Self-organization of molecules via non-covalent interactions is able to generate complex matters and structures[1] that function at both supramolecular and cellular levels. For example, proteins self-assemble into a variety of nanofibers (or filaments), which are critical components for many essential cellular activities. Inside cells, actins, lamins and tubulins self-assemble to afford microfilaments, intermediate filaments, and microtubules, respectively;[2] outside cells, the self-assembly of collagen and elastin provides nanofibers that serve as the extracellular matrix to support and regulate differentiation, proliferation, and interaction of cells.[2] While the nanofibers of normal proteins are essential for normal cellular functions, the aggregation of aberrant proteins also results in aggregates (e.g., β-amyloid oligomers)[3] that are toxic to cells. Like proteins, certain small organic molecules also self-assemble in water to afford supramolecular nanofibers in which aromatic-aromatic interactions and hydrophobicity play critical roles for guiding hydrogen bonding, promoting the self-assembly, and stabilizing the resulted nanofibers.[4] While the successful formation of supramolecular nanofibers of small molecules has resulted in hydrogels[5] as a new class of biomaterials for tissue engineering,[6] drug delivery,[7] protein arrays,[8] electrophoresis,[9] bacteria typing,[10] and protocells,[11] there is little exploration on the protein targets[12] and cytotoxicity of the supramolecular nanofibers of small molecules,[10, 13] especially at the concentration below the critical gelation concentration (cgc).
Recently, reported by Wells et al.,[13] nanofibrils formed by aggregation of small molecules bind to procaspase-3 and activate its transformation to generate caspase-3 and to initiate the apoptosis cascade. Encouraged by this work and our previous results on cell death induced by intracellular formation of molecular nanofibers,[14] we choose to evaluate the cytotoxicity of a versatile self-assembly motif, naphthalen-2-yl-acetamido-Phe-Phe (Nap-FF, 1 (Scheme 1a)). We select 1 for three reasons: (i) 1 exhibits remarkable ability to self-assemble in water and to form nanofibers at low concentration; (ii) 1 has a known crystal structure[15] that allows us to infer plausible molecular arrangement in the nanofibers formed by 1 (Scheme 1B); (iii) a structurally close analogue consisted of 1 exhibits specific interactions with certain protein targets (e.g., tubulins).[12] After determining the threshold of 1 to form molecular nanofibers, we incubate cells with 1 at different concentrations and use cell-based tubulin assay to elucidate the possible molecular targets of the nanofibers of 1. Our results suggest that the molecules of 1 self-assemble at 320-340 μM to form soluble nanofibers that disrupt the dynamics of microtubule, leading to apoptosis of glioblastoma cells (Scheme 1c). Notably, the nanofibers of 1 exhibit little acute toxicity towards a neuronal cell line (PC12). As the first example of supramolecular nanofiber formed by self-assembly of small molecules to disrupt self-organization of functional proteins, this work not only promises potential therapeutic application of supramolecular nanofibers as anticancer agent to treat glioblastoma, a devastating disease without effective cure, but also provides mechanism for inherent cytotoxicity of hydrophobic supramolecular nanofibers.
Scheme 1.

a) The chemical structure of 1 and b) a possible molecular arrangement in the nanofibers of 1 based on its crystal structure (Enlarged shows the hydrogen bonding (cyan dashed line) and aromatic-aromatic interaction (orange dashed line) between the molecules. c) Illustration of supramolecular nanofibers of 1 to selectively inhibit growth of glioblastoma cells.
We first characterize the morphology of nanofibers formed by 1 and verify its threshold concentration of self-assembly. Molecules of 1 self-assemble efficiently to form nanofibers, which entangle and hold water to form hydrogel (Figure S1) due to physical cross-linking. This observation suggests that 1 is able to self-assemble in water to form nanofibers below the cgc of 1 (0.4 wt% (0.83mM)). Using transmission electron microscopy (TEM), we analyze the solutions of 1 at different concentrations in PBS buffer (pH 7.4) below the cgc of 1. Negative stained[16] TEM images of 1 at 400 μM show nanofibers with uniform width of 24 ± 2 nm (Figure 1a) as dominant morphology. Measuring all nanofibers in the TEM images, we find a relatively wide distribution of the lengths of the nanofibers with the average value of about 181 nm (Figure 1b). On the contrary, the TEM images of solution of 1 at 300 μM only displays amorphous feature (Figure S2), suggesting that 1 exists primarily in monomeric state at the concentration of 300 μM and has a threshold concentration of the formation of nanofibers between 300 to 400 μM. As shown in Scheme 1b, the crystal structure of 1 suggests that, being driven by aromatic-aromatic interactions and hydrogen bonding, 1 self-assembles into β strand-like structures. The circular dichroism (CD) spectrum of 1 at 400 μM, indeed, correlates to a typical β-sheet structure according to CD simulation of a negative peak at 190 nm followed by a positive peak at 200 nm (Figure 1c).[17] These results confirm that nanofibers of 1 formed by self-assembly of 1 adopting β-sheet like structure, which lead us to use thioflavin T (ThT), a benzothiazole dye exhibiting enhanced fluorescence upon binding to β-sheet containing fibrils,[18] for quantifying the self-assembly of 1. As shown in Figure 1d, at concentration of 1 below 320 μM, the ThT emission has little change; at and above 340 μM of 1, the ThT emission increases linearly with the increase of concentration of 1, indicating the threshold concentration for 1 to form the nanofibers in the range of 320-340 μM. In addition, the fluorescence of the solution of 1 at the concentration of 200 or 300 μM, before and after the filtration, exhibit essentially the same spectra (Figure S3) as the monomeric naphthalene in water,[19] indicating that 1 mainly exist as monomers at those concentrations.
Figure 1.
Characterization of molecular nanofibers of 1 in PBS buffer. a) Negative-stained TEM images of the solution of 1 at 400 μM in PBS buffer and b) histogram of length distribution of the nanofibers according to the TEM images of 1 at 400 μM. c) Circular dichroism spectra of 1 at different concentrations. d) ThT emission at 484 nm (λex = 440 nm) of ThT (20 μM) with increased concentrations of 1.
After determined the threshold of 1 to form the nanofibers, we examine the cytotoxicity of monomeric 1 and nanofiber of 1 by incubation of mammalian cells with 1 at concentrations below and above the threshold. We first test the cytotoxicity of 1 towards HeLa cells:[20] the most widely studied malignant cancer cell line.[21] While monomers of 1 (i.e., 1 at 200 and 300 μM) exhibit little cytotoxicity, 400 or 500 μM of 1 significantly decreases the viability of the HeLa cells to less than 20% within 48 h (Figure 2a). Above its threshold concentration of self-assembly, 1 exists as a mixture of nanofibers and monomers of 1 in the solution. To verify that the nanofibers of 1 result in the cytotoxicity, we slowly pass the culture medium containing 1 through 0.22 μm nylon membrane. Due to their sizes (Figure 1b), some of the nanofibers of 1 are removed by the filtration, but the monomers of 1 remain in the filtrate. As shown in Figure 2b, filtrates of 400 or 500 μM of 1 exhibits less cytotoxicity towards HeLa cells (over 50% of cells are viable at 48 h) than without filtration. This result agrees with that the concentrations of 1 in the solution, after the filtration, are below the threshold concentration of aggregation (Table S1). The above results suggest that the monomers of 1 are innocuous to cells while the nanofibers of 1 inhibit the growth of HeLa cells.
Figure 2.
Cell viability assay (MTT) of HeLa cells after 24 and 48 h of treatment with a) as prepared or b) filtered medium with different initial concentrations of 1. MTT assay of c) T98G and d) PC12 cell lines incubated with 1.
To test whether the nanofibers of 1 inhibit the growth of other cancer cell lines in vitro, we incubate 1 with T98G cells,[22] a cancer cell line that causes glioblastoma, the most aggressive malignant primary brain tumor in human.[23] As shown in Figure 2c, the nanofibers of 1 also efficiently inhibit growth of T98G cells while monomers of 1 show little cytotoxicity to T98G. In the brain, glial cells (from which glioblastoma derives) and neuronal cells together produce functional nervous systems.[24] We then test the cytotoxicity of 1 on a neuronal cell line PC12,[25] a frequently used model of neuronal cells.[26] While being acutely cytotoxic to T98G, both monomers and nanofibers of 1 exhibit little toxicity towards PC12 (Figure 2d). Prolong the incubation of PC12 with nanofibers of 1 for 7 days, over 60% of the PC12 cells still remain viable (Figure S4), suggesting that PC12 cells, unlike T98G, tolerant nanofibers of 1 to a certain degree.
As an insidious facet of malignant cancer cells, which makes them different with benign tumor cells, is that they ignore tissue boundaries and infiltrate or migrate into surrounding or distant tissues.[27] So we test the influence of nanofibers of 1 on the migration of HeLa cells. We first create a gap with width of 0.9 mm by a plastic insert on fully confluent layer of HeLa cells, (Figure 3a), then apply the media containing 1 at different concentrations. After 18 h of culture, the untreated cells and the cells incubated with 300 μM of 1 exhibit similar rates of migration and cover 47% of the gaps (Figure 3b, c). Being incubated with 400 μM of 1, the cells only cover 27% of the gaps (Figure 3d), indicating that the nanofibers of 1 delay the migration of cells. Since microtubule and filamentous actin are essential for establishing and maintaining cell migration,[28] we speculate that nanofibers of 1 may affect function of either microtubule or filamentous actin to delay cell migration. This result, in fact, agrees with an in vitro experiment in our previous study, which shows the nanofibers of a photo-reactive hydrogelator, which is structurally similar with that of 1, selectively bind tubulin.[12]
Figure 3.
Cell migration assay of HeLa cells treated with 1. a) The gaps were created on HeLa cells of 100% confluence in 24 well plates, and measured after 18 h of incubation with b) 0, c) 300, or d) 400 μM of 1.
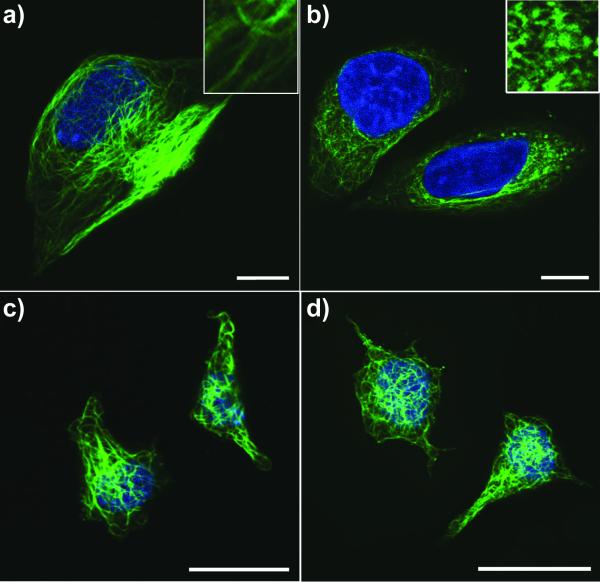
To verify the speculation that the nanofibers of 1 interact with tubulin, we use tubulin tracker to visualize microtubule in T98G and PC12 cells incubated with or without nanofibers of 1 for 24 h. As shown in Figure 4a, the untreated T98G cells displays long microtubule network that stretch through the cell body. Treated with 400 μM of 1 (i.e., the nanofibers of 1), tubulins in T98G cells aggregate into clusters with a few number of short microtubules (Figure 4b), illustrating the nanofibers of 1 disrupts the dynamics of microtubules. Based on the time dependent cytotoxicity curve of nanofibers of 1 (Figure S5), cancer cells enter death phase after 38 h of incubation, which place the disruption of microtubules prior to cell death. As it is known that malfunction of microtubules can lead to apoptosis,[29] we use apoptosis assays (FITC-annexin V, PI and TUNEL)[30] to evaluate T98G cells treated with 400 μM of 1 for 24 h. Result of the assays confirm that the T98G cells enter early apoptosis stage (Figure S6). These data together suggests that the nanofibers of 1 disrupt the dynamics of microtubules and consequently induce apoptosis of glioblastoma cells (e.g., T98G). Moreover, the HeLa cells enter death phase after 38h regardless the concentration (to be 400 or 500 μM), which also agrees with that the aggregates of 1 cause the cell death and deviate from the conventional dosage response based on the concentration of the monomers. Unlike in T98G cells, microtubules in PC12 cells treated by nanofibers of 1 have little change comparing with the untreated PC12 cells (Figure 4c, d), indicating that microtubule in PC12 cells tolerant the treatment of 1. The reasons T98G cells experience much obvious microtubule disruption and consequently higher cytotoxicity than PC12 cells lie in the accelerated metabolic rate of glioblastoma cells due to Warburg effect[31] and that small molecule tend to accumulate in cell once forms aggregates.[14b] Since 1 accumulates much slower in PC12 cell than that in T98G cells (Figure S7), the amount of nanofibers of 1 in PC12 cells is much less than that in T98G cells. Thus 1 induces little microtubule disruption and little cytotoxicity on PC12 cells.
Figure 4.
Confocal images of tubulin stained T98G (a, b) and PC12 (c, d) cells treated with medium containing 0 (a, c) or 400 μM (b, d) of 1 for 24 h. DNA was counterstained blue by DAPI. Insets are 3X enlarged images. Scale bar = 10 μm.
In conclusions, this work establishes that, as a form of supramolecular assemblies, the nanofibers of small molecules exhibit properties drastically different from the individual molecules when they interact with cells. Moreover, hydrophobicity of 1 appears to be critical to result in the observed cytotoxicity because 2, obtained by using tyrosine (Y) to replace the phenylalanine (F) in 1, exhibits relatively high critical aggregation concentration (1.0 mM) and considerably low cytotoxicity (Figure S8). Considering the relative high hydrophobicity of 1, which has higher logP value (4.57) than naphthalene (logP = 3.03), the result from cell-based tubulin assay implies inherent toxicity of other nanofibers with high local density of hydrophobicity might also arise from a similar mechanism. To elucidate molecular mechanism of these nanofibers with high hydrophobicity, cell-based assay is necessary, as shown in the elucidation of the molecular mechanism of nanofibers of 1. Moreover, the successful inhibition of growth of glioblastoma cells without detrimental effects to neuronal cells promises potential therapeutic application of supramolecular nanofibers of small hydrophobic molecules in treating this most aggressive malignant brain tumor. Of course, one has to engineer the temporal profiles of the supramolecular nanofibers to achieve the therapeutic effect, which is our ongoing study.
Experimental Section
Materials and Methods
All chemicals and reagents were obtained from FisherSci. Circular dichroism was performed on a JASCO J-810 spectrometer, transmission electron microscopy on Morgagni 268 transmission electron microscope, MTT viability assay and tubulin polymerization assay on DTX 880 multimode detector, flow cytometry on FACSCalibur flow cytometer, isothermal titration calorimetry on TA instrument NANO ITC Low volume, fluorescence spectra on RF-5301PC spectrofluorophotometer, confocal images on Leica SP2 microscope.
Tubulin staining[32]
Cells in exponential growth phase were seeded in glass bottomed culture chamber at 10,000 cell/mL. The cells were allowed for attachment for 24 h at 37 °C, 5% CO2. The culture medium was removed, and new culture medium containing 1 at 0 or 400 μM was added. After 24 h of incubation, cells were washed with PBS buffer for 3 times and stained by Tubulin Tracker™ Green at 100 nM and DAPI 300 nM in PBS for 30 min at 37 °C in dark. The sample was rinsed three times in PBS, and the cells were kept in PBS for imaging.
Supplementary Material
Acknowledgments
[**] This work was partially supported by NIH (R01CA142746) and startup fund from Brandeis University. We acknowledge the help of EM facility in Brandeis University. BX thanks Prof. N. Grigorieff for advices on TEM.
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- [1]a).Lehn JM. Science. 2002;295:2400–2403. doi: 10.1126/science.1071063. [DOI] [PubMed] [Google Scholar]; b) Whitesides GM, Mathias JP, Seto CT. Science. 1991;254:1312–1319. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]; c) Whitesides GM, Grzybowski B. Science. 2002;295:2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- [2].Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J. Mol. Cell Biol. St. Martin’s Press; 2000. [Google Scholar]
- [3].Cherny I, Gazit E. Angew. Chem. Int. Ed. 2008;47:4062–4069. doi: 10.1002/anie.200703133. [DOI] [PubMed] [Google Scholar]
- [4]a).Brizard A, Stuart M, van Bommel K, Friggeri A, de Jong M, van Esch J. Angew. Chem. Int. Ed. 2008;47:2063–2066. doi: 10.1002/anie.200704609. [DOI] [PubMed] [Google Scholar]; b) Ma ML, Kuang Y, Gao Y, Zhang Y, Gao P, Xu B. J. Am. Chem. Soc. 2010;132:2719–2728. doi: 10.1021/ja9088764. [DOI] [PubMed] [Google Scholar]; c) Martin-Gago P, Gomez-Caminals M, Ramon R, Verdaguer X, Martin-Malpartida P, Aragon E, Fernandez-Carneado J, Ponsati B, Lopez-Ruiz P, Cortes MA, Colas B, Macias MJ, Riera A. Angew. Chem. Int. Ed. 2012;51:1820–1825. doi: 10.1002/anie.201106406. [DOI] [PubMed] [Google Scholar]
- [5].Estroff LA, Hamilton AD. Chem. Rev. 2004;104:1201–1217. doi: 10.1021/cr0302049. [DOI] [PubMed] [Google Scholar]
- [6].Smith AM, Williams RJ, Tang C, Coppo P, Collins RF, Turner ML, Saiani A, Ulijn RV. Adv. Mater. 2008;20:37–41. [Google Scholar]
- [7]a).Zhao F, Ma ML, Xu B. Chem. Soc. Rev. 2009;38:883–891. doi: 10.1039/b806410p. [DOI] [PubMed] [Google Scholar]; b) Tiller JC. Angew. Chem. Int. Ed. 2003;42:3072–3075. doi: 10.1002/anie.200301647. [DOI] [PubMed] [Google Scholar]
- [8].Kiyonaka S, Sada K, Yoshimura I, Shinkai S, Kato N, Hamachi I. Nature Mater. 2004;3:58–64. doi: 10.1038/nmat1034. [DOI] [PubMed] [Google Scholar]
- [9].Yamamichi S, Jinno Y, Haraya N, Oyoshi T, Tomitori H, Kashiwagi K, Yamanaka M. Chem. Commun. 2011;47:10344–10346. doi: 10.1039/c1cc13826j. [DOI] [PubMed] [Google Scholar]
- [10].Yang ZM, Ho PL, Liang GL, Chow KH, Wang QG, Cao Y, Guo ZH, Xu B. J. Am. Chem. Soc. 2007;129:266–267. doi: 10.1021/ja0675604. [DOI] [PubMed] [Google Scholar]
- [11].Kumar RK, Yu XX, Patil A, Li M, Mann S. Angewandte Chemie-International Edition. 2011;50:9343–9347. doi: 10.1002/anie.201102628. [DOI] [PubMed] [Google Scholar]
- [12].Gao Y, Long MJC, Shi J, Hedstrom L, Xu B. Chem. Commun. 2012;48:8404–8406. doi: 10.1039/c2cc33631f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zorn JA, Wille H, Wolan DW, Wells JA. J. Am. Chem. Soc. 2011;133:19630–19633. doi: 10.1021/ja208350u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]a).Yang ZM, Xu KM, Guo ZF, Guo ZH, Xu B. Adv. Mater. 2007;17:3152–3156. [Google Scholar]; b) Yang ZM, Liang GL, Guo ZF, Guo ZH, Xu B. Angew. Chem. Int. Ed. 2007;46:8216–8219. doi: 10.1002/anie.200701697. [DOI] [PubMed] [Google Scholar]
- [15].Zhang Y, Kuang Y, Gao YA, Xu B. Langmuir. 2011;27:529–537. doi: 10.1021/la1020324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Frado LL, Craig R. J. Mol. Biol. 1992;223:391–397. doi: 10.1016/0022-2836(92)90659-8. [DOI] [PubMed] [Google Scholar]
- [17].Greenfield NJ. Nat. Protoc. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Levine H. Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lee S, Winnik MA. Can. J Chem. 1993;71:1216–1224. [Google Scholar]
- [20].Rafferty KA. Virchows Archiv. B. 1986;50:167–180. doi: 10.1007/BF02889899. [DOI] [PubMed] [Google Scholar]
- [21].Skloot R. The Immortal Life of Henrietta Lacks. Crown; New York: 2010. [Google Scholar]
- [22].Schlenska-Lange A, Knüpfer H, Lange TJ, Kiess W, Knüpfer M. Anticancer Res. 2008;28:1055–1060. [PubMed] [Google Scholar]
- [23].Rubenstein M, Shaw M, Mirochnik Z, Slobodskoy L, Glick R, Lichtor T, Chou P, Guinan P. Methods Find Exp. Clin. Pharmacol. 1999;21:391–393. doi: 10.1358/mf.1999.21.6.541918. [DOI] [PubMed] [Google Scholar]
- [24].Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DM, Cavenee WK, DePinho RA. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- [25].Cheng Y, Zhizhin I, Perlman RL, Mangoura D. J. Biol. Chem. 2000;275:23326–23332. doi: 10.1074/jbc.M001837200. [DOI] [PubMed] [Google Scholar]
- [26].Troy CM, Shelanski ML. Proc. Nat. Acad. Sci. USA. 1994;91:6384–6387. doi: 10.1073/pnas.91.14.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Giese A, Bjerkvig R, Berens ME, Westphal M. J. Clin. Oncol. 2003;21:1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- [28].Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Nat. Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- [29].Sherwood SW, Sheridan JP, Schimke RT. Exp. Cell Res. 1994;215:373–379. doi: 10.1006/excr.1994.1354. [DOI] [PubMed] [Google Scholar]
- [30].Del Bino G, Darzynkiewicz Z, Degraef C, Mosselmans R, Fokan D, Galand P. Cell Prolif. 1999;32:25–37. doi: 10.1046/j.1365-2184.1999.3210025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hsu PP, Sabatini DM. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- [32].Koss DJ, Hindley KP, Riedel G, Platt B. J. Neurochem. 2007;102:1009–1023. doi: 10.1111/j.1471-4159.2007.04579.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.