Abstract
The regulation of the protein synthesis has a crucial role in governing the eukaryotic cell growth. Subtle changes of proteins involved in the translation process may alter the rate of the protein synthesis and modify the cell fate by shifting the balance from normal status into a tumoral or apoptotic one. The largest eukaryotic initiation factor involved in translation regulation is eIF3. Amongst the 13 factors constituting eIF3, the f subunit finely regulates this balance in a cell-type-specific manner. Loss of this factor causes malignancy in several cells, and atrophy in normal muscle cells. The intracellular interacting partners which influence its physiological significance in both cancer and muscle cells are detailed in this review. By delineating the global interaction network of this factor and by clarifying its intracellular role, it becomes apparent that the f subunit represents a promising candidate molecule to use for biotherapeutic applications.
Keywords: Apoptosis, Atrophy, Cancer, eIF3f, Hypertrophy
Introduction
Protein synthesis is one of the most complex and energy-consuming processes in eukaryotic cells, by which the genetic information is translated from a nucleic acid code into an amino acidic language to make proteins. Global protein synthesis rates have a key role in governing cell growth, the control of cell size, and cell proliferation. Changes in the levels of specific regulatory proteins involved in the translation process may affect the translation rates and the amount of produced proteins. Thus, the regulation of gene expression is a crucial step in cellular physiology considering that subtle defects in the mechanism of translational control may lead to either cell malignancy or cell death. Protein synthesis is mainly regulated at the initiation step. In eukaryotes, initiation can be divided into two steps: formation of a 48S initiation complex and its joining with a 60S subunit [1]. First, the eukaryotic initiation factors eIF3, eIF1, eIF1A and eIF2-GTP-Met-tRNAi bind to the 40S ribosomal subunit to form a 43S preinitiation complex. This complex subsequently attaches to the 5′-proximal region of mRNAs, after which eIF4A, 4B and 4F unwind their secondary structure. The 43S complex then scans the 5′ untranslated region in the 5′–3′ direction until the initiation codon, where it stops and forms a 48S complex with codon–anticodon base-pairing through eIF5. Second, eIF5B promotes the displacement of the eukaryotic initiation factors and the subsequent joining of 48S complex with the 60S subunit [1].
The largest and more complex eukaryotic initiation factor, which plays a role in translation regulation, cell growth and cancer, is eIF3. This review is focused on the function of the f-subunit of eIF3 (eIF3f) in the translation regulation process. An analysis of its physiological significance and its intracellular interacting partners involved in translational regulation both in cancer and muscle cells is sequentially detailed. The aim is to delineate the global eIF3f interaction network and to clarify its intracellular role in order to validate eIF3f as a lead candidate to use for biotherapeutic applications.
Eukaryotic initiation factor 3
Translation initiation in eukaryotes is an intricate process requiring at least nine eukaryotic initiation factors [1, 2]. Among these factors, eIF3 is the largest as it comprises up to 13 non-identical subunits. Several functions have been ascribed to eIF3, including the interaction and stabilization of the eIF2-GTP-Met-tRNAi ternary complex, binding to the 40S ribosomal subunit, facilitating the binding of mRNA to the 40S ribosome, and promoting dissociation of the 40S and 60S ribosomal subunits. Other initiation factors (e.g., eIF4G, eIF4B, eIF5 and eIF1) are known to interact with eIF3, suggesting that this factor might have a role in ‘organizing’ proteins on the surface of the 40S ribosome [3–5].
Structure and nomenclature
Mammalian eIF3 is an 800-kDa molecular mass assembly of 13 subunits that are designated eIF3a–eIF3m [3–11]. In the past, ad hoc nomenclatures for eIF3 subunits have been derived from subunit molecular weights or gene names. In 2001, to eliminate confusion surrounding cross-species comparisons of eIF3 subunits, a unified nomenclature was proposed by Browning et al. [7]. Each subunit is identified by a letter designation based on the decreasing order of the mammalian subunit masses determined from SDS-PAGE mobility (Table 1). For two novel subunits, −l and −m, the nomenclature has been adapted as they have arisen.
Table 1.
Summary of eukaryotic eIF3 subunits
| Unified nomenclature | S. cerevisiae + MW | H. sapiens | S. pombe | A. thaliana | Consensus motif |
|---|---|---|---|---|---|
| eIF3a | Tif32—110 kDa | p170 | p107 | p114 | PCI |
| eIF3b | Prt1—90 kDa | p116 | p84 | p82 | RRM |
| eIF3c | Nip1—93 kDa | p110 | p104 | p105 | PCI |
| eIF3d | – | p66 | Moe1 a | p66 | – |
| eIF3e | – | p48 | Int6 a | p51 | PCI |
| eIF3f | – | p47 | Csn6 | p32 | MPN |
| eIF3g | Tif35—33 kDa | p44 | Tif35 | p33 | RBD |
| eIF3h | p40 | p40 b | p38 | MPN | |
| eIF3i | Tif34—39 kDa | p36 | Sum1 | p36 | WD repeats |
| eIF3j | Hcr1—35 kDa | p35 | – | – | – |
| eIF3k | – | p28 | – | p25 | PCI |
| eIF3l | – | p67 | – | p60 | PCI |
| eIF3m | – | GA17 | Csn7B b | – | PCI |
The ‘conserved’ core subunits are highlighted in bold type, the ‘functional’ core subunits are underlined, and the ‘dispensable’ ones are in italics
MPN Mpr1p and Pad1p N-terminal conserved domain; PCI 26S proteasome, COP9 signalosome and eukaryotic initiation factor eIF3 conserved domain; RBD RNA-binding domain; RRM RNA-recognition motif; S6K1 ribosomal protein S6 kinase 1; WD conserved regions of approximately 40 amino acids typically bracketed by Trp–Asp
aSubunits contained in S. pombe Int6 eIF3 complex
bSubunits contained in S. pombe Csn7B eIF3 complex
Initial characterizations of mammalian eIF3 were based on biochemical methods applied to purified proteins from rabbit reticulocytes and HeLa cells [12, 13]. Due to its large size and complexity, a detailed understanding of eIF3 structure and function has been achieved more recently from experiments based on the cloning and characterization of human cDNAs encoding the eIF3 subunits [5, 14–16]. By comparing the cDNA sequences of the mammalian eIF3 subunits to the entire genome of the budding yeast Saccharomyces cerevisiae, it appears that only five mammalian subunits eIF3-a, -b, -c, -g and -i have identifiable homologues encoded in yeast (Table 1). A sixth ortholog, eIF3j, is a nonessential subunit of the budding yeast eIF3 that enhances interactions with other eIFs [17], promotes binding of eIF3 to the 40S subunit [18], and has an independent function in 40S ribosome biogenesis [19]. Biochemical interactions have been detected between these five conserved subunits in both mammalian and yeast systems [14, 15, 20]. The S. cerevisiae eIF3 complex also contains 3 additional proteins, p135, p62 and p16, for which corresponding homologs are not found in the mammalian complex [21–23], whereas 7 subunits (eIF3-d, -e, -f, -h, -k, -l and -m) identified in the mammalian eIF3 complex are absent in the yeast complex (Table 1). Most of these 7 mammalian subunits of eIF3 appear to be highly conserved in Drosophila melanogaster, Caernorhabditis elegans, Arabidopsis thaliana and the fission yeast Schizosaccharomyces pombe [4, 5]. Nevertheless, the biochemical function of this complex is conserved inasmuch as eIF3 purified from S. cerevisiae can replace mammalian eIF3 in the in vitro methionyl-puromycin synthesis assay [21]. Accordingly, the mammalian eIF3 complex consists of five subunits (eIF3-a, -b, -c, -g, -i) that represent a ‘conserved’ core essential for translation initiation in vivo, and the remaining ‘noncore’ subunits that appear to be evolutionarily divergent and serve to modulate its activity [5, 14, 20].
The function of each subunit of eIF3 has first been investigated in S. cerevisiae. Deletion or mutation of eIF3a [24, 25], eIF3b [15, 26], eIF3c [27], eIF3g [28], or eIF3i [20, 28, 29] in yeast leads to a vast reduction in protein synthesis, suggesting that each of these five subunits is required for eIF3 integrity. The role of each subunit and the minimum combination of components required for the formation of an active and functional core have also been investigated in mammalian eIF3 [30]. By using a baculovirus-based coexpression system, recombinant eIF3 complexes lacking different individual subunits have been reconstituted. Remarkably, deletion studies showed that the rate of the protein synthesis does not change by knockdown of the two evolutionarily conserved subunits, eIF3g and eIF3i, in HeLa cells, and the eIF3 activity remains unaltered after deletion of the three non-conserved subunits eIF3-d, -k, -l [30]. Therefore, eIF3-g, -i, -d, -k, -l subunits are not essential for eIF3 activity but might be involved in translational control of specific mRNAs or in particular cellular conditions [30]. Hence, the functional core of mammalian eIF3 consists of three conserved subunits, eIF3-a, -b, -c, and three non-conserved subunits, eIF3-e, -f and -h. These three last subunits might serve to stabilize the eIF3-abc complex which directly participates in translation initiation [30]. In vitro stepwise reconstitution of human eIF3 subunits has revealed the architectural features of this factor. The dimer of subunits eIF3a and eIF3c serves as a central scaffold to which most of the other subunits bind. This dimer directly interacts with the subunits eIF3e, -k, -l and -m containing the PCI domain (the 26S proteasome, COP9 signalosome and eukaryotic initiation factor eIF3 conserved domain) and with the subunits eIF3-f and-h containing a MPN (Mpr1p and Pad1p N-terminal conserved domain) to form a highly stable PCI/MPN octamer. Subunits -d, -b, -g, -i, and -j then finalize the assembly of the eIF3 complex [11].
eIF3 function in cell cycle regulation, translational regulation, and cancer
The global protein synthesis in mammals occurs mainly in the G1 phase of a cell cycle and is reduced during mitosis. This process is modulated by several translation initiation factors [31]. The first studies that associated two eIF3 subunits, eIF3a and eIF3b, to cell cycle regulation were performed in S. cerevisiae. Both subunits are essential for the G1-S phase transition in yeast [32, 33]. A study on the eIF3a expression in mammalian cells has shown that its levels oscillate during the cell cycle and its maximum of expression is detected in the S phase, indicating that eIF3a may be a translational regulator for proteins important for S phase entrance [34]. Additionally, the subunits eIF3a [35], eIF3e [36, 37], eIF3k [38] and eIF3f [39] have been found in both cytoplasm and nucleus, indicating that these factors may shuttle between the two subcellular localizations according to cell cycle progression.
Proper execution of the cell cycle requires the synthesis and activation of key proteins at specific times. The most prevalent mechanism for regulating the overall rate of protein synthesis involves the phosphorylation of the initiation factors. Besides phosphorylation, the rate of protein synthesis is regulated by other mechanisms and post-translational modifications of the translational machinery, such as methylation of lysine or arginine residues and O-glycosylation [40]. Recent reports confirm the role of the eIF3 factor in regulation of translation initiation rate [40–42]. However, the molecular mechanisms by which the function of eIF3 is regulated are poorly understood. A mass spectrometric approach has been used to determine post-translational modifications that regulate the activity of eIF3 during the translation initiation process (Table 2). A total of 29 phosphorylation sites and several other post-translational modifications (loss of N-terminal methionine and/or N-terminal protein acetylation) have recently been identified [10].
Table 2.
Identification of eIF3 protein subunits and their corresponding post-translational modifications using a mass spectrometric analysis [10]
| Protein name | UniProt accession number | Molecular massa (Da) | Sequence coverage (%) | Post-translational modifications |
|---|---|---|---|---|
| eIF3a | Q14152 | 166,758.3 | 86 | Loss of Met-1, phosphorylation (Ser-881, Ser-1198, Ser-1336b, Ser-1364b) |
| eIF3b | P55884 | 93,093.7 | 77 | Acetylation (Met-1), phosphorylation (Ser-83, Ser-85, Ser-119, Ser-125b, Ser-152, Ser-154, Ser-164) |
| eIF3c | Q99613 | 106,143.8 | 65 | Phosphorylation (Ser-9, Ser-11, Ser-13, Ser-15, Ser-16, Ser-18, Ser-39, Ser-166c, Thr-524b, Ser-909c) |
| eIF3d | O15371 | 63,972.9 | 74 | Not found |
| eIF3e | P60228 | 52,131.8 | 84 | Loss of Met-1, acetylation (Ala-2) |
| eIF3f | O00303 | 37,554.8 | 79 | Loss of Met-1, acetylation (Ala-2), phosphorylation (Ser-258b) |
| eIF3g | O75821 | 35,639.8 | 83 | Loss of Met-1, phosphorylation (Thr-41, Ser-42) |
| eIF3h | O15372 | 40,010.4 | 89 | Phosphorylation (Ser-183b) |
| eIF3i | Q13347 | 36,501.9 | 93 | Not found |
| eIF3j | O75822 | 29,293.2 | 81 | Loss of Met-1, acetylation (Ala-2), phosphorylation (Ser-11, Ser-13, Ser-20, Thr-109c) |
| eIF3k | Q9UBQ5 | 24,970.6 | 75 | Loss of Met-1, acetylation (Ala-2) |
| eIF3l | Q9Y262 | 66,637.9 | 70 | Loss of Met-1, acetylation (Ser-2) |
| eIF3m | Q7L2H7 | 42,413.8 | 74 | Loss of Met-1, acetylation (Ser-2) |
aCalculated from the theoretical average mass of the corresponding eIF3 protein subunit plus any post-translational modifications identified
bFound only after TiO2 phosphopeptide enrichment
cFound only after Ga(III) IMAC or TiO2 phosphopeptide enrichment
Alternatively, an indirect regulation of the activity of eIF3 is performed by association of its subunits with other proteins involved in the regulation of protein synthesis. For example, the subunit eIF3e binds p56 in interferon-treated or virus-infected mammalian cells, and inhibits the translation in vitro and in vivo [43, 44]. The subunit eIF3g interacts with Paip1, a Poly (A)-binding protein and stimulates translation initiation [45], whereas the subunits eIF3h and eIF3f interact with TRC8, a ubiquitin E3 ligase, and inhibit protein synthesis, possibly through ubiquitilation of eIF3 or some other translational components [46]. These mechanisms and interacting partners render eIF3 a pivotal player in controlling the protein synthesis and degradation.
A proper level of translation initiation is necessary to regulate cell proliferation, since the hyperactivation or the down-regulation of the rate of the protein synthesis contributes importantly to cell malignancy. The contribution of eIF3 to oncogenesis and maintenance of the cancer state has been demonstrated in several studies [47–49]. Amongst the 13 subunits constituting eIF3, overexpression of subunits eIF3-a, -b, -c, -h, -i, and -m has been detected in several different solid tumors and in several different cancer cell lines (Table 3; for literature citations, see [42, 47, 48]. The correlation of abnormal eIF3 subunit levels and cancer indicates that eIF3 has an important role in determining the balance between cell proliferation and apoptosis. A study performed on NIH3T3 fibroblasts, stably transfected with five individual eIF3 subunit cDNAs, has confirmed that their overexpression contributes to malignant phenotype [48]. Ectopic overexpression of eIF3-a, -b, -c, -h or -i induces an increase in protein synthesis rate, the resistance to apoptosis, and the oncogenic transformation of NIH3T3 cells. Hence, eIF3 may play a causal role in neoplasia, and the misregulation of translation may be a contributory factor to cancer pathology [48]. It has been demonstrated that the overexpression of a single eIF3 subunit causes malignant transformation and induces an increase in eIF3 activity, which in turn stimulates the translation of mRNAs involved in cell proliferation. Zhang et al. [48] have suggested that transforming subunit overexpression leads to the preferential stimulation of so-called “weak” mRNAs, which are translated more efficiently considering the abundant presence of secondary structures in their 5′-UTRs. Generally, “weak” mRNAs encode proteins involved in promoting cell growth and proliferation, such as c-myc, cyclin D1 and growth factors. Thus, the increase of “weak” mRNA products stimulates the malignant transformation of cells.
Table 3.
Translational factor alterations in cancers
| Subunit | Observed modification | Cancer association |
|---|---|---|
| eIF3a | Increased expression | Mouse melanoma, human breast, cervical, esophageal, lung, and gastric cancers |
| eIF3b | Increased expression | Human breast carcinoma |
| eIF3c | Increased expression | Human testicular seminomas |
| eIF3e | Decreased expression | Human breast and lung carcinomas |
| eIF3f | Decreased expression | Pancreas, vulva, ovary, breast, small intestine tumors, and melanoma |
| eIF3h | Increased expression | Human breast, prostate, hepatocellular carcinomas |
| eIF3i | Increased expression | Cadmium transformed NIH3T3 cell lines |
| eIF3m | Increased expression | Human colon cancer |
Two other eIF3 subunits, eIF3e and eIF3f, are downregulated in human breast/lung carcinomas and melanoma/pancreatic cancer, respectively [50–54]. Low eIF3e expression is considered as a significant factor in predicting poor prognosis for nonsmall cell lung cancer [52]. It may be likely that these factors have a causal role in cancer development inasmuch as their decrease removes the restraints that block the protein synthesis and stimulates the selective translation of oncogenic proteins involved in cell proliferation [53, 54].
The f subunit of eIF3
In 2003, Shi et al. [39] showed, for the first time, that in human cells the noncore f subunit of the mammalian eIF3 multiprotein complex acts as a negative regulator of translation. Furthermore, in mammalian cells, eIF3f inhibits protein synthesis at the translational level and not by affecting mRNA levels [55]. Translation initiation can be cap-dependent or cap-independent/IRES-dependent. It is not clearly known whether eIF3f specifically inhibits one of these two types of translation. However, the silencing of eIF3f increased both cap-dependent and IRES-dependent translation, indicating a suppressive role of eIF3f on both translation initiation mechanisms [56]. Furthermore, it has been recently demonstrated that the f subunit plays a role in the inhibition of HIV-1 replication. Overexpression of N-terminal 91 amino acids of eIF3f or full-length eIF3f drastically restricts HIV-1 replication by interfering with the 3′-end processing of HIV-1 mRNAs and reducing their nuclear and cytoplasmic levels [57, 58].
Intracellular localization
Under normal conditions, most of the endogenous eIF3f is bound to 40S ribosomal subunit [55]. However, confocal microscopy images of human melanoma cells show that eIF3f is localized at both cytoplasmic and nuclear levels [39]. Other eIF3 subunits, such as eIF3a, eIF3e and eIF3k, have been reported to have nuclear localization [35–38]. In fact, there are at least two different types of eIF3 complexes localized in both the cytoplasm and the nucleus of cells. Hence, eIF3 is considered as a ‘dynamic’ complex, where ‘dynamic’ means that it may contain a different and variable number of subunits [59, 60]. The cytoplasmic eIF3 complex contains the subunits eIF3a-c and f, whereas the nuclear one lacks the subunits eIF3a and f [60]. What correlates this finding and the microscopy results?
It has been demonstrated that the phosphorylation of eIF3f may specifically increase its association with other eIF3 core complex subunits. In particular, during apoptosis, eIF3f is directly phosphorylated by the cyclin-dependent kinase CDK11p46 in living cells [60], and is incorporated into a micro-complex containing eIF3b and eIF3c, but not eIF3a. By cell fractionation, it has been observed that this complex precipitates with the nucleus in the insoluble fraction of the cells. Nevertheless, eIF3f can also associate in a smaller complex consisting of eIF3a, eIF3b and eIF3c in the soluble fraction of both normal and apoptotic cells [60]. Therefore, under normal conditions, the nuclear eIF3 contains eIF3b and eIF3c and, during apoptosis, the phosphorylated eIF3f joins the nuclear complex. The model currently suggested is that the translation initiation can be regulated during apoptosis by the phosphorylated eIF3f binding to different sub-fractions of the eIF3 complex [60]. This finding, coupled with the fact that eIF3f is a Mov34 family member, led Shi et al. [39] to elaborate the hypothesis that this factor may have a nuclear function in mRNA metabolism such as splicing or transport. It is also possible that the nuclear eIF3 complex has functions other than translation initiation. For example, the synthesis of ribosomes in eukaryotes takes place in the nucleus and then ribosomes are exported to the cytoplasm. Shi et al. [60] suggest that CDK11p46 may regulate the function of the nuclear eIF3 complex by phosphorylating eIF3f, considering that cyclin-dependent kinases are involved in ribosome biogenesis and nucleolar organization.
Regulation of eIF3f activity
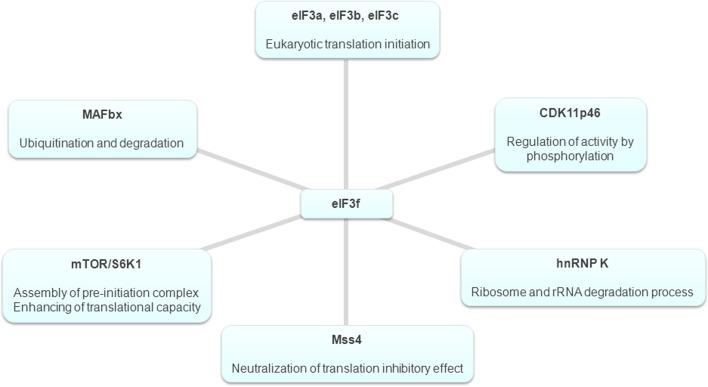
The eIF3f subunit is a Mov34 family member, containing an MPN motif, which is found in two other macromolecular complexes homologous to eIF3, the COP9 signalosome and the 19S proteasome [5, 6, 61, 62]. The presence of a MPN domain in complexes with apparently divergent functions, such as protein synthesis, signal transduction, and protein degradation, has been explained by the fact that this domain is probably necessary to promote complex assembly and is required for proper interactions between subunits of these complexes [63]. Recent crystallographic studies have revealed that there are two classes of MPN domains, one possessing a metalloprotease activity and a second possessing a structural function [64, 65]. The subunits eIF3f and eIF3h contain the second type of MPN domain, which is responsible for protein–protein interactions and for protein stability [64, 65]. The crucial role of this domain in determining the cellular function and the intracellular behaviour of eIF3f has been confirmed by several studies. In 2006, Shi et al. [55] showed that this domain, and more precisely the amino acid region spanning residues 170–248, is responsible for the translation inhibition function of eIF3f. Furthermore, it has been demonstrated that an eIF3f activating phosphorylation site is located in the Mov34 domain [60]. The phosphorylation status of this subunit can significantly influence its association with eIF3 core complex, its intracellular localization, and its function in the regulation of protein synthesis rate and in the control of the subtle balance between cell growth and cell death (see below). Moreover, several intracellular interacting partners of eIF3f have been identified (Fig. 1) using yeast two-hybrid screens. In most identified partners, the interactions are mediated by the Mov domain, suggesting that it may play a central role in regulating the activity of this subunit.
Fig. 1.
Schematic representation of the intracellular interacting partners of the eIF3f subunit
Apoptosis versus cell growth: the importance of eIF3f rate
Phosphorylation of eIF3f during apoptosis
Using a yeast two-hybrid system, Shi et al. [39] showed that endogenous cyclin-dependent kinase 11 (CDK11) and eIF3f protein can interact and that their interaction can be strengthened by the stimulation of apoptosis.
The CDK11 kinases belong to a large family of p34cdc2-related kinases [66] containing at least 20 CDK11 isoforms that are differentially expressed in mammalian tissues and regulate diverse cellular functions. The largest CDK11p110 isoform is associated with cyclin L and various splicing factors, and is involved in the regulation of transcription and RNA splicing in proliferating cells [67, 68]. Expression of the CDK11p110 is ubiquitous and constant throughout the cell cycle [69]. During Fas- and tumor necrosis factor-induced cell death, CDK11p110 is activated by proteolytic cleavage by caspase-3. Caspase-3 processing removes the N-terminal part of the protein, which contains nuclear translocation signals and is responsible for protein stability, and generates a smaller 46- to 50-kDa protein (CDK11p46) containing the catalytic kinase domain [70, 71].
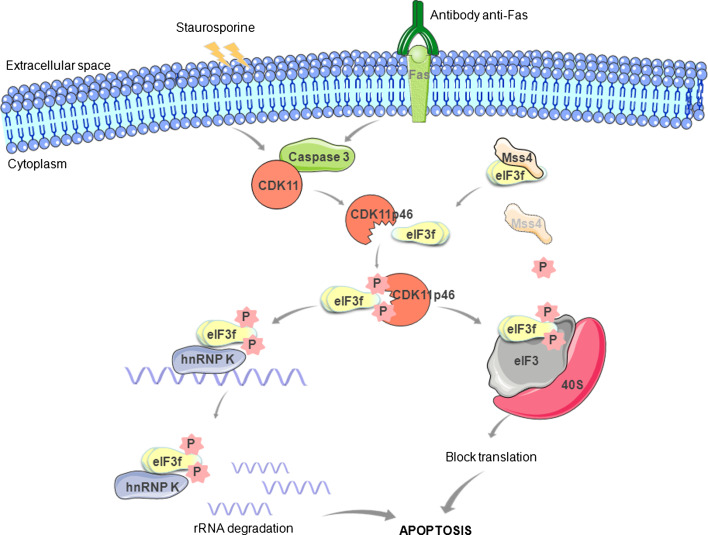
After anti-Fas or staurosporine treatments in human melanoma cells, the caspase-processed C-terminal fragment CDK11p46 strongly interacts with eIF3f via its Mov34 domain [39] and, due to its kinase activity, phosphorylates eIF3f inducing the inhibition of translation (Fig. 2). The mutation of the phosphate transfer site in CDK11p46 abrogates the phosphorylation of eIF3f protein. Two different phosphorylation sites have been identified in eIF3f during apoptosis: Ser46 [39] and Thr119 [60]. CDK11p46 phosphorylates both sites. Thr119 is located in the Mov34 domain of eIF3f, which is important for both the translational inhibitory function and the eIF3f interaction with CDK11p46. The biological consequence of the phosphorylation of eIF3f during apoptosis is not completely known. The phosphorylation status of eIF3f can significantly influence its association with the eIF3 core complex and its function in the regulation of translation and apoptosis [60]. Considering that the phosphorylations on Ser46 and Thr119 have been detected in cells undergoing apoptosis, it has been postulated that eIF3f may be involved in the inhibition of the protein synthesis when phosphorylated, acting as a downstream death executer.
Fig. 2.
Schematic representation of eIF3f interactions during apoptosis. Upon apoptotic stimulation, CDK11p110 is cleaved by caspase 3 to generate a CDKp46 that interacts strongly with the Mov34 domain of eIF3f and phosphorylates it at Ser46 and Thr119. According to Wen et al.’s [56] hypothesis (left side), phosphorylated eIF3f interacts with hnRNP K and promotes rRNA degradation by interfering with rRNA protective function of hnRNP. According to Walter et al.’s [73] hypothesis, under normal conditions (right side), Mss4 is tightly bound to eIF3f, inhibiting its phosphorylation and subsequent association with eIF3 protein complex and pro-apoptotic functions. After prolonged stress-induced apoptosis, Mss4 is downregulated, leading to a release of eIF3f, which is phosphorylated by the CDK11p46 kinase and the translation initiation results inhibited
eIF3f expression levels in cancer and apoptotic cells
The biological role of eIF3f in translation and apoptosis in tumor cells has been investigated by Shi et al. [39, 53–56, 60]. Transcriptome analysis has shown that endogenous eIF3f expression level changes from one cell type to another and from normal to tumor cells [55]. Furthermore, the eIF3f protein level also increases during apoptosis with maximal expression at 36 h after stimulation [55].
Using a cancer profiling array and real-time reverse transcription PCR, eIF3f transcript levels have been shown to be downregulated in most human tumors relative to matched normal tissues [55]. In particular, 100 % of pancreas and vulva tumors, 90 % of breast tumors, 71 % of melanomas, and 70 % of ovary and small intestine tumors showed a significant decrease of eIF3f expression [55]. Using loss of heterozygosity and gene copy number analyses, it has been demonstrated that there is an allelic loss of eIF3f gene in human melanoma and in pancreatic cancer cells [53, 54]. No mutations are responsible for the decreased eIF3f expression in these two cancer types [53, 54].
The molecular mechanism by which eIF3f protein expression decreases, contributing to cancer development, is unclear. One possible explanation is that the decrease of a negative regulator of translation like eIF3f may lead to an increased eIF3 activity, which in turn stimulates translation of specific mRNAs encoding for proteins involved in cell proliferation [53, 54]. eIF3f was also found to interact with mTOR and S6K1. The decrease of eIF3f expression may deregulate the function of the mTOR pathway, which in turn leads to increased translation of oncogenic proteins and malignant transformation [53, 54]. Finally, eIF3f is ubiquitously expressed in all tissues and is associated with both subpolysomal particles and polysomes. eIF3f is localized at both cytoplasmic and nuclear levels according to the cell cycle phase and is able to associate to different sub-fractions of the eIF3 complex. Probably, these various multiproteins complexes are implicated in the role of eIF3f in discriminating the mRNAs to be translated into normal cells, and may also reflect the deregulation of the translational pathway in tumor cells.
eIF3f overexpression in cancer inducing apoptosis
Enforced expression of eIF3f inhibits translation, cellular growth and proliferation, and induces apoptosis in melanoma and pancreatic cancer cells [53–55]. Ectopic expression of eIF3e and eIF3f subunits in stably transfected NIH3T3 fibroblasts also inhibits cell growth and proliferation [48]. Western blot analyses have demonstrated that eIF3f induces apoptosis in caspase 3/7 and in a Bcl-2-, Bax- or Bcl-XL-independent manner [55]. It is still unclear whether decreased eIF3f expression is the cause, rather than the consequence, of malignant transformation. Knockdown of endogenous eIF3f by siRNA attenuates apoptosis in melanoma cells after treatment with staurosporine as apoptotic agent [55]. Wen et al. [56] have stably knocked down endogenous eIF3f in normal human pancreatic ductal epithelial cells (HDPE) and have observed increased cell proliferation, clonogenicity, apoptotic resistance, survival, resistance to chemotherapy drug, mesenchymal morphology, and migration. eIF3f-silenced cells also show an increased cell size, nuclear pleiomorphism, aneuploidy, and cell cycle abnormality. By using an ex vivo 3D-cell culture system, they have also shown that eIF3f-silenced HPDE cells form more irregular masses with abnormal architecture and polarity (recapitulating malignant tumors in vivo), while control cells develop into a single-layer epithelial hollow spheres (resembling normal pancreatic ductal structure in vivo). eIF3f-silenced HPDE cells proliferate in an anchorage-independent manner. These observations confirm that eIF3f is an important negative regulator of cell growth and proliferation, and the decrease of its expression contributes to tumor cells’ evading apoptosis via upregulation of protein synthesis [56].
A link between translation initiation and rRNA degradation
Besides the hypotheses described above to explain how decreased eIF3f may contribute to cancer development, a further model linking the translation initiation factor and rRNA degradation has been proposed by Wen et al. [56]. This model finds its basis in the observation that eIF3f-transfected cancer cells show a significant decrease of 60S and 80S ribosomes and 60S/40S ribosomal subunit ratio, and an increase in RNA absorbance during apoptosis [55]. It is still unclear whether the reduced 60S ribosomal subunit contributes to or is a result of apoptosis. The increasing of the absorbance is correlated to an increased amount of rRNA degradation, which contributes to the apoptotic process [55]. The major rRNA degraded in eIF3f-transfected cells is the 28S rRNA followed by the 18S rRNA in a minor rate [55]. How exactly eIF3f induces rRNA degradation is unclear. However, as shown for other eIF3 subunits, the translation may be inhibited by specific eIF3f binding to RNAs and their consequent degradation [29]. The discovery that eIF3f can interact with the heterogenous nuclear ribonucleoprotein K (hnRNP K) during apoptosis in melanoma and HPDE cells corroborates this model [56]. hnRNP K is a RNA-binding protein responsible for the maintenance of RNA stability by binding to the 3′-UTR of the mRNAs. Yeast three-hybrid screens and RNA pull-down assays have shown that hnRNP binds to 18S and 28S rRNA [72]. In human cells, dissociation of hnRNP from rRNA contributes to rRNA degradation during apoptosis [56]. However, whether hnRNP K regulates rRNA degradation is not known. Considering that hnRNP K protects rRNA from degradation whereas eIF3f promotes this process, Wen et al. [56] have proposed that eIF3f may interact with hnRNP K during apoptosis and promote rRNA degradation by interfering with its rRNA protective function (Fig. 2). In other words, eIF3f sequesters hnRNP K and inhibits its binding to rRNA, which leads to increased rRNA degradation and attenuated translation.
The rRNA synthesis and degradation in normal cells needs to be perfectly balanced in order to maintain homeostasis of protein synthesis. Under physiological conditions, eIF3f competes with rRNA for hnRNP K binding. This contributes to the maintenance of the homeostasis of rRNA level and translation in cells. Increased eIF3f expression contributes to apoptosis via hnRNP K sequestration and increased rRNA degradation. Loss of eIF3f contributes to tumorigenesis via the increased binding of hnRNP K to rRNA and an increased rRNA level. Currently, it is still unclear where and how normal human rRNA is degraded during various physiologic or pathologic cellular processes; however, eIF3f might play a role in mRNA and rRNA degradation.
Mss4: the neutralizing agent of eIF3f translation inhibitory effect
Starting from an investigation into the regulatory properties of the mammalian suppressor of Sec4 (Mss4) in response to cellular stress, Walter et al. [73] have recently demonstrated that this protein can act as an anti-apoptotic agent by direct binding to eIF3f. High expression levels of Mss4 protein are beneficial for cells, as they protect them from stress-induced apoptosis. The expression of Mss4 is strongly regulated by stress stimuli at both transcription and post-transcriptional levels: it is upregulated at early stages of stress stimulation, but it declines with prolonged stimulation. Yeast two-hybrid screens have indicated that the most abundant Mss4 interaction partner is eIF3f, and studies on A7 melanoma cells have shown that Mss4 neutralizes the translation inhibitory effect of eIF3f [73]. Thus, Mss4 is probably involved in the regulation of protein translation, and its association to eIF3f protects cells from apoptosis. In other words, eIF3f is a negative regulator of translation: its overexpression makes apoptosis easier, whereas its downregulation has the opposite effect. Under normal conditions, Mss4 is tightly bound to eIF3f, inhibiting its phosphorylation and subsequent association with eIF3 complex. At early stages of stress stimulation, increasing amounts of Mss4 protein efficiently bind the eIF3f preventing its interaction with CDK11p46. When Mss4 proteins in cells drop-down owing to ongoing stress, the CDK11p46 can phosphorylate the released eIF3f. The translation initiation is inhibited and apoptosis is activated (Fig. 2).
Atrophy versus hypertrophy: the importance of eIF3f
Hypertrophy
In muscle cells, the regulation of protein synthesis rate also has an important role in the control of cell growth. Muscular hypertrophy and atrophy are two opposite and mechanistically linked phenomena regulating muscle cell size, finely determined throughout a balance between new protein accumulation and degradation of pre-existing proteins [74]. A major mediator of skeletal muscle hypertrophy is the serine/threonine-specific protein kinase Akt1. Two major downstream branches of the Akt pathway, which are relevant to muscle hypertrophy, are the mTOR pathway, which is activated by Akt, and glycogen synthase kinase 3β, which is blocked by Akt. Both pathways control the protein synthesis [74].
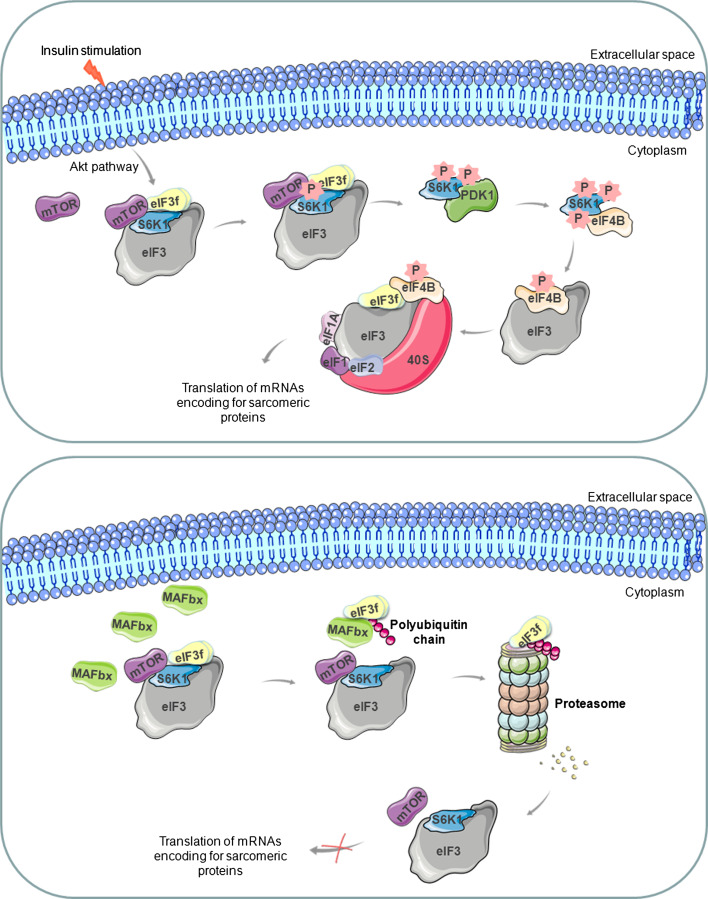
The kinase mammalian target of rapamycin (mTOR) has recently emerged as a key regulator of mammalian cell growth that integrates signals from growth factors, nutrients, and energy status to control protein synthesis and other cell functions [75, 76]. As the name implies, mTOR is selectively inhibited by rapamycin, a drug used as an immunosuppressant in organ transplantation. The role of mTOR in muscle growth has been demonstrated by in vivo studies showing that rapamycin blocks overload hypertrophy and regenerating muscle growth [77, 78]. The effect of mTOR on the translation machinery and protein synthesis is executed by phosphorylation of the ribosomal protein S6 kinases and of 4E-BP1, a repressor of the cap-binding protein eIF4E [79, 80]. mTOR and S6K1 maneuver on and off the eIF3 translation initiation complex in a signal-dependent fashion [79]. In fact, the eIF3 complex acts as a scaffolding platform that associates with mTOR and S6K1 in a growth-factor and rapamycin-sensitive manner. Under normal conditions, S6K1 is bound to the eIF3 complex, whereas mTOR association is greatly reduced. Following an activating signal, such as insulin stimulation, mTOR is recruited to the eIF3-preinitiation complex, leading to phosphorylation of the bound and inactive S6K1. Phosphorylated S6K1 is released from the eIF3 complex, and interacts with PDK1, which promotes a second phosphorylation of S6K1. Activated S6K1 then phosphorylates eIF4B, which is then recruited to the eIF3-preinitiation complex and promotes the recruitment of other initiation factors and the translation of mRNAs encoding proteins involved in muscle hypertrophy (Fig. 3, upper panel) [79]. Thus, eIF3, mTOR, and S6K1 coordinate the assembly of a translation initiation complex with enhanced translational capacity under conditions of nutrient and energy sufficiency. The understanding of how this assembly is controlled is still incomplete. However a physical and functional link exists between these proteins. Yeast two-hybrid screens have revealed that both mTOR [80] and S6K1 [79] physically interact with the f subunit of eIF3. Particularly, the inactive hypophosphorylated form of S6K1 physically associates with the Mov34 domain of eIF3f [81]. Thus, mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein/protein interchange and might control the function of eIF3 [80].
Fig. 3.
Schematic representation of the intracellular signals characterizing the hypertrophic (upper panel) and atrophic (lower panel) pathways in muscle cells. During hypertrophy, mTOR phosphorylates S6K1, which is released from eIF3 complex, and phosphorylated again by PDK1. Activated S6K1 phosphorylates eIF4B, which promotes the recruitment of other initiation factors and the translation of mRNAs encoding proteins involved in muscle growth. During atrophy, the ubiquitin ligase MAFbx is upregulated and S6K1 is accumulated in its inactive hypophosphorylated form. By binding the Mov34 domain, MAFbx transfers polyubiquitin chains on eIF3f and promotes its degradation by the proteasome and block the synthesis of proteins involved in muscle cell growth
Translational regulation of eIF3f
The mRNA encoding for the eIF3f factor contains at the 5′ end a relatively short stretch of 6–12 pyrimidines, called the terminal oligopyrimidine (TOP) sequence, and consequently its mRNA is known as TOP mRNA [82]. The TOP sequence serves as a cis-acting element necessary for a growth-associated translation regulation [83]. The translational control of TOP mRNAs is used by cells to coordinate the synthesis of different ribosomal proteins, elongation factors, and several other proteins associated with the assembly or function of the translational apparatus [83]. Interestingly, among the initiation factors, only eIF3e, eIF3f and eIF3h exhibit the characteristics of TOP mRNAs [82]. A polysome mRNA analysis on HeLa, HEK293 and NIH3T3 cells has revealed that eIF3e, eIF3f and eIF3h mRNAs are mainly associated with subpolysomal particles in quiescent or growth-arrested cells, but mostly associated with polysomes in growing cells [82]. Consequently, the fact that these three eIF3 subunits are encoded by TOP mRNAs is considered relevant for their role in the growth-associated translation regulation.
It is classically known that mTOR complex 1 (mTORC1) regulates cell growth and proliferation through modulation of protein synthesis in response to growth factors, hormones and amino acids [84]. In a recent study, it has been demonstrated that the mTORC1 kinase specifically controls the translation of TOP mRNAs through the eIF4E-binding proteins (4E-BPs) and the eIF4G1 initiation factor [85]. In eIF4E-dependent initiation, mTORC1 phosphorylates the 4E-BPs promoting their dissociation from the cap-binding protein eIF4E. Active eIF4E then associates to eIF4G1, which interacts with eIF3 resulting in the assembly of the 43S pre-initiation complex on the mRNA [75]. When mTORC1 is inactivated by rapamycin or Torin1, dephosphorylated 4E-BPs bind eIF4E and thereby prevent its association with eIF4G1 and the consequent activation of TOP mRNA translation [85].
In 2010, Csibi et al. [86] reported that the decreased activity of mTORC1 kinase is correlated with the degradation of eIF3f and the accumulation of unphosphorylated forms of S6K1 during muscle atrophy. In contrast, during terminal muscle differentiation, the amount of eIF3f increases as well as mTORC1, leading to an increase in S6K1 activation and phosphorylation of rpS6 and 4E-BP1 [86]. They also observed that rapamycin treatment destabilizes the mTORC1/eIF3f interaction in differentiated myotubes, and that ablation of eIF3f in muscle cells prevents mTORC1 activity and phosphorylation of S6K1, rpS6 and 4E-BP1 [86].
Thus, in muscle cells, the eIF3f translation may be regulated in an mTORC1-dependent manner.
Atrophy
Atrophy is a decrease in cell size mainly caused by a loss of organelles, cytoplasm and proteins. A major contribution in understanding muscle atrophy comes from the pioneering studies on gene expression profiling performed independently by Gomes and Bodine [87, 88]. The idea to compare gene expression in different models of muscle atrophy leads to the identification of a subset of genes that are commonly up- or down-regulated. Since all the diseases used for the experiments of microarray (i.e., diabetes, cancer cachexia, chronic renal failure, fasting, and denervation) have muscle atrophy in common, the up or down genes are believed to regulate the loss of muscle components and are called atrophy-related genes or atrogenes [89]. Together, these findings indicate that muscle atrophy is an active process controlled by specific signaling pathways and transcriptional programs [74]. Furthermore, the two most induced genes encode for two muscle-specific ubiquitin ligases, atrogin-1/MAFbx and MuRF1, which are upregulated in different models of muscle atrophy and are responsible for the increased protein degradation through the ubiquitin–proteasome system [87, 88]. These two genes are actually the best markers for muscle atrophy and could be considered as master genes for muscle wasting.
Ectopic expression of MAFbx in myotubes leads to atrophy and appears to be essential for accelerated muscle protein loss [90]. Interestingly, during atrophy, MAFbx physically interacts with the f subunit of eIF3 via its Mov34 domain [91], leading to its ubiquitination and its degradation by the proteasome (Fig. 3, lower panel) [90]. The silencing of MAFbx expression by small hairpin RNA interference prevents eIF3f degradation in myotubes undergoing atrophy [91]. In C2C12 myotubes that undergo atrophy, MAFbx accumulates in the nucleus and triggers the translocation of eIF3f from the cytoplasm to the nucleus. The translocation of the f subunit between the two compartments may represent a mechanism for regulating its activity during atrophy.
Interestingly, eIF3f can itself act as an enzyme by deubiquinating the transmembrane receptor Notch1 [92]. Notch 1 is involved in a highly conserved signaling pathway essential for development and is monoubiquitinated during its activation. To enter the nucleus and fulfill its transcriptional functions, this receptor needs to be deubiquitinated. The f subunit acts as a positive regulator of Notch signaling. The activated form of Notch is able to interact with eIF3f only in the presence of the E3 ubiquitin ligase Deltex [92]. It remains to be elucidated whether eIF3f works as a deubiquinating enzyme outside the translation complex or in association with the whole translation initiation complex.
The role of eIF3f in atrophy and hypertrophy
By using an inducible protein expression system, it has been demonstrated that the genetic activation of eIF3f is sufficient to block atrophy and induces a massive hypertrophy. This cellular growth is associated with an increase of sarcomeric proteins but not the muscle regulatory factors such as MyoD and/or myogenin [91]. eIF3f is upregulated during terminal differentiation of skeletal muscle and absent in the undifferentiated embryonic rhabdomyosarcoma. Hence, in skeletal muscle, eIF3f can act as a ‘translational enhancer’ that increases the efficiency of the structural muscle protein synthesis leading to muscle hypertrophy in vitro and in vivo. These findings are consistent with previous suggestions that overload-induced hypertrophy is due to an increase in translational capacity and/or translational efficiency [93, 94]. Thus, eIF3f plays an important role in controlling muscle mass and size.
In constrast, the loss of eIF3f, due to MAFbx-mediated ubiquitination, induces atrophy in normal myotubes. Proteins destined for degradation are marked by covalent linkage with a chain of ubiquitin molecules on lysine residues for further degradation into short peptides by the 26 S proteasome. Deletion studies, focused on the identification of the lysine residues marked by MAFbx, have shown that the target for proteasomal degradation is the C-terminal domain of eIF3f. This domain contains 6 C-terminal lysine residues that serve as ubiquitination sites. The mutation of these 6 lysine into arginine residues (mutant K5-10R) does not suppress the MAFbx-eIF3f interaction nor its activity. Thus, the C-terminal domain finely regulates eIF3f activity in skeletal muscle [90]. When levels of eIF3f are maintained by electroporation of the eIF3f K5-10R mutant into adult fibers, muscles are protected from atrophy [90]. The eIF3f mutant K5-10R induces a strong hypertrophic phenotype in cellulo and in vivo, and its overexpression provokes an enhanced ability in avoiding skeletal muscle atrophy [90].
The ubiquitination of eIF3f on C-terminal lysine residues may serve two functions: addressing eIF3f degradation specifically by MAFbx during skeletal muscle atrophy and regulating the translation of specific pools of mRNAs of key sarcomeric proteins [90].
Hence, atrophy and hypertrophy processes communicate with each other via the translation regulator eIF3f [81]. Its Mov34 domain is necessary for both S6K1 and MAFbx binding. In particular, muscles undergoing atrophy accumulate the inactive hypophosphorylated form of S6K1, stimulating the MAFbx-mediated degradation of eIF3f (Fig. 3, lower panel). Whether MAFbx interacts with free eIF3f or with eIF3f molecules bound to S6K1 still remains unclear.
Conclusion
In the light of the eIF3f properties illustrated above, a global analysis of its interactome is necessary. Two opposite features of eIF3f have emerged by analyzing the data available in the literature. This factor is able to act as both a ‘negative repressor’ of translation initiation [39] and a ‘translational enhancer’ [91], and as both a ‘downstream death executer’ in cancer cells [55] and a ‘cell growth enhancer’ in muscle cells [91]. The loss of eIF3f seems to contribute to tumorigenesis whereas its ectopic expression is sufficient to restore apoptosis in cancer cells. Interestingly, in normal myotubes, the genetic repression of eIF3f induces atrophy whereas genetic activation of eIF3f is sufficient to induce hypertrophy.
The same protein seems to act in two different ways according to the cell type considered. It is not clear whether the endogenous eIF3f protein rate is the cause or the effect of this behavior. However, a cell-type-specific role is believed to exist for eIF3f.
Several eIF3f interacting partners have been identified. During the apoptotic pathway, eIF3f is activated by CDK11p46, interacts with hnRNP K for RNA degradation, and is neutralized by Mss4. During the muscle cell growth, eIF3f interacts with mTOR/S6K1 to promote the assembly of the translation initiation complex and is degraded by MAFbx. A positive role in the activation of Notch signaling pathway has also been reported.
Moreover, two different types of eIF3 complexes (nuclear and cytoplasmic) have been found in the cell, suggesting that eIF3f could shuttle between the cytoplasm and the nucleus in a cell cycle-dependent manner.
All these data confirm that eIF3f has a multileveled control of multiple functions in the cells, outside its usual function in translation. Keeping that in mind, targeting eIF3f may be a strategy to reorganize different intracellular pathways and alter the basis of the balance between cell proliferation and apoptosis. Thus, eIF3f represents a lead candidate to use for biotherapeutic applications for both inhibiting the growth of cancer cells or muscle atrophy and, thus, preventing its progression into irreversible cachexia.
References
- 1.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pestova TV, Hellen CU. The structure and function of initiation factors in eukaryotic protein synthesis. Cell Mol Life Sci. 2000;57(4):651–674. doi: 10.1007/PL00000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31(10):553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Asano K, Kinzy TG, Merrick WC, Hershey JW. Conservation and diversity of eukaryotic translation initiation factor eIF3. J Biol Chem. 1997;272(2):1101–1109. doi: 10.1074/jbc.272.2.1101. [DOI] [PubMed] [Google Scholar]
- 5.Asano K, Vornlocher HP, Richter-Cook NJ, Merrick WC, Hinnebusch AG, Hershey JW. Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits. Possible roles in RNA binding and macromolecular assembly. J Biol Chem. 1997;272(43):27042–27052. doi: 10.1074/jbc.272.43.27042. [DOI] [PubMed] [Google Scholar]
- 6.Hershey JW, Asano K, Naranda T, Vornlocher HP, Hanachi P, Merrick WC. Conservation and diversity in the structure of translation initiation factor EIF3 from humans and yeast. Biochimie. 1996;78(11–12):903–907. doi: 10.1016/S0300-9084(97)86711-9. [DOI] [PubMed] [Google Scholar]
- 7.Browning KS, Gallie DR, Hershey JW, Hinnebusch AG, Maitra U, Merrick WC, Norbury C. Unified nomenclature for the subunits of eukaryotic initiation factor 3. Trends Biochem Sci. 2001;26(5):284. doi: 10.1016/S0968-0004(01)01825-4. [DOI] [PubMed] [Google Scholar]
- 8.Mayeur GL, Fraser CS, Peiretti F, Block KL, Hershey JW. Characterization of eIF3 k: a newly discovered subunit of mammalian translation initiation factor elF3. Eur J Biochem/FEBS. 2003;270(20):4133–4139. doi: 10.1046/j.1432-1033.2003.03807.x. [DOI] [PubMed] [Google Scholar]
- 9.Morris-Desbois C, Rety S, Ferro M, Garin J, Jalinot P. The human protein HSPC021 interacts with Int-6 and is associated with eukaryotic translation initiation factor 3. J Biol Chem. 2001;276(49):45988–45995. doi: 10.1074/jbc.M104966200. [DOI] [PubMed] [Google Scholar]
- 10.Damoc E, Fraser CS, Zhou M, Videler H, Mayeur GL, Hershey JW, Doudna JA, Robinson CV, Leary JA. Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol Cell Proteomics. 2007;6(7):1135–1146. doi: 10.1074/mcp.M600399-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Sun C, Todorovic A, Querol-Audi J, Bai Y, Villa N, Snyder M, Ashchyan J, Lewis CS, Hartland A, Gradia S, Fraser CS, Doudna JA, Nogales E, Cate JH. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3) Proc Natll Acad Sci USA. 2011;108(51):20473–20478. doi: 10.1073/pnas.1116821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benne R, Hershey JW. Purification and characterization of initiation factor IF-E3 from rabbit reticulocytes. Proc Natll Acad Sci USA. 1976;73(9):3005–3009. doi: 10.1073/pnas.73.9.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown-Luedi ML, Meyer LJ, Milburn SC, Yau PM, Corbett S, Hershey JW. Protein synthesis initiation factors from human HeLa cells and rabbit reticulocytes are similar: comparison of protein structure, activities, and immunochemical properties. Biochemistry. 1982;21(18):4202–4206. doi: 10.1021/bi00261a002. [DOI] [PubMed] [Google Scholar]
- 14.Asano K, Phan L, Anderson J, Hinnebusch AG. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273(29):18573–18585. doi: 10.1074/jbc.273.29.18573. [DOI] [PubMed] [Google Scholar]
- 15.Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol. 1998;18(8):4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Block KL, Vornlocher HP, Hershey JW. Characterization of cDNAs encoding the p44 and p35 subunits of human translation initiation factor eIF3. J Biol Chem. 1998;273(48):31901–31908. doi: 10.1074/jbc.273.48.31901. [DOI] [PubMed] [Google Scholar]
- 17.Valasek L, Phan L, Schoenfeld LW, Valaskova V, Hinnebusch AG. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 2001;20(4):891–904. doi: 10.1093/emboj/20.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen KH, Valasek L, Sykes C, Jivotovskaya A, Hinnebusch AG. Interaction of the RNP1 motif in PRT1 with HCR1 promotes 40S binding of eukaryotic initiation factor 3 in yeast. Mol Cell Biol. 2006;26(8):2984–2998. doi: 10.1128/MCB.26.8.2984-2998.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valasek L, Hasek J, Nielsen KH, Hinnebusch AG. Dual function of eIF3j/Hcr1p in processing 20 S pre-rRNA and translation initiation. J Biol Chem. 2001;276(46):43351–43360. doi: 10.1074/jbc.M106887200. [DOI] [PubMed] [Google Scholar]
- 20.Verlhac MH, Chen RH, Hanachi P, Hershey JW, Derynck R. Identification of partners of TIF34, a component of the yeast eIF3 complex, required for cell proliferation and translation initiation. EMBO J. 1997;16(22):6812–6822. doi: 10.1093/emboj/16.22.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naranda T, MacMillan SE, Hershey JW. Purified yeast translational initiation factor eIF-3 is an RNA-binding protein complex that contains the PRT1 protein. J Biol Chem. 1994;269(51):32286–32292. [PubMed] [Google Scholar]
- 22.Garcia-Barrio MT, Naranda T, Vazquez de Aldana CR, Cuesta R, Hinnebusch AG, Hershey JW, Tamame M. GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes Dev. 1995;9(14):1781–1796. doi: 10.1101/gad.9.14.1781. [DOI] [PubMed] [Google Scholar]
- 23.Naranda T, MacMillan SE, Donahue TF, Hershey JW. SUI1/p16 is required for the activity of eukaryotic translation initiation factor 3 in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16(5):2307–2313. doi: 10.1128/mcb.16.5.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valasek L, Trachsel H, Hasek J, Ruis H. Rpg1, the Saccharomyces cerevisiae homologue of the largest subunit of mammalian translation initiation factor 3, is required for translational activity. J Biol Chem. 1998;273(33):21253–21260. doi: 10.1074/jbc.273.33.21253. [DOI] [PubMed] [Google Scholar]
- 25.Vornlocher HP, Hanachi P, Ribeiro S, Hershey JW. A 110-kilodalton subunit of translation initiation factor eIF3 and an associated 135-kilodalton protein are encoded by the Saccharomyces cerevisiae TIF32 and TIF31 genes. J Biol Chem. 1999;274(24):16802–16812. doi: 10.1074/jbc.274.24.16802. [DOI] [PubMed] [Google Scholar]
- 26.Danaie P, Wittmer B, Altmann M, Trachsel H. Isolation of a protein complex containing translation initiation factor Prt1 from Saccharomyces cerevisiae. J Biol Chem. 1995;270(9):4288–4292. doi: 10.1074/jbc.270.9.4288. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg JR, Phan L, Gu Z, deSilva A, Apolito C, Sherman F, Hinnebusch AG, Goldfarb DS. Nip1p associates with 40 S ribosomes and the Prt1p subunit of eukaryotic initiation factor 3 and is required for efficient translation initiation. J Biol Chem. 1998;273(36):23485–23494. doi: 10.1074/jbc.273.36.23485. [DOI] [PubMed] [Google Scholar]
- 28.Hanachi P, Hershey JW, Vornlocher HP. Characterization of the p33 subunit of eukaryotic translation initiation factor-3 from Saccharomyces cerevisiae. J Biol Chem. 1999;274(13):8546–8553. doi: 10.1074/jbc.274.13.8546. [DOI] [PubMed] [Google Scholar]
- 29.Naranda T, Kainuma M, MacMillan SE, Hershey JW. The 39-kilodalton subunit of eukaryotic translation initiation factor 3 is essential for the complex’s integrity and for cell viability in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17(1):145–153. doi: 10.1128/mcb.17.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masutani M, Sonenberg N, Yokoyama S, Imataka H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26(14):3373–3383. doi: 10.1038/sj.emboj.7601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pyronnet S, Sonenberg N. Cell-cycle-dependent translational control. Curr Opin Genet Dev. 2001;11(1):13–18. doi: 10.1016/S0959-437X(00)00150-7. [DOI] [PubMed] [Google Scholar]
- 32.Kovarik P, Hasek J, Valasek L, Ruis H. RPG1: an essential gene of saccharomyces cerevisiae encoding a 110-kDa protein required for passage through the G1 phase. Curr Genet. 1998;33(2):100–109. doi: 10.1007/s002940050314. [DOI] [PubMed] [Google Scholar]
- 33.Evans DR, Singer RA, Johnston GC, Wheals AE. Cell-cycle mutations among the collection of Saccharomyces cerevisiae dna mutants. FEMS Microbiol Lett. 1994;116(2):147–153. doi: 10.1111/j.1574-6968.1994.tb06693.x. [DOI] [PubMed] [Google Scholar]
- 34.Dong Z, Liu Z, Cui P, Pincheira R, Yang Y, Liu J, Zhang JT. Role of eIF3a in regulating cell cycle progression. Exp Cell Res. 2009;315(11):1889–1894. doi: 10.1016/j.yexcr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chudinova EM, Ivanov PA, Nadezhdina ES. Large subunit of translation initiation factor–3 p170 contains potentially functional nuclear localization signals. Mol Biol (Mosk) 2004;38(4):684–691. doi: 10.1023/B:MBIL.0000037010.48936.af. [DOI] [PubMed] [Google Scholar]
- 36.Morris-Desbois C, Bochard V, Reynaud C, Jalinot P. Interaction between the Ret finger protein and the Int-6 gene product and co-localisation into nuclear bodies. J Cell Sci. 1999;112(Pt 19):3331–3342. doi: 10.1242/jcs.112.19.3331. [DOI] [PubMed] [Google Scholar]
- 37.Watkins SJ, Norbury CJ. Cell cycle-related variation in subcellular localization of eIF3e/INT6 in human fibroblasts. Cell Prolif. 2004;37(2):149–160. doi: 10.1111/j.1365-2184.2004.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen X, Yang Y, Liu W, Sun M, Jiang J, Zong H, Gu J. Identification of the p28 subunit of eukaryotic initiation factor 3(eIF3 k) as a new interaction partner of cyclin D3. FEBS Lett. 2004;573(1–3):139–146. doi: 10.1016/j.febslet.2004.07.071. [DOI] [PubMed] [Google Scholar]
- 39.Shi J, Feng Y, Goulet AC, Vaillancourt RR, Sachs NA, Hershey JW, Nelson MA. The p34cdc2-related cyclin-dependent kinase 11 interacts with the p47 subunit of eukaryotic initiation factor 3 during apoptosis. J Biol Chem. 2003;278(7):5062–5071. doi: 10.1074/jbc.M206427200. [DOI] [PubMed] [Google Scholar]
- 40.Hershey JW. Regulation of protein synthesis and the role of eIF3 in cancer. Braz J Med Biol Res. 2010;43(10):920–930. doi: 10.1590/S0100-879X2010007500098. [DOI] [PubMed] [Google Scholar]
- 41.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10(4):254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 42.Dong Z, Zhang JT. Initiation factor eIF3 and regulation of mRNA translation, cell growth, and cancer. Crit Rev Oncol/Hematol. 2006;59(3):169–180. doi: 10.1016/j.critrevonc.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 43.Guo J, Hui DJ, Merrick WC, Sen GC. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 2000;19(24):6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hui DJ, Bhasker CR, Merrick WC, Sen GC. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. J Biol Chem. 2003;278(41):39477–39482. doi: 10.1074/jbc.M305038200. [DOI] [PubMed] [Google Scholar]
- 45.Martineau Y, Derry MC, Wang X, Yanagiya A, Berlanga JJ, Shyu AB, Imataka H, Gehring K, Sonenberg N. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol Cell Biol. 2008;28(21):6658–6667. doi: 10.1128/MCB.00738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JP, Brauweiler A, Rudolph M, Hooper JE, Drabkin HA, Gemmill RM. The TRC8 ubiquitin ligase is sterol regulated and interacts with lipid and protein biosynthetic pathways. Mol Cancer Res MCR. 2010;8(1):93–106. doi: 10.1158/1541-7786.MCR-08-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goh SH, Hong SH, Lee BC, Ju MH, Jeong JS, Cho YR, Kim IH, Lee YS. eIF3 m expression influences the regulation of tumorigenesis-related genes in human colon cancer. Oncogene. 2011;30(4):398–409. doi: 10.1038/onc.2010.422. [DOI] [PubMed] [Google Scholar]
- 48.Zhang L, Pan X, Hershey JW. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem. 2007;282(8):5790–5800. doi: 10.1074/jbc.M606284200. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L, Smit-McBride Z, Pan X, Rheinhardt J, Hershey JW. An oncogenic role for the phosphorylated h-subunit of human translation initiation factor eIF3. J Biol Chem. 2008;283(35):24047–24060. doi: 10.1074/jbc.M800956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyazaki S, Imatani A, Ballard L, Marchetti A, Buttitta F, Albertsen H, Nevanlinna HA, Gallahan D, Callahan R. The chromosome location of the human homolog of the mouse mammary tumor-associated gene INT6 and its status in human breast carcinomas. Genomics. 1997;46(1):155–158. doi: 10.1006/geno.1997.4996. [DOI] [PubMed] [Google Scholar]
- 51.Marchetti A, Buttitta F, Pellegrini S, Bertacca G, Callahan R. Reduced expression of INT-6/eIF3-p48 in human tumors. Int J Oncol. 2001;18(1):175–179. doi: 10.3892/ijo.18.1.175. [DOI] [PubMed] [Google Scholar]
- 52.Buttitta F, Martella C, Barassi F, Felicioni L, Salvatore S, Rosini S, D’Antuono T, Chella A, Mucilli F, Sacco R, Mezzetti A, Cuccurullo F, Callahan R, Marchetti A. Int6 expression can predict survival in early-stage non-small cell lung cancer patients. Clin Cancer Res. 2005;11(9):3198–3204. doi: 10.1158/1078-0432.CCR-04-2308. [DOI] [PubMed] [Google Scholar]
- 53.Doldan A, Chandramouli A, Shanas R, Bhattacharyya A, Cunningham JT, Nelson MA, Shi J. Loss of the eukaryotic initiation factor 3f in pancreatic cancer. Mol Carcinog. 2008;47(3):235–244. doi: 10.1002/mc.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doldan A, Chandramouli A, Shanas R, Bhattacharyya A, Leong SP, Nelson MA, Shi J. Loss of the eukaryotic initiation factor 3f in melanoma. Mol Carcinog. 2008;47(10):806–813. doi: 10.1002/mc.20436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi J, Kahle A, Hershey JW, Honchak BM, Warneke JA, Leong SP, Nelson MA. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene. 2006;25(35):4923–4936. doi: 10.1038/sj.onc.1209495. [DOI] [PubMed] [Google Scholar]
- 56.Wen F, Zhou R, Shen A, Choi A, Uribe D, Shi J. The tumor suppressive role of eIF3f and its function in translation inhibition and rRNA degradation. PLoS ONE. 2012;7(3):e34194. doi: 10.1371/journal.pone.0034194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valente ST, Gilmartin GM, Mott C, Falkard B, Goff SP. Inhibition of HIV-1 replication by eIF3f. Proc Natll Acad Sci USA. 2009;106(11):4071–4078. doi: 10.1073/pnas.0900557106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valente ST, Gilmartin GM, Venkatarama K, Arriagada G, Goff SP. HIV-1 mRNA 3′ end processing is distinctively regulated by eIF3f, CDK11, and splice factor 9G8. Mol Cell. 2009;36(2):279–289. doi: 10.1016/j.molcel.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunand-Sauthier I, Walker C, Wilkinson C, Gordon C, Crane R, Norbury C, Humphrey T. Sum1, a component of the fission yeast eIF3 translation initiation complex, is rapidly relocalized during environmental stress and interacts with components of the 26S proteasome. Mol Biol Cell. 2002;13(5):1626–1640. doi: 10.1091/mbc.01-06-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi J, Hershey JW, Nelson MA. Phosphorylation of the eukaryotic initiation factor 3f by cyclin-dependent kinase 11 during apoptosis. FEBS Lett. 2009;583(6):971–977. doi: 10.1016/j.febslet.2009.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aravind L, Ponting CP. Homologues of 26S proteasome subunits are regulators of transcription and translation. Protein Sci. 1998;7(5):1250–1254. doi: 10.1002/pro.5560070521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23(6):204–205. doi: 10.1016/S0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- 63.Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 2001;20(24):7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanches M, Alves BS, Zanchin NI, Guimaraes BG. The crystal structure of the human Mov34 MPN domain reveals a metal-free dimer. J Mol Biol. 2007;370(5):846–855. doi: 10.1016/j.jmb.2007.04.084. [DOI] [PubMed] [Google Scholar]
- 65.Zhang H, Gao ZQ, Wang WJ, Liu GF, Shtykova EV, Xu JH, Li LF, Su XD, Dong YH. The crystal structure of the MPN domain from the COP9 signalosome subunit CSN6. FEBS Lett. 2012;586(8):1147–1153. doi: 10.1016/j.febslet.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 66.Bunnell BA, Heath LS, Adams DE, Lahti JM, Kidd VJ. Increased expression of a 58-kDa protein kinase leads to changes in the CHO cell cycle. Proc Natll Acad Sci USA. 1990;87(19):7467–7471. doi: 10.1073/pnas.87.19.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loyer P, Trembley JH, Lahti JM, Kidd VJ. The RNP protein, RNPS1, associates with specific isoforms of the p34cdc2-related PITSLRE protein kinase in vivo. J Cell Sci. 1998;111(Pt 11):1495–1506. doi: 10.1242/jcs.111.11.1495. [DOI] [PubMed] [Google Scholar]
- 68.Dickinson LA, Edgar AJ, Ehley J, Gottesfeld JM. Cyclin L is an RS domain protein involved in pre-mRNA splicing. J Biol Chem. 2002;277(28):25465–25473. doi: 10.1074/jbc.M202266200. [DOI] [PubMed] [Google Scholar]
- 69.Xiang J, Lahti JM, Grenet J, Easton J, Kidd VJ. Molecular cloning and expression of alternatively spliced PITSLRE protein kinase isoforms. J Biol Chem. 1994;269(22):15786–15794. [PubMed] [Google Scholar]
- 70.Ariza ME, Broome-Powell M, Lahti JM, Kidd VJ, Nelson MA. Fas-induced apoptosis in human malignant melanoma cell lines is associated with the activation of the p34(cdc2)-related PITSLRE protein kinases. J Biol Chem. 1999;274(40):28505–28513. doi: 10.1074/jbc.274.40.28505. [DOI] [PubMed] [Google Scholar]
- 71.Beyaert R, Kidd VJ, Cornelis S, Van de Craen M, Denecker G, Lahti JM, Gururajan R, Vandenabeele P, Fiers W. Cleavage of PITSLRE kinases by ICE/CASP-1 and CPP32/CASP-3 during apoptosis induced by tumor necrosis factor. J Biol Chem. 1997;272(18):11694–11697. doi: 10.1074/jbc.272.18.11694. [DOI] [PubMed] [Google Scholar]
- 72.Paziewska A, Wyrwicz LS, Ostrowski J. The binding activity of yeast RNAs to yeast Hek2p and mammalian hnRNP K proteins, determined using the three-hybrid system. Cell Mol Biol Lett. 2005;10(2):227–235. [PubMed] [Google Scholar]
- 73.Walter BM, Nordhoff C, Varga G, Goncharenko G, Schneider SW, Ludwig S, Wixler V. Mss4 protein is a regulator of stress response and apoptosis. Cell Death Dis. 2012;3:e297. doi: 10.1038/cddis.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sandri M. Signaling in muscle atrophy and hypertrophy. Physiology. 2008;23:160–170. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- 75.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 76.Teleman AA, Hietakangas V, Sayadian AC, Cohen SM. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 2008;7(1):21–32. doi: 10.1016/j.cmet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 77.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3(11):1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 78.Pallafacchina G, Calabria E, Serrano AL, Kalhovde JM, Schiaffino S. A protein kinase B-dependent and rapamycin-sensitive pathway controls skeletal muscle growth but not fiber type specification. Proc Natll Acad Sci USA. 2002;99(14):9213–9218. doi: 10.1073/pnas.142166599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123(4):569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 80.Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence JC., Jr mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J. 2006;25(8):1659–1668. doi: 10.1038/sj.emboj.7601047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Csibi A, Tintignac LA, Leibovitch MP, Leibovitch SA. eIF3-f function in skeletal muscles: to stand at the crossroads of atrophy and hypertrophy. Cell Cycle. 2008;7(12):1698–1701. doi: 10.4161/cc.7.12.6090. [DOI] [PubMed] [Google Scholar]
- 82.Iadevaia V, Caldarola S, Tino E, Amaldi F, Loreni F. All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA. 2008;14(9):1730–1736. doi: 10.1261/rna.1037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem/FEBS. 2000;267(21):6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 84.Avruch J, Lin Y, Long X, Murthy S, Ortiz-Vega S. Recent advances in the regulation of the TOR pathway by insulin and nutrients. Curr Opin Clin Nutr Metab Care. 2005;8(1):67–72. doi: 10.1097/00075197-200501000-00010. [DOI] [PubMed] [Google Scholar]
- 85.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Csibi A, Cornille K, Leibovitch MP, Poupon A, Tintignac LA, Sanchez AM, Leibovitch SA. The translation regulatory subunit eIF3f controls the kinase-dependent mTOR signaling required for muscle differentiation and hypertrophy in mouse. PLoS ONE. 2010;5(2):e8994. doi: 10.1371/journal.pone.0008994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA. 2001;98(25):14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 89.Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21(1):140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 90.Csibi A, Leibovitch MP, Cornille K, Tintignac LA, Leibovitch SA. MAFbx/Atrogin-1 controls the activity of the initiation factor eIF3-f in skeletal muscle atrophy by targeting multiple C-terminal lysines. J Biol Chem. 2009;284(7):4413–4421. doi: 10.1074/jbc.M807641200. [DOI] [PubMed] [Google Scholar]
- 91.Lagirand-Cantaloube J, Offner N, Csibi A, Leibovitch MP, Batonnet-Pichon S, Tintignac LA, Segura CT, Leibovitch SA. The initiation factor eIF3-f is a major target for atrogin1/MAFbx function in skeletal muscle atrophy. EMBO J. 2008;27(8):1266–1276. doi: 10.1038/emboj.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moretti J, Chastagner P, Gastaldello S, Heuss SF, Dirac AM, Bernards R, Masucci MG, Israel A, Brou C. The translation initiation factor 3f (eIF3f) exhibits a deubiquitinase activity regulating Notch activation. PLoS Biol. 2010;8(11):e1000545. doi: 10.1371/journal.pbio.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Carson JA. The regulation of gene expression in hypertrophying skeletal muscle. Exerc Sport Sci Rev. 1997;25:301–320. doi: 10.1249/00003677-199700250-00013. [DOI] [PubMed] [Google Scholar]
- 94.Baar K, Esser K. Phosphorylation of p70(S6 k) correlates with increased skeletal muscle mass following resistance exercise. Am J Physiol. 1999;276(1 Pt 1):C120–C127. doi: 10.1152/ajpcell.1999.276.1.C120. [DOI] [PubMed] [Google Scholar]