA repair. Furthermore, we show that UL97 inhibits aggregate deposition in cellular models of HD and SCA3. UL97 prevents the deposition of aggregates of the mutant huntingtin exon 1 containing 82 glutamine repeats (HttExon1-Q82) or full length ataxin-3 containing a 72 polyQ track (AT3-72Q). The kinase activity of UL97 appears critical, as the kinase-dead UL97 mutant (K335M) fails to prevent aggregate formation. We further show that UL97 disrupts nuclear PML bodies and decreases p53-mediated transcription. The universality of the antiaggregation effect of UL97 suggests that UL97 targets a key cellular factor that regulates cellular aggregation mechanisms. Our results identify UL97 as a novel means to modulate polyQ aggregation and suggest that UL97 can serve as a novel tool to probe the cellular mechanisms that contribute to the formation of aggregates in polyglutamine disorders.
Keywords: UL97, Polyglutamine, Aggresome, Ataxin 3, Huntingtin, PML bodies, p53
Introduction
The expansion of trinucleotide CAG repeats encoding glutamines within specific cellular proteins is the cause of inherited neurodegenerative diseases termed polyglutamine disorders. Huntington’s disease, spinobulbar muscular atrophy (SBMA), dentatorubral–pallidoluysian atrophy (DRPLA), and spinocerebellar ataxias (SCA) 1, 2, 3, 6, 7, and 17 are all caused by an abnormally expanded polyQ domain (reviewed in Cummings and Zoghbi, 2000; McCampbell et al., 2001; Orr and Zoghbi, 2007). A striking neuropathological hallmark of these polyQ diseases is the presence of neuronal insoluble cytoplasmic aggregates and nuclear insoluble inclusions (NIIs) formed by mutant polyQ proteins (reviewed in Ross, 2002). The role NIIs and cytoplasmic aggregates in the pathological processes of polyQ diseases remains contentious. While some consider the NIIs/cytoplasmic aggregates as direct toxic intermediates, some have proposed that NIIs/cytoplasmic aggregates are a point of sequestration of a toxic product and thereby beneficial to a neuron (Arrasate et al., 2004; Ross et al., 1999).
Multiple lines of evidence support the view that protein aggregation is a complex process that is initiated by the accumulation of misfolded polyQ-containing proteins into a variety of higher-order intermediate conformational assemblies that ultimately form insoluble inclusion bodies. Conformational rearrangements of these mutated proteins likely change their biological activities and contribute to their toxicities. Importantly, recent reports suggest that the toxicity of mutant polyQ-containing proteins might be not related to the insoluble inclusion bodies but rather to soluble oligomeric or other intermediate conformations (Kayed et al., 2003, 2004). In support of this idea, polyQ-induced neurodegeneration can be prevented by pharmacological and molecular manipulations that target processes leading to oligomerization and aggregation. For instance, inhibition of polyglutamine oligomerization in a transgenic mouse model of HD has a marked protective effect on survival, weight loss, and motor function (Sanchez et al., 2003), supporting the idea that oligomerization of expanded polyglutamine may play a pivotal role in the protein’s toxicity. Similarly, the green tea polyphenol epi-gallocatechin-3-gallate (EGCG) inhibits aggregation of polyQ Httex1 in vitro and reduces polyQ-induced cytotoxicity in yeast cells and Drosophila models of the disease (Ehrnhoefer et al., 2006).
In addition to pharmacological approaches, modulations of cellular pathways that prevent aggregation also reverse neurotoxicity. Molecular chaperones are the first line of defense against protein aggregation, and numerous studies have shown that overexpression of chaperones prevents polyQ aggregation and reduces the pathology of neurodegenerative diseases (Muchowski and Wacker, 2005). In cultured cells, the chaperones heat shock protein 40 kDa (Hsp40) and 70 kDa (Hsp70) have been found to prevent the formation of spherical and annular oligomeric structures by a polyQ Httex1 (Wacker et al., 2004). Significantly, overexpression of Hsp70 in SCA1 transgenic mice decreases neurodegeneration and improves motor coordination (Cummings et al., 2001). Similarly, inhibiting polyQ aggregation by overexpressing Hsp70 (Warrick et al., 1999, 2005) and Hsp40 (Chan et al., 2000) rescues neurodegeneration in Drosophila models of SCA3. Altogether, these findings strongly suggest that specific polyQ conformational structures confer the cellular toxicity of the mutated proteins and that the identification of novel pharmacological, molecular, or genetic means to prevent polyQ aggregation is likely to have direct impact on developing therapeutic strategies for neurodegenerative diseases such as HD and SCA3.
We have recently documented that the UL97 kinase of HCMV prevents the aggregation of viral components during infection (Prichard et al., 2008). UL97 is a ~80-kDa tegument protein composed of 707 amino acids that is expressed during HCMV infection (Michel et al., 1998). UL97 contains a nuclear localization signal within its N-terminal domain and is targeted to the nucleus in infected or transfected cells (Prichard et al., 2005). UL97 is a kinase and shows homology to cellular serine/threonine kinases within conserved subdomains involved in substrate and ATP binding (Michel et al., 1998). The kinase activity of UL97 requires the invariant lysine at position 355, and the K355M mutant is inactive (Marschall et al., 2001). UL97 is important for viral replication, and a recombinant virus with a large deletion in UL97 replicates poorly and contains abnormal aggregates of viral proteins within the nuclei (Prichard et al., 1999, 2005). One of the aggregated proteins is the pp65 viral tegument protein, which also forms nuclear aggregates when expressed in transfected mammalian cells. Importantly, UL97, but not the catalytically inactive UL97 K355M, prevents pp65 aggregation (Prichard et al., 2005), suggesting that UL97 has antiaggregation activity and that the antiaggregation effect of UL97 is dependent on its kinase activity.
In addition to viral proteins, UL97 also prevents aggregation of GFP170*, a protein chimera formed by fusing an internal segment (amino acids 566–1375) of the Golgi protein GCP-170 to the C-terminus of GFP (Misumi et al., 2001; Hicks and Machamer, 2002; Prichard et al., 2008). We have shown previously that GFP170* forms nuclear aggregates similar in structure to those formed by the viral pp65 and also deposits in large ribbon-like aggregates within the cytoplasm (Fu et al., 2005a). UL97 prevents the formation of both the nuclear and the cytoplasmic GFP170* aggregates (Prichard et al., 2008). As with pp65, the catalytically inactive UL97/K355M mutant is unable to prevent GFP170* aggregation. Thus, UL97 prevents the aggregation of both a viral and a cellular protein. Herein, we examined the possibility that UL97 may possess a general antiaggregation activity and may serve as a tool for understanding and inhibiting the mechanisms that contribute to aggregation in polyQ diseases.
We report that UL97 has a strong antiaggregation effect on non-polyQ proteins as well as polyQ-expanded proteins associated with HD and SCA3. We show that UL97 prevents the deposition of aggregates of the non-polyQ Werner protein (WRN) that causes the premature aging disease Werner syndrome. We also show that UL97 prevents aggregation of a pathogenic construct that encodes the full-length ataxin-3 containing a 72-glutamine expansion (AT3-72Q), and of a pathogenic N-terminal huntingtin domain corresponding to the exon1 of this protein and containing an expanded track of 82 glutamine residues (HttExon1-82Q). In all cases, the catalytically inactive UL97/ K355M mutant does not prevent aggregation. The similarity of the UL97 effect on the viral pp65 protein, the non-polyQ GFP170* and WRN proteins, and the polyQ AT3-72Q and HttExon1-82Q proteins suggests that UL97 has general antiaggregation effect. This similarity is also consistent with the hypothesis that aggregation of diverse proteins may occur through a common mechanism that is targeted by the UL97 kinase. In agreement, we show that UL97 disperses nuclear PML bodies and causes a decrease in p53-mediated transcription.
Our results identify UL97 as a novel means to inhibit the aggregation of polyQ proteins. They also designate UL97 as a new molecular tool to further examine the cellular mechanisms that lead to polyQ aggregation and neurodegeneration in HD and SCA3.
Materials and methods
Antibodies and reagents
Monoclonal antibody to the V5-epitope was purchased from Invitrogen (catalogue no. R960-25; Carlsbad, CA). Polyclonal anti-GFP antibody was purchased from Abcam (catalogue no. ab290-50; Cambridge, MA). Anti-myc monoclonal antibody was purchased from Covance (catalogue no. PRB-150B; Denver, PA). Anti-myc (A-14) (catalogue no. sc-789) polyclonal antibody, anti-PML (PG-M3) (catalogue no. sc-966) monoclonal antibody, and anti-PML (H-238) (sc-5621) polyclonal antibody were from Santa Cruz (Santa Cruz, CA). Fugene 6 transfection reagent was purchased from Roche (catalogue no. 11814443001; Indianapolis, IN), and was used in luciferase experiments, Mirus IT-LTI transfection reagent (catalogue no. MIR2300; Madison,WI) was used for transfection of cells for immunoflourescence microscopy. BCA protein assay kit was purchased from Thermo Scientific (catalogue no. 23225; Rockford, IO). Alexa Fluor 594-labeled goat anti-rabbit, Alexa Fluor 488-labeled goat anti-mouse, and Hoechst 33258 were from Invitrogen Molecular Probes, Inc (Eugene, OR).
DNA constructs
The construction of the GFP-GCP170* chimera has been previously described (Fu et al., 2005b). UL97 and K355M V5-epitope tagged plasmids have been previously described (Prichard et al., 2005). Plasmids encoding pGL2-p21A luciferase have been previously described (Chinery et al., 1997) and were a generous gift from Dr. Xinbin Chen (UC Davis School of Veterinary Medicine). HttExon1-8299Q-GFP plasmid has been described in Chun et al. (2001). The AT3-72Q plasmid was generously provided by Dr. Randall Pittman (University of Pennsylvania School of Medicine). The mRFP-Werner construct has been described previously (Vaitiekunaite et al., 2007) and was a gift from Dr. Marek Rusin (Maria Sklodowska-Curie Memorial Institute, Gliwice, Poland).
Cell culture and transfections
HeLa cells were purchased from ATCC (Manassas, VA). HeLa cells were cultured in minimum essential medium (MEM) supplemented with glucose, and glutamine (Mediatech, Comprehensive Cancer Center of the University of Alabama at Birmingham, AL). Media were supplemented with 10% fetal bovine serum (FBS, Life Technologies, Grand Island, NY), 100 U/ml of penicillin and streptomycin (Invitrogen Corporation, Grand Island, NY), nonessential amino acids, and 1 mM sodium pyruvate. HT1080 cells were a gift from Dr. Susan Nozell (University of Alabama at Birmingham). HT1080 cells were maintained in MEM plus L-glutamine and 10% fetal bovine serum. These cells express endogenous wild-type p53 and were used for luciferase assays. All cells were grown in incubator with 95% air in the atmosphere and 5% carbon dioxide (CO2) at 37 °C.
HeLa cells were grown in monolayers on coverslips in six-well plates, and cells were transfected with TransLTI (Mirus) or FuGENE 6 (Roche) reagents or with Lipofectin (Invitrogen), according to the manufacturers’ protocols.
Cell viability assays were performed as follows: cells were cotransfected with AT3-72Q and either a vector lacking UL97, a vector encoding UL97, or a vector encoding UL97/K355M. After 24 or 48 h of culture, cells were treated with 15 mg/ml of fluorescein diacetate for 5 min, washed 1× in phosphate-buffered saline (PBS), and processed for immunofluorescence microscopy.
Immunofluorescence microscopy and quantification
At 48 to 72 h after transfection, cells were washed three times in PBS, fixed in 3% paraformaldehyde for 10 min and then quenched with 10 mM ammonium chloride. The cells were permeabilized in 0.2% Triton X-100 in PBS for 7 min and washed 3 times in PBS. The coverslips were blocked in PBS,2.5% goat serum,0.2% Tween-20 for 5 min followed by blocking in PBS, 0.4% fish skin gelatin, and 0.2% Tween-20. Cells were incubated with primary antibody for 1 h at room temperature. Cover-slips were washed with PBS, 0.2% Tween-20 5 times for 5 min and incubated with secondary antibodies for 45 min. Coverslips were washed as described above and the nucleus was stained with Hoechst 33258. The coverslips were mounted on slides in 9:1 glycerol/PBS with 0.1% p-phenylenediamine (Sigma-Aldrich; St. Louis, MO).
Fluorescence patterns were visualized with a Leitz Orthoplan microscope with epifluorescence and Hoffman Modulation Contrast optics from Chroma Technology (Brattleboro, VT, USA). Optical sections were captured with a CCD high-resolution camera from Roper Scientific (Tucson, AZ, USA) equipped with a camera/computer interface. Images were analyzed with a power Mac using IPLab Spectrum software (Scanalytics, Fairfax, VA, USA). A Perkin Elmer ERS6FE FRAP Spinning Disc Confocal was used to capture images to visualize the association of polyQ aggregates with PML Bodies. Images were analyzed in Volocity software.
For quantitation of aggresome patterns (diffuse staining versus aggresome localization), 100 cells were counted from randomly selected fields. Counting was performed in a “blind” manner by different members of the Sztul laboratory for GCP170*, mRFP-Werner, AT3-72Qmyc, and HttExon1-82Q-GFP. Data are presented as mean± SD. The Student’s t-test was used to determine statistical significance, and a p value less than 0.05 was considered significant.
Luciferase reporter assays
HT1080 cells were seeded in two 12-well plates and were grown for 24 h before transfection. The cells were cotransfected in triplicate with 500 ng of pGL2-p21A luciferase reporter construct and 350 ng of pcDNA3 (empty vector), or UL97-V5, or K355M–V5. Cells were washed once in PBS and lysed in 200 µl of luciferase lysis buffer (25 mM Tris-phosphate (pH 7.8), 2 mM DTT, 2 mM 1,2-diaminocyclohexane-N,N,N´, N´-tetraacetic acid, 10% glycerol,1%Triton X-100) with rocking for 5 min at room temperature. Then, 40 µl of lysate was added to 100 µl of luciferin substrate (20 mM Tricine, 1 mM MgCO3, 2.67 mM MgSO4, 0.1 mMEDTA,33.3 mMDTT,270 µMcoenzymeA,470 µMLuciferin,and 530 µM ATP). The lucifirase activity in each sample was measured in a Luminometer from Promega. The settings for the luminometer were a delay time of 3 sec with an integration time of 10 sec. Each experiment was done in triplicate.
Results
UL97 prevents aggregation of the non-polyQ proteins GFP170* and WRN
We have shown previously that GFP170* is an aggregation-prone protein that can be used to explore cellular mechanisms involved in the formation of aggresomes (Fu et al., 2005b). To test whether UL97 might affect deposition of GFP170* aggregates, we compared GFP170* localization in cells transfected with GCP170* in the presence of either an empty plasmid, a plasmid encoding UL97 or a plasmid encoding the catalytically inactive UL97/K355M. Consistent with our previous findings, cellular expression of GFP170* in the presence of an empty plasmid resulted in the presence of large ribbon-like aggregates within the cytoplasm, and large spherical inclusions within the nucleus (Supplemental Fig. 1A). In clear contrast with these observations, when GFP170* was expressed in the presence of UL97, large cytoplasmic and nuclear GFP170* aggregates were rarely observed, and instead, GFP170* appeared diffuse throughout the cytoplasm and the nucleoplasm (Supplemental Fig. 1B). To determine whether the effect of UL97 on the formation of GFP170* aggregates was dependent on its kinase activity, HeLa cells were transfected with plasmids encoding GFP170* and the kinase-dead UL97/K355M. Expression of the kinase-dead UL97 did not prevent the formation of cytoplasmic or nuclear GFP170* aggregates (Supplemental Fig. 1C). Interestingly, UL97/K355M colocalizes with cytoplasmic GCP170* aggregates but does not associate with nuclear inclusions. This is consistent with the predominantly cytoplasmic localization of UL97/K355M observed in transfected cells (Prichard et al., 2005 and Supplemental Fig. 2). It is unknown why the kinase-dead UL97/K355M remains within the cytoplasm while the wild-type UL97 shuttle into the nucleus.
Quantitative analysis revealed that the frequency of cells with large nuclear GFP170* aggregates is significantly decreased in cells transfected with UL97 (~10%) as compared to cells transfected with the empty plasmid (~79%) (Supplemental Fig. 1D). Expression of the kinase-dead UL97/K355M did not significantly affect the frequency of cells with aggregates (~73%) when compared to cells transfected with the empty plasmid (Supplemental Fig. 1D). These observations suggest that UL97 prevents GFP170* aggregation by a mechanism dependent on its kinase activity.
To further examine UL97 effects on protein aggregation, we tested whether UL97 can prevent aggregation of the WRN protein that when mutated causes the adult progeria disease Werner syndrome. WRN is a RecQ helicase and exonuclease (Gray et al., 1997; Huang et al., 1998) involved in DNA repair and telomere maintenance (Kamath-Loeb et al., 2000, 2004). Endogenous WRN exhibits a diffuse nuclear pattern, sometimes interspersed with nuclear or nucleolar foci (Marciniak et al., 1998; Opresko et al., 2003). When WRN is tagged at the N-terminus with monomeric red fluorescent protein (WRN–RFP), the protein forms multiple nuclear aggregates (Vaitiekunaite et al., 2007).
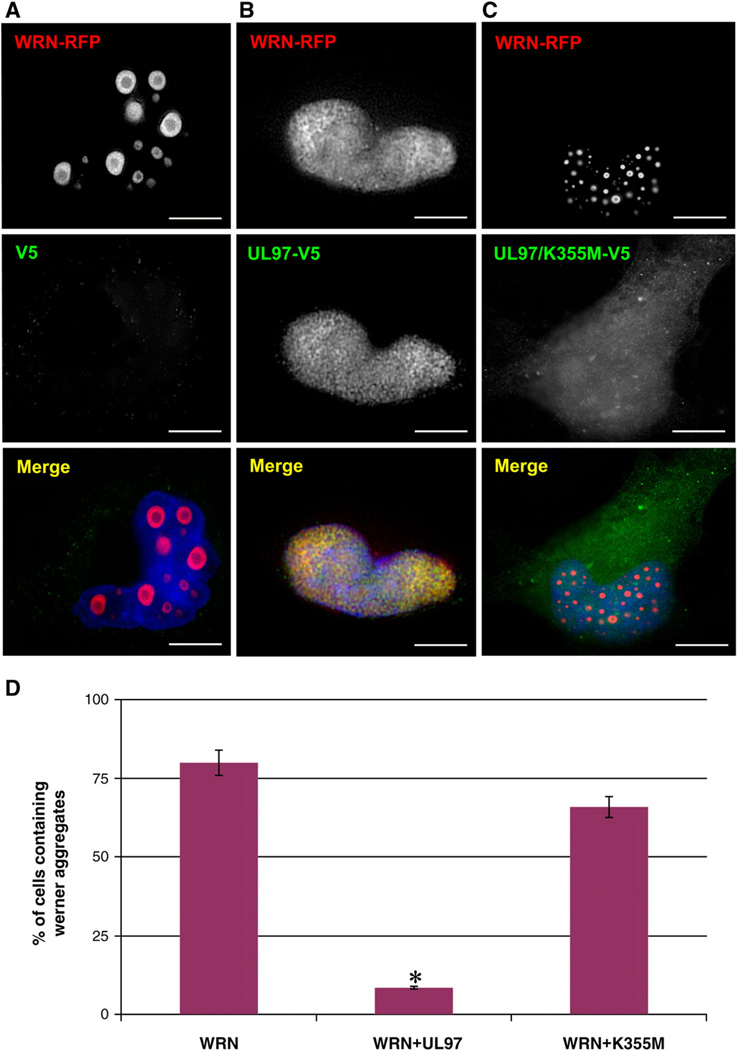
To test the effect of UL97 on the formation of WRN aggregates, cells were transfected with WRN–RFP in the presence of either an empty plasmid, a plasmid encoding UL97 or a plasmid encoding the kinase-dead UL97/K355M. Consistent with previous studies (Vaitiekunaite et al., 2007), expression of the WRN–RFP protein resulted in the presence of numerous “donut-shaped” nuclear inclusions (Fig. 1A). In clear contrast to these observations, when WRN–RFP was expressed in the presence of UL97, nuclear WRN–RFP aggregates were rarely observed but instead WRN–RFP fluorescence appeared diffuse throughout the nucleoplasm (Fig. 1B). Expression of UL97 had no effect on the nuclear localization of WRN–RFP, suggesting that the mechanism preventing the formation of WRN–RFP aggregates does not involve its nuclear import and/or retention. Expression of the kinase-dead UL97/K355M does not prevent the formation of nuclear WRN–RFP aggregates (Fig. 1C). Thus, UL97 prevents the formation of WRN–RFP nuclear aggregates in a kinase-dependent manner.
Fig. 1.
UL97 prevents deposition of nuclear aggregates of WRN protein. HeLa cells were cotransfected with mRFP-WRN and a vector either lacking UL97 (A), encoding UL97-V5 (B), or encoding UL97/K355M–V5 (C). After 24 h, cells were processed for IF with anti-GFP antibodies to detect WRN and anti-V5 antibodies to detect UL97. Cells expressing WRN were scored for aggregates in three independent experiments, and the graph represents the percentage of cells containing aggregates (D). mRFP-WRN forms large donut-shaped nuclear aggregates in control cells and in the presence of catalytically inactive UL97/K355M. Aggregates are not detected in cells expressing UL97. The difference is significant (p<0.0004). Bars, 19 µm.
Quantitative analysis revealed that in the absence of UL97 a majority (~80%) of cells expressing WRN–RFP contained nuclear aggregates, whereas expression of UL97 resulted in significantly decreased frequency of cells with aggregates (~7%) (Fig. 1D). Expression of the kinase-dead UL97/K355M did not significantly affect the frequency of cells with aggregates (~70%) when compared to cells transfected with the empty plasmid (Fig. 1D). However, we did notice that the size of WRN–RFP aggregates was consistently reduced in the presence of the UL97/K355M kinase-dead mutant.
The finding that UL97 exerts antiaggregation effect on the pp65 and pp71 viral proteins (Prichard et al., 2008) and on two nonviral proteins, GFP170* and WRN (this study), suggests that UL97 has a general antiaggregation property. To test this hypothesis, we next explored the possibility that UL97 prevents the aggregation of polyQ-expanded proteins linked to the neurodegenerative diseases SCA3 and HD.
UL97 prevents the aggregation of Ataxin-3 containing an expanded polyQ domain
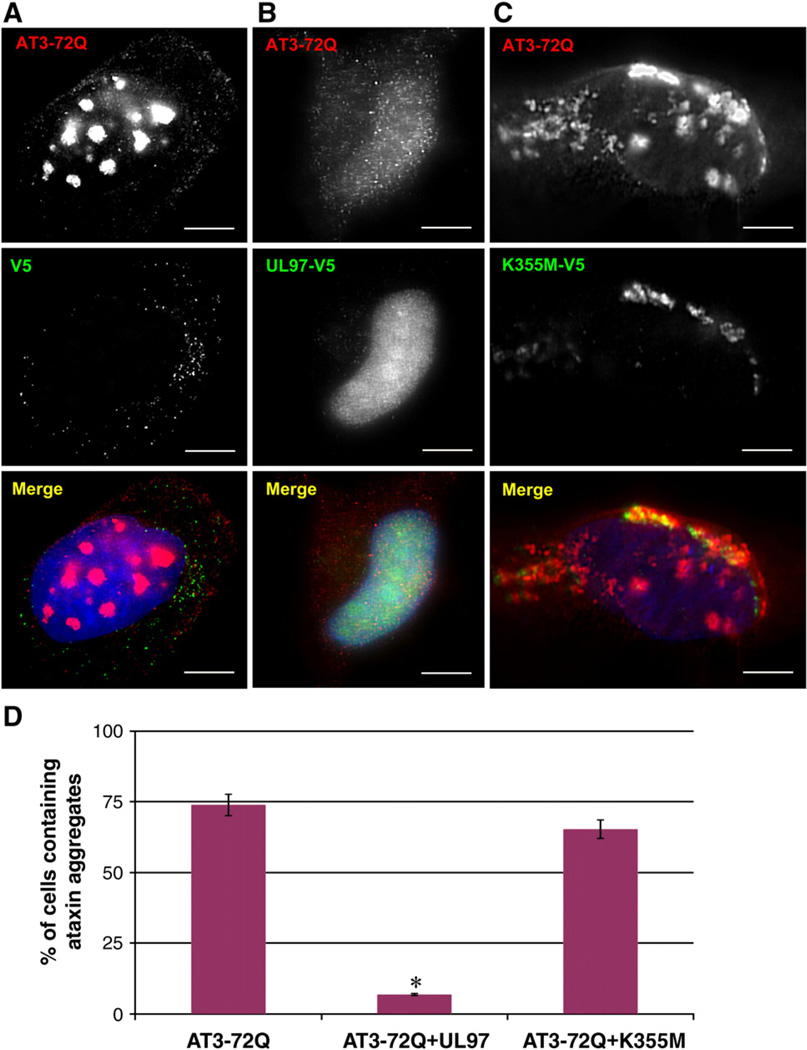
We used a HeLa cell-based assay to examine the formation of aggregates after transfecting cells with a plasmid encoding full-length ataxin-3 containing 72 glutamine residues (AT3-72Q) and tagged at its C-terminus domain with myc (Zhong and Pittman, 2006).To assess the effect of UL97 on AT3-72Q aggregation, cells were transfected with AT3-72Q in the presence of either an empty plasmid, a plasmid encoding UL97, or a plasmid encoding the catalytically inactive UL97/ K355M. In agreement with previous studies (Burnett et al., 2003; Burnett and Pittman, 2005), expression of AT3-72Q and the empty vector resulted in the presence of large aggregates that were localized within the nucleus (Fig. 2A). In contrast, expression of AT3-72Q in the presence of UL97 prevented the formation of aggregates, and AT3-72Q localized in small punctate structures scattered throughout the cytoplasm and the nucleoplasm (Fig. 2B). Expression of AT3-72Q in the presence of the kinase-dead UL97/K355M did not prevent the formation of large AT3-72Q aggregates (Fig. 2C). The kinase-dead mutant UL97/K355M colocalizes with cytoplasmic AT3-72Q aggregates but not with the nuclear aggregates. This is consistent with the cytoplasmic localization of UL97/K355M (Supplemental Figs. 1 and 2).
Fig. 2.
UL97 prevents aggregation of ataxin-3 containing expanded polyQ track. HeLa cells were cotransfected with AT3-72Q-myc and either a vector lacking UL97 (A), a vector encoding UL97-V5 (B), or a vector encoding UL97/K355M–V5 (C). After 72 h, cells were processed for IF with anti-myc antibodies to detect AT3-72Q and anti-V5 antibodies to detect UL97. Cells expressing AT3-72Q were scored for aggregates in three independent experiments, and the graph represents the percentage of cells containing aggregates (D). AT3-72Q-myc forms large cytoplasmic and nuclear aggregates in control cells and in cells expressing the catalytically inactive UL97/K355M. Aggregates are not detected or greatly reduced in cells that express UL97. The difference is significant (p<0.0007). Bars, 19 µm.
Quantitative analysis reveals that the frequency of cells with AT3-72Q aggregates is significantly decreased in cells transfected with UL97 (~6%) as compared to cells transfected with the empty plasmid (~71%) (Fig. 2D). Expression of the kinase-dead UL97/K355M did not significantly affect the frequency of cells with aggregates (~73%) when compared to cells transfected with AT3-72Q and an empty plasmid (Fig. 2D). Thus, UL97 prevents the formation of AT3-72Q aggregates in a kinase-dependent manner.
UL97 prevents the aggregation of the huntingtin exon-1 fragment containing an expanded polyQ domain
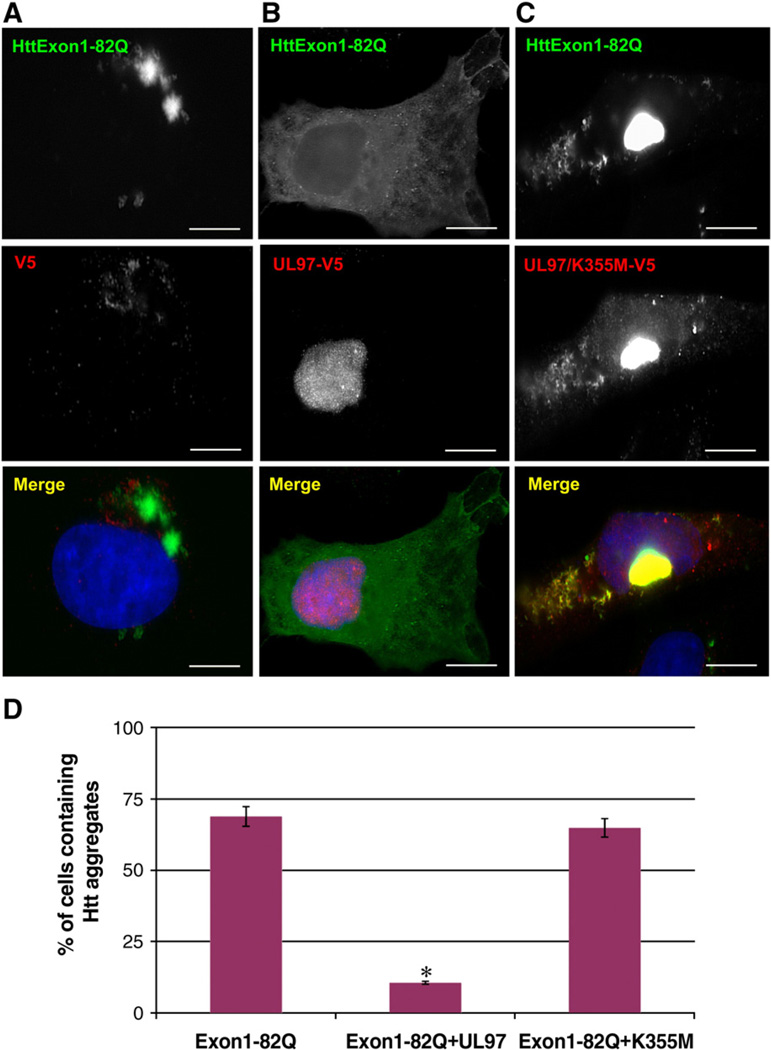
We next examined the effects of UL97 on the formation of polyQ aggregates in an HD cellular model. HeLa cells were transfected with a plasmid encoding the huntingtin exon1 containing 82 glutamines residues and a GFP tag at its N-terminus domain (HttExon1-82Q) (Chun et al., 2001). In agreement with our previous studies (Chun et al., 2001), expression of HttExon1-82Q resulted in the formation of large aggregates in the perinuclear area of cells expressing HttExon1-82Q and the empty vector (Fig. 3A). In contrast, aggregates were not detected in cells expressing HttExon1-82Q in the presence of UL97, and instead HttExon1-82Q presented a diffuse cytoplasmic staining (Fig. 3B). UL97 prevents the formation of HttExon1-82Q in a kinase dependent manner as the dead kinase UL97/K355M did not prevent the formation of Htt Exon1–82Q aggregates (Fig. 3C). In cells coexpressing Htt Exon1–82Q aggregates and UL97/K355M, the mutant kinase colocalize with Htt Exon1–82Q cytoplasmic aggregates (analysis of multiple confocal planes indicate all structures containing UL97/K355M are outside the nucleus). This is consistent with the cytoplasmic distribution of UL97/K355M and the preferential association of this mutant with cytoplasmic GCP170* and AT-372Q aggregates (Supplemental Figs. 1 and 2; Fig. 2C).
Fig. 3.
UL97 prevents aggregation of the polyQ expanded HttExon1-82Q. HeLa cells were cotransfected with HttExon1-82Q-YFP and either a vector lacking UL97 (A), a vector expressing UL97-V5 (B), or a vector expressing UL97/K355M–V5. After 48 h, cells were processed for IF with anti-GFP antibodies to detect HttExon1-82Q and anti-V5 antibodies to detect UL97. Cells expressing HttExon1-82Q were scored for aggregates in three independent experiments, and the graph represents the percentage of cells containing aggregates (D). Bars, 19 µm. HttExon1-82Q-YFP forms large cytoplasmic and nuclear aggregates in the absence of UL97 or in the presence of catalytically inactive UL97/K355M. Aggregates are not detected or greatly reduced in cells that express UL97. The difference is significant (p<0.004).
Quantification of these results reveals that the number of HttExon1-82Q aggregates is significantly lower in cells transfected with UL97 (~10%) when compared to cells transfected with the empty vector (~69%) (Fig. 3D). In contrast, expression of the kinase-dead UL97/K355M did not significantly affect the frequency of cells with aggregates (~67%) when compared to cells transfected with HttExon1-82Q and an empty plasmid (Fig. 3D).
UL97 coordinately disrupts PML bodies and prevents polyQ aggregation
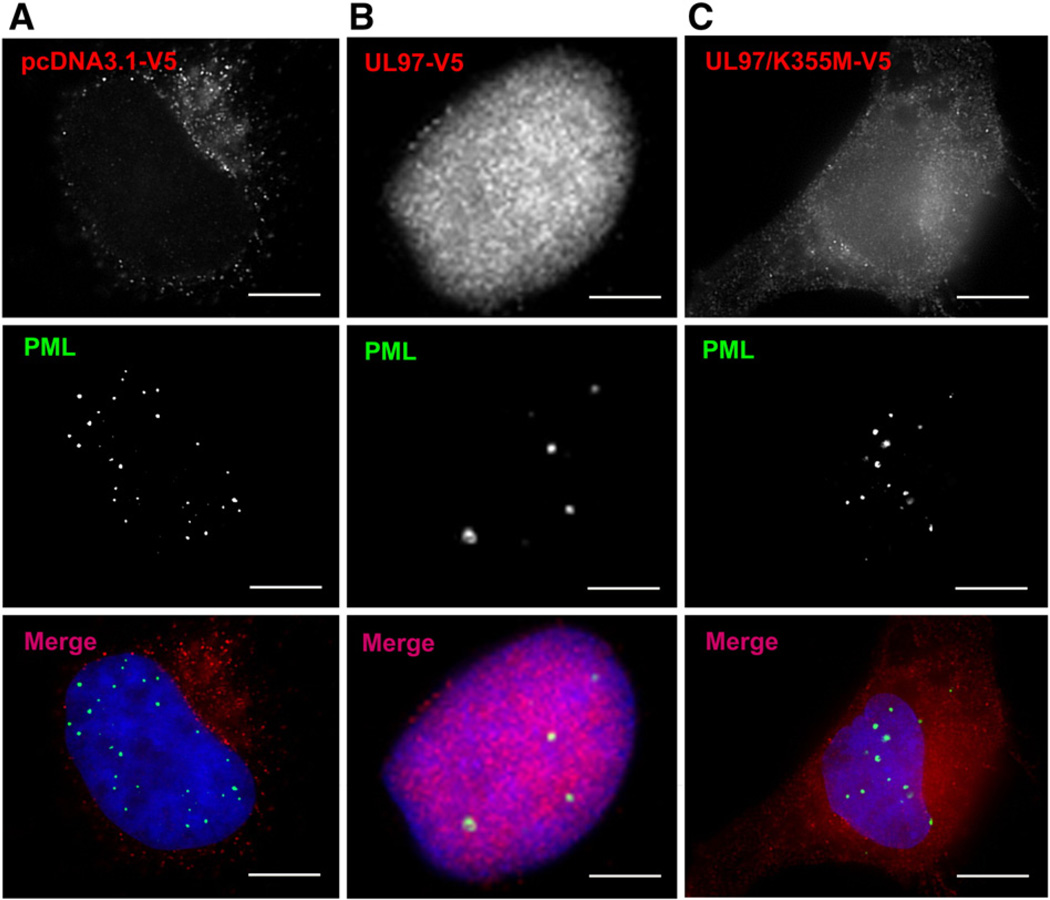
PML bodies have been linked to the aggregation of various viral and cellular proteins (Bernardi and Pandolfi, 2007; Bonilla et al., 2002; Nichol et al., 2009). We have reported recently that UL97 disrupt PML bodies, as monitored by the dispersion of the Sp100 component of PML bodies (Prichard et al., 2008). This suggests that the UL97 antiaggregation activity may be linked to PML disruption. To test this possibility, we examined the effect of UL97 on the integrity of PML bodies by following the localization of the defining component of PML bodies, the PML protein (pPML). pPML is required for nucleating and maintaining PML bodies, and cells devoid of pPML do not assemble PML bodies (Shen et al., 2006). In control cells, pPML is detected in 15– 30 discrete nuclear foci (Fig. 4A). Expression of UL97 has a dramatic effect and causes the disappearance of PML bodies, with the number of PML bodies decreasing to only few (sometimes larger) structures (Fig. 4B). UL97 localizes in a diffuse nuclear pattern in transfected cells, as previously reported (Prichard et al., 1999, 2008). Interestingly, although UL97 localizes to the nucleus, it does not appear to target to PML bodies, even when they become enlarged. In contrast, the inactive UL97/K355M mutant does not cause pPML mislocalization (Fig. 4C). UL97/K355M is detected in the cytoplasm and appears largely restricted from the nucleus. It is likely that the mutant cannot enter or be retained within the nucleus. This might explain the inability of UL97/K355M to disrupt PML bodies.
Fig. 4.
UL97 disrupts PML bodies. HeLa cells were transfected with an empty vector (A), transfected with V5-tagged UL97 (B), or with V5-tagged inactive UL97/K355M (C). After 24 h, cells were processed for IF with anti-V5 antibodies to detect UL97 and anti-pPML antibodies to detect PML bodies. In control cells, ~20 PML bodies are visible. Cells expressing UL97 show reduced number of PML bodies. A normal PML pattern is visible in cells expressing the inactive UL97/K355M.
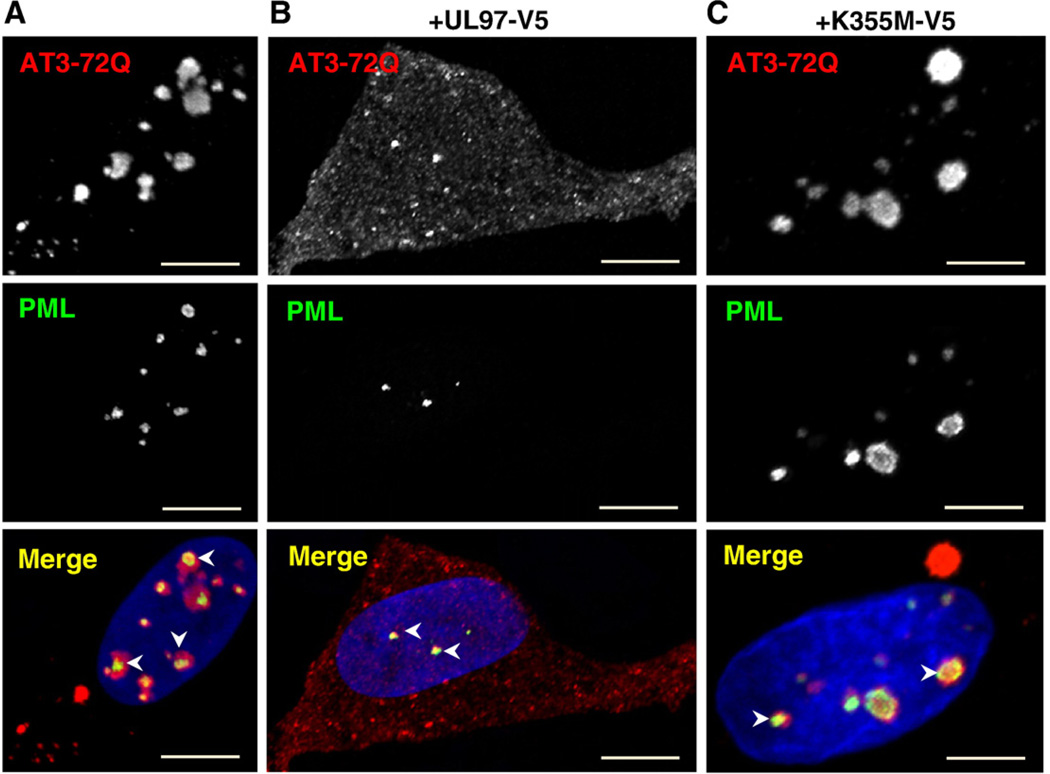
The disruptive effect of UL97 on PML bodies might be linked to its antiaggregation property. To test this hypothesis, we explored the relationship between UL97 effects on PML bodies and UL97 antiaggregation property in cells expressing AT3-72Q or HttExon1-82Q. The integrity of PML bodies was assessed in cells expressing mutant AT3-72Q and either a vector lacking UL97, a vector encoding UL97, or a vector encoding the catalytically inactive UL97/K355M. Cells expressing AT3-72Q and the empty vector contain multiple nuclear and cytoplasmic AT3-72Q aggregates (Fig. 5A). The cells also contain multiple PML bodies within the nucleus. There appears to be a close spatial association between the AT3-72Q aggregates and PML bodies, as most PML bodies lie adjacent to an aggregate (arrowheads). Cells coexpressing AT3-72Q and UL97 show diffuse cytoplasmic and nuclear staining of AT3-72Q, confirming the antiaggregation effect of UL97 (Fig. 5B). The cells contain drastically reduced number of PML bodies. The few remaining PML bodies colocalize with the small AT3-72Q aggregates detected in the nucleus (arrowheads). Cells coexpressing AT3-72Q and the inactive UL97/K355M contain a number of large nuclear and cytoplasmic AT3-72Q aggregates (Fig. 5C). PML bodies are present in these cells and appear to be extensively remodeled. The PML bodies are recruited to the aggregates to form a “beaded necklace” pattern around the aggregates (arrowheads). Thus, UL97 appears to coordinately disrupt PML bodies and prevents aggregation of AT3-72Q.
Fig. 5.
Disruption of PML bodies by UL97 is linked to inhibition in AT3-72Q aggregation. HeLa cells were cotransfected with AT3-72Q-myc and either a vector lacking UL97 (A), encoding UL97-V5 (B), or encoding UL97/K355M–V5 (C). At 72 h later, cells were processed for IF with anti-myc antibodies to detect AT3-72Q and anti-pPML antibodies to detect PML bodies. Cells transfected with an empty vector or the inactive UL97/K355M contain large cytoplasmic and nuclear AT3-72Q aggregates. In such cells, PML bodies are associated with the AT3-72Q aggregates (arrowheads in A and C). Cells transfected with UL97 lack AT3-72Q aggregates and have a reduced number of PML bodies. Sometimes small AT3-72Q aggregates are detected adjacent to the dispersed PML bodies (arrowheads in B). Bars, 19 µm.
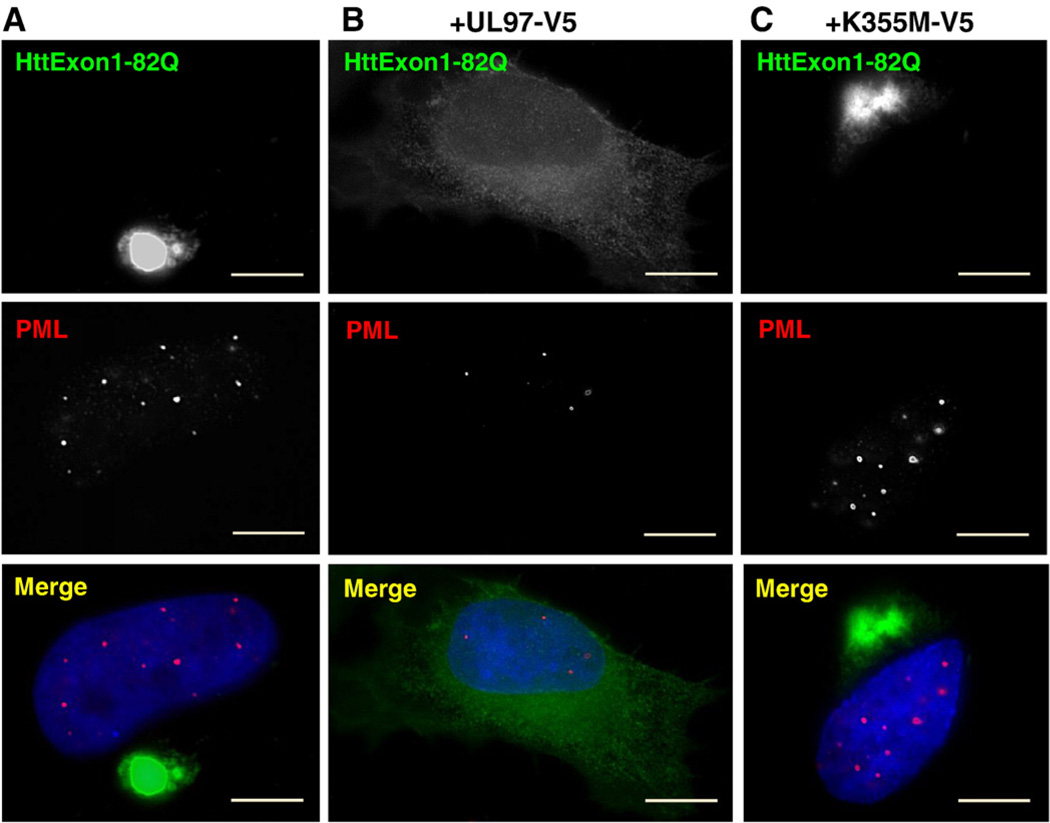
We also explored the relationship between PMLs and inhibition of HttExon1-82Q aggregation. Cells were cotransfected with HttExon1-82Q and either a vector lacking UL97, a vector encoding UL97, or a vector encoding the catalytically inactive UL97/K355M. Cells expressing HttExon1-82Q and the empty vector contain large cytoplasmic HttExon1-82Q aggregates (Fig. 6A). The cells show multiple PML bodies within the nucleus in a pattern indistinguishable from that in control cells (compare to Fig. 4A). In contrast, cells coexpressing HttExon1-82Q and UL97 show diffuse staining of HttExon1-82Q throughout the cytoplasm (Fig. 6B). The lack of HttExon1-82Q aggregates confirms the antiaggregation effect of UL97. The cells contain dramatically reduced number of PML bodies, and the remaining PMLs appear smaller than in control cells. Cells expressing HttExon1-82Q and the inactive UL97/K355M contain large, irregularly shaped cytoplasmic aggregates of HttExon1-82Q (Fig. 6C). The cells contain a normal complement of PML bodies within the nucleus. Thus, expression of UL97 causes the disruption of PML bodies and prevents aggregation of HttExon1-82Q.
Fig. 6.
Disruption of PML bodies by UL97 is linked to inhibition in HttExon1-82Q aggregation. HeLa cells were cotransfected with HttExon1-82Q-myc and either a vector lacking UL97 (A), encoding UL97-V5 (B), or encoding UL97/K355M–V5 (C). After 72 h, cells were processed for IF with anti-myc antibodies to detect HttExon1-82Q and anti-pPML antibodies to detect PML bodies. Cells transfected with an empty vector or the inactive UL97/K355M contain large HttExon1-82Q aggregates and normal PML bodies. Cells transfected with UL97 lack HttExon1-82Q aggregates and have dispersed PML bodies. Bars, 19 µm.
The commonality of the regulatory effect of UL97 on PML integrity suggests that the aggregation processes of polyQ proteins could be functionally coupled to the status of PML bodies and that the dispersion of PML bodies by UL97 could be linked to antiaggregation. The coordinated actions of UL97 in dispersal of PML bodies and prevention of aggregation both require the kinase activity of UL97.
UL97 inhibits p53-dependent transcription
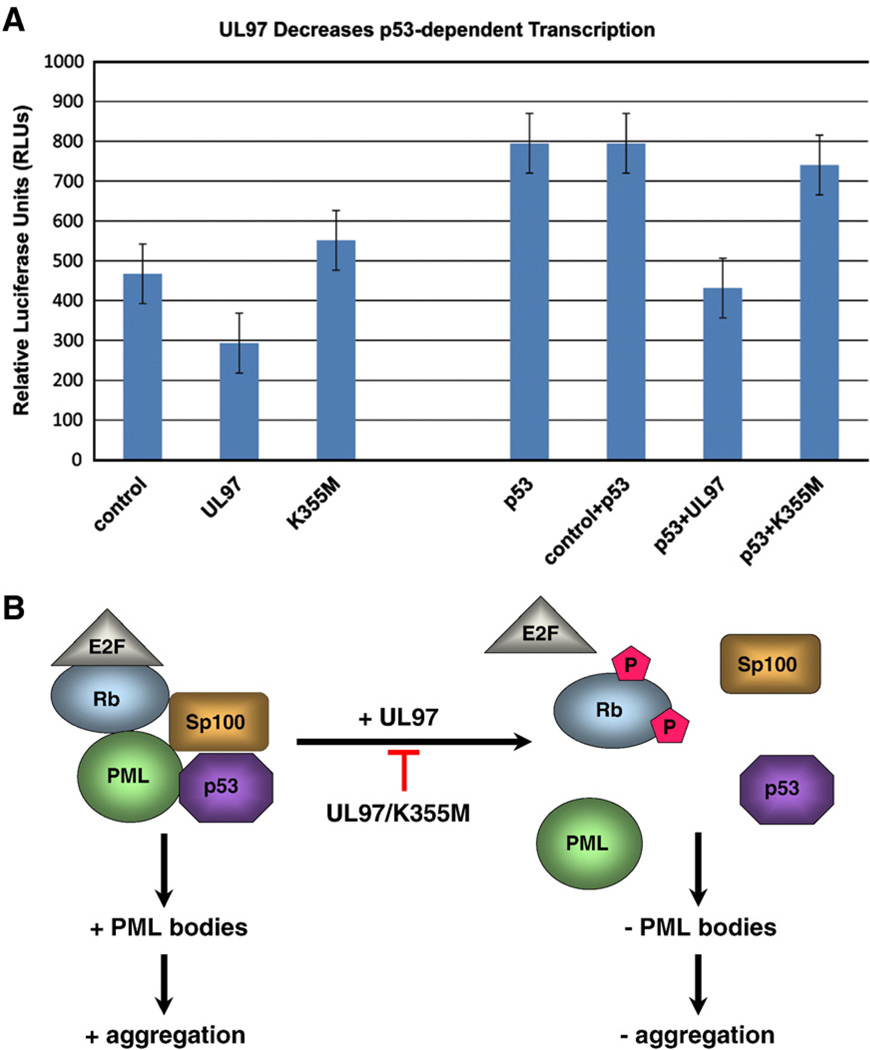
PML bodies have been implicated in transcription, and they contain the transcriptional regulators Sp100, pPML, CBP, and p53 (de Stanchina et al., 2004; Hofmann et al., 2002; Moller et al., 2003; Pampin et al., 2006; Salomoni et al., 2006; reviewed in Bernardi and Pandolfi, 2007). Thus, we explored the link between UL97 effects on PML bodies and p53-mediated transcription. p53 activity was assessed in HT1080 fibrosarcoma cells by measuring transcription from a reporter construct composed of firefly luciferase fused to the p21 promoter that contains two p53 response elements (pGL2-p21A). Comparisons were made between cells cotransfected with pGL2-p21A and either the empty vector (control), a vector containing UL97 (UL97), or a vector containing UL97/K355M (UL97/K355M). Basal transcription levels were quantified in cells transfected with pGL2-p21A and the empty vector (Fig. 7A, control), and these are consistent with previously reported results. Cells cotransfected with pGL2-p21A and UL97 showed a 30% reduction in p53 activity (Fig. 7A, UL97). The inhibitory effect was dependent upon UL97 kinase activity, as cells cotransfected with pGL2-p21A and the inactive UL97/K355M showed no changes in the levels of p53-dependent transcription (Fig. 7A, UL97/K355M).
Fig. 7.
UL97 decreases p53-mediated transcription. (A) A set of HT1080 cells was cotransfected with pGL2-p21A (a p21-responsive luciferase reporter) and either a vector lacking UL97, encoding UL97-V5, or encoding UL97/K355M–V5. Another set of HT1080 cells was cotransfected with pGL2-p21A and p53, and either a vector lacking UL97, encoding UL97-V5, or encoding UL97/K355M–V5. At 24 h after transfection, cells were assayed for luciferase activity. Each transfection was done in triplicate and an average value of relative luciferase units (RLUs) is reported. (B) A model for UL97 action in modulating aggregation. Control cells contain PML bodies and form aggregates. In cells expressing the UL97 kinase, the Rb component of PML bodies is phosphorylated, PML bodies are dispersed and p53-mediated transcription is decreased. These changes correlate with inhibition in the formation of aggregates.
The HT1080 cells used in these experiments contain endogenous p53. To increase p53-dependent transcription and thus increase the experimental signal, we performed additional experiments in HT1080 cells transfected with exogenous p53. Comparisons were made between cells cotransfected with p53, pGL2-p21A and either a vector lacking UL97 (p53+control), a vector encoding UL97 (p53+UL97), or a vector encoding UL97/K355M (p53+UL97/K355M). As expected, cells expressing exogenous p53 showed a significantly higher basal level of transcription (Fig. 7A, p53 and p53+control). Coexpression of UL97 decreased p53-mediated transcription by 50% (Fig. 7A, p53+ UL97). Coexpression of the inactive UL97/K355M had no effect on p53-mediated transcription (Fig. 7A, p53+UL97/K355M). These results indicate that UL97 represses the transcriptional activity of p53 in a kinase-dependent manner and suggest a potential mechanism that links the effects of UL97 on aggregation, integrity of PML bodies, and p53-mediated transcription.
We assessed whether the UL97 kinase might rescue the cytotoxicity associated with AT3-72Q polyQ aggregates. In these experiments, cells were cotransfected with AT3-73Q and either a vector lacking UL97, a vector encoding UL97, or a vector encoding UL97/K355M. After 24 or 48 h of culture, cell viability was measured by the uptake of fluorescein diacetate. UL97 appears to increase the percentage of viable cells expressing AT3-72Q (data not shown).
Discussion
The results of this study provide strong evidence for an antiaggregation effect of the UL97 viral kinase. UL97 prevents the aggregation of GFP170* and WRN proteins, and of two polyQ-containing proteins, AT3-72Q and HttExon1-82Q that are associated with SCA3 and Huntington disease, respectively. Remarkably, UL97 is able to prevent the formation of both cytoplasmic and nuclear aggregates formed by these proteins. Consistent with previous studies revealing that UL97 possesses a nuclear localization signal (Prichard et al., 2005), we find that UL97 localizes preferentially within the nucleus. This suggests that the antiaggregation properties of UL97 are likely to be mediated through events occurring within the nucleus. Within the nucleus,the UL97 kinase may interact and phosphorylate nuclear substrates to prevent aggregation. This model is consistent with our findings that the UL97/K355M mutant defective in kinase activity is essentially restricted from the nucleus and is unable to prevent protein aggregation. Further work will be required to dissect the relative contributions of the nuclear localization and the kinase activity towards the antiaggregation function of UL97. Together, our studies document that UL97 localizes within the nucleus and prevents protein aggregation in a kinase-dependent manner.
UL97 shows kinase activity towards a number of cellular substrates. For example, UL97 phosphorylates the myelin basic protein; histones H1, H2B, and H3; lamins A, B, and C; and the largest subunit of RNA polymerase II in vitro (Baek et al., 2002a,b, 2004; Marschall et al., 2005). Whether any of these known UL97 substrates are linked to the antiaggregating property of UL97 remains to be elucidated. However, recent studies suggest a role for the retinoblastoma (Rb) protein, a recently identified UL97 substrate, in UL97-mediated antiaggregation. Rb is a tumor-suppressor that controls cell cycle progression through G1 phase (Hume et al., 2008; Prichard et al., 2008). Importantly, Rb localizes to PML bodies, where it forms stable complexes with unphosphorylated pPML (Alcalay et al., 1998). The identification of Rb as UL97 substrate seems especially relevant in the light of the apparent relationship between PML bodies and aggregate formation, and the coordinate ability of UL97 to disrupt PML bodies and prevent aggregation. The mechanisms by which UL97 causes disruption of PML bodies remains to be further characterized. However, our findings are consistent with a model (Fig. 7B) in which UL97 causes changes in the architecture of PML bodies through a phosphorylation dependent mechanism. Remodeling and disruption of PML bodies would then lead to changes in the transcription of p53-responsive genes and affect the formation of aggregates. In support of this model, we provide evidence that GFP170*, WRN, and AT3-72Q deposit adjacent to PML bodies and cause the rearrangement of PML bodies around the aggregates. Further, we show that UL97 disrupts PML bodies. The UL97-mediated disruption of PML is likely to have important consequences on transcriptional activity. In support, our study provides clear evidence that UL97-mediated disruption of PML bodies results in the inhibition of p53-mediated transcription. Whether the antiaggregation action of UL97 is linked to alterations in a larger transcriptional repertoire remains to be explored. Importantly, UL97-induced disruption of PML bodies and alteration in p53-dependent transcription are linked to UL97 ability to prevent aggregation of AT3-72Q and HttExon1-82Q in cellular models of SCA3 and HD, respectively.
Huntington’s disease, spinocerebellar ataxia 3, and several other polyglutamine disorders are characterized by the accumulation of cytoplasmic and/or nuclear aggregates within the brain of affected individuals (Davies et al., 1997; DiFiglia et al., 1997). In these diseases, a mutated protein misfolds to undergo an alternative conformation that in most cases results in its aggregation and accumulation as inclusion bodies in neurons. The role of these aggregates in the etiology of misfolded diseases has been the subject of intense debate, with models that consider the aggregates as harmful, benign, or beneficial (Ross and Poirier, 2005; Sisodia, 1998). A body of emerging evidence suggests that aggregates might be cytoprotective. Initial clues came from postmortem studies that revealed a relatively poor correlation between the neurons in which inclusion bodies are present and the neurons preferentially vulnerable in specific neurodegenerative disorders (Kuemmerle et al., 1999; reviewed in Ross, 2002). Further temporal studies of the appearance of aggregates and the onset of clinical pathology suggest that tissue damage and pathology can manifest before detection of aggregates (Saudou et al., 1998), raising the possibility that aggregates may actually be protective. Additional studies by Arrasate et al. (2004) provided evidence that the formation of inclusion bodies reduces the risk of neuronal death. A potential explanation is that inclusion bodies and visible aggregates constitute a point of sequestration of intermediate conformational assemblies that correlate more directly to the neuronal vulnerability than the aggregates (Poirier et al., 2002, 2005; Ross et al., 2003). Indeed, accumulating evidence supports the view that protein aggregation is a complex process, thought to be initiated by the interactions between the misfolded proteins to generate a variety of higher-order intermediate assemblies that ultimately form insoluble inclusion bodies (Poirier et al., 2002; Wetzel, 2006).
Using recombinant mutant huntingtin exon1 with an expanded polyQ track in in vitro studies, a number of studies have characterized the morphological and structural features of aggregation and provided evidence of globular and protofibrillar intermediates rich in β-structure before the formation of fibrils and aggregates (Chen et al., 2001, 2002; Poirier et al., 2002, 2005). Similar intermediate conformational assemblies have been identified for other proteins capable of forming aggregates in various neurodegenerative disorders, suggesting common mechanism of aggregation and potentially toxicity (Kayed et al., 2003). In line with this model, accumulating evidence suggests that the toxicity of amyloidogenic proteins involved in several neurodegenerative disorders may not be related to the insoluble protein aggregates but rather to the soluble oligomeric or other intermediate assemblies. For instance, soluble oligomeric or protofibrillar forms of amyloids are more potent toxins than the mature fibrils of Aβ (Dahlgren et al., 2002), α-synuclein (Conway et al., 2000), IAPP (Anguiano et al., 2002), and polyQ (Kayed et al., 2003, 2004, 2009). In effect, the formation of aggregates might be a protective mechanism to sequester and isolate these toxic intermediate conformational species (Ross and Poirier, 2005).
In this study, we used cellular models of aggregation to show the efficacy of the UL97 kinase in preventing the deposition of non-polyQ and polyQ aggregates. Although it is clear that UL97 targets cellular pathways that control aggregation, it remains to be determined whether UL97 affects the formation and accumulation of potentially toxic soluble oligomeric or protofibrillar conformations of the mutated proteins. The ability of UL97 to inhibit aggregation of polyQ proteins in cellular models of diseases as diverse as HD and SCA3 raises the possibility that UL97 might be a tool to dissect the aggregation mechanisms in diseases associated with polyQ protein deposits. Thus, characterizing the molecular mechanisms of UL97 action in preventing protein aggregation may reveal a novel rational approach to prevent polyQ-mediated neurodegeneration.
Supplementary Material
Acknowledgments
We thank Dr. Susan Nozelle for assisting with luciferase assays and thoughtful insight into transcriptional aspects of this work. We are also grateful to Dr. Melanie Styers for critically reviewing the manuscript.
Footnotes
Supplementary materials related to this article can be found online at doi:10.1016/j.nbd.2010.08.013.
References
- Alcalay M, Tomassoni L, Colombo E, Stoldt S, Grignani F, Fagioli M, Szekely L, Helin K, Pelicci PG. The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol. Cell. Biol. 1998;18:1084–1093. doi: 10.1128/mcb.18.2.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguiano M, Nowak RJ, Lansbury PT., Jr. Protofibrillar islet amyloid polypeptide permeabilizes synthetic vesicles by a pore-like mechanism that may be relevant to type II diabetes. Biochemistry. 2002;41:11338–11343. doi: 10.1021/bi020314u. [DOI] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Baek MC, Krosky PM, Coen DM. Relationship between autophosphorylation and phosphorylation of exogenous substrates by the human cytomegalovirus UL97 protein kinase. J. Virol. 2002a;76:11943–11952. doi: 10.1128/JVI.76.23.11943-11952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek MC, Krosky PM, He Z, Coen DM. Specific phosphorylation of exogenous protein and peptide substrates by the human cytomegalovirus UL97 protein kinase. Importance of the P+5 position. J. Biol. Chem. 2002b;277:29593–29599. doi: 10.1074/jbc.M202312200. [DOI] [PubMed] [Google Scholar]
- Baek MC, Krosky PM, Pearson A, Coen DM. Phosphorylation of the RNA polymerase II carboxyl-terminal domain in human cytomegalovirus-infected cells and in vitro by the viral UL97 protein kinase. Virology. 2004;324:184–193. doi: 10.1016/j.virol.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Bonilla WV, Pinschewer DD, Klenerman P, Rousson V, Gaboli M, Pandolfi PP, Zinkernagel RM, Salvato MS, Hengartner H. Effects of promyelocytic leukemia protein on virus-host balance. J. Virol. 2002;76:3810–3818. doi: 10.1128/JVI.76.8.3810-3818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett BG, Pittman RN. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc. Natl Acad. Sci. USA. 2005;102:4330–4335. doi: 10.1073/pnas.0407252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett B, Li F, Pittman RN. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol Genet. 2003;12:3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- Chan HY, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum. Mol. Genet. 2000;9:2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- Chen S, Berthelier V, Yang W, Wetzel R. Polyglutamine aggregation behavior in vitro supports a recruitment mechanism of cytotoxicity. J. Mol. Biol. 2001;311:173–182. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]
- Chen S, Berthelier V, Hamilton JB, O’Nuallain B, Wetzel R. Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry. 2002;41:7391–7399. doi: 10.1021/bi011772q. [DOI] [PubMed] [Google Scholar]
- Chinery R, Brockman JA, Peeler MO, Shyr Y, Beauchamp RD, Coffey RJ. Antioxidants enhance the cytotoxicity of chemotherapeutic agents in colorectal cancer: a p53-independent induction of p21WAF1/CIP1 via C/EBPbeta. Nat. Med. 1997;3:1233–1241. doi: 10.1038/nm1197-1233. [DOI] [PubMed] [Google Scholar]
- Chun W, Lesort M, Tucholski J, Faber PW, MacDonald ME, Ross CA, Johnson GV. Tissue transglutaminase selectively modifies proteins associated with truncated mutant huntingtin in intact cells. Neurobiol. Dis. 2001;8:391–404. doi: 10.1006/nbdi.2001.0390. [DOI] [PubMed] [Google Scholar]
- Conway KA, Harper JD, Lansbury PT., Jr. Fibrils formed in vitro from alpha-synuclein and two mutant forms linked to Parkinson’s disease are typical amyloid. Biochemistry. 2000;39:2552–2563. doi: 10.1021/bi991447r. [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Zoghbi HY. Trinucleotide repeats: mechanisms and pathophysiology . Annu. Rev. Genomics Hum. Genet. 2000;1:281–328. doi: 10.1146/annurev.genom.1.1.281. [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Sun Y, Opal P, Antalffy B, Mestril R, Orr HT, Dillmann WH, Zoghbi HY. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum. Mol. Genet. 2001;10:1511–1518. doi: 10.1093/hmg/10.14.1511. [DOI] [PubMed] [Google Scholar]
- Dahlgren KN, Manelli AM, Stine WB, Jr., Baker LK, Krafft GA, LaDu MJ. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J. Biol. Chem. 2002;277:32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW. PML is a direct p53 target that modulates p53 effector functions. Mol. Cell. 2004;13:523–535. doi: 10.1016/s1097-2765(04)00062-0. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Ehrnhoefer DE, Duennwald M, Markovic P, Wacker JL, Engemann S, Roark M, Legleiter J, Marsh JL, Thompson LM, Lindquist S, Muchowski PJ, Wanker EE. Green tea (−)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington’s disease models. Hum. Mol. Genet. 2006;15:2743–2751. doi: 10.1093/hmg/ddl210. [DOI] [PubMed] [Google Scholar]
- Fu L, Gao YS, Sztul E. Transcriptional repression and cell death induced by nuclear aggregates of non-polyglutamine protein. Neurobiol. Dis. 2005a;20:656–665. doi: 10.1016/j.nbd.2005.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Gao YS, Tousson A, Shah A, Chen TL, Vertel BM, Sztul E. Nuclear aggresomes form by fusion of PML-associated aggregates. Mol. Biol. Cell. 2005b;16:4905–4917. doi: 10.1091/mbc.E05-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nat. Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- Hicks SW, Machamer CE. The NH2-terminal domain of Golgin-160 contains both Golgi and nuclear targeting information. J. Biol. Chem. 2002;277:35833–35839. doi: 10.1074/jbc.M206280200. [DOI] [PubMed] [Google Scholar]
- Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, Will H, Schmitz ML. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′-N5′ exonuclease. Nat. Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume AJ, Finkel JS, Kamil JP, Coen DM, Culbertson MR, Kalejta RF. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science. 2008;320:797–799. doi: 10.1126/science.1152095. [DOI] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Johansson E, Burgers PM, Loeb LA. Functional interaction between the Werner syndrome protein and DNA polymerase delta. Proc. Natl Acad. Sci. USA. 2000;97:4603–4608. doi: 10.1073/pnas.97.9.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Welcsh P, Waite M, Adman ET, Loeb LA. The enzymatic activities of the Werner syndrome protein are disabled by the amino acid polymorphism R834C. J. Biol. Chem. 2004;279:55499–55505. doi: 10.1074/jbc.M407128200. [DOI] [PubMed] [Google Scholar]
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- Kayed R, Sokolov Y, Edmonds B, McIntire TM, Milton SC, Hall JE, Glabe CG. Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J. Biol. Chem. 2004;279:46363–46366. doi: 10.1074/jbc.C400260200. [DOI] [PubMed] [Google Scholar]
- Kayed R, Pensalfini A, Margol L, Sokolov Y, Sarsoza F, Head E, Hall J, Glabe C. Annular protofibrils are a structurally and functionally distinct type of amyloid oligomer. J. Biol. Chem. 2009;284:4230–4237. doi: 10.1074/jbc.M808591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuemmerle S, Gutekunst CA, Klein AM, Li XJ, Li SH, Beal MF, Hersch SM, Ferrante RJ. Huntington aggregates may not predict neuronal death in Huntington’s disease. Ann. Neurol. 1999;46:842–849. [PubMed] [Google Scholar]
- Marciniak RA, Lombard DB, Johnson FB, Guarente L. Nucleolar localization of the Werner syndrome protein in human cells. Proc. Natl Acad. Sci. USA. 1998;95:6887–6892. doi: 10.1073/pnas.95.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall M, Stein-Gerlach M, Freitag M, Kupfer R, van Den Bogaard M, Stamminger T. Inhibitors of human cytomegalovirus replication drastically reduce the activity of the viral protein kinase pUL97. J. Gen. Virol. 2001;82:1439–1450. doi: 10.1099/0022-1317-82-6-1439. [DOI] [PubMed] [Google Scholar]
- Marschall M, Marzi A, aus dem Siepen P, Jochmann R, Kalmer M, Auerochs S, Lischka P, Leis M, Stamminger T. Cellular p32 recruits cytomegalo-virus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 2005;280:33357–33367. doi: 10.1074/jbc.M502672200. [DOI] [PubMed] [Google Scholar]
- McCampbell A, Taye AA, Whitty L, Penney E, Steffan JS, Fischbeck KH. Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc. Natl Acad. Sci. USA. 2001;98:15179–15184. doi: 10.1073/pnas.261400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel D, Schaarschmidt P, Wunderlich K, Heuschmid M, Simoncini L, Muhlberger D, Zimmermann A, Pavic I, Mertens T. Functional regions of the human cytomegalovirus protein pUL97 involved in nuclear localization and phosphorylation of ganciclovir and pUL97 itself. J. Gen. Virol. 1998;79(Pt 9):2105–2112. doi: 10.1099/0022-1317-79-9-2105. [DOI] [PubMed] [Google Scholar]
- Misumi Y, Sohda M, Tashiro A, Sato H, Ikehara Y. An essential cytoplasmic domain for the Golgi localization of coiled-coil proteins with a COOH-terminal membrane anchor. J. Biol. Chem. 2001;276:6867–6873. doi: 10.1074/jbc.M010121200. [DOI] [PubMed] [Google Scholar]
- Moller A, Sirma H, Hofmann TG, Staege H, Gresko E, Ludi KS, Klimczak E, Droge W, Will H, Schmitz ML. Sp100 is important for the stimulatory effect of homeodomain-interacting protein kinase-2 on p53-dependent gene expression. Oncogene. 2003;22:8731–8737. doi: 10.1038/sj.onc.1207079. [DOI] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- Nichol JN, Petruccelli LA, Miller WH., Jr Expanding PML’s functional repertoire through post-translational mechanisms. Front. Biosci. 2009;14:2293–2306. doi: 10.2741/3380. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Cheng WH, von Kobbe C, Harrigan JA, Bohr VA. Werner syndrome and the function of the Werner protein; what they can teach us about the molecular aging process. Carcinogenesis. 2003;24:791–802. doi: 10.1093/carcin/bgg034. [DOI] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Pampin M, Simonin Y, Blondel B, Percherancier Y, Chelbi-Alix MK. Cross talk between PML and p53 during poliovirus infection: implications for antiviral defense. J. Virol. 2006;80:8582–8592. doi: 10.1128/JVI.00031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier MA, Li H, Macosko J, Cai S, Amzel M, Ross CA. Huntingtin spheroids and protofibrils as precursors in polyglutamine fibrilization. J. Biol. Chem. 2002;277:41032–41037. doi: 10.1074/jbc.M205809200. [DOI] [PubMed] [Google Scholar]
- Poirier MA, Jiang H, Ross CA. A structure-based analysis of huntingtin mutant polyglutamine aggregation and toxicity: evidence for a compact beta-sheet structure. Hum. Mol. Genet. 2005;14:765–774. doi: 10.1093/hmg/ddi071. [DOI] [PubMed] [Google Scholar]
- Prichard MN, Gao N, Jairath S, Mulamba G, Krosky P, Coen DM, Parker BO, Pari GS. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J. Virol. 1999;73:5663–5670. doi: 10.1128/jvi.73.7.5663-5670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN, Britt WJ, Daily SL, Hartline CB, Kern ER. Human cytomegalovirus UL97 kinase is required for the normal intranuclear distribution of pp 65 and virion morphogenesis. J. Virol. 2005;79:15494–15502. doi: 10.1128/JVI.79.24.15494-15502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN, Sztul E, Daily SL, Perry AL, Frederick SL, Gill RB, Hartline CB, Streblow DN, Varnum SM, Smith RD, Kern ER. Human cytomegalovirus UL97 kinase activity is required for the hyperphosphorylation of retinoblastoma protein and inhibits the formation of nuclear aggresomes. J. Virol. 2008;82:5054–5067. doi: 10.1128/JVI.02174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington’s disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Opinion: what is the role of protein aggregation in neurodegeneration? Nat. Rev. Mol. Cell Biol. 2005;6:891–898. doi: 10.1038/nrm1742. [DOI] [PubMed] [Google Scholar]
- Ross CA, Wood JD, Schilling G, Peters MF, Nucifora FC, Jr., Cooper JK, Sharp AH, Margolis RL, Borchelt DR. Polyglutamine pathogenesis. Philos. Trans. R. Soc. Lond. B BiolSci. 1999;354:1005–1011. doi: 10.1098/rstb.1999.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Poirier MA, Wanker EE, Amzel M. Polyglutamine fibrillogenesis: the pathway unfolds. Proc. Natl Acad. Sci. USA. 2003;100:1–3. doi: 10.1073/pnas.0237018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni P, Guernah I, Pandolfi PP. The PML-nuclear body associated protein Daxx regulates the cellular response to CD40. Cell Death Differ. 2006;13:672–675. doi: 10.1038/sj.cdd.4401820. [DOI] [PubMed] [Google Scholar]
- Sanchez I, Lin H, Scaglioni P. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 2003;421:373–379. doi: 10.1038/nature01301. [DOI] [PubMed] [Google Scholar]
- Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- Shen T, Lin H, Scaglioni P, Yung T, Pandolfi P. The mechanisms of PML-nuclear body formation. Mol. Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisodia SS. Nuclear inclusions in glutamine repeat disorders: are they pernicious, coincidental, or beneficial? Cell. 1998;95:1–4. doi: 10.1016/s0092-8674(00)81743-2. [DOI] [PubMed] [Google Scholar]
- Vaitiekunaite R, Butkiewicz D, Krzesniak M, Przybylek M, Gryc A, Snietura M, Benedyk M, Harris CC, Rusin M. Expression and localization of Werner syndrome protein is modulated by SIRT1 and PML. Mech. Ageing Dev. 2007;128:650–661. doi: 10.1016/j.mad.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat. Struct. Mol. Biol. 2004;11:1215–1222. doi: 10.1038/nsmb860. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Morabito LM, Bilen J, Gordesky-Gold B, Faust LZ, Paulson HL, Bonini NM. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol. Cell. 2005;18:37–48. doi: 10.1016/j.molcel.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Wetzel R. Kinetics and thermodynamics of amyloid fibril assembly. Acc. Chem. Res. 2006;39:671–679. doi: 10.1021/ar050069h. [DOI] [PubMed] [Google Scholar]
- Zhong X, Pittman RN. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum. Mol. Genet. 2006;15:2409–2420. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.