Abstract
Many histone co-valent modifications have been identified and shown to play key regulatory roles in eukaryotic transcription, DNA damage repair and replication. In vitro experiments designed to understand the mechanistic role of individual modifications require the availability of substantial quantities of pure histones, homogeneously modified at specific residues. We have applied the amber stop codon/suppressor tRNA strategy to the production of histone H4 acetylated at lysine 16, a particularly important isoform of this histone. Our success relies on adapting the H4 DNA sequence to the codon preference of E. coli and on preventing the premature decay of the H4 mRNA. These modifications to the original procedure render it easily applicable to the generation of any co-valently modified histone H4 isoform.
In the nuclei of eukaryotic cells, the basic repeating subunit of chromatin is the nucleosome, an octamer of four different histone molecules around which approximately 147 base pairs of DNA are wrapped. Nucleosomes interact, and attract a variety of non-histone proteins thereby condensing chromatin and rendering it refractory to transcription, replication and DNA damage repair. In order for these fundamental processes to occur, the architecture of chromatin must be modified. This is achieved by a variety of epigenetic mechanisms, among which the co-valent modification of the histones plays a critical role. These modifications either alter the topology of the chromatin fiber directly, or serve as platforms for the docking of remodeling complexes. Histones can be acetylated, methylated, phosphorylated, ubiquitinated, ADP-ribosylated, sumoylated and glycosylated at various positions, largely but not exclusively along their N-terminal tails. Combinations of these modifications determine the local chromatin state. One of the better studied among histone modifications is the acetylation of histone H4 at lysine16 (H4K16ac), a modification that is laid down by the histone acetyl transferase hMOF (MYST1/KAT8) [1, 2]. Loss of H4K16 acetylation has been reported in a variety of human cancers. In addition, H4K16 hypoacetylation is associated with defective DNA repair and premature senescence [3-5]. Recently, H4K16 acetylation was shown to be essential for the renewal of pluripotent stem cells [6].
The chromatin modifications that regulate the DNA processes mentioned above have been studied using all of the modern tools of molecular biology, from biochemistry and immunochemistry to mutagenesis and bioinformatics. The realization that a greater understanding of the regulatory mechanisms that underlie these processes would greatly benefit from the study of chromatin architecture at the biophysical level, has led to in-vitro experiments with individual nucleosomes, reconstituted nucleosomal arrays, or single chromatin fibers [7-9]. Understanding the structural role played by histone modifications, individually or in combination, requires the ability to reconstitute histone octamers and nucleosomal arrays that are uniformly modified. Histone acetyl transferase complexes are difficult to purify and the acetylation reactions that they mediate are not easily driven to completion. These considerations necessitate alternative strategies for the synthesis of substantial quantities of specifically modified histones.
Modified histones have been generated by native ligation or chemical modification. Native ligation requires joining by trans-thioesterification, an N-terminal fragment ending in a C-terminal thioester, with a C-terminal fragment bearing an N-terminal cysteine [10, 11]. Chemical synthesis of modified histones can also be obtained by converting a selected lysine to cysteine followed by alkylation of this residue to produce a methyl lysine analog [12, 13]. An alternate approach involves replacing the codon of the amino acid that needs to be modified with an amber codon that will be read by a tRNA preloaded with the modified amino acid [7, 14].
There are problems that can arise in attempting to implement these different approaches. Native chemical ligation is challenging for molecular biology laboratories that are not equipped to handle the use of hydrofluoric acid to generate the required thioester. Purchasing N-terminal peptides with a C-terminal thioester is not a solution as the synthesis of such peptides appears to be problematic for most commercial companies. Importantly, although amino acid-codon mutagenesis was used to generate co-valently modified histone H3, attempts to use this procedure to express modified H4 in bacterial cells have failed (J. Chin, personal communication). Here we present a protocol that has resulted in the abundant production of H4K16ac by the amber codon suppressor tRNA system, which can be applied to the other co-valently modified isoforms of this histone.
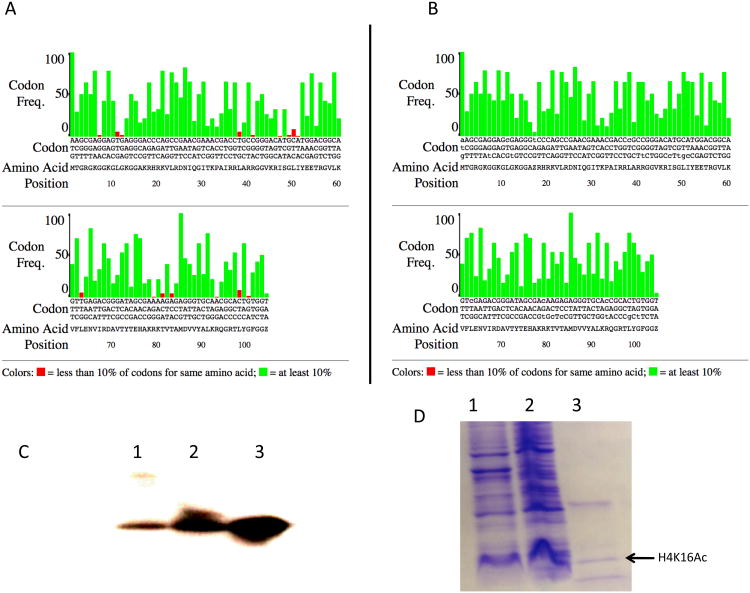
To induce the synthesis of H4K16ac, BL21 competent E. coli cells, grown on LB medium containing kanamycin (50ug/ml) and spectinomycin (50ug/ml), were transformed with a pBK-AckRS-3 plasmid carrying an engineered, orthogonal acetyl-lysyl-tRNA synthetase/tRNACUA, and a pCDF PylT-1 plasmid carrying the open reading frame for histone H4 with an amber codon at the K16 site. A single colony was inoculated in Y2T broth with 0.2% sucrose (v/w) and grown at 37°C in a shaker incubator until 0.6 OD600 was attained. At this point the culture was supplemented with 20mM nicotinamide and 10mM acetyl-lysine and allowed to grow for 30 minutes. Protein expression was induced by the addition of 0.5mM IPTG at 37°C for 2 hours, and purification using Ni-NTA beads was performed following the protocol described in reference [15]. As determined by western blot, the protein yield was extremely low, presumably due to the extensive difference in codon bias between H4 and bacterial proteins (Fig. 1C). An additional possibility was the instability of the mRNA. To address both considerations, we repeated the experiment with the following two modifications. Reducing the codon bias led to a modest increase in the level of expression (Fig. 1B and C) and we were able to purify approximately 0.1 ug of protein per litre of E. coli culture. Switching to an RNaseE mutant BL21 strain (Invitrogen #C6020-03) substantially improved the expressed protein yield (approximately 200 ug per liter of culture) and allowed its visualization in Coomassie-stained gels (Fig. 1D).
Figure 1.
(A) E.coli codon usage analysis for wildtype Drosophila melanogaster histone H4 reading frame. (B) Modified reading frame. Codons are indicated vertically. Drosophila codons with a frequency of less than 10% in E. coli are indicated in red.
(C) Western blot of recombinant histone H4k16Ac. Lane 1: uninduced expression; lane 2: induced expression of the unmodified reading frame; lane 3: induced epression of the reading frame after codon optimization. (D) Coomassie stained gel of H4K16ac expression in RNaseE mutant BL21 cells. Total extract from uninduced cells (lane 1) and from induced cells (lane 2). Lane 3: Ni-NTA bound protein.
We confirmed the identity of the purified protein by mass spectrometry (MS). Histones contain many arginine and lysine residues in their N-terminal tails, resulting in very small tryptic products that are not retained on reversed phase high performance liquid chromatography (RP-HPLC) columns for MS analysis. For this reason we resorted to chemical derivatization of the protein [16]. 3 ug of the purified histone H4 (in 50mM NaH2PO4, 500mM NaCl, 500mM Imidazole, 6M Urea) was desalted on a mini C8 reverse phase liquid chromatography (LC) column packed with Sep-Pak material (Waters). The eluted protein was dried and resuspended in deionized water at a concentration of 0.5 μg/μl. All primary amines were derivatized twice at 51°C with propionic anhydride and the protein was digested in solution with trypsin (20:1 wt/wt) overnight at 37°C. The reaction was stopped with glacial acetic acid (1:15 v/v), frozen at −80°, and dried in a vacuum concentrator. Derivatization with propionic anhydride was repeated twice to cancel the charge on amino termini of the peptides. Aliquots of 1-1.5 μg of peptides were analyzed in two technical replicates on a hybrid LTQ Orbitrap XL mass spectrometer (Thermo) run in positive ion mode with a nano-ESI source (Thermo) for peptide ionization with 2.0 kV electrospray. Each peptide sample was resuspended in LC loading buffer (0.1% formic acid, 0.03% trifluoroacetic acid, 1% acetonitrile) and loaded onto a 15 cm or longer nano-HPLC column (internal diameter 100 Å) packed with Reprosil-Pur 120 C18-AQ 1.9 μm beads (Dr. Maisch Corporation) and eluted over an 80 min 4-80% buffer B reverse phase gradient (Buffer A: 0.1% formic acid, 1% acetonitrile in water; Buffer B: 0.1% formic acid in acetonitrile) generated by a NanoAcquity UPLC system (Waters Corporation). Data dependent acquisition of centroid MS spectra at 60,000 resolution following collision induced dissociation (collision energy 35%, activation Q 0.25, activation time 30 ms) for the top 10 precursor ions with charge determined by the acquisition software to be z≥2. Dynamic exclusion of peaks already sequenced was for 15s with early expiration for 2 count events with signal-to-noise > 2. Automatic gating control was set to 150 ms maximum injection time or 106 counts.
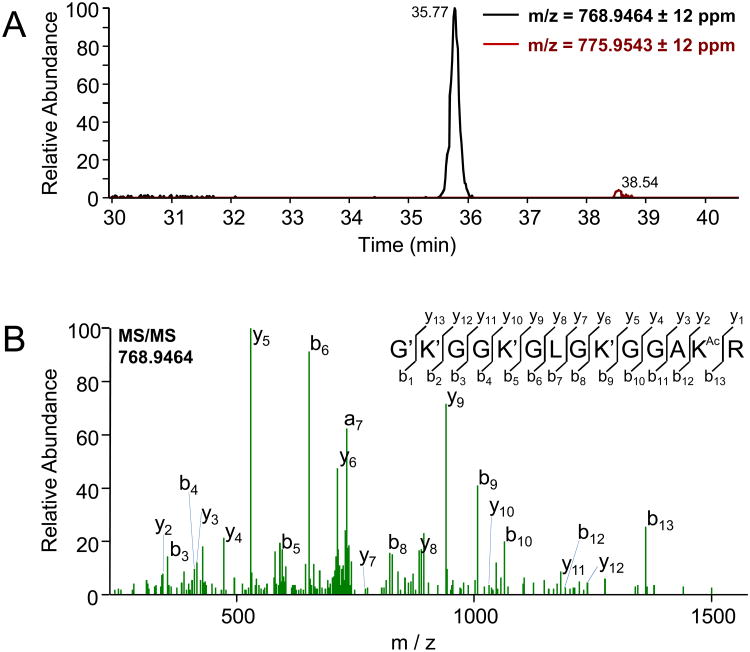
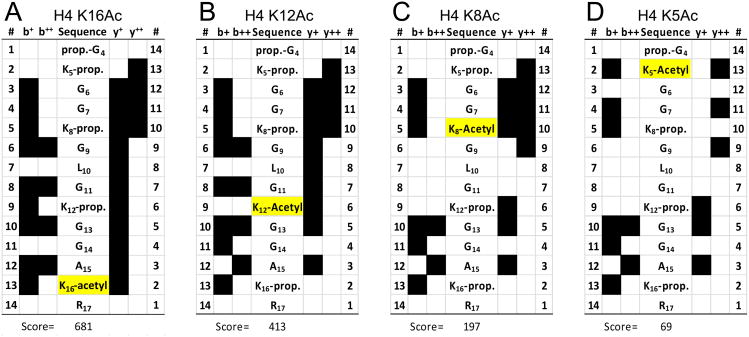
MS/MS events were searched against eukaryotic and E. coli databases using Mascot with 20 ppm mass accuracy. The major eukaryotic protein identified was histone H4, by 16 MS/MS events for at least 13 distinct peptides. E coli proteins identified included Hfq, glutamyl tRNA synthase, and universal stress protein G (supplementary materials). Analysis of raw data on histone H4 was more thoroughly performed manually in the Thermo Xcalibur Qual Browser. The precursor mass for fully propionylated, singly acetylated G4-to-R17 peptide from Drosophila melanogaster histone H4 (768.9464±12 ppm) was located as a single LC elution peak at 35-36 min in each replicate (Fig. 2A); no other isobaric chromatographic peaks were found. The MS/MS for this precursor was then analyzed in the Peptide Sequence Fragmentation Modelling module of Molecular Weight Calculator 6.48 (Matthew Monroe, Pacific Northwest National Laboratory), which identified at least 11 b and y ions from each representative MS/MS spectrum, indicating that the single acetyl modification was on K16 (Fig. 2B,C). PNNL Molecular Weight Calculator match scores were used to confirm specificity of acetylation site assignment by Mascot. A mascot search with fixed propionylation on lysine and N-termini and variable acetylation on lysine identified 3 spectral counts with both +2 and +3 charge states, and a maximum Mascot Ion score of 78 for the conserved G3-R17 peptide with acetylation assigned to K16 in each case. Scoring in the software with acetylation manually assigned to any other position gave a lower number of matched ions and consequently, a lower matching score (Fig. 3). To allow us to conclude that K16 was the major site of acetylation, we also performed a manual search for the unmodified, fully propionylated peptide for G4-to-R17 (775.9543±12 ppm) for each replicate, to compare the peak intensity (<2%) to that of the monoacetylated peptide. Additionally, we searched for an elution peak at 866.0061 m/z ±12 ppm, representing the derivatized, acetylated K91 peptide [KTVTALDVVYALK(Ac)R]. Given the absence of this peak, the near absence of the unmodified G4-to-R17 peptide, and a single, homogeneous chromatographic peak for the acetylated K16 peptide, we were able to conclude that the vast majority of histone H4 recovered was acetylated at K16.
Figure 2.
(A) Liquid chromatography indicates a single uniformly modified chemical species at the expected mass/charge (m/z) ratio, and only 2% of combined signal is seen from the expected m/z peak for the unmodified peptide. (B) MS/MS spectrum of the derivatized H4K16ac protein PropionylGKpropionylGGKpropionylGLGKpropionylGGAKAcetylR. MS/MS Spectrum with b (N-terminal) and y (C-terminal) fragment ions labeled. Best manual fragmentation pattern match assigns acetylation to K16.
Figure 3.
MS/MS ion coverage and specificity. Scoring algorithm and peak matching for the MS/MS event representing the sole chromatographic peak at the expected precursor mass indicate that K16 is the sole acetylation site.
In summary, we have succeeded in using the amber stop codon/suppressor tRNA strategy developed by Jason Chin and colleagues [14] to create and purify histone H4 acetylated at lysine 16 in substantial amounts. Our results can be applied to the purification of this histone bearing any of the plethora of co-valent modifications that native chromatin exhibits by using the modified H4 sequence presented in Fig, 1B and introducing an amber codon at the appropriate position.
Codon usage analysis was done using a free online software available at http://www.biology.ualberta.ca/pilgrim.hp/links/usage2.0c.html The two technical replicate runs and method specification files are in a dropbox available via the following link https://www.dropbox.com/sh/up8n6fo1dlcj6x3/czFsdEWSXD The Peptide Sequence Fragmentation Modelling software is freely available from the PNNL website: http://omics.pnl.gov/software/MWCalculator.php
Supplementary Material
Acknowledgments
This study was supported by grant GM015691 from the National Institutes of Health to JCL We are grateful to Dr. Jason W. Chin for the pBK-AckRS-3 and pCDF PylT-1 plasmids. SK planned and executed the expression of the protein; ED, DD and NS carried out the MS-MS analysis; JCL supervised the research and writing of the manuscript.
Footnotes
The authors declare no conflicts of interest.
References
- 1.Smith ER, Cayrou C, Huang R, Lane WS, et al. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taipale M, Rea S, Richter K, Vilar A, et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25:6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X, Corsa CA, Pan PW, Wu L, et al. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol. 2010;30:5335–5347. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma GG, So S, Gupta A, Kumar R, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30:3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krishnan V, Chow MZ, Wang Z, Zhang L, et al. Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc Natl Acad Sci U S A. 2011;108:12325–12330. doi: 10.1073/pnas.1102789108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Li L, Pandey R, Byun JS, et al. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012;11:163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neumann H, Hancock SM, Buning R, Routh A, et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Molecular cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunlap D, Yokoyama R, Ling H, Sun HY, et al. Distinct contributions of MSL complex subunits to the transcriptional enhancement responsible for dosage compensation in Drosophila. Nucleic Acids Res. 2012;40:11281–11291. doi: 10.1093/nar/gks890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grigoryev SA, Woodcock CL. Chromatin organization - the 30 nm fiber. Experimental cell research. 2012;318:1448–1455. doi: 10.1016/j.yexcr.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 10.Shogren-Knaak MA, Peterson CL. Creating designer histones by native chemical ligation. Methods Enzymol. 2004;375:62–76. doi: 10.1016/s0076-6879(03)75004-6. [DOI] [PubMed] [Google Scholar]
- 11.Allahverdi A, Yang R, Korolev N, Fan Y, et al. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011;39:1680–1691. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon MD. Installation of site-specific methylation into histones using methyl lysine analogs. Curr Protoc Mol Biol. 2010;Chapter 21:21–10. doi: 10.1002/0471142727.mb2118s90. Unit 21 18. [DOI] [PubMed] [Google Scholar]
- 13.Simon MD, Chu F, Racki LR, de la Cruz CC, et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 15.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 16.Garcia BA, Mollah S, Ueberheide BM, Busby SA, et al. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc. 2007;2:933–938. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.