Summary
Despite the apparent absence of genes coding for the known pathways for biosynthesis, the monosaccharide rhamnose was detected in the D configuration in Mycoplasma pneumoniae and Mycoplasma pulmonis, and in both the D and L configurations in Mycoplasma arthritidis. Surprisingly, the monosaccharide glucose was not a precursor for rhamnose biosynthesis and was not incorporated at detectable levels in glucose-containing polysaccharides or glycoconjugates. In contrast, carbon atoms from starch, a polymer of glucose, were incorporated into rhamnose in each of the three species examined. When grown in a serum-free medium supplemented with starch, M. arthritidis synthesized higher levels of rhamnose, with a shift in the relative amounts of the D and L configurations. Our findings suggest the presence of a novel pathway for rhamnose synthesis that is widespread in the genus Mycoplasma.
Keywords: butanolysis, 13C-saccharides, gas chromatography, mass spectrometry, mollicutes
Introduction
Mycoplasmas (Class Mollicutes) are significant pathogens of humans and animals, causing chronic diseases of the respiratory system, genital tract, and joints. Factors that contribute to disease chronicity include polysaccharides that protect the mycoplasma from host defenses and have roles in biofilm formation (Bolland et al., 2012, Shaw et al., 2013, Shaw et al., 2012, Daubenspeck et al., 2009). The mycoplasmas lack a cell wall but have descended from A+T-rich Firmicutes through a process that resulted in a streamlined genome containing about 600–800 protein-coding regions. Extensive bioinformatic analysis of each gene in Mycoplasma arthritidis and Mycoplasma pulmonis suggests that these species have limited machinery for synthesis of monosaccharides and glycoconjugates (Dybvig et al., 2008, French et al., 2008, Daubenspeck et al., 2014). These murine pathogens have only a single annotated gene (Marth-_orf849 and MYPU_7700) coding for a glycosyltransferase and no strong candidate for a nucleotidyltransferase gene. Nevertheless, the mycoplasmas produce glycolipids, polysaccharides, and glycoproteins (Klement et al., 2007, Razin et al., 1970, Chandler et al., 1989, Buttery and Plackett, 1960, Plackett and Buttery, 1958, Daubenspeck et al., 2009, Demina et al., 2009). M. pulmonis and probably Mycoplasma pneumoniae produce a polysaccharide that contains glucose (Daubenspeck et al., 2009, Simmons et al., 2013).
The pathways for synthesis of D- and L-rhamnose have been extensively studied in bacteria and generally involve nucleotide sugar intermediates (Giraud and Naismith, 2000). Of the twenty or so mycoplasma genomes that have been sequenced, no rhamnose synthesis genes have been identified. Nevertheless, we find rhamnose in several species of mycoplasma including the human pathogen M. pneumoniae and the murine pathogens M. arthritidis and M. pulmonis. The presence of rhamnose was unexpected. These mycoplasmas are unlikely to acquire rhamnose from the environment because their mammalian hosts neither synthesize rhamnose nor incorporate it into glycoconjugates. Rhamnose was in the rare D configuration in all three species, and M. arthritidis also contained L-rhamnose. When grown in the presence of [U-13C]starch, the carbon atoms of rhamnose were labeled with 13C, demonstrating rhamnose synthesis. Surprisingly, when the medium was supplemented with D-[U-13C]glucose, no labeling of rhamnose was detected. Hence, rhamnose is not synthesized through a traditional pathway beginning with the conversion of glucose to glucose-6-phosphate (G6P), G6P to glucose-1-phosphate (G1P), and then G1P to UDP-glucose as generally found for L-rhamnose. Similarly, D-rhamnose is not synthesized by the usual pathway in which glucose is converted to G6P and eventually to GDP-mannose via mannose-6-phosphate and mannose-1-phosphate (M1P) (Giraud and Naismith, 2000). Because starch, a polymer of glucose, supports rhamnose synthesis but glucose monosaccharide does not, we propose that the energy from the glycosidic bonds between the glucose residues of starch is a requirement for synthesis.
Results
Mycoplasmas contain D-rhamnose
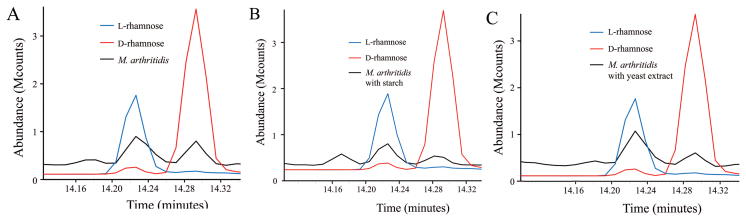
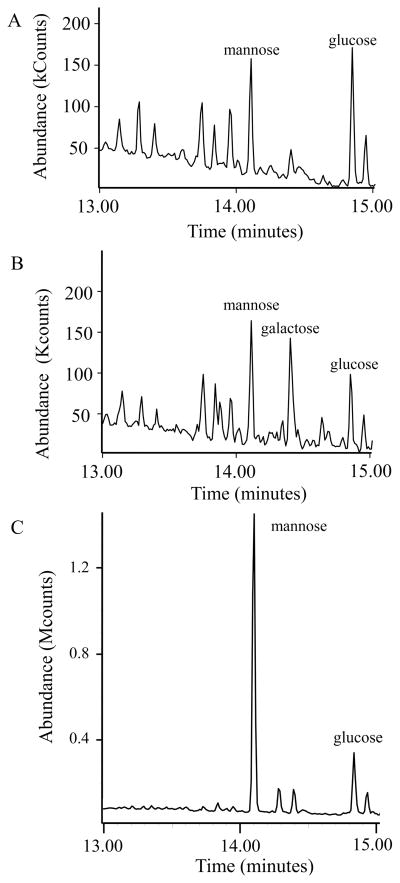
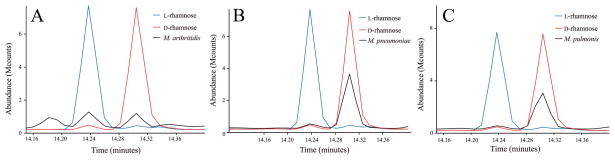
The monosaccharide composition of the glycomoieties (glycolipids, glycoproteins, and polysaccharides) of mycoplasmas was examined. In pilot experiments, lysates of 11 species of Mollicutes were dialyzed extensively to remove monosaccharides, and the remaining glycomoieties were analyzed by GC/MS. The dialyzed lysates of each species (Acholeplasma laidlawii, Mycoplasma arginini, M. arthritidis, Mycoplasma capricolum, Mycoplasma fermentans, Mycoplasma genitalium, Mycoplasma gallisepticum, Mycoplasma hyorhinis, Mycoplasma mycoides subsp. capri, M. pneumoniae, and M. pulmonis) contained the 6-deoxyhexose rhamnose. M. arthritidis was chosen for further study because initial experiments indicated that this species had a relatively high level of rhamnose. In addition to rhamnose, this species also had significant levels of glucose and mannose (Fig. 1). Standard curves of known amounts of these sugars were used to quantitate the amount of each sugar in the lysates (Fig. S1). Most of the rhamnose in nature is in the L configuration, but D-rhamnose has been reported in a few organisms such as Pseudomonas (Rocchetta et al., 1998). The trimethylsilyl (TMS) derivatives of the methyl glycosides of D- and L-rhamnose cannot be resolved by GC/MS, but the TMS derivatives of the butyl glycosides can be. Hence, lysates were subjected to butanolysis and compared to the butyl glycosides generated from standards of D- and L-rhamnose. Both D- and L-rhamnose were detected in M. arthritidis (Fig. 2A). Because the finding of both enantiomers of rhamnose was surprising, the TMS derivatives of the butyl glycosides of M. pneumoniae and M. pulmonis were also analyzed. The rare D configuration predominated in these two species (Fig. 2B and C).
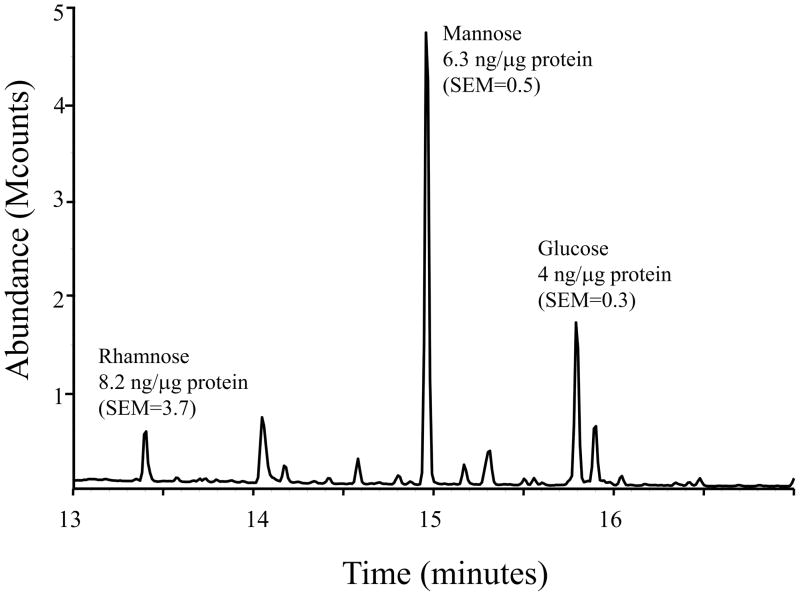
Fig. 1.
Representative chromatogram of dialyzed lysates of M. arthritidis indicating the amount of each sugar per μg protein with standard error of the mean (SEM). Lysates (n =3) contained 20-μg protein.
Fig. 2.
Gas chromatograms of dialyzed lysates of M. arthritidis (A), M. pneumoniae (B), and M. pulmonis (C), subjected to butanolysis to resolve and L-rhamnose. The enantiomers of rhamnose were identified by comparison to standards of each configuration.
13C labeling
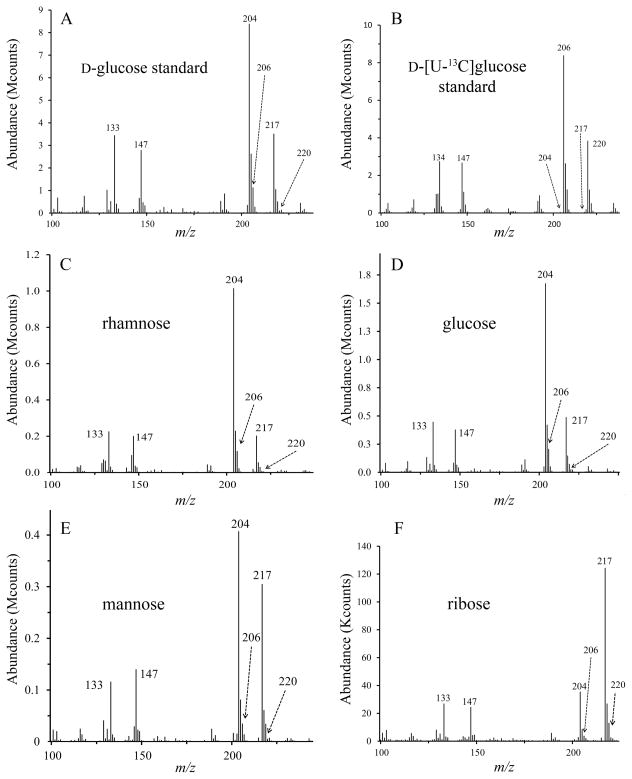
To confirm rhamnose biosynthesis, mycoplasmas were grown in medium supplemented with carbohydrates containing 13C isotopes (universally labeled). When the TMS derivatives of the methyl glycosides of 12C glucose, mannose and rhamnose are fragmented by electron ionization, ions at m/z 133 containing one carbon atom (C1), 204 containing two carbon atoms (C2—C3 or C3—C4) and 217 containing three carbon atoms (C2—C3—C4) are detected by MS (DeJongh et al., 1969). The increased mass of 13C isotopes shifts the more abundant ions to m/z 134, 206 and 220. Also abundant are ions at m/z 147, a known adduct of the TMS reagent used for methanolysis (DeJongh et al., 1969). The 13C labeling of mycoplasmal glycomoieties was examined by GC/MS analysis of dialyzed lysates. The results are summarized in Table 1. When grown in medium supplemented with D-[U-13C]glucose monosaccharide, the abundant ions from glucose, mannose and rhamnose are found at m/z 133, 204 and 217, not m/z 134, 206 and 220 for the three species M. arthritidis, M. pneumoniae and M. pulmonis (Figs. 3, S2 and S3). Thus, the glucose, mannose and rhamnose residues of the glycomoieties were not labeled with 13C. The mass spectra of TMS derivatives of the methyl glycosides of ribose standards were analyzed and had ions at m/z 133, 204 and 217, as expected for fragmentation of pentose sugars (Kochetkov and Chizhov, 1965, Lawson et al., 1971). Ribose was detected in the mycoplasmal samples grown in D-[U-13C]glucose, suggesting that some nucleic acids were not removed by nuclease digestion and dialysis. The ions from the TMS derivatives of the methyl glycosides of ribose were found at m/z 133, 204 and 217 in the case of M. arthritidis but at 134, 206 and 220 for M. pneumoniae and M. pulmonis. Thus, ribose was labeled with 13C in the glycolytic species of mycoplasma only, indicating that glucose was imported and metabolized in these species but not in the non-glycolytic M. arthritidis.
Table 1.
13C labeling (ratio of 13C to 12C ions) of the indicated sugars in M. arthritidis, M. pulmonis, and M. pneumoniae when medium is supplemented with 13C-glucose or 13C-starch.
| M. arthritidis | M. pulmonis | M. pneumoniae | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||||
| Ion ratios |
|
|
|
|
|
|
|
|
|
|||||||||
| 13C-glucose supplement | Rhamnose | 1a | 1 | 1 | 2 | 1 | 3b | 2 | 1 | 7b | ||||||||
| Glucose | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||
| Mannose | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||
| Ribose | 1 | 1 | 1 | 4 | 6 | 17 | 17 | 6 | 17 | |||||||||
| Galactose | NAc | NA | NA | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||
|
| ||||||||||||||||||
| 13C-starch supplement | Rhamnose | 9 | 290 | 600 | 7 | 25 | 467 | 7 | 21 | 43 | ||||||||
| Glucose | 27 | 2,300 | 16,000 | 26 | 820 | 6,000 | 26 | 1,100 | 8,300 | |||||||||
| Mannose | 2 | 3 | 7 | 1 | 1 | 1 | 1 | 1 | 2 | |||||||||
| Ribosed | 1 | 2 | 1 | 2 | 2 | 3 | 2 | 5 | 2 | |||||||||
| Galactose | NA | NA | NA | 5 | 5 | 13 | 5 | 27 | 7 | |||||||||
Each ion ratio for the indicated sugar was divided by the ion ratio of the 12C standard to normalize. For both 12C glucose and 12C starch, the ratio of 134 to 133 ions, the ratio of 206 to 204 ions, and the ratio of 220 to 217 ions was 0.1, 0.1, and 0.03, respectively. For comparison, the ratio of 134 to 133 ions, 206 to 204 ions, and 220 to 217 ions was 2.7, 220, and 530, respectively, for 13C glucose. The ratio of 134 to 133 ions, 206 to 204 ions, and 220 to 217 ions was 2.9, 160, and 300, respectively, for 13C starch.
Apparent increase in ion ratio attributed to labeled ribose (major peak) overlapping the major rhamnose peak in the chromatogram.
NA, Not applicable, galactose not detected in this species.
There is no issue of labeled rhamnose overlapping the ribose peak because the ribose minor peak was used for this calculation.
Fig. 3.
Mass spectra of TMS derivatives of the methyl glycosides from M. arthritidis cells grown in medium supplemented with D-[U-13C]glucose. (A) 12C glucose standard, (B) D-[U-13C]glucose standard, (C) rhamnose, (D) glucose, (E) mannose, (F) ribose.
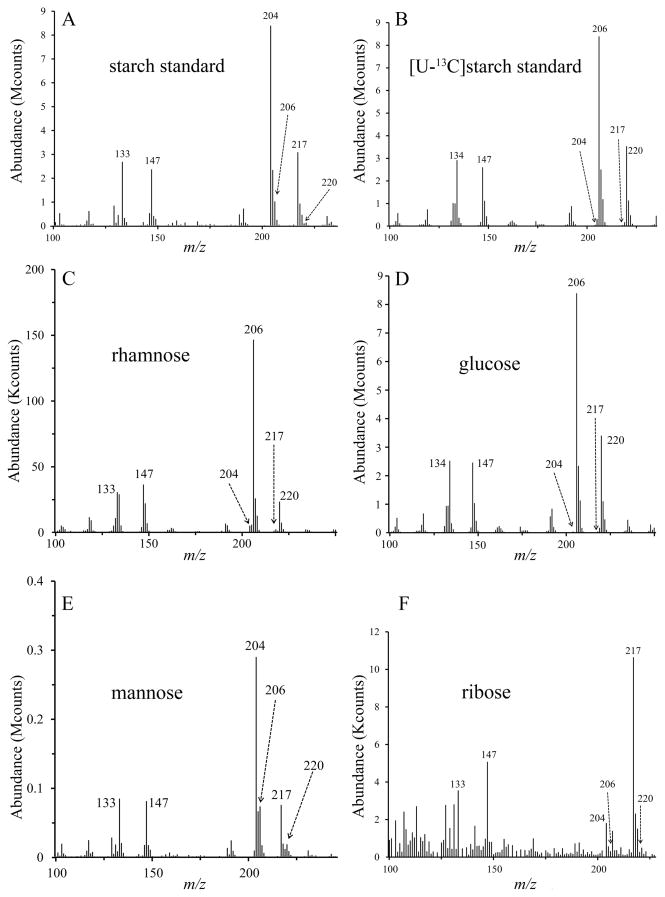
The failure of even the glucose residues of the glycomoieties of these species to become labeled when the medium was supplemented with D-[U-13C]glucose demonstrated that monosaccharides were not utilized for synthesis of these macromolecules. The commercial availability of [U-13C]starch provided a resource to determine whether glucose polymers would support the synthesis of rhamnose. In contrast to the results obtained with D-[U-13C]glucose monosaccharide, cells grown in the presence of [U-13C]starch yielded 13C-labeled rhamnose as evidenced by the abundant ions at m/z 134, 206 and 220 (Fig. 4C, and panel C of Figs. S4 and S5). We believe that [U-13C]starch also labeled glucose (Fig. 4D, and panel D of Figs. S4 and S5) but no firm conclusion could be reached because some of the starch molecules did not remain in solution during the course of the experiment and were harvested along with the mycoplasma cells. [U-13C]starch did not label mannose in M. pneumoniae and M. pulmonis but perhaps slightly did so in M. arthritidis (Fig. 4E, and panel E of Figs. S4 and S5). In contrast to D-[U-13C]glucose, ribose was not labeled appreciably by [U-13C]starch (Fig. 4F, and panel F of Figs. S4 and S5), suggesting that monosaccharides and polysaccharides are metabolized distinctly. Galactose was absent in lysates of M. arthritidis but found in M. pneumoniae and M. pulmonis. Galactose in both M. pneumoniae and M. pulmonis was labeled in cultures supplemented with [U-13C]starch but not D-[U-13C]glucose (panel G of Figs. S2–S5).
Fig. 4.
Mass spectra of TMS derivatives of the methyl glycosides from M. arthritidis cells grown in medium supplemented with [U-13C]starch. (A) 12C starch standard, (B) [U-13C]starch standard, (C) rhamnose, (D) glucose, (E) mannose, (F) ribose.
Relative amounts of D- and L-rhamnose are dependent on medium composition
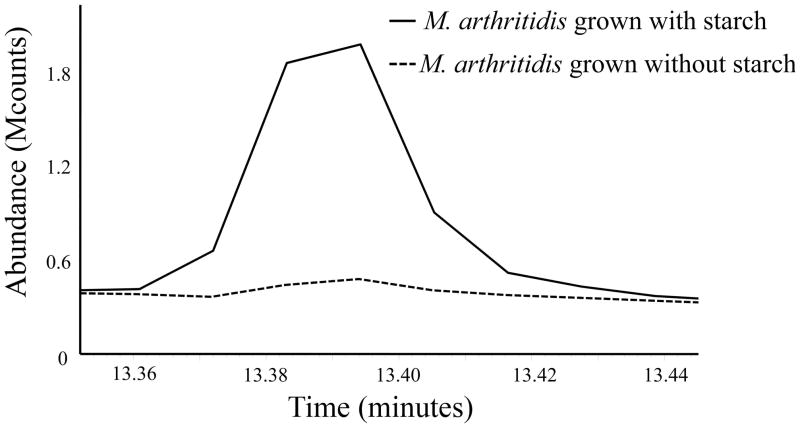
The amount of rhamnose in M. arthritidis increased by about 10-fold when serum-free medium (SFM) was supplemented with starch (Fig. 5), with an increase in the relative abundance of L-rhamnose (Fig. 6). These results are in contrast to the finding of nearly equal amounts of D- and L-rhamnose in M. arthritidis cells grown in medium without starch (Figs. 2A and 6A). A medium supplement commonly used for growth of mycoplasmas is yeast extract. When yeast extract was added to SFM, there was a similar increase in the relative amount of L-rhamnose (Fig. 6C).
Fig. 5.
Representative chromatogram showing the abundance of rhamnose from cells grown in medium with and without starch. Dialyzed lysates containing 20-μg protein were analyzed by GC/MS.
Fig. 6.
Gas chromatograms of dialyzed lysates of M. arthritidis grown in SFM (A), SFM supplemented with starch (B), or SFM supplemented with yeast extract (C) and subjected to butanolysis to resolve D- and L-rhamnose. The enantiomers of rhamnose were identified by comparison to standards of each configuration.
Medium glycomoieties
Rhamnose was detected in all species of mycoplasma examined, even when starch was absent from the growth medium. Because the mycoplasmas did not convert glucose monosaccharide into rhamnose, the growth medium must supply oligosaccharides or glycoconjugates that would support rhamnose biosynthesis. Some of the media formulations used in this study contained serum and yeast extract, which is a component of mycoplasma broth base. Serum and yeast extract have an abundance of glycoproteins that could serve as starting material for rhamnose synthesis. SFM would lack these glycoproteins. Peptone and bovine serum albumin (BSA) were major ingredients of the SFM. All SFM components were dialyzed to remove monosaccharides and analyzed by GC/MS to examine the composition of any glycoconjugates that might be present. Dialyzed peptone and BSA were the only reagents that contained significant levels of glucose and mannose, and the peptone also contained an abundant level of galactose (Fig. 7A and B). Hence, the SFM did contain glycoconjugates of mammalian origin that might support a minimal level of rhamnose synthesis in the absence of a glucose polymer (starch). Also analyzed by GC/MS was the yeast extract that had been used in some experiments to supplement SFM. Yeast extract contained substantial quantities of mannose and glucose (Fig. 7C). We note that no rhamnose whatsoever was detected in BSA, peptone, yeast extract, or any of the other reagents used in the media formulations for study.
Fig. 7.
Gas chromatograms of dialyzed (A) BSA (1 mg), (B) peptone (2.5 mg), and (C) yeast extract (3 mg). Due to a column replacement, the retention times of the TMS derivatives of the glycosides in this figure vary from other figures.
Rhamnose partitioning in chloroform-methanol
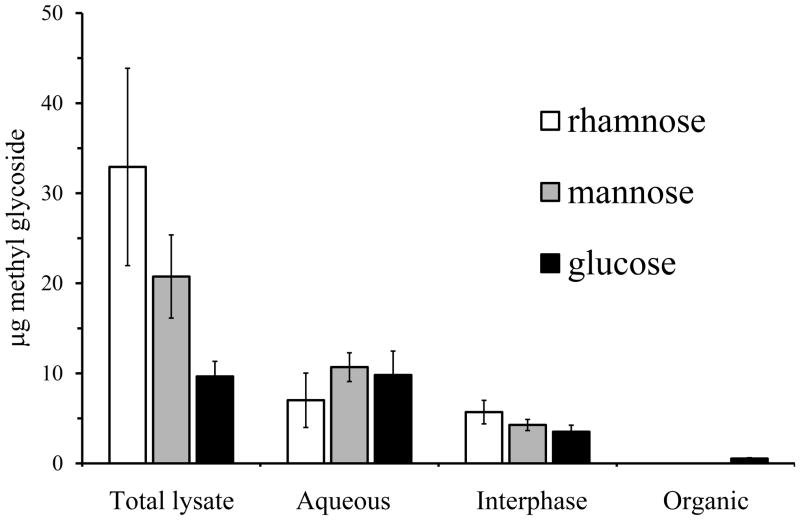
Dialyzed lysates of M. arthritidis were extracted with chloroform-methanol. GC analysis was performed on the material that partitioned into the aqueous and organic phases as well as the insoluble material at the interphase. With the extraction protocol, glycolipids partition into the organic phase and polysaccharide into the aqueous phase. Some protein should be found in the aqueous phase but most of it is in the interphase (Wessel and Flugge, 1984). No rhamnose was detected in the organic phase, indicating that rhamnose is not a component of glycolipid (Fig. 8). Glucose was found in the organic phase, consistent with a previous report of glucolipid in this species (Li et al., 1997). Glucose, mannose and rhamnose were found in both the aqueous phase and the interphase.
Fig. 8.
Phase partitioning of M. arthritidis lysate in chloroform-methanol (n = 4). Error bars are standard error of the mean.
Discussion
Over 40 species of mycoplasma are listed in the NCBI genome projects database (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/microbial_taxtree.html). There are few genes annotated as coding for nucleotidyltransferases, glycosyltransferases, and other enzymes associated with sugar synthesis (Daubenspeck et al., 2014). However, it is clear that mycoplasmas synthesize polysaccharides and glycolipids. M. mycoides synthesizes a galactan (Plackett and Buttery, 1958), M. pulmonis synthesizes a capsule that is antiphagocytic and protects from complement (Daubenspeck et al., 2009, Bolland et al., 2012, Shaw et al., 2013), and M. pneumoniae synthesizes an adhesive polysaccharide necessary for robust biofilm formation (Simmons et al., 2013). Possibly all species of mycoplasma produce glycolipids. For M. pneumoniae, the glycolipids contain galactose or glucose (Klement et al., 2007). These glycolipids are thought to be important for pathogenesis through their involvement in molecular mimicry (Ang et al., 2002, Yuki, 2007, Kitazawa et al., 1998, Kusunoki et al., 2001).
Rhamnose biosynthesis is well studied in bacteria (Giraud and Naismith, 2000). This 6-deoxy monosaccharide is common in bacteria and also found in some plants and eukaryotic microbes but not in mammals. The stereoisomers D- and L-rhamnose are mirror images, with the L-form being significantly more common. Generally, these enantiomers are synthesized by two distinct pathways. G6P is converted to G1P, which is used for production of dTDP-glucose to initiate the pathway for synthesis of L-rhamnose. Synthesis of D-rhamnose involves the production of GDP-mannose from M1P. A hydrolase converts the nucleotide sugar to a 4-keto-6-deoxyhexose. For L-rhamnose synthesis, the RmlC enzyme catalyzes an isomerase reaction that generates dTDP-L-lyxo-6-deoxy-4-hexulose. The final step in synthesis is shared between the two pathways and is a NADPH-dependent reaction that reduces the ketone at the C4 position, giving rise to dTDP-L-rhamnose or GDP-D-rhamnose. None of the species of mycoplasma for which the genome sequence is available contain any homologs of known rhamnose synthesis machinery.
Although rhamnose was previously reported to be present in M. pneumoniae (Allen and Prescott, 1978), it was unexpected to find rhamnose throughout the Mycoplasma genus because of the lack of recognizable rhamnose synthesis genes in any species, including M. pneumoniae. A bigger surprise was that the mycoplasmas produce D-rhamnose, an exceedingly rare carbohydrate. The glycolytic species M. pneumoniae and M. pulmonis had the D-form only while the non-glycolytic M. arthritidis had both D- and L-rhamnose. The failure of carbon isotopes from D-[U-13C]glucose monosaccharide to be incorporated into rhamnose suggested a novel pathway for synthesis that did not involve the phosphorylation of glucose to G6P followed by its conversion to G1P or M1P. The glycolytic species do import glucose monosaccharide, converting it to G6P (Pollack et al., 1983). In these species, ribose was labeled by D-[U-13C]glucose monosaccharide, demonstrating that hexoses can be converted to pentoses as suggested by bioinformatic analysis. In contrast, ribose was not labeled with D-[U-13C]glucose monosaccharide in M. arthritidis, and this species might not import monosaccharides.
[U-13C]starch labeled rhamnose and galactose but not mannose and ribose in M. pneumoniae and M. pulmonis. Bioinformatic analysis indicates that these species are missing phosphoglucomutase and glucose phosphatases. Hence, the mycoplasmas should not be able to interconvert G6P and G1P or generate glucose from either hexose phosphate. The mycoplasmas presumably hydrolyze starch to oligosaccharides that are imported. Perhaps G1P is produced in mycoplasmas only by transfer of phosphate to the hexose as the glycosidic bond between the sugar residues of an imported oligosaccharide is broken, similar to the proposed production of galactose-1-P in bifidobacteria (Yamamoto, 2012). The resulting G1P is then used to support synthesis of rhamnose and glycoconjugates. Because ribose was not labeled with [U-13C]starch, glycolysis and synthesis of glycoconjugates may occur by separate pathways with no interconversion of G6P and G1P as predicted from bioinformatics. It is also possible that the mycoplasmas transfer the energy from the glycosidic bond of oligosaccharides to something other than G1P, such as a lipid-linked sugar (Lairson et al., 2008). The failure of mannose to be labeled by D-[U-13C]glucose or [U-13C]starch suggests that there is no interconversion of glucose and mannose, the two primary sugars found in the glycomoieties of SFM. The requirement for oligosaccharides as substrate for glycosyltransferase reactions instead of nucleotide sugars would explain the failure of homology searches to identify the synthesis machinery (Henrissat et al., 2008).
BSA and peptone are the only possible sources of mannose and glucose glycoconjugates in the SFM and must contain the appropriate precursors to synthesize both L- and D-rhamnose in M. arthritidis. The ability of medium supplements such as yeast extract and starch to vary the relative amounts of D- and L-rhamnose in M. arthritidis suggests that some glycomoieties in the medium support synthesis of L-rhamnose and others support D-rhamnose. Perhaps oligosaccharides, such as those derived from α-linked polymers of starch, primarily support the synthesis of L-rhamnose via a pathway that does not involve nucleotide sugars but nevertheless includes dehydratase, epimerase and reductase reactions similar to the conversion of UDP-glucose to dTDP-L-rhamnose in other organisms. D-rhamnose in mycoplasmas may be synthesized from a different glycan, such as a mannose glycoconjugate, similar to the conversion of GDP-mannose to GDP-D-rhamnose in other organisms. In this scenario, one might have predicted that yeast extract, which contains high levels of mannose, would primarily support the synthesis of D-rhamnose, which was not the case. The mannose-containing oligosaccharides in yeast extract might have linkages that are not used or used only inefficiently by the mycoplasma.
Although bioinformatic analysis suggests M. arthritidis and M. pulmonis cannot convert G1P to UDP-glucose, M. pneumoniae has the required nucleotidyltransferase for this reaction. M. pneumoniae, but not the other two species, also has the GalE enzyme that can interconvert UDP-glucose and UDP-galactose. M. pneumoniae has three annotated glycosyltransferases while the other species have only one. One of the glycosyltransferases of M. pneumoniae is known to catalyze synthesis of glycolipids and can use UDP-glucose and UDP-galactose as a substrate (Klement et al., 2007). The pathway for glycolipid synthesis in species of mycoplasma that lack recognizable nucleotidyltransferases has not been investigated.
Chloroform-methanol partitioning of M. arthritidis indicates that rhamnose is not a component of glycolipid. We have no evidence to support rhamnose as being a residue of polysaccharides produced by M. pneumoniae or M. pulmonis (Simmons et al., 2013, Daubenspeck et al., 2009). There is evidence for glycoproteins in mycoplasmas (Demina et al., 2009), and the finding of rhamnose at the interphase after extraction with chloroform-methanol suggests that rhamnose may be attached to protein. We are currently investigating protein glycosylation in M. arthritidis to determine whether rhamnose, glucose and mannose are associated with glycoproteins.
M. pneumoniae and M. pulmonis are considered to be primarily respiratory pathogens of humans and murine animals, respectively, while M. arthritidis is a murine pathogen that persists in joints and causes arthritis. These species are found only in a mammalian host, which provides sterols and other nutrients that are essential for growth. These mycoplasmas have evolved to acquire sugars from host molecules such as glycoproteins and glycosaminoglycans. Bioinformatic analysis indicates that the mycoplasmas produce several glycosidases, which might generate the oligosaccharides to support glycoconjugate synthesis. The bovine pathogen Mycoplasma dispar reportedly has a capsule that is induced by coculture with host cells (Almeida and Rosenbusch, 1991), perhaps because the host cells provided glycans that supported polysaccharide synthesis. The strategy of using host oligosaccharides for glycoconjugate synthesis would be favorable from the standpoint of energy conservation and have pathogenic consequences as glycans are stripped from host molecules.
Experimental procedures
Strains and cultures
For initial pilot experiments, GC/MS analysis was performed on Mycoplasma fermentans strain PG18 (ATCC 19989), M. mycoides subsp. capri strain GM9 (Voelker et al., 1995), M. capricolum ATCC 27343, A. laidlawii strain JA1 (Liss and Maniloff, 1973), M. gallisepticum strain PG31 (ATCC 19610), M. pneumoniae strain M129 (ATCC 29342), M. genitalium strain G37 (ATCC 33530), M. arginini ATCC 23838, M. hyorhinis strain GDL (ATCC 23839), M. arthritidis strain 158 (Dybvig and Khaled, 1990), and M. pulmonis strain CTG (Daubenspeck et al., 2009). The identity of the mycoplasma species was confirmed by amplifying and sequencing the 16S rRNA gene of each strain. For these pilot studies only, the culture media was mycoplasma broth (MB), prepared as described elsewhere (Dybvig et al., 2010, Dybvig et al., 2008). For growth of the glycolytic species, the MB contained 0.5% dextrose and had a starting pH of 7.8. For growth of the two non-glycolytic species, M. arginini and M. arthritidis, the only change in the preparation of the MB was that the medium contained 0.5% arginine-HCl in place of dextrose and the pH was adjusted to 7.4.
Other than the pilot experiments, mycoplasmas were propagated in SFM similar to that described by Yus et al. (Yus et al., 2009). The following were mixed in 1 liter of water: 25 ml 20% arginine-HCl for non-glycolytic species of mycoplasma (M. arthritidis) or 10 ml 50% dextrose for glycolytic species (M. pneumoniae and M. pulmonis), 13.5 g Dulbecco’s Modified Eagle’s Medium (DMEM, Sigma), 6 ml Isovitalex (Becton Dickinson), 1.2 ml 20% DNA (degraded herring sperm, Sigma), 50 mg ampicillin, 2.5 g Select Peptone 140 (Life Technologies), 0.2 mg α-lipoic acid (Sigma), 20 mg uracil (Sigma), 20 mg spermine, 0.5 g glycerol, and 6.8 g amino acid mixture. The amino acid mixture was prepared by mixing 7.1 g L-alanine, 12.5 g L-arginine, 10.6 g L-asparagine, 5.4 g glycine, 11.8 g L-histidine, 8.4 g L-isoleucine, 8.4 g L-leucine, 9.3 g L-lysine, 23.2 g L-methionine, 2.0 g L-phenylalanine, 9.2 g L-proline, 1.3 g L-serine, 7.6 g L-threonine, 2.0 g L-tryptophan, 0.4 g L-tyrosine, and 16.8 g L-valine (all amino acids supplied by Sigma). The SFM mixture was adjusted to pH 7.4 for growth of M. arthritidis or pH 7.8 for growth of glycolytic species and sterilized by filtration. SFM was completed by adding lipid and cholesterol suspensions. The lipid suspension was made by dissolving 11 mg palmitic acid, 11 mg oleic acid, 11 mg linoleic acid, 11 mg glyceryl tripalmitate, 11 mg glyceryl trioleate, and 11 mg glyceryl trilinoleate in 0.5 ml 100% ethanol, heating to 80°C, and injecting it into 1 ml 10 mM HEPES buffer (pH was 7.4) also heated to 80°C. The cholesterol suspension was made by dissolving 20 mg cholesterol in 0.5 ml ethanol, heating to 80°C, and injecting it into 1 ml 10 mM HEPES heated to 80°C. The lipid and cholesterol suspensions were each added to filter-sterilized 20 ml HEPES (pH 7.4) with 1 g BSA. Both of the suspensions were incubated at room temperature for 30 minutes with occasional mixing with a vortex before adding to the SFM mixture. All lipids and cholesterol were supplied by Sigma. Injection is defined as pipetting vigorously at the bottom of the solvent tube. It is important for growth that the lipids and cholesterol be in a suspended state. If the suspensions do not remain in a suspended state after the 30-minute incubation, repeat the process. These suspensions were made fresh each time for optimal growth. For some experiments SFM was supplemented with starch (3.3 mg per ml culture medium) or yeast extract (Invitrogen, 6.6 mg per ml medium).
For labeling with 13C glucose or 13C starch, a low-glucose SFM (LG-SFM) was prepared as described for SFM except that Low Glucose DMEM was used instead of DMEM and a vitamin supplement was devised for use in place of glucose-containing Isovitalex. A 167x stock solution of vitamin supplement was prepared in 10 mM HEPES buffered at pH 7.2 in a total volume of 10 ml with 0.1 mg cobalamine, 0.13 mg para-aminobenzoic acid, 2.5 mg nicotinamide adenine dinucleotide, 10 mg L-cystine, 6.7 mg pyridoxine, 184 mg sodium pyruvate, 0.03 mg thiamine and 1 mg thiamine phosphate (all supplied by Sigma). The mixture was filter sterilized and stored in 1ml aliquots at 20°C until use.
Lysate preparation
Cultures were harvested (late-logarithmic to stationary growth phase) and washed three times with phosphate-buffered saline (PBS). The cells were suspended in 1 ml digestion buffer (100 mM Tris-Hcl, 10 mM MgCl2, 5 mM NaN3, pH 7) and sonicated at full power at 90% duty cycle on a Branson Sonifier 450 for 30 seconds. For experiments employing nuclease and protease digestions, lysates were heated to 80°C for 15 minutes to denature protein, digested overnight with 25 μg RNaseA and 25 μg DNaseI, and then digested overnight with 25 μg proteinase K.
GC/MS
Lysates were subjected to methanolysis to generate methyl glycosides that were analyzed by GC/MS. To accomplish this, lysates were first dialyzed with 5,000 volumes of water (Millipore) in 2,000 molecular-weight cutoff dialysis cassettes (Pierce) and then dessicated. The samples were subjected to methanolysis by treatment with 0.4 ml acidic methanol (1:10 acetyl chloride in methanol) at 80°C for at least 16 hours and dried again. The resulting methyl glycosides were dissolved in 150 μl methanol, transferred to polyspring inserts in glass tubes, evaporated and sealed under argon gas, and injected with 50 μl of reagent from the HMDS + TMCS + Pyridine, 3:1:9 (Sylon™ HTP) Kit (Sigma). The resulting TMS derivatives were analyzed by GC/MS with an Agilent Technologies 6890N Network GC System and a 5973 Network Mass Selective Detector with MSD Productivity Chemstation Software. The instrument injected 0.20 μl of sample into the 30-meter column holding a temperature of 70°C and after 5 minutes the temperature was increased to 220°C at 20°C/minute, and then increased to 275°C at 10°C/minute where it was maintained for 10 minutes. For GC/MS identification, standards consisted of D-arabinose, D-galactose, D-glucose, D-fucose, D-mannose, D-rhamnose, L-rhamnose, D-ribose, and D-xylose. All standards were obtained from Sigma, except D-rhamnose, which was obtained from Carbosynth Ltd.
Butanolysis
Butanolysis of methyl glycosides was performed as previously described using (R)-(-)-2-butanol (Gerwig et al., 1978). Lysates containing 125-μg protein were subjected to methanolysis as described above. The methyl glycosides were dried in a new ampule and 0.5 ml acidic butanol (1:10 acetyl chloride in butanol) was added and the ampule was sealed and kept at 80° C for 8 hours. The sample was dried again and analyzed by GC/MS as described above. Controls consisted of standards of D- and L-rhamnose.
Isotopes
D-[U-13C]glucose and [U-13C]starch were obtained from Cambridge Isotopes Laboratories, Inc. Six mg D-[U-13C]glucose in 1 ml water was added to 10 ml LG-SFM. In other experiments, 33 mg [U-13C]starch in 1 ml water was added to 10 ml LG-SFM cultures that were also supplemented with 6 mg unlabeled glucose.
Acid hydrolysis of starch
For solubilization, 3.3 mg/ml starch was treated in 1N HCl at 80°C and vortexed every 15 minutes until it remained in solution. The pH was then adjusted to 7.2 with NaOH.
Lipid extraction
Lysates were extracted with chloroform and methanol by the method of Bligh and Dyer (Bligh and Dyer, 1959) to separate lipids, which partition into the organic phase, from hydrophilic material that partitioned into the aqueous phase.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (AI63909, AR44252 and AI93750). We thank Portia Caldwell for technical assistance and Warren Simmons for helpful suggestions.
References
- Allen PZ, Prescott B. Immunochemical studies on a Mycoplasma pneumoniae polysaccharide fraction: cross-reactions with type 23 and 32 antipneumococcal rabbit sera. Infect Immun. 1978;20:421–429. doi: 10.1128/iai.20.2.421-429.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida RA, Rosenbusch RF. Capsulelike surface material of Mycoplasma dispar induced by in vitro growth in culture with bovine cells is antigenically related to similar structures expressed in vivo. Infect Immun. 1991;59:3119–3125. doi: 10.1128/iai.59.9.3119-3125.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang CW, Tio-Gillen AP, Groen J, Herbrink P, Jacobs BC, Van Koningsveld R, et al. Cross-reactive anti-galactocerebroside antibodies and Mycoplasma pneumoniae infections in Guillain–Barré syndrome. J Neuroimmunol. 2002;130:179–183. doi: 10.1016/s0165-5728(02)00209-6. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bolland JR, Simmons WL, Daubenspeck JM, Dybvig K. Mycoplasma polysaccharide protects against complement. Microbiology. 2012;158:1867–1873. doi: 10.1099/mic.0.058222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery SH, Plackett P. A specific polysaccharide from Mycoplasma mycoides. J Gen Microbiol. 1960;23:357–368. doi: 10.1099/00221287-23-2-357. [DOI] [PubMed] [Google Scholar]
- Chandler DK, Olson LD, Kenimer JG, Probst PG, Rottem S, Grabowski MW, et al. Biological activities of monoclonal antibodies to Mycoplasma pneumoniae membrane glycolipids. Infect Immun. 1989;57:1131–1136. doi: 10.1128/iai.57.4.1131-1136.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubenspeck JM, Bolland JR, Luo W, Simmons WL, Dybvig K. Identification of exopolysaccharide-deficient mutants of Mycoplasma pulmonis. Mol Microbiol. 2009;72:1235–1245. doi: 10.1111/j.1365-2958.2009.06720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubenspeck JM, Jordan DS, Dybvig K. The glycocalyx of Mollicutes. In: Browning GF, Citti C, editors. Mollicutes: Molecular Biology and Pathogenesis. Norfolk, UK: Caister Academic Press; 2014. pp. 131–147. [Google Scholar]
- DeJongh DC, Radford T, Hribar JD, Hanessian S, Bieber M, Dawson G, et al. Analysis of trimethylsilyl derivatives of carbohydrates by gas chromatography and mass spectrometry. J Am Chem Soc. 1969;91:1728–1740. [Google Scholar]
- Demina IA, Serebryakova MV, Ladygina VG, Rogova MA, Zgoda VG, Korzhenevskyi DA, et al. Proteome of the bacterium Mycoplasma gallisepticum. Biochem (Moscow) 2009;74:165–174. doi: 10.1134/s0006297909020072. [DOI] [PubMed] [Google Scholar]
- Dybvig K, Khaled M. Isolation of a second cryptic plasmid from Mycoplasma mycoides subsp. mycoides. Plasmid. 1990;24:153–155. doi: 10.1016/0147-619x(90)90018-8. [DOI] [PubMed] [Google Scholar]
- Dybvig K, Lao P, Jordan DS, Simmons WL. Fewer essential genes in mycoplasmas than previous studies suggest. FEMS Microbiol Lett. 2010;311:51–55. doi: 10.1111/j.1574-6968.2010.02078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybvig K, Zuhua C, Lao P, Jordan DS, French CT, Tu AHT, et al. Genome of Mycoplasma arthritidis. Infect Immun. 2008;76:4000–4008. doi: 10.1128/IAI.00516-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CT, Lao P, Loraine AE, Matthews BT, Yu H, Dybvig K. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol Microbiol. 2008;69:67–76. doi: 10.1111/j.1365-2958.2008.06262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwig GJ, Kamerling JP, Vliegenthart JFG. Determination of the D and L configuration of neutral monosaccharides by high-resolution capillary G.L.C. Carbohydr Res. 1978;62:349–357. [Google Scholar]
- Giraud MF, Naismith JH. The rhamnose pathway. Curr Opin Struct Biol. 2000;10:687–696. doi: 10.1016/s0959-440x(00)00145-7. [DOI] [PubMed] [Google Scholar]
- Henrissat B, Sulzenbacher G, Bourne Y. Glycosyltransferases, glycoside hydrolases: surprise, surprise! Curr Opin Struct Biol. 2008;18:527–533. doi: 10.1016/j.sbi.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Kitazawa K, Tagawa Y, Honda A, Yuki N. Guillain–Barré syndrome associated with IgG anti-GM1b antibody subsequent to Mycoplasma pneumoniae infection. J Neurol Sci. 1998;156:99–101. doi: 10.1016/s0022-510x(98)00020-3. [DOI] [PubMed] [Google Scholar]
- Klement ML, Ojemyr L, Tagscherer KE, Widmalm G, Wieslander A. A processive lipid glycosyltransferase in the small human pathogen Mycoplasma pneumoniae: involvement in host immune response. Mol Microbiol. 2007;65:1444–1457. doi: 10.1111/j.1365-2958.2007.05865.x. [DOI] [PubMed] [Google Scholar]
- Kochetkov NK, Chizhov OS. Mass spectrometry of methylated methyl glycosides: Principles and analytical application. Tetrahedron. 1965;21:2029–2047. [Google Scholar]
- Kusunoki S, Shiina M, Kanazawa I. Anti-Gal-C antibodies in GBS subsequent to mycoplasma infection: Evidence of molecular mimicry. Neurology. 2001;57:736–738. doi: 10.1212/wnl.57.4.736. [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- Lawson AM, Stillwell RN, Tacker MM, Tsuboyama K, McCloskey JA. Mass spectrometry of nucleic acid components. Trimethylsilyl derivatives of nucleotides. J Amer Chem Soc. 1971;93:1014–1023. doi: 10.1021/ja00733a039. [DOI] [PubMed] [Google Scholar]
- Li JL, Matsuda K, Takagi M, Yamamoto N. Detection of serum antibodies against phosphocholine-containing aminoglycoglycerolipid specific to Mycoplasma fermentans in HIV-1 infected individuals. J Immunol Methods. 1997;208:103–113. doi: 10.1016/s0022-1759(97)00135-x. [DOI] [PubMed] [Google Scholar]
- Liss A, Maniloff J. Infection of Acholeplasma laidlawii by MVL51 virus. Virology. 1973;55:118–126. doi: 10.1016/s0042-6822(73)81013-x. [DOI] [PubMed] [Google Scholar]
- Plackett P, Buttery SH. A galactan from Mycoplasma mycoides. Nature. 1958;182:1236–1237. doi: 10.1038/1821236a0. [DOI] [PubMed] [Google Scholar]
- Pollack JD, V, Tryon V, Beaman KD. The metabolic pathways of Acholeplasma and Mycoplasma: an overview. Yale J Biol Med. 1983;56:709–716. [PMC free article] [PubMed] [Google Scholar]
- Razin S, Prescott B, Caldes G, James WD, Chanock RM. Role of glycolipids and phosphatidylglycerol in the serological activity of Mycoplasma pneumoniae. Infect Immun. 1970;1:408–416. doi: 10.1128/iai.1.4.408-416.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchetta HL, Pacan JC, Lam JS. Synthesis of the A-band polysaccharide sugar D-rhamnose requires Rmd and WbpW: identification of multiple AlgA homologues, WbpW and ORF488, in Pseudomonas aeruginosa. Mol Microbiol. 1998;29:1419–1434. doi: 10.1046/j.1365-2958.1998.01024.x. [DOI] [PubMed] [Google Scholar]
- Shaw BM, Daubenspeck JM, Simmons WL, Dybvig K. EPS-I polysaccharide protects Mycoplasma pulmonis from phagocytosis. FEMS Microbiol Lett. 2013;338:155–160. doi: 10.1111/1574-6968.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw BM, Simmons WL, Dybvig K. The Vsa shield of Mycoplasma pulmonis is antiphagocytic. Infect Immun. 2012;80:704–709. doi: 10.1128/IAI.06009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons WL, Daubenspeck JM, Osborne JD, Balish MF, Waites KB, Dybvig K. Type 1 and type 2 strains of Mycoplasma pneumoniae form different biofilms. Microbiology. 2013;159:737–747. doi: 10.1099/mic.0.064782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker LL, Weaver KE, Ehle LJ, Washburn LR. Association of lysogenic bacteriophage MAV1 with virulence of Mycoplasma arthritidis. Infect Immun. 1995;63:4016–4023. doi: 10.1128/iai.63.10.4016-4023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. Biological analysis of the microbial metabolism of hetero-oligosaccharides in application to glycotechnology. Biosci Biotechnol Biochem. 2012;76:1815–1827. doi: 10.1271/bbb.120401. [DOI] [PubMed] [Google Scholar]
- Yuki N. Ganglioside mimicry and peripheral nerve disease. Muscle Nerve. 2007;35:691–711. doi: 10.1002/mus.20762. [DOI] [PubMed] [Google Scholar]
- Yus E, Maier T, Michalodimitrakis K, van Noort V, Yamada T, Chen WH, et al. Impact of genome reduction on bacterial metabolism and its regulation. Science. 2009;326:1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.