Abstract
Malignant neuroblastomas contain stem-like cells. These tumors also overexpress the Forkhead box transcription factor FoxM1. In this study, we investigated the roles of FoxM1 in the tumorigenicity of neuroblastoma. We showed that depletion of FoxM1 inhibits anchorage-independent growth and tumorigenicity in mouse xenografts. Moreover, knockdown of FoxM1 induces differentiation in neuroblastoma cells, suggesting that FoxM1 plays a role in the maintenance of the undifferentiated progenitor population. We showed that inhibition of FoxM1 in malignant neuroblastoma cells leads to the downregulation of the pluripotency genes sex determining region Y box 2 (Sox2) and Bmi1. We provided evidence that FoxM1 directly activates expression of Sox2 in neuroblastoma cells. By using a conditional deletion system and neurosphere cultures, we showed that FoxM1 is important for expression of Sox2 and Bmi1 in the mouse neural stem/progenitor cells and is critical for its self-renewal. Together, our observations suggested that FoxM1 plays an important role in the tumorigenicity of the aggressive neuroblastoma cells through maintenance of the undifferentiated state.
Introduction
Neuroblastoma, a malignancy derived from neural crest of the sympathetic nervous systems, is the second most common solid tumor in childhood and the most common tumor of infancy with an incidence of 10.2 cases per million children under age of 15 (1, 2). The origin of neuroblastoma is thought to be incompletely committed precursor cells derived from neural crest tissues (2). Neuroblastoma is unique in terms of its clinical bipolarity. Although tumors found in patients younger than 1 year are highly curable and undergo regression with minimal treatments, tumors diagnosed in older patients often grow relentlessly despite intensive and multimodal treatments with only 30% to 40% long-term survival rate (1-3). The unfavorable prognosis has been associated with several factors including MYCN and TrkB gene amplification, chromosome 1p losses, and so on (4-6). However, the molecular pathways mediating tumorigenicity of aggressive neuroblastoma remain largely unclear.
Consistent with its clinical bipolarity, neuroblastomas are heterogeneous in terms of pathologic features, ranging from tumors containing predominantly undifferentiated neuroblast cells to those that are mainly well-differentiated neurons surrounded by Schwann stroma cells (1, 7). This heterogeneous feature is manifested in the cell lines established in vitro. The less malignant S-type cells (substrate adherent and non-neuronal) are usually flattened and attach strongly to the substrate. The N-type cells (neuroblastic), which grow as poorly attached aggregates of small and rounded cells, are tumorigenic and rapidly proliferating. The I-type cells (intermediate), which are less differentiated than the N type, represent malignant, multipotent neural crust stem cells (8-10). The I-type neuroblastoma cells possess self-renewal ability and have significantly higher tumor-forming capacity, as determined by soft agar colony formation and tumor growth in immunodeficient mice (7, 9, 10).
Studies from both patient samples and in vitro cell culture system suggested that neuroblastoma contains pluripotent tumor initiating cells (TIC; refs. 11-14). The existence of TICs may account for both the heterogeneity nature of neuroblastoma as well as the tumor relapse (11, 13, 14). It is also consistent with the observation that the I-type neuroblastoma cells, the most aggressive type of neuroblastoma cells, are malignant neural crest stem cells that possess the ability to self-renewal (10). High frequency of the I-type cells in tumor is associated with increased recurrence (9). A better understanding of the tumorigenicity mechanism of the neuroblastoma possessing stem cell properties will be critical to improve therapeutic outcomes.
Sox2 (sex determining region Y box 2) is a transcription factor that is essential for the maintenance of self-renewal and growth of both embryonic and adult stem cells (15). Recent evidences imply that Sox2 is involved in promoting tumorigenicity in malignant tissues. Sox2 functions as a lineage survival oncogene in lung and esophageal squamous cell carcinoma, where it promotes oncogenic function of tumor cells (16). Consistently, Sox2 silencing in glioma leads to inhibition of proliferation and loss of tumorigenicity (17). Its expression is also detectable in several other types of malignant tumors including neuroblastoma (18-22).
FoxM1, also known as HFH-11B, Trident, WIN, or MPP2, belongs to the Forkhead box (Fox) transcriptional factor family, which consists of more than 50 members sharing homology in the winged-helix DNA binding domains (23, 24). FoxM1 is a proliferation-specific transcriptional factor, given the fact that its expression is strongly correlated with the proliferation capacity of the cells. It is expressed ubiquitously in all proliferating cells, including many tumor-derived cell lines. In normal tissues, FoxM1 is detectable in progenitors with extensive proliferating capacity, whereas its expression is depleted in differentiated or resting cells (24, 25). Numerous transgenic studies in mouse systems showed that FoxM1 is crucial for the development and progression of tumors of different origins, including liver, prostate, colon, breast, lung, brain, and so on (26-30). However, the involvement of FoxM1 in neuroblastoma has not been characterized.
Here, we showed that depletion of FoxM1 inhibits tumorigenicity of neuroblastoma, which is associated with the induction of differentiation. Furthermore, we found FoxM1 is able to directly activate the expression of pluripotency gene Sox2 in neuroblastoma. Also, we showed that deletion of FoxM1 impairs the self-renewal of mouse neural stem/progenitor cells.
Materials and Methods
Cell culture
SK-N-BE(2) cells and BE(2)-C cells were obtained from the American Type Culture Collection. Both SK-N-BE(2) cells and BE(2)-C cells were cultured in MEM/F12 medium supplemented with 15% FBS and penicillin–streptomycin.
Plasmids and siRNA
The pCMV-FoxM1b vector was constructed as previously described (31). The Sox2 expression construct was made by amplifying the Sox2 cDNA fragment sequence from pMSCV-Flag-hSox2 (Addgene; ref. 32) and ligating it into the pcDNA3 construct (Invitrogen). The siRNA oligonucleotide sequence specific for human FoxM1 was 5′ GGACCACUUUCCCUACUUUUU-3′ (33), and for human Sox2 was 5′GGAAUGGACCUUGUAUAGAUU-3′ (34). Oligonucleotides were synthesized by Dharmacon Research. The plasmids and siRNA duplexes were transfected into cells by using Lipofectamine 2000 reagent (Invitrogen) in serum-free tissue culture medium following the manufacturer's protocol.
Neural stem/progenitor cell isolation, culture, and neurosphere frequency assay
Neural stem/progenitor cells were generated from 14.5-day-old embryo cerebral cortical tissue and cultured in serum-free DMEM/F12 medium supplemented with N2 supplement (Invitrogen), 20 ng/mL epidermal growth factor and fibroblast growth factor (Peprotech), 2 mmol/L glutamine (Invitrogen), 6 mg/mL glucose, 14 mmol/L NaHCO3, and 5 mmol/L HEPES (Invitrogen). Neurospheres were dissociated by using chemical dissociation kit following the manufacture's protocol (Stemcell Technologies). Dissociated cells were seeded in culture dish with grid (Nunc) at a clonal density. After 6 to 8 days, the newly generated neurospheres were counted under microscope.
Antibodies and immunoblots
Rabbit polyclonal antibody against FoxM1 was described (31). The following antibodies were also used: Sox2 (Abcam ab15830 and Cell Signaling #3579), Bmi1 (Cell Signaling #2830 and Millipore clone F6 05–637), cleaved caspase-3 (Cell Signaling #9661), β-catenin (BD 610153), neurofilament medium (NF-M; Zymed 13–0900), tubulin-β III (Millipore Mab1637), and Nestin (BD Pharmingen 560393). Horseradish peroxidase-conjugated secondary antibodies were used to amplify the signal from primary antibody (Bio-rad). Protein lysates were prepared in NP-40 lysis buffer consisted of 1% NP-40, 5% glycerol, 20 nmol/L β-glycerophosphate, 2 mmol/L NaF, 5 mmol/L EDTA, 5 mmol/L EGTA, and freshly added protease inhibitor cocktail (Roche).
Cell viability assay
Cells were trypsinized after siRNA transfection, counted, and seeded at a density of 2 × 103 cells per well in triplicate in 48-well plate (Corning). The growth of the cell was monitored by measuring the luminescent signal by using the CellTiter-Glo Kit (Promega) every other day following manufacture's protocol.
Soft agar and in vivo tumorigenicity assay
For soft agar assay, cells were counted and plated in 6-well plates in 0.35% agarose on a 0.7% agarose bed in triplicate. Colonies were stained with crystal violet and counted after 3 weeks. Pictures were taken under dissecting microscope. For in vivo tumorigenicity assay, cells were treated with control or FoxM1 siRNA for 24 hours. A total of 1 × 106 cells were injected subcutaneously into nude mice (Nu/Nu strain; Charles River). Picture of the mice were taken 4 weeks after injection.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were conducted as previously described (33). FoxM1 antibody was used for immunoprecipitation. The following primers were used: Sox2 —15k,a (−14,964 to −14,802) 5′-ACTACTGGTTCCTGATTCCCTCATC-3′ and 5′-GCAAGTCCGCAAAAGTTGTCTC-3′; Sox2 —15k,b (−15,149 to −15,046) 5′-TTCCCAACCCCGTGAGAAAG-3′ and 5′-GCAGAAACTGAGGTGACTGACCAG-3′; and Sox2 —2.5k (−2,668 to −2,517) 5′-CCACCCTTATCCACACCAATTCC-3′ and 5′-TGATTGTCCAGACGCCACAAAG-3′.
Promoter reporters and dual luciferase assay
BE(2)-C cells were plated at 8 × 104 cells per well in a 24-well plate and transfected via Lipofectamine 2000 with different combinations of 100 ng of either CMV-FoxM1B expression construct or empty vector and 0.5 μg of either wild type or mutant −15,178/−14,836 Sox2-firefly luciferase reporter as indicated. In all treatments, 3 ng of CMV-Renilla luciferase was cotransfected as an internal control. Cells were harvested 24 hours after transfection, and protein extracts were subjected to dual luciferase assays (Promega) with firefly luciferase activity normalized to Renilla luciferase activity. Promoter activity was expressed as fold induction of transcription by the FoxM1b expression vector, where the promoter activity resulting from transfection with empty vector was set at 1.
Real-time—PCR
Quantitative (q) real-time (RT)–PCR was carried out as described previously (33). The following primers were used: human FoxM1 5′-GGAGGAAATGCCACACTTAGCG-3′ and 5′-TAGGACTTCTTGGGTCTTGGGGTG-3′; human Sox2 5′-TGAATGCCTTCATGGTGTGGTC-3′ and 5′-CCGTCTCCGACAAAAGTTTCC-3′; human Bmi1 5′-TGATGTGTGTGCTTTGTGGAGG-3′ and 5′-GTGGTCTGGTCTTGTGAACTTGG-3′; human cyclophilin 5′-GCAGACAAGGTCCCAAAGACAG-3′ and 5′-CACCCTGACACATAAACCCTGG-3′; mouse Foxm1, 5′AGCGTTAAGCAGGAACTGGA-3′ and 5′-GGAAGTGGTCCTCAATCCAA-3′; mouse Sox2 5′-AACGGCTCGCCCACCTACAGC-3′ and 5′-CAGGGGCAGTGTGCCGTATTTGG-3′; mouse Bmi1 5′-AGAGGGATGGACTACGAATGC-3′ and 5′-AACAGGAAGAGGTGGAGGGAAC-3′; and mouse cyclophilin 5′-GGCAAATGC-TGGACCAAACAC-3′ and 5′-TTCCTGGACCCAAAACGCTC-3′. For semiquantitative RT-PCR experiments, the linear ranges for amplicon of each PCR primers were determined to allow semiquantitative comparisons. The primers used in semiquantitative RT-PCR were human FoxM1 5′-GGAGGAAATGCCACACTTAGCG-3′ and 5′-TAGGACTTCTTGGGTCTTGGGGTG-3′; human Sox2 5′-TGAATGCCTTCATGGTGTGGTC-3′ and 5′-CCGTCTCCGACAAAAGTTTCC-3′; human Oct4 5′-GGGGTT-CTATTTGGGAAGGTATTC-3′ and 5′-GGTTCGCTTTCTCT-TTCGGG-3′; human Nanog 5′-CCAG-TCCCAAAGGCAAACAAC-3′ and 5′-TGGAGGCTGAGGTATT-TCTGTCTC-3′; human Bmi1 5′-TGATGTGTGTGCTTTGTGGAGG-3′ and 5′-GTGGTCTGGTCTTGTGAACTTGG-3′; human Ezh2 5′AGTTGGTGAATGCCCTTGGTC-3′ and 5′-TGCTGTGCCCTTAT-CTGGAAAC-3′; human Suz12 5′- GCCAACCTGGATTT-GCTTTTAGTC′ and ′TCTTTGCTGT-TCTACTTCCCCATC-3′; and human cyclophilin 5′-GCAGACAAGGTCCCAAAGACAG-3′ and 5′-CACCCTGACACATAAACCCTGG-3′.
Statistical analysis
Statistical significance was calculated by the Student's t test (2 tailed). Statistically significant changes were indicated with asterisks (*, P < 0.05; **, P < 0.001).
Results
FoxM1 is critical for the tumorigenicity of neuroblastoma cells
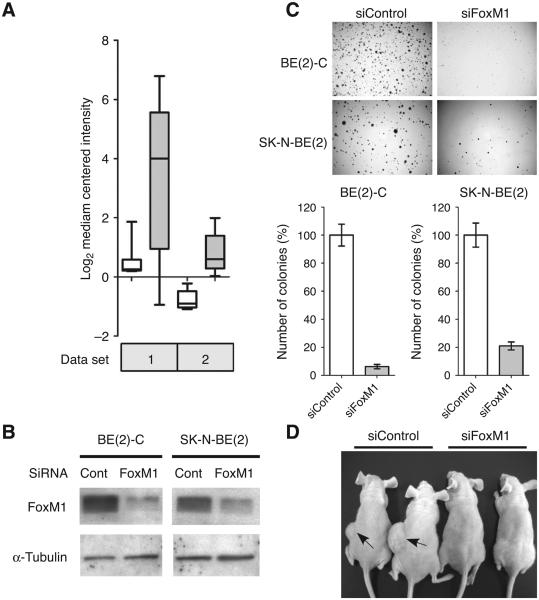
Expression studies with patient samples by several groups had revealed that FoxM1 mRNA is significantly upregulated in neuroblastoma tissue samples compared with noncancerous ganglioneuroma or less aggressive ganglioneuroblastoma (35-37). A box plot representing 2 different data sets from publicly available database is shown (Fig. 1A; refs .35, 36). However, the biological function of FoxM1 in neuroblastoma had not been elucidated. To evaluate the role of FoxM1 in neuroblastoma, we first investigated the effects of FoxM1 on anchorage-independent growth, which is a hallmark of tumorigenicity. We reduced FoxM1 expression by siRNA in 2 different types of aggressive neuroblastoma cell lines: SK-N-BE(2) and BE(2)-C. SK-N-BE(2) belongs to N-type neuroblastoma cells, whereas BE(2)-C belongs to the most malignant I-type neuroblastoma cells (8-10). After 72 hours siRNA transfection, the protein level of FoxM1 was reduced significantly in these 2 cell lines, as evidenced by the Western blot (Fig. 1B). The anchorage-independent growth capacity of neuroblastoma cells was checked by carrying out soft agar colony formation assay. Ablation of FoxM1 by siRNA led to a profound decrease in the number of colonies formed in both cell lines (Fig. 1C). The SKN-BE(2) cells formed about 80% less colonies in FoxM1 siRNA treated cells compared with control siRNA-treated cells. For the BE(2)-C line, the reduction was even more severe. FoxM1 knockdown led to about 90% reduction in the number of colonies by quantification (Fig. 1C).
Figure 1.
FoxM1 is critical for the tumorigenicity of neuroblastoma. A, normalized FoxM1 mRNA level in 2 publically available mRNA expression profile data sets. White box, benign ganglioneuroma or ganglioneuroblastoma. Grey box, neuroblastoma. P values for the 2 data sets are 2.06E-10 and 3.82E-6. B, immunoblot showing depletion of FoxM1 by siRNA in BE(2)-C and SK-N-BE(2)-C cells. Cell lysates were collected at 72 hours after transfection. C, representative pictures and quantification of anchorage-independent growth on soft agar plates. Twenty-four hours after FoxM1 or control siRNA silencing, cells were plated at a density of 8 × 103 cells per well in a 6-well plate. Colonies were stained and counted after 3 weeks. D, picture of nude mice after 4 weeks subcutaneous injection of BE(2)-C cells treated with control or FoxM1 siRNA.
Similar effect was observed in vivo when BE(2)-C cells (1 × 106 cells) were injected subcutaneously in nude mice. The control group, expressing control siRNA, formed tumors within 2 weeks of injection, whereas the FoxM1 siRNA expressing cells failed to form tumors in 4 weeks (Fig. 1D). The strong inhibition of tumor growth showed that FoxM1 is critical for the tumorigenicity of the neuroblastoma cells. The loss of tumorigenicity could not be explained sufficiently by the inhibition of cell growth. A growth curve analysis following depletion of FoxM1 indicated only a partial retardation of growth at the initial time points (Supplementary Fig. S1A). At later time points, cell counts on FoxM1 silencing increased, most likely because of reexpression of FoxM1. Moreover, we did not see any significant increase in apoptosis based on changes in the sub-G1 population and caspase-3 activation following depletion of FoxM1 in the BE(2)-C cells (Supplementary Fig. S1B and C). The differential effect of transient FoxM1 knockdown on growth curve versus anchorage-independent growth or growth in xenografts suggests that a continuous presence of FoxM1 is critical for the tumorigenicity of the neuroblastoma cells.
Transient loss of FoxM1 leads to spontaneous differentiation of the neuroblastoma cells
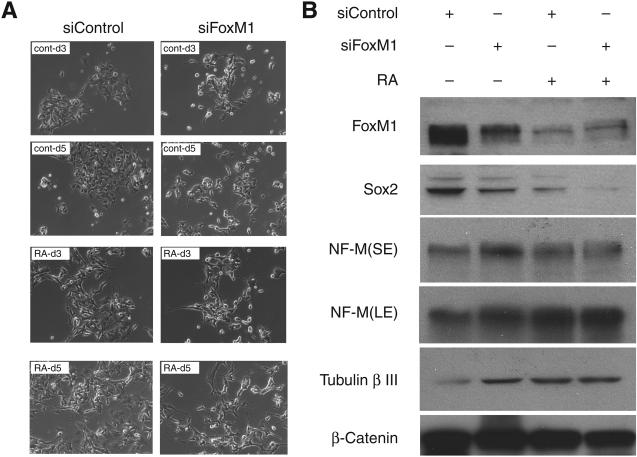
Several recent studies, in other tumor models, indicated a link between the state of differentiation of the tumor cells and their tumorigenicity. For example, in human liver cancer, it was shown that the tumorigenicity correlated with the presence of stem cell-like cancer cells (38). Moreover, there is a strong association between poor differentiation and aggressiveness of breast cancers (39). Because the neuroblastoma cells used in our tumorigenicity studies contain stem-like progenitor cells, we considered the possibility that the depletion of FoxM1 inhibits tumorigenicity by inducing differentiation. We investigated the effects of FoxM1 depletion on differentiation of the BE(2)-C cells, which belong to I-type neuroblastoma cells representing the neural crust stem cell. Retinoic acid is able to induce differentiation of these cells toward neuronal lineage (10). We observed that the following 5 days after retinoic acid treatment, BE(2)-C cells started to exhibit morphology of differentiated neurons with neurite extension (Fig. 2A). Interestingly, the level of FoxM1 was significantly decreased in the differentiated cells (Fig. 2B). Moreover, we observed that FoxM1 knockdown alone was able to induce a significant increase of the neuronal differentiation phenotype in BE(2)-C cells (Fig. 2A). Furthermore, depletion of FoxM1 alone resulted in a significant increase in the levels of neuronal differentiation markers, NF-M and β-tubulin III (Fig. 2B). Interestingly, the pluripotency gene Sox2 was downregulated by both retinoic acid and FoxM1 siRNA (Fig. 2B). Together these results clearly indicate that FoxM1 is important for maintaining the undifferentiated state of the I-type neuroblastoma cells.
Figure 2.
BE(2)-C cells with reduced FoxM1 undergo differentiation. A, BE(2)-C cells were transfected with control siRNA or FoxM1 siRNA. Retinoic acid (RA) was added to induce differentiation 72 hours after transfection. Representative pictures of these cells were taken 3 days (d3) and 5 days (d5) after retinoic acid treatment under microscope. Cells without retinoic acid treatment were also pictured at the same time point (CONT). B, immunoblot of cell lysates collected 5 days after retinoic acid treatment. FoxM1, Sox2, NF-M, and tubulin β-III were detected by Western blot. β-catenin was used as loading control. SE, short exposure; LE, long exposure.
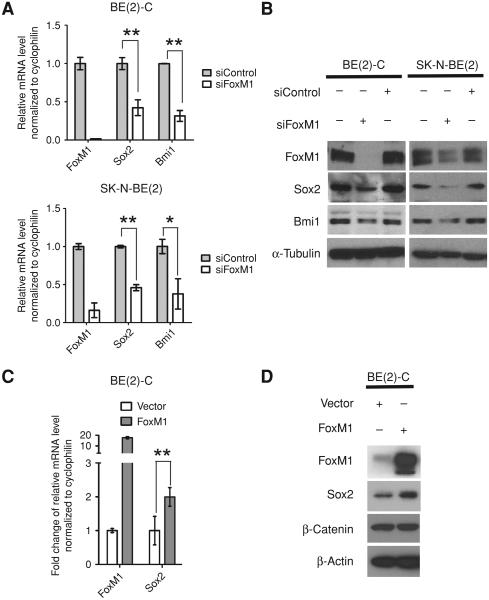
FoxM1 directly activates expression of the pluripotency gene Sox2 that is required also for the tumorigenicity of neuroblastoma cells
The pluripotency gene Sox2 had been implicated in the maintenance of neural stem cell pool (15) and it had been found to be involved in mediating tumorigenicity of several types of human malignancies by impacting the anchorage-independent growth (16, 17). Therefore, we tested the hypothesis that FoxM1 is critical for the expression of Sox2 in the neuroblastoma cells. One study reported that overexpression of FoxM1 in P19 teratocarcinoma cells increases expression of Sox2 (40). However, it is unclear whether the regulation is direct. To elucidate the connection between FoxM1 and Sox2 in neuroblastoma, we checked the expression level of Sox2 after FoxM1 silencing. The mRNA level of Sox2 was remarkably reduced in both SKN-BE(2) and BE(2)-C cells following FoxM1-silencing (Fig. 3A). Furthermore, the protein level of Sox2 was decreased in FoxM1 siRNA treated samples compared with control siRNA treated or nontreated samples, evidenced by immunoblot (Fig. 3B). These results indicated that FoxM1 might regulate Sox2 at the transcriptional level. We assayed for the expression of polycomb family member Bmi1, which has been implicated in promoting tumorigenicity of neuroblastoma cells (41, 42). Interestingly, it was observed that, similar to Sox2, Bmi1 expression level also tightly correlated with FoxM1 in neuroblastoma, which indicated that it might also be involved in FoxM1-mediated tumorigenicity of neuroblastoma cells (Fig. 3A and B).
Figure 3.
Sox2 expression correlates with FoxM1 in neuroblastoma cells. A, mRNA levels of FoxM1, Sox2, and Bmi1 detected by qRT-PCR. BE(2)-C and SK-N-BE(2)-C cells were transfected with control siRNA or FoxM1 siRNA for 72 hours. The mRNA levels were normalized to human cyclophilin mRNA, and the control groups were set to 1. B, immunoblot showing the protein level of FoxM1, Sox2, and Bmi1 from the same samples described in A. α-Tubulin was used as loading control. C, mRNA levels of FoxM1 and Sox2 detected by qRT-PCR 24 hours after transfecting BE(2)-C cells with empty vector or pCMV-FoxM1 plasmid. The mRNA levels were normalized to human cyclophilin mRNA, and the control groups were set to 1-fold. D, immunoblot showing the expression of FoxM1 and Sox2 from the samples described in C. β-Catenin and β-actin were used as loading controls.
Bmi-1 had been previously reported to function downstream of FoxM1 through activation of c-Myc (43). Therefore, we focused on revealing the relationship between FoxM1 and Sox2. To test whether FoxM1 is able to stimulate Sox2 expression in neuroblastoma cells, we transiently transfected FoxM1 expression plasmid into BE(2)-C cells. Expression of FoxM1 led to an increase in the Sox2 mRNA level (Fig. 3C). In addition, by immunoblot, we observed that the Sox2 protein also responded to FoxM1 upregulation compared with the control transfection (Fig. 3D). Similar result was observed by stably overexpressing FoxM1 in S-type neuroblastoma SK-NAS cells (Supplementary Fig. S2) and the ectopic expression of FoxM1 led to increased colony formation on plate and anchorage-independent growth in soft agar (Supplementary Fig. S2A and B).
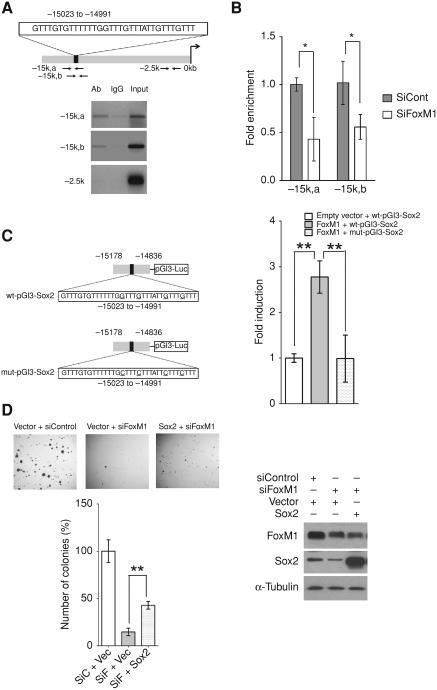
Next, we tested the possibility that FoxM1 directly stimulates Sox2 expression by binding to its promoter region. Analysis of human Sox2 upstream promoter region by using MacVector software revealed 1 putative FoxM1 binding motif 15 kb upstream of the transcriptional start site (−15,023 to −14,991; Fig. 4A). To determine whether FoxM1 binds to this site, we utilized quantitative ChIP. We first tested whether FoxM1 binds to Sox2 promoter region at endogenous level. BE(2)-C cells were cross-linked and sonicated. The chromatins were immunoprecipitated with either FoxM1 specific antibody (Ab) or rabbit serum (control). The amount of endogenous Sox2 DNA bound by FoxM1 was determined by PCR by using 2 different sets of primers flanking the DNA sequences near the potential binding sites (−14,963 to −14,801 and −15,148 to −15,040). Compared with serum control, FoxM1 antibody was able to enrich the DNA fragments upstream 15 kb region of the Sox2 gene with both sets of primers and there was no enrichment by using primers flanking nonspecific region around −2.5 kb upstream (Fig. 4A). To confirm the specificity of the endogenous binding of FoxM1 to the Sox2 promoter, we investigated whether knockdown of FoxM1 would disrupt the interaction. Chromatin samples were collected from both FoxM1 siRNA treated BE(2)-C cells and control siRNA treated BE(2)-C cells. The amount of Sox2 promoter DNA enriched by FoxM1 antibody in both samples was quantified by RT-PCR with 2 different sets of primers specific to upstream 15 kb region of the Sox2 gene. FoxM1 knockdown led to a half-fold reduction in the immunoprecipitation of the −15 kb region amplicons (Fig. 4B) which indicated the binding is FoxM1 specific. These results showed that, in the BE(2)-C cells, endogenous FoxM1 binds to the Sox2 upstream region (Fig. 4A) and activates expression of Sox2 (Fig. 3A).
Figure 4.
FoxM1 activates Sox2 by binding to its upstream promoter region. A, schematic diagram of human Sox2 upstream promoter region. Predicted FoxM1 binding region was shown in the box(−15,023 to −14,991). Arrows indicated positions for designed ChIP primers −15k,a (−14,963 to −14,801), −15k,b (−15,148 to −15,040), and −2.5k (−2,668 to −2,517). Endogenous binding of FoxM1 to Sox2 upstream region as determined by ChIP assay. Crosslinked and sonicated chromatin fragments from BE(2)-C cells were precipitated with either FoxM1 antibody or rabbit IgG. Semiquantitative PCR was carried out to determine the amount of DNA that was precipitated by FoxM1 antibody or IgG control by using indicated primers targeting predicted binding region (−15k,a and −15k,b) or nonbinding region (−2.5k). B, BE(2)-C cells were transfected with control or FoxM1 siRNA for 72 hours. Chromatins were crosslinked, sonicated, and precipitated with FoxM1 antibody. The amount of Sox2 upstream region precipitated by FoxM1 antibody was determined by qRT-PCR by using primers targeting predicted binding region (−15k,a and −15k,b). C, left, schematic diagram of wild type and mutated human Sox2 promoter luciferase constructs. Four canonical FoxM1 binding motif GTTTs were mutated into CTTTs. Right, fold induction of human Sox2 promoter luciferase activity by overexpression of FoxM1 is shown. The amount of wide-type Sox2 promoter luciferase activity by empty vector stimulation was set at 1-fold. D, representative pictures and quantification of anchorage-independent growth on soft-agar plates. BE(2)-C cells were cotransfected with different combination of expression vector and siRNA as indicated. Twenty-four hours after transfection, cells were plated at a density of 2 × 103 cells per well in a 6-well plate. Colonies were stained and counted after 3 weeks.
To test the transcriptional activity of FoxM1 on human Sox2 promoter, we cloned the human Sox2 promoter sequence (−15,178 to −14,836) encompassing the predicted binding motif into pGl3 construct (wt-pGl3-Sox2) as well as the mutated human Sox2 promoter sequence where 4 canonical FoxM1 binding motif GTTTs were mutated into CTTTs (mut-pGl3-Sox2; Fig. 4C). We carried out dual luciferase assay by cotransfecting BE(2)-C cells with FoxM1b expression construct and either wild-type Sox2 reporter plasmid or mutated reporter plasmid. Overexpression of FoxM1b resulted in a 3-fold increase in wild-type Sox2 promoter activation relative to empty vector transfection, whereas it failed to stimulate the mutated Sox2 promoter luciferase (Fig. 4C). Similar results were obtained in U2OS cells (data not shown).
In addition, we found that expression of Sox2 in BE(2)-C cells treated with FoxM1-siRNA caused a partial, but significant, reversal of the anchorage-independent growth (Fig. 4D). An incomplete reversal is consistent with the possibility that FoxM1 activates expression of other genes required for the anchorage-independent growth. It is likely that Bmi1, the expression of which decreased following FoxM1 silencing, is involved (Fig. 3A and B). Also, we assayed for other pluripotency genes in BE(2)-C cells for their dependence on FoxM1. Depletion of FoxM1 resulted in a significant loss of Oct4, Ezh2, and Suz12 expression (Supplementary Fig S3A). To further investigate the role of Sox2 in the anchorage-independent growth of the BE(2)-C cells, we employed Sox2 siRNA, which caused a significant loss of the anchorage-independent growth in soft agar colony formation assay compared with control siRNA treatment, indicating a critical role of Sox2 in neuroblastoma cells (Supplementary Fig. S4A–C). More interestingly, silencing of Sox2 largely compromised the increased number of soft agar colonies caused by overexpression of FoxM1 in the BE(2)-C cells (Supplementary Fig. S4C and D), suggesting that Sox2 is one of the key downstream mediators of FoxM1 in inducing anchorage-independent growth of the neuroblastoma cells.
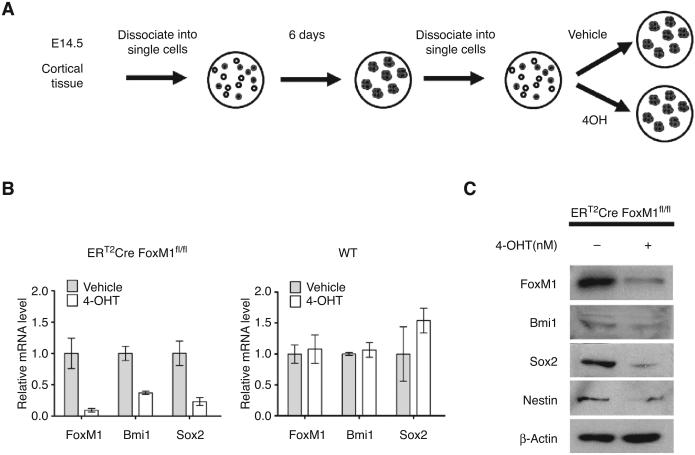
FoxM1 deletion results in impaired self-renewal of the E14.5 neural stem/progenitor cells
Both Sox2 and Bmi1 are critical for self-renewal of the neural cortical stem/progenitor cells (15, 44). If FoxM1 is important for expression of these genes in those cell type, we hypothesized that deletion of FoxM1 will inhibit self-renewal of the neural cortical stem/progenitor cells. To test this possibility, we took advantage of the well-established protocol to culture neural cortical stem/progenitor cell in vitro (45; also see schematic in Fig. 5A). The embryonic cortical tissue from day 14.5 ERT2-Cre FoxM1fl/fl embryo was dissected and digested to generate neurospheres in serum-free medium. The ERT2-Cre FoxM1fl/fl strain was generated by crossing ERT2-Cre strain with FoxM1 fl/fl strain. ERT2-Cre allele allows the activation of Cre-recombinase upon tamoxifen administration, which in turn excises the FoxM1 alleles from the genome. To check whether FoxM1 regulates Sox2 and Bmi1 in neural stem/progenitor cells, same number of dissociated neural stem/progenitor cells was kept in serum-free neural stem cell medium containing either 4-OH tamoxifen or ethanol as vehicle control. After 4 days, FoxM1 mRNA was remarkably decreased along with decreases in Sox2 and Bmi1 expression, as evidenced by RT-PCR and immunoblot (Fig. 5B and C). The level of Nestin, another neural stem cell marker, also went down (Fig. 5C). To exclude the possible side effect from tamoxifen, in parallel, wild-type neurospheres were generated and cultured in the same setting. Tamoxifen treatment did not lead to any significant reduction of Sox2 and Bmi1, which indicated the effect of Sox2 and Bmi1 reduction is because of FoxM1 ablation (Fig. 5B).
Figure 5.
Decreased Sox2 expression in neural stem/progenitor cells following FoxM1 depletion. A, schematic diagram describing the experimental procedure. B, ERT2-Cre FoxM1fl/fl neural stem/progenitor cells were treated with vehicle or 50 nmol/L 4OH-tamoxifen treatment for 4 days after being dissociated into single cells. mRNA levels of FoxM1, Sox2, and Bmi1 were detected by qRT-PCR. Wild-type neural stem/progenitor cells with same treatment were used as control. The mRNA levels were normalized to mouse cyclophilin mRNA, and the vehicle control groups were set to 1-fold. C, immunoblot showing the expression of FoxM1, Bmi1, Sox2, and Nestin of ERT2-Cre FoxM1fl/fl neural stem/progenitor cells treated with vehicle or 50 nmol/L 4OH-tamoxifen. β-actin was used as loading control.
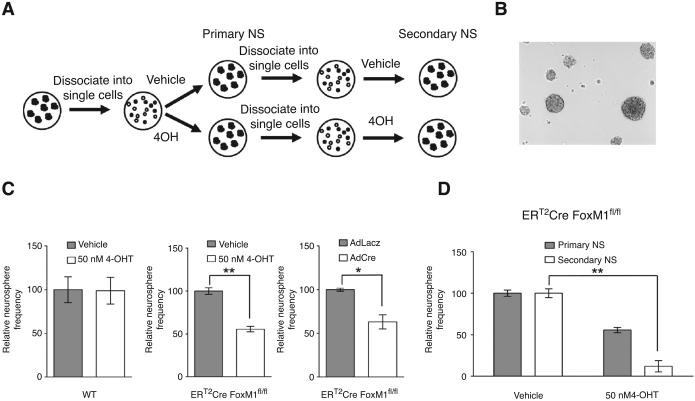
In addition, we carried out the neurosphere assay, which measures the capacity of a primary neurosphere to form new multipotent neurosphere after dissociation, as an indicatior of self-renewal capacity (see schematic in Fig. 6A). The ERT2-Cre FoxM1fl/fl neurospheres were dissociated into single cells. Same number of cells was kept in serum-free neural stem cell medium containing either 4-OH tamoxifen or ethanol as vehicle control. After 6 days, number of neurosphere formed was counted. A representative picture of the neurospheres counted was shown (Fig. 6B). We found that FoxM1 deletion by tamoxifen treatment led to a significant decrease in the frequency of newly-formed neurosphere compared with vehicle control (Fig. 6C), indicating that loss of FoxM1 impairs the self-renewal of previous neural stem/progenitor cell population. The same procedure was repeated with the newly formed primary neurospheres to obtain secondary neurospheres in either 4-OHT tamoxifen–treated or control-treated samples (Fig. 6A). After 6 days, we found the difference in neurosphere frequency was even more dramatic (Fig. 6D). We observed no obvious difference in primary neurosphere frequency in the wild-type neurospheres. Similar effects on primary neurosphere frequency were observed when the ERT2Cre FoxM1fl/fl neurospheres were treated with adenovirus expressing lacZ gene or Cre recombinase. The primary frequency of neurosphere was decreased by adeno-Cre treatment (Fig. 6C). Together, these data suggested that FoxM1 is necessary to maintain the self-renewal of cortical neural stem/progenitor cells.
Figure 6.
Loss of FoxM1 impairs the self-renewal of neural stem/progenitor cells. A, schematic diagram describing the experimental procedure of neurosphere assay. B, representative picture of neurospheres. C, quantification of primary neurosphere frequency. Neural stem/progenitor cells were dissociated into single cells and same amount of cells was plated in triplicate for each treatment. Wild-type neural stem/progenitor cells were treated with vehicle or 50 nmol/L 4OH-tamoxifen as controls. ERT2-Cre FoxM1fl/fl neural stem/progenitor cells were treated with vehicle or 50 nmol/L 4OH-tamoxifen in 1 set and AdlacZ or AdCre virus in another set. Six days later, the number of neurospheres was counted under microscope. The frequency of neurosphere was calculated by dividing the number of neurospheres formed by the number of initial single neural stem/progenitor cells plated. The neurosphere frequency was normalized to the vehicle control which was set to 100%. D, ERT2-Cre FoxM1fl/fl neurosphere formed from the set treated with vehicle or 50 nmol/L 4OH-tamoxifen described in C were dissociated again and the same amount of cells was plated and treated with another round of vehicle or 50 nmol/L 4OH-tamoxifen. Six days later, the secondary neurosphere frequency was obtained in the same way.
Discussion
Studies described here are significant as they provide new molecular insights into the aggressive nature of neuroblastoma. Although FoxM1 is overexpressed in the aggressive forms of neuroblastoma, its involvement in neuroblastoma has not been investigated. We showed a dual role of FoxM1 in positively regulating tumorigenicity and in maintenance of the progenitor population in neuroblastoma. First, we showed that FoxM1 serves as a critical activator of tumorigenic properties of the aggressive forms of the neuroblastoma cells. In addition, we discovered a direct connection between FoxM1 and pluripotency-associated gene Sox2 in mediating the anchorage-independent growth of the neuroblastoma cells. Finally, we observed that neuroblastoma cells with reduced FoxM1 expression undergo spontaneous differentiation with diminished levels of Sox2. Furthermore, in mouse cortical neural stem/progenitor cells, loss of FoxM1 largely impaired the self-renewal ability.
The aggressive forms of neuroblastoma still remain a challenge in the clinic, largely because of limited knowledge of biological and prognostic characteristic of this childhood disease. Our results revealed that FoxM1 is crucial for the tumorigenicity of aggressive neuroblastoma cells, which is often related to the metastatic potential of the tumors. These observations make FoxM1 an attractive therapeutic target for treating neuroblastoma patients. That is particularly significant because several groups are actively involved in characterizing inhibitors of FoxM1 that also inhibits tumor progression (46, 47).
Mounting evidences suggested that the existence of TIC within neuroblastoma might be responsible for its clinical relapse (11, 13, 14). The striking clinical bipolarity of its pathologic feature also indicated that neuroblastoma may be a disease of stem cells (2,3). Expression of the pluripotency genes, which are critical for normal stem cell maintenance, have been detected in neuroblastoma cells (11, 12, 14, 22). However, very little is known about the molecular basis of how the pluripotency genes get activated. Here, we discovered that one of the core pluripotent genes Sox2 is directly activated by FoxM1. In that regard, it is noteworthy that several pluripotency genes have been shown to be activated by FoxM1 (40, 43). Our observation suggests that the pluripotency gene Sox2 is critical for the tumorigenic activity of FoxM1. Thus, FoxM1 might be involved in altering the cellular characteristic favoring oncogenic growth of tumor cells by potentially upregulating pluripotency-associated genes.
Sox2 has been found to be involved in tumorigenicity of several types of tumors and, it has been regarded as a cancer stem cell marker. For example, loss of Sox2 was reported to impair the anchorage-independent growth of glioma, Ewing's sarcoma, and lung tumors (16-18, 20). In this study, we discovered similar phenotype in Sox2 silenced neuroblastoma cells. In addition, we found Sox2 functions downstream of FoxM1 because silencing of Sox2 abolished the induction of soft agar colony formation observed with neuroblastoma cells overexpressing FoxM1. Although the downstream effectors in the pathways that impact anchorage-independent growth or tumorigenicity are unclear, we think that they are related to the state of differentiation as well.
Two gene expression studies in tumors showed a link between expression of embryonic stem (ES) cell pluripotency signatures genes and aggressiveness of the tumors, including increased tumorigenicity (39, 48). The study with breast cancer gene expression identified 9 transcription factor genes (Core 9 genes) among the ES signature transcriptional factors that were mostly overexpressed in high-grade breast cancers (39). The study then ranked all putative transcription factor genes whose expression correlated with the Core 9 genes. Interestingly, FoxM1 ranked number 2 in that list (39). Also, we compiled a list of 538 genes that were shown to be activated by FoxM1 from the literature and compared them with the 338 genes identified by Wong and colleagues as core embryonic stem cell–like module expressed in epithelial cancer stem cells (48). In that comparison, we observed about 10% of the embryonic stem cell–like module (33 genes) overlapped with the FoxM1 target genes (Supplementary Fig. S3B; P = 9.5 × 10−5). Also, there was a significant overlap between the FoxM1 and Sox2 target genes (49; Supplementary Fig. S3B; P = 2.0 × 10−15). These observations further support the notion that FoxM1 plays a role in the expression of the pluripotency genes in cancer stem cells.
The I-type neuroblastoma retains the ability to differentiate in vitro upon addition of different stimuli (10, 42). FoxM1 loss alone is sufficient to induce spontaneous differentiation of BE (2)-C cells, indicating that the expression of FoxM1 is important for maintaining the cells in a relatively undifferentiated state. We also observed that upon retinoic acid-induced differentiation, the expression of FoxM1 is gradually diminished (data not shown; Fig. 2) which further supports our hypothesis. This is consistent with our finding that FoxM1 is necessary for the expression of pluripotency genes, such as Sox2 and Bmi1, which are important for maintenance of an undifferentiated state of the I-type neuroblastoma cells. These observations were also validated by using neural cortical precursors. The possibility that the neural cortical progenitor cells depend upon FoxM1 is supported by our observation on the neurosphere frequency assay. We observed that the ability of the progenitor cells to undergo self-renewal was compromised significantly in cells depleted of FoxM1, which also correlated with loss of Sox2 and Bmi1 expression. The studies in Figures 5 and 6, on neural stem/progenitor cell pool, are incomplete, but they support the studies with the neuroblastoma cells. The observations suggest that FoxM1 might be involved in expression of the Sox2 and Bmi1 genes in multiple progenitor types and potentially plays a role in the aggressiveness of multiple tumor types in which FoxM1 is overexpressed.
Supplementary Material
Supplementary figure 1
FoxM1 moderately affects the growth of neuroblastoma cells. A, Twenty-four hours after FoxM1 or control siRNA transfection, cells were trypsinized and seeded at a density of 2×103 cells per 48-well plate for proliferation assay. Viable cell number was measured by using CellTiter-Glo luminescent cell viability assay kit (Promega) every two days. The fold change in cell count was presented by luminescence unit and was normalized by the value at day zero. B, Seventy-two hours after FoxM1 or control siRNA transfection, cells were fixed and stained with propidium iodide (PI) and analyzed by flow cytometry for sub-G1 population. C, Seventtwo hours after FoxM1 or control siRNA transfection, cell lysates were collected and assayed for cleaved caspase-3 by immunoblot. β-catenin was used as loading control.
Supplementary figure 2
FoxM1 promotes tumorigenicity in SK-N-AS cells. A, Colony formation is induced in cells stably expressing FoxM1. 2×103 SK-N-AS cells stabling expressing empty vector or FoxM1 were plated per well in a six-well plate in triplicate. Colonies were stained and quantified after 2 weeks. B, Representative pictures and quantification of anchorage-independent growth of SK-N-AS cells stabling expressing empty vector or FoxM1. Cells were plated a density of 1.6×104 cells per well in a six-well plate. Colonies were stained and counted after three weeks. C, Immunoblot showing expression of FoxM1 and Sox2 of SK-N-AS cells stabling expressing empty vector for FoxM1. α−tubulin was used as the loading control.
Supplementary figure 3
FoxM1 and ESC-like gene signature. A, Seventy-two hours after FoxM1 and control siRNA treatment, semiquantitative RT-PCR was performed to analyze the mRNA level of the pluripotency genes including Sox2, Oct4, Nanog, Bmi1, Ezh2, Suz12. Cyclophilin was used as a loading control. The intensity of the bands was determined by ImageJ software and the intensity of control samples were set at one. B, Venn diagram of overlapping genes in three groups: FoxM1 target genes, Sox2 target genes and core ESC-like module. FoxM1 target genes were summarized from 201 published FoxM1 related literature where FoxM1 regulated genes were reported and from differentially expressed genes from one publicly available FoxM1 array of breast carcinoma (NCBI GDS1477) (50). Sox2 target genes were reported from chip array study (49, 39, table S1)) and core ESC-like module was previously purposed (49(table S6)). P value for both FoxM1 with Sox2 targets overlap (F/S) and FoxM1 with core module overlap (F/E) were obtained using hypergeometric calculation. C, List of 33 overlapping genes of FoxM1 targets and core ESC-like module. Among them, genes that were also Sox2 targets were highlighted in red.
Supplementary figure 4
FoxM1 mediated anchorage-independent growth requires expression of Sox2. A Immunoblot showing depletion of Sox2 by siRNA in BE(2)-C cells. Cell lysates were collected at four days after transfection. B, 24 hours after Sox2 or control siRNA transfection, cells were trypsinized and seeded at a density of 2×103 cells per well in a 48-well plate for proliferation assay. Viable cell number was measured by using CellTiter-Glo luminescent cell viability assay kit (Promega) every two days. The fold change in cell count was presented by luminescence unit and was normalized by day zero. C, BE(2)-C cells were first transfected with empty or pCMV-FoxM1 plasmid for 24 hours, and then transfected with either control siRNA or Sox2 siRNA for another 24 hours. Cells were plated at a density of 8×103 cells per well in a six-well plate in soft agar plate. Colonies were stained and quantified after three weeks. D, Quantification of colonies numbers derived from C. Colony large than 100μm in diameter was defined as large colony and colony which has diameter between 25-100μm was defined as small colony.
Acknowledgments
We thank Dr. Xao X. Tang for sharing reagents and Michael Demars and Joel Bernard Fontanarosa for technical support. We dedicate this work to the memory of Dr. Robert H. Costa.
Grant Support
This work was supported by U.S. Public Health Service (PHS) grants CA 124488 and by a Merit Review grant (IO1BX000131) from the Veteran's Administration to P. Raychaudhuri. A.L. Tyner is supported by the PHS grants DK 44525 and DK068503. S. Bagchi is supported by the PHS grant CA156164.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–16. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 2.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 3.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–11. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attiyeh EF, London WB, Mosse YP, Wang Q, Winter C, Khazi D, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–53. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 5.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–4. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 6.Jaboin J, Kim CJ, Kaplan DR, Thiele CJ. Brain-derived neurotrophic factor activation of TrkB protects neuroblastoma cells from chemotherapy-induced apoptosis via phosphatidylinositol 3′-kinase pathway. Cancer Res. 2002;62:6756–63. [PubMed] [Google Scholar]
- 7.Ross RA, Biedler JL, Spengler BA. A role for distinct cell types in determining malignancy in human neuroblastoma cell lines and tumors. Cancer Lett. 2003;197:35–9. doi: 10.1016/s0304-3835(03)00079-x. [DOI] [PubMed] [Google Scholar]
- 8.Ciccarone V, Spengler BA, Meyers MB, Biedler JL, Ross RA. Phenotypic diversification in human neuroblastoma cells: expression of distinct neural crest lineages. Cancer Res. 1989;49:219–25. [PubMed] [Google Scholar]
- 9.Walton JD, Kattan DR, Thomas SK, Spengler BA, Guo HF, Biedler JL, et al. Characteristics of stem cells from human neuroblastoma cell lines and in tumors. Neoplasia. 2004;6:838–45. doi: 10.1593/neo.04310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross RA, Spengler BA, Domenech C, Porubcin M, Rettig WJ, Biedler JL. Human neuroblastoma I-type cells are malignant neural crest stem cells. Cell Growth Differ. 1995;6:449–56. [PubMed] [Google Scholar]
- 11.Hansford LM, McKee AE, Zhang L, George RE, Gerstle JT, Thorner PS, et al. Neuroblastoma cells isolated from bone marrow metastases contain a naturally enriched tumor-initiating cell. Cancer Res. 2007;67:11234–43. doi: 10.1158/0008-5472.CAN-07-0718. [DOI] [PubMed] [Google Scholar]
- 12.Mahller YY, Williams JP, Baird WH, Mitton B, Grossheim J, Saeki Y, et al. Neuroblastoma cell lines contain pluripotent tumor initiating cells that are susceptible to a targeted oncolytic virus. PLoS One. 2009;4:e4235. doi: 10.1371/journal.pone.0004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A. 2004;101:14228–33. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith KM, Datti A, Fujitani M, Grinshtein N, Zhang L, Morozova O, et al. Selective targeting of neuroblastoma tumor-initiating cells by compounds identified in stem cell-based small molecule screens. EMBO Mol Med. 2010;2:371–84. doi: 10.1002/emmm.201000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–65. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 16.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–42. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, et al. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–8. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Pinilla SM, Sarrio D, Moreno-Bueno G, Rodriguez-Gil Y, Martinez MA, Hernandez L, et al. Sox2: a possible driver of the basal-like phenotype in sporadic breast cancer. Mod Pathol. 2007;20:474–81. doi: 10.1038/modpathol.3800760. [DOI] [PubMed] [Google Scholar]
- 19.Phi JH, Park SH, Kim SK, Paek SH, Kim JH, Lee YJ, et al. Sox2 expression in brain tumors: a reflection of the neuroglial differentiation pathway. Am J Surg Pathol. 2008;32:103–12. doi: 10.1097/PAS.0b013e31812f6ba6. [DOI] [PubMed] [Google Scholar]
- 20.Riggi N, Suva ML, De Vito C, Provero P, Stehle JC, Baumer K, et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010;24:916–32. doi: 10.1101/gad.1899710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y, Lin Q, Zelterman D, Yun Z. Hypoxia-regulated delta-like 1 homologue enhances cancer cell stemness and tumorigenicity. Cancer Res. 2009;69:9271–80. doi: 10.1158/0008-5472.CAN-09-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melone MA, Giuliano M, Squillaro T, Alessio N, Casale F, Mattioli E, et al. Genes involved in regulation of stem cell properties: studies on their expression in a small cohort of neuroblastoma patients. Cancer Biol Ther. 2009;8:1300–6. doi: 10.4161/cbt.8.13.8890. [DOI] [PubMed] [Google Scholar]
- 23.Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- 24.Ye H, Kelly TF, Samadani U, Lim L, Rubio S, Overdier DG, et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17:1626–41. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25:1715–9. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–50. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV, et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–20. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–61. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 29.Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida Y, Wang IC, Yoder HM, Davidson NO, Costa RH. The fork-head box M1 transcription factor contributes to the development and growth of mouse colorectal cancer. Gastroenterology. 2007;132:1420–31. doi: 10.1053/j.gastro.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 31.Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24:2649–61. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–84. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 33.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–94. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otsubo T, Akiyama Y, Yanagihara K, Yuasa Y. SOX2 is frequently downregulated in gastric cancers and inhibits cell growth through cell-cycle arrest and apoptosis. Br J Cancer. 2008;98:824–31. doi: 10.1038/sj.bjc.6604193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albino D, Scaruffi P, Moretti S, Coco S, Truini M, Di Cristofano C, et al. Identification of low intratumoral gene expression heterogeneity in neuroblastic tumors by genome-wide expression analysis and game theory. Cancer. 2008;113:1412–22. doi: 10.1002/cncr.23720. [DOI] [PubMed] [Google Scholar]
- 36.Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–70. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Diskin S, Rappaport E, Attiyeh E, Mosse Y, Shue D, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–62. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- 38.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90 +cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–66. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie Z, Tan G, Ding M, Dong D, Chen T, Meng X, et al. Foxm1 transcription factor is required for maintenance of pluripotency of P19 embryonal carcinoma cells. Nucleic Acids Res. 2010;38:8027–38. doi: 10.1093/nar/gkq715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui H, Hu B, Li T, Ma J, Alam G, Gunning WT, et al. Bmi-1 is essential for the tumorigenicity of neuroblastoma cells. Am J Pathol. 2007;170:1370–8. doi: 10.2353/ajpath.2007.060754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui H, Ma J, Ding J, Li T, Alam G, Ding HF. Bmi-1 regulates the differentiation and clonogenic self-renewal of I-type neuroblastoma cells in a concentration-dependent manner. J Biol Chem. 2006;281:34696–704. doi: 10.1074/jbc.M604009200. [DOI] [PubMed] [Google Scholar]
- 43.Li SK, Smith DK, Leung WY, Cheung AM, Lam EW, Dimri GP, et al. FoxM1c counteracts oxidative stress-induced senescence and stimulates Bmi-1 expression. J Biol Chem. 2008;283:16545–53. doi: 10.1074/jbc.M709604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–7. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992;12:4565–74. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gusarova GA, Wang IC, Major ML, Kalinichenko VV, Ackerson T, Petrovic V, et al. A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J Clin Invest. 2007;117:99–111. doi: 10.1172/JCI27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gartel AL. FoxM1 inhibitors as potential anticancer drugs. Expert Opin Ther Targets. 2008;12:663–5. doi: 10.1517/14728222.12.6.663. [DOI] [PubMed] [Google Scholar]
- 48.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–44. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1
FoxM1 moderately affects the growth of neuroblastoma cells. A, Twenty-four hours after FoxM1 or control siRNA transfection, cells were trypsinized and seeded at a density of 2×103 cells per 48-well plate for proliferation assay. Viable cell number was measured by using CellTiter-Glo luminescent cell viability assay kit (Promega) every two days. The fold change in cell count was presented by luminescence unit and was normalized by the value at day zero. B, Seventy-two hours after FoxM1 or control siRNA transfection, cells were fixed and stained with propidium iodide (PI) and analyzed by flow cytometry for sub-G1 population. C, Seventtwo hours after FoxM1 or control siRNA transfection, cell lysates were collected and assayed for cleaved caspase-3 by immunoblot. β-catenin was used as loading control.
Supplementary figure 2
FoxM1 promotes tumorigenicity in SK-N-AS cells. A, Colony formation is induced in cells stably expressing FoxM1. 2×103 SK-N-AS cells stabling expressing empty vector or FoxM1 were plated per well in a six-well plate in triplicate. Colonies were stained and quantified after 2 weeks. B, Representative pictures and quantification of anchorage-independent growth of SK-N-AS cells stabling expressing empty vector or FoxM1. Cells were plated a density of 1.6×104 cells per well in a six-well plate. Colonies were stained and counted after three weeks. C, Immunoblot showing expression of FoxM1 and Sox2 of SK-N-AS cells stabling expressing empty vector for FoxM1. α−tubulin was used as the loading control.
Supplementary figure 3
FoxM1 and ESC-like gene signature. A, Seventy-two hours after FoxM1 and control siRNA treatment, semiquantitative RT-PCR was performed to analyze the mRNA level of the pluripotency genes including Sox2, Oct4, Nanog, Bmi1, Ezh2, Suz12. Cyclophilin was used as a loading control. The intensity of the bands was determined by ImageJ software and the intensity of control samples were set at one. B, Venn diagram of overlapping genes in three groups: FoxM1 target genes, Sox2 target genes and core ESC-like module. FoxM1 target genes were summarized from 201 published FoxM1 related literature where FoxM1 regulated genes were reported and from differentially expressed genes from one publicly available FoxM1 array of breast carcinoma (NCBI GDS1477) (50). Sox2 target genes were reported from chip array study (49, 39, table S1)) and core ESC-like module was previously purposed (49(table S6)). P value for both FoxM1 with Sox2 targets overlap (F/S) and FoxM1 with core module overlap (F/E) were obtained using hypergeometric calculation. C, List of 33 overlapping genes of FoxM1 targets and core ESC-like module. Among them, genes that were also Sox2 targets were highlighted in red.
Supplementary figure 4
FoxM1 mediated anchorage-independent growth requires expression of Sox2. A Immunoblot showing depletion of Sox2 by siRNA in BE(2)-C cells. Cell lysates were collected at four days after transfection. B, 24 hours after Sox2 or control siRNA transfection, cells were trypsinized and seeded at a density of 2×103 cells per well in a 48-well plate for proliferation assay. Viable cell number was measured by using CellTiter-Glo luminescent cell viability assay kit (Promega) every two days. The fold change in cell count was presented by luminescence unit and was normalized by day zero. C, BE(2)-C cells were first transfected with empty or pCMV-FoxM1 plasmid for 24 hours, and then transfected with either control siRNA or Sox2 siRNA for another 24 hours. Cells were plated at a density of 8×103 cells per well in a six-well plate in soft agar plate. Colonies were stained and quantified after three weeks. D, Quantification of colonies numbers derived from C. Colony large than 100μm in diameter was defined as large colony and colony which has diameter between 25-100μm was defined as small colony.