SUMMARY
The extremely thermoacidophilic archaea are a particularly intriguing group of microorganisms that must simultaneously cope with biologically extreme pHs (≤ 4) and temperatures (Topt ≥ 60°C) in their natural environments. Their expandi ng biotechnological significance relates to their role in biomining of base and precious metals and their unique mechanisms of survival in hot acid, at both the cellular and biomolecular levels. Recent developments, such as advances in understanding of heavy metal tolerance mechanisms, implementation of a genetic system, and discovery of a new carbon fixation pathway, have been facilitated by availability of genome sequence data and molecular genetic systems. As a result, new insights into the metabolic pathways and physiological features that define extreme thermoacidophily have been obtained, in some cases suggesting prospects for biotechnological opportunities.
Keywords: extreme thermoacidophile, bioleaching, CO2 fixation, archaeal genetics
INTRODUCTION
Over the past 20 years much has been written about the biotechnological potential of microorganisms from extreme environments, primarily focusing on individual enzymes capable of withstanding the otherwise harsh conditions required for long-term efficacy in bioprocessing environments [1–5]. However, as genome sequence data have become available for extremophiles (The UCSC Archaeal Genome Browser; http://archaea.ucsc.edu) and molecular genetics tools have begun to emerge [6,7], there exists the possibility to go beyond single biocatalytic steps to take advantage of the novel pathways and physiological characteristics that are intrinsic to these unique microorganisms. By incorporating these features into less extreme organisms and cells and by metabolically engineering extremophiles directly, a new horizon in microbial biotechnology can emerge. Here, we consider the extremely thermoacidophilic archaea, microorganisms that thrive in hot acid.
Extremely thermoacidophilic archaea and their physiological characteristics
For the purposes of this review, an “extreme thermoacidophile” is a microorganism with both an optimal growth temperature ≥ 60°C and an optimal pH of ≤ 4.0. A majority of the extremely thermoacidophilic species studied to date [8] belong to the archaeal orders Sulfolobales and Thermoplasmatales (Figure 1). From what is currently known, it is interesting that the most heat-tolerant extreme thermoacidophiles are not the most acid-tolerant and vice versa. The most thermophilic of the extreme thermoacidophiles, crenarcheon Acidianus infernus, grows at temperatures up to 95°C (Topt of 85–90°C) but at pHs only as low as 1.0 (pHopt 2.0) [9]. In contrast, Picrophilus species of the euryarchaeal order Thermoplasmatales are the most acidophilic, growing at pHs as low as 0 (pHopt 0.7), but at temperatures up to only 65°C (Topt of 60°C) [10]. Insights into life in hot acid may be soon forthcoming, since genome sequences exist or are underway for many extreme thermoacidophiles (see Figure 1, Table 1) (www.genomesonline.org). Furthermore, several new species in known genera of Sulfolobales (Acidianus, Metallosphaera) have been reported, as well as a new member of the Thermoplasmatales, Thermogymnomonas acidicola [11–14]. We may have only scratched the surface with respect to extreme thermoacidophile diversity, because new environments not previously known to harbor these microorganisms have recently been identified. For example, mathematical modeling and 16S rRNA data suggest that conditions conducive to thermoacidophilic growth exist in deep-sea hydrothermal vents, a hypothesis supported by reports of the first euryarchaeon from the order Deep-sea Hydrothermal Vent Euryarchaeotic 2 (DHVE2), Aciduliprofundum boonei [15••]. Although A. boonei (Topt 70°C) grows best at a pH slightly above 4.0, 16S rRNA indicates that this microorganism comprises 10–15% of selected vent-associated archaeal populations, suggesting that there are extreme thermoacidophiles from these sites yet to be isolated [15].
Figure 1.
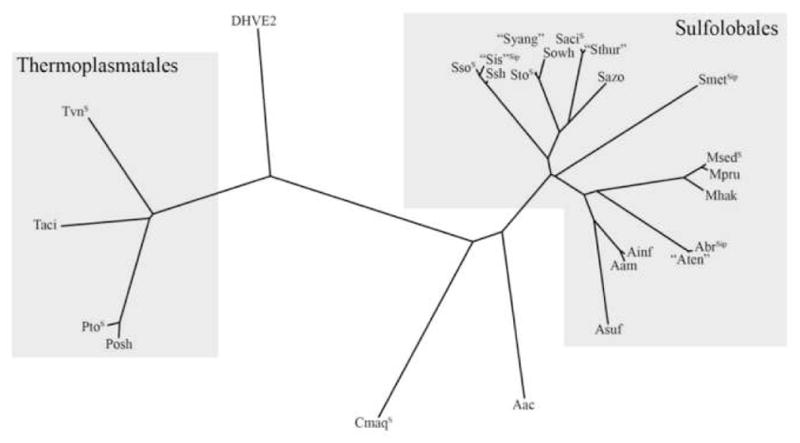
Unrooted 16S phylogenentic tree (constructed using ClustalW and Phylip 3.5c available on http://mobyle.pasteur.fr) of extremely thermoacidophilic archaea (Topt ≥60°C, pHopt <4), compiled from [8,9,11,14,72,73]. Genomes are denoted as Ssequenced or Sipsequencing in progress, according to www.genomesonline.org. Organisms denoted in “ ” have not yet been described in full detail. Accession numbers for 16S rRNA are listed in parentheses. Sso = S. solfataricus (SSOr03), “Sis”=”S. islandicus” strain M14A (AY247895), Ssh=S. shibatae (M32504), Sto=S. tokodaii (ABO22438), “Syang”= ”Sulfolobus yangmingensis” (AB010957), Sowh= Sulfurisphaera ohwakuensis (D85507), Saci= S. acidocaldarius (D14876), “Sthur” = “Sulfolobus thuringensis” (X90485), Sazo=Stygiolobus azoricus (D85520), Smet= S. metallicus (U40813), Msed=M. sedula (Msed_R0026), Mpru= Metallosphaera prunae (X90482), Mhak=Metallosphaera hakonensis (D86414), Abr=Acidianus brierleyi (X90477), “Aten”= ”Acidianus tengchongensis” (AF226987), Ainf=A. infernus (X89852), Aam=Acidianus ambivalens (D85506), Asuf=Acidianus sulfidivorans (AY907891), Aac=Acidolobus aceticus (AF191225), Cmaq=Caldivirga maquilingensis (ABO13926), Posh=Picrophilus oshimae (X84901), Pto=Picrophilus torridus (PTOr02), Taci=Thermogymnomonas acidicola (AB269873), Tvn=Thermoplasma volcanium (Tvnr04), DHVE2 represented by 16S of A. boonei (DQ451875). No 16S sequences are available for extreme thermoacidophiles Sulfurococcus yellowsonensis and Sulfurococcus mirabilis.
Table 1.
Selected features of extreme thermoacidophiles with sequenced genomes.
Compiled from NCBI May 2008 genome project links (http://www.ncbi.nlm.nih.gov/) and references listed for this article.
| Sequenced Thermoacidophile | Growth Temp (°C) | Growth pH | Growth Optimum (°C, pH) | Isolated from | Genome Sequence reported | GC content (%) | Genome Size (Mb) | Estimated # of encoded proteins | IS elements1 | toxin-like COGs2 | “hypo-thetical proteins” |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Crenarchaea-Thermoprotei | |||||||||||
| Thermoproteales | |||||||||||
| Caldivirga maquilingensis | 60–92 | 2.3–6.4 | 85, 3.7–4.2 | acidic hot spring, Philippines | 2007 | 43.1 | 2.1 | 1963 | 7 | 15 | 549 |
| Sulfolobales | |||||||||||
| Metallosphaera sedula | 50–80 | 1–4.5 | 75, 2–3 | solfataric thermal pond drainage, Italy | 2007 | 46.2 | 2.2 | 2256 | 11 | 18 | 673 |
| Sulfolobus acidocaldarius | 55–85 | 1–6 | 70–75, 2–3 | solfataric hot spring, Italy | 2005 | 36.7 | 2.2 | 2223 | 14 | 19 | 1002 |
| Sulfolobus solfataricus | 50–87 | 2–5.5 | 85, 3–4.5 | solfataric hot spring, Italy | 2001 | 35.8 | 3 | 2977 | 159 | 26 | 1340 |
| Sulfolobus tokodaii | 70–85 | 2–5.5 | 80, 2.5–3 | hot spring, Japan | 2001 | 32.8 | 2.7 | 2825 | 12 | 32 | 1874 |
| Euryarchaea- Thermoplasmata | |||||||||||
| Thermoplasmatales | |||||||||||
| Picrophilus torridus | 47–65 | 0–3.5 | 60, 0.7 | volcanic solfataric field, Japan | 2004 | 36 | 1.5 | 1535 | 4 | 4 | 291 |
| Thermoplasma acidophilum | 45–63 | 0.5–4 | 59, 1–2 | burning coal refuse pile, USA | 2001 | 46 | 1.6 | 1482 | 4 | 6 | 583 |
| Thermoplasma volcanium | 33–67 | 1–4 | 60, 2 | volcanic solfataric field, Italy | 2001 | 39.9 | 1.6 | 1499 | 56 | 6 | 261 |
annotation contains “transposase”, “integrase”, or “resolvase”
ORFs similar to COGs 1412, 1439, 1487, 1848, 3413, or 4113 containing nucleic acid binding and/or PIN (PilT N terminus) domains
Mechanisms of resistance to and survival in hot acid
The mechanisms by which microbial life thrives in hot acid have been investigated in some detail in recent years, triggered by the availability of genome sequence data, functional genomics tools, and molecular genetics. While the intrinsic basis for this novel growth physiology is not clear, clues are emerging as to how these microorganisms survive in the face of hot, acidic, and often metal-laden conditions which are typically associated with DNA damage, protein denaturation, and other disruptions in cellular processes.
DNA damage and repair
High temperatures and the potential for cytosol acidification heighten the possibility of DNA damage or modification in extreme thermoacidophiles relative to mesophilic neutrophiles. Thus, clues to DNA damage repair may emerge from examination of this cellular function in hot acid biotopes. It is surprising that basal mutation rates for extreme thermoacidophiles are not particularly high. For example, Sulfolobus acidocaldarius has a spontaneous mutation rate similar to that of E. coli [16]. Furthermore, when Sulfolobus solfataricus and S. acidocaldarius were exposed to UV-irradiation, no significant increase in transcription of known DNA repair proteins was noted [17•,18]; however, it is possible that these genes are constitutively transcribed at higher levels than in mesophiles. Following irradiation, aggregates resembling those formed during plasmid-mediated conjugation were found, spurring speculation that Sulfolobus species may use conjugational DNA exchange and homologous recombination to repair mutated DNA [17]. In a related study, S. solfataricus infected with the Sulfolobus spindle-shaped virus (SSV1) exhibited a similar, but heightened, response to UV-induced DNA damage, suggesting that viruses may be an evolutionary component of stress management systems [19]. Another spindle-shaped virus, SSV2 from native host “Sulfolobus islandicus” REY15/4, sent infected “S. islandicus” REY15A cells into a metabolically inactive state upon encountering unfavorable environmental conditions and then played a role in re-starting metabolic activity once favorable growth conditions emerged [20]. Up-regulation of two S. solfataricus recA/rad51 homologs (radA, SSO0250; radA-like, SSO0777) in response to a DNA-damaging antibiotic led to discovery of the first regulatory protein involved in archaeal DNA damage repair (Sta1, SSO0048) [21]. Neither the radA-like SSO0777, its sta1 activator, nor radA itself were induced by UV-irradiation, indicating that the nature of DNA damage may drive the specific type of repair response [21].
Heat shock
Though extreme thermoacidophiles thrive at temperatures up to 95°C, they are still susceptible to thermal stresses such that they exhibit both cold shock and heat shock responses. Extremely thermoacidophilic archaea react to supraoptimal temperatures in much the same way as other microorganisms [22–24]. Most work to date has focused on the archaeal thermosome, or rosettasome, a heat-shock responsive HSP60-like molecular chaperone that has been implicated in many cellular roles [25]. However, recent efforts have shown that heat shock response in extreme thermoacidophiles is extensive, involving much more than chaperones or other proteins involved in protein refolding. When exponentially growing S. solfataricus was shifted from its growth temperature optimum (80°C) to 90°C, approximately 1/3 of the transcriptome responded within 5 minutes [26]. Included in this set of genes were many insertion elements and chromosomally encoded toxin-antitoxin (TA) loci - 22 TA pairs and 1 solitary toxin – all from the VapBC family [26]. Chromosomally-encoded TA loci in bacteria are thought to be stress response elements [27], although the role of these tandem protein complexes in archaea has not been examined. Since PIN domain-containing VapCs, the “toxin” component of TA loci, are putative ribonucleases [28,29], these proteins could play an important role in post-transcriptional regulation in archaea, especially during heat shock.
Metal resistance
Extreme thermoacidophiles have developed mechanisms for tolerating heavy metals that are physiologically toxic to most microorganisms (Figure 2). These mechanisms involve their capacity to recover from metal-induced damage (similar to oxidative stress) [30] and to limit the effective concentration of the toxic metal itself. In some cases, enzymes reduce or oxidize metals to less toxic forms - for example, the mercuric reductase in S. solfataricus reduces soluble intracellular Hg2+ to volatile elemental Hg0 [31••]. In other cases, metal chelation or complexation can accomplish the same objective. In Sulfolobus metallicus, a polyphosphate (polyP)-based mechanism is believed to underlie cellular tolerance to high levels of copper; greater accumulation of polyP granules was observed in S. metallicus (considered to have higher levels of Cu tolerance) compared to S. solfataricus, and granule size was noted to decrease as Cu levels were increased [32•]. Other strategies, however, do not involve metal transformation, direct or indirect, and instead are based on exporting toxic metal ions via P-type ATPases [33]. Evidence to date suggests that multiple systems can operate in parallel in extreme thermoacidophiles to provide cumulative tolerance, with particular strategies useful for multiple metals [31–33]. For example, copper tolerance in Sulfolobus species involves efflux ATPases in addition to the polyP pathway, but the same ATPases also contribute to cadmium tolerance [32,33]. There may also be some intrinsic redundancy in protecting against heavy metal toxicity. Disruption mutants lacking mercuric reductase and its regulator (merAR) were found to still exhibit some mer operon transcription [31]. In fact, creation of a mutant with a disrupted regulator resulted in increased merA expression and consequently increased Hg2+ tolerance [31], underscoring the importance of understanding the regulation of resistance mechanisms with respect to engineering characteristics for bioprocesses. While traditional acclimation and/or spontaneous mutant generation approaches are still useful, direct genetic manipulation offers the possibility of conferring similar levels of metal tolerance increase in a systematic manner.
Figure 2.
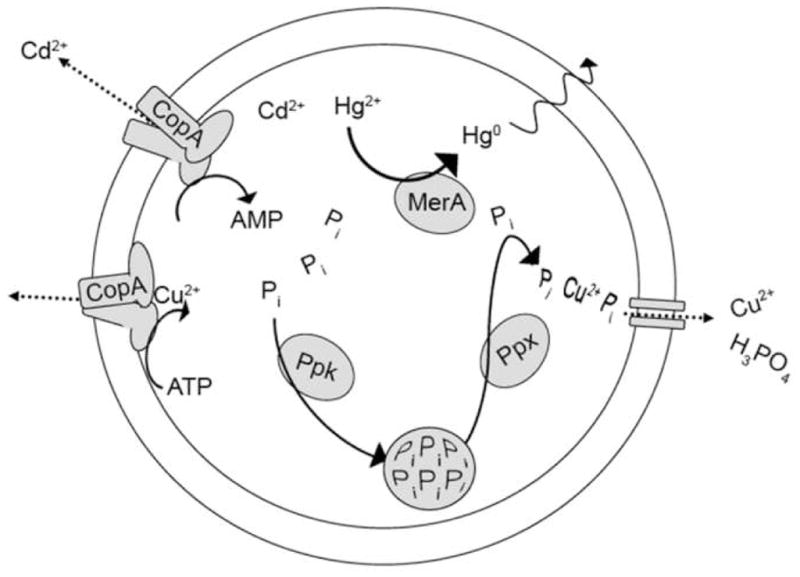
Metal resistance mechanisms in extreme thermoacidophiles. CopA is the P-type ATPase shown to be involved in copper and cadmium cation efflux in S. solfataricus [33]. MerA is the mercuric reductase which reduces soluble Hg2+ to volatile elemental Hg and is constitutively expressed in S. solfataricus [31]. Ppk (polyphosphate kinase) and Ppx (exopolyphosphatase) which comprise the polyP system described in S. metallicus [32].
Molecular genetics of extreme thermoacidophiles
Versatile genetic systems for extreme thermoacidophiles are a critical need for many reasons. Recombinant expression of genes encoding extreme thermoacidophile proteins in commonly used bacterial hosts can be problematic [34,35], likely reflecting intrinsic differences between archaea and bacteria. Also, molecular genetic systems could provide the basis for investigating biological mechanisms enabling life in hot acid. Fortunately, promising developments along these lines have been reported, including successful efforts with gene disruption [36,37] and protein tagging [6]. For a comprehensive review of the development of molecular genetics for archaea see [7].
Viruses and plasmids
Virus-based plasmids have been tailored for specific needs to support development of genetics systems in extreme thermoacidophiles. The first virus from an extreme thermoacidophile, SSV1, was isolated over 20 years ago from Sulolobus shibatae (B12) [38] and is the one most developed for use in studying Sulfolobus species [7]. There are some useful features of this virus for molecular genetics. For example, removal of the integrase gene from SSV1 demonstrated that the integrase protein is essential only for integration into the host genome, and not for virus infection and replication [39]. This characteristic could be exploited for novel vector development, where non-homologous recombination of the vector into the host genome is undesirable. Recently constructed Sulfolobus-E. coli fusion vectors have potential as easily modifiable genetic elements for extreme thermoacidophiles. The fusion shuttle vector pSSVrt, based on “S. islandicus” REY15/4 pSSVx and E. coli pUC19, accommodated insertion of foreign sequences up to ~11 Kb with efficient propagation and vector stability at high-copy numbers with no integration [40]. Furthermore, the pRN1-E. coli transposon fusion, noted for its relatively small size (5.4 Kb) and stable copy numbers of 10–20 in mid-log phase, is stable in S. acidocaldarius and S. solfataricus, as well as in E. coli. [41]. By adding the pyrE gene to this plasmid and using pyrE mutants, a selectable marker could be introduced into the host. An SSV1-E. coli pUC18 reporter gene system was developed with selectable marker genes pyrEF, both heat- and arabinose-inducible promoters, and convenient restriction sites [42]. The improved vector (modified from pMJ03) was used for heterologous and homologous production of tagged proteins in S. solfataricus. All of these developments are exciting and offer promise for expanding molecular genetics capabilities in extreme thermoacidophiles.
Gene disruption
Construction of directed gene deletion mutants in extremely thermoacidophilic archaea is a significant challenge, but progress is being made on this front. The focus has been on S. solfataricus PBL2025, a constructed mutant of S. solfataricus 98/2, which lacks about 50 genes, including lacS [43]. Thus, the inability to grow on lactose-based minimal media provides a selectable marker. A protocol for efficient integration of exogenous DNA into the S. solfataricus PBL2025 genome has been described [36]. A similar method was used to construct a deficient mutant to study α-amylase function and regulation in S. solfataricus [37]. Markerless exchange using a plasmid that encodes a cloned copy of a modified DNA sequence and a selectable marker gene, again lacS for natural deletion mutant PBL2025, has also been used for development of mercury reductase (merA) deficient Sulfolobus solfataricus [31].
Recombinant production of extremely thermoacidophilic proteins
Production of extremely thermoacidophilic proteins in mesophilic hosts (e.g., E. coli) can take advantage of overexpression and simplified purification methods (e.g., heat treatment), but codon usage and inclusion body problems often temper the enthusiasm for this approach [34,35]. Some solutions to existing problems have been recently proposed. S. solfataricus genes have rare (compared to E. coli) codon clustering at the 5′ transcript end which specifically inhibits target translation. But, this can be relieved by adding rare codon tRNAs (utilizing strains like BL21(DE3) CodonPlus-RIL or Rosetta(DE3)) or by changing rare codons (via primer design) to those more frequently translated by the host [34]. Yields of active S. tokodaii and P. torridus proteins produced in E. coli (Rosetta(DE3)) were increased by growth and expression at elevated temperatures up to 46°C, where it was hypothesized that protein synthesis was slowed, contributing to an increase in rate of proper folding [35].
Bioleaching
The biomining industry has a longstanding interest in the use of extreme thermoacidophiles for metals recovery from ores [44,45]. These organisms, as is the case with certain mesophilic chemolithotrophic bacteria such as Acidothiobacillus ferrooxidans [46•], can liberate precious (e.g., gold) and base (e.g., copper) metals trapped in, and as, metal sulfides (e.g., iron pyrite and chalcopyrite) through dissimilatory oxidative processes. Biological regeneration of Fe3+ from Fe2+ is key to chemical attack of metal sulfides. However, biooxidation of reduced inorganic sulfur compounds (RISCs) is also important to prevent the accumulation of passivating sulfur compounds on metal surfaces that can limit metal mobilization rates. Extreme thermoacidophiles grow at temperatures where mesoacidophilic biocatalysts (or contaminants from non-sterile substrates) cannot, and where passivation from RISCs is nearly eliminated, leading to higher effective leaching rates [46]. Efficacy in biomining environments also requires tolerance of high levels of toxic heavy metals as well as the ability to assimilate inorganic carbon, as organic sources can be scarce in this environment. A full complement of the aforementioned desirable traits is not typically resident in a single native microorganism, but may be in a consortium [46]. Alternatively, extreme thermoacidophile genomes [47–49], including that of a biomining organism [50•], can be examined for pathways responsible for conferring these desirable biomining traits. Taken together with the emerging molecular genetic tools for extreme thermoacidophiles, metabolic engineering of biomining organisms with enhanced properties may soon be a reality.
Iron-sulfur oxidation
Several terminal components of respiratory electron transport chains (ETC) in extreme thermoacidophiles have been known for years [51–54]. However, it was recently shown that the relative membrane concentration of these components depends on the electron donating substrate [55], suggesting involvement in dissimilatory Fe2+ and RISC oxidation. Substrate-dependent ETC expression led to identification of a Fe2+ oxidation- (fox) induced gene cluster in the autotrophic biominer, S. metallicus [56••]. Comparative genomics subsequently revealed that homologs to this gene cluster are also present in Metallosphaera sedula [50] and Sulfolobus tokodaii, the latter of which was not previously shown to have iron-oxidizing capabilities [56]. This cluster contains not only ORFs similar to previously recognized terminal components (foxABCD), but also ferredoxins and other putative iron-sulfur binding proteins typically involved in electron transfer (foxFGHJ). This implies that more than terminal components, possibly entire ETCs, are differentially expressed in response to certain substrates. Although the sulfur oxygenase reductase (SOR) does not appear to be directly connected to an ETC [57•], it was also up-regulated in S° compared to Fe2+ grown cells [56]. SOR is believed to participate in the early steps of cytoplasmic sulfur oxidation in extremely thermoacidophilic archaea. Surprisingly, M. sedula, a putative sulfur oxidizer [58•], does not encode SOR [50], indicating that either some extremely thermoacidophilic archaea have alternative (yet unknown) sulfur oxidation enzymes, or that M. sedula has lost (or never possessed) the capacity to oxidize sulfur (similar to S. solfataricus and S. acidocaldarius) [58].
CO2 fixation
With the identification of a 5th pathway for inorganic carbon fixation [59••], new perspectives on autotrophy have emerged. The new cycle model starts with a 2-carbon acetyl-CoA molecule and assimilates two bicarbonates. The key bicarbonate-incorporating enzyme is a heterotrimeric acetyl-/propionyl-CoA carboxylase [60]. One way in which this new pathway is distinguished from the 3-hydroxypropionate cycle is by a putative homotetrameric 4-hydroxybutyryl-CoA dehydratase (Figure 3). While activities have been detected for all steps in this pathway [59], only a few enzymes have been characterized biochemically [59–62]. Comparative genomics suggests multiple candidates (all having similar sequences) for the final enzymatic reactions of the cycle; experimental work is required to identify which of these candidates supports the function of the final three steps of the pathway, involving re-arrangement of a 4-carbon molecule (crotonyl-CoA) prior to splitting into two molecules of acetyl-CoA. Understanding and controlling regulation of this new inorganic carbon fixation pathway could lead to improvements in bioleaching through enhanced biomass/biocalatyst levels, and even new CO2 sequestration strategies.
Figure 3.
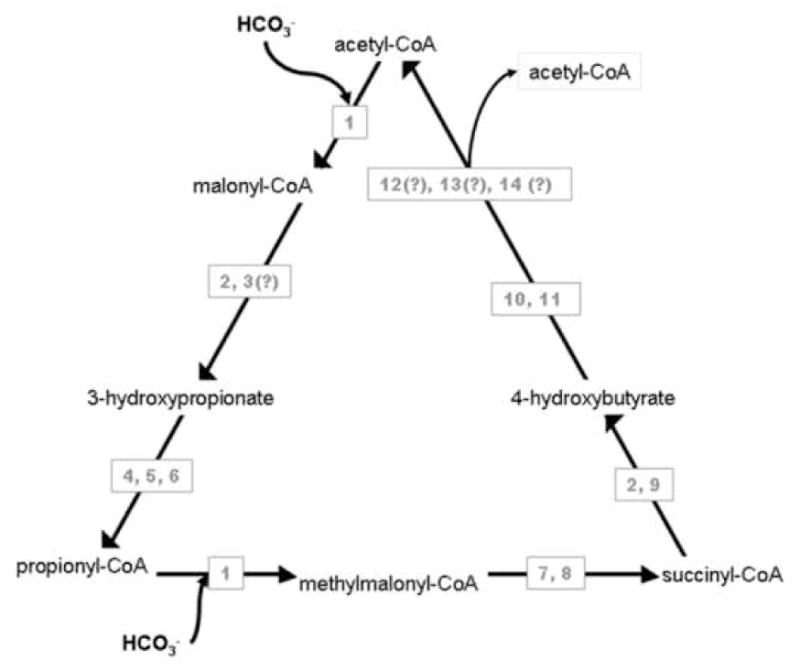
The recently proposed 5th cycle of autotrophic carbon fixation, adapted from[59]. 1. acetyl-CoA/propionyl-CoA carboxylase, 2. malonyl-CoA/succinyl-CoA reductase, 3. malonate semialdehyde reductase, 4. 3-hydroxypropionyl-CoA synthetase, 5. 3-hydroxypropionyl-CoA dehydratase, 6. acryloyl-CoA reductase 7. methylmalonyl-CoA epimerase, 8. metylmalonyl-CoA mutase, 9. succinate semialdehyde reductase, 10. 4-hydroxybutyryl-CoA synthetase, 11. 4-hydroxybutyryl-CoA dehydratase, 12. crotonyl-CoA hydratase, 13. 3-hydroxybutaryl-CoA dehydrogenase, 14. acetoacetyl-CoA β-keothiolase. ORFs encoding enzymes with (?) have not been finalized in the extreme thermoacidophile in which the cycle was studied. Many of the intermediates of this cycle are the same as found in the 3-hydroxypropionate cycle (left side and base of triangle). The right side of triangle represents rearrangement after the second carbon addition, concluding with a split into two molecules of acetyl-CoA, which distinguishes the 3-hydroxpropionate/4-hydroxybutyrate cycle from the 3-hydroxypropionate cycle.
OTHER DEVELOPMENTS
ncRNA
Non-coding RNAs (ncRNAs) in extremely thermoacidophilic archaea, particularly in Sulfolobus species [63–66], could be used for transient control of biocatalytic steps in a bioprocess via dosing of small interfering RNA (siRNA). While many ncRNAs are also small RNAs (sRNA) of 60 nucleotides or less, ncRNAs as long as 500 nucleotides have been reported [63]. Most ncRNAs are characterized by a K-turn motif and appear to possess a post-transcriptional modification or regulation (“silencing”) function. Many ncRNAs recognize their targets via full (“anti-sense”) or partial (“antisense-box motif”) complementarity to transposons, other ORFs, or non-coding RNA (rRNA, tRNA, sRNA, etc.) [63,64]. ncRNAs are thought to interact with proteins to influence structure and function. For example, in S. solfataricus, the Rbp18 protein of the 30S ribosomal subunit has been shown to bind free sRNA (in vitro and in vivo) with some degree of selectivity [65]. In S. acidocaldarius, the anti-sense-box motifs (and their location) in sRNA are important for both proper structural complexation with ribosomal core proteins and complex activity [64]. sRNA produced from clustered regularly interspaced short palindromic repeats (CRISPRs) are believed to interact with catalytically active CRISPR-associated sequences (Cas proteins), which are often encoded immediately up- (crenarchaea) or downstream (euryarchaea) of CRISPRs [63,66]. Some CRISPRs contain non-repetitive spacer sequences with similarity to viruses or genomic ORFs. The Cas proteins may be involved with adding/removing spacer sequences to/from CRISPR regions (i.e. Cas1, Cas2), processing of long ncRNA to sRNA form (i.e. Cas3, Cas5), and possibly complexing with resulting sRNAs to form a microbial equivalent of the eukaryotic RNA-induced silencing (RISC) complex (i.e. Cas4) [66,67].
S-layers and extreme thermoacidophile membranes
S-layers of extremely thermoacidophilic archaea are of interest in nanobiotechnology, because their self-assembled periodicity and uniform properties, as well as thermoacid stability, make them attractive for use in ultrafiltration, immobilization matrix, and coating capacities [68]. Most work to date with these S-layers has focused on structure and properties of the purified self-assembled proteins [69]. Models suggest that archaeal S-layers, with their membrane anchors, help maintain cell shape and stabilize the membrane against environment-induced osmotic pressure changes [70]. Study and use of tetraetherlipids themselves have been somewhat hampered due to high costs/low purification yields. However, recent efforts to defray bioleaching costs by processing by-products have resulted in a lower-cost, higher yield purification process for extremely thermoacidophilic tetraetherlipid, calditoglycerocaldarchaeol [71]. The next significant advances in functional understanding may come from studies of the natural environment of S-layers, in which they are bound to the tetraetherlipid cell membrane [69]. A search through GenBank (July 2008) shows that although S-layer domain proteins are annotated in sequenced extreme thermoacidophiles (COG 1361), genes related to their modification and assembly are not well known/annotated. Future work in this area will most likely begin with analysis of gene neighborhoods (many containing transporters and/or transcriptional regulators). Manipulation via genetic tools will be invaluable in study of the formation and regulation of these proton influx barriers with long-term potential to produce tunable acid stability.
SUMMARY
Recently available extreme thermoacidophile genome sequences are revealing novel pathways and strategies that contribute to survival in hot, acidic environments. With the emerging availability of molecular genetics for these microorganisms, metabolic engineering efforts to realize biotechnological opportunities are within reach. Also promising is the prospect of finding novel extreme thermoacidophiles in yet untapped acidic niches, such as deep sea hydrothermal biotopes.
Acknowledgments
KSA and CRC acknowledge NIH T32 Biotechnology Traineeships for support. This work was funded in part by a grant to RMK from the US National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adams MW, Perler FB, Kelly RM. Extremozymes: expanding the limits of biocatalysis. Biotechnology (N Y) 1995;13:662–668. doi: 10.1038/nbt0795-662. [DOI] [PubMed] [Google Scholar]
- 2.Antranikian G, Vorgias CE, Bertoldo C. Extreme environments as a resource for microorganisms and novel biocatalysts. Adv Biochem Eng Biotechnol. 2005;96:219–262. doi: 10.1007/b135786. [DOI] [PubMed] [Google Scholar]
- 3.Hough DW, Danson MJ. Extremozymes. Curr Opin Chem Biol. 1999;3:39–46. doi: 10.1016/s1367-5931(99)80008-8. [DOI] [PubMed] [Google Scholar]
- 4.Tindall KR, Kunkel TA. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988;27:6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- 5.Atomi H. Recent progress towards the application of hyperthermophiles and their enzymes. Curr Opin Chem Biol. 2005;9:166–173. doi: 10.1016/j.cbpa.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Sowers KR, DasSarma S, Blum PH. Gene Transfer in Archaea. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf GA, Schmidt TM, editors. Methods for General and Molecular Microbiology. American Society for Microbiology; 2007. [Google Scholar]
- 7.Allers T, Mevarech M. Archaeal genetics - the third way. Nat Rev Genet. 2005;6:58–73. doi: 10.1038/nrg1504. [DOI] [PubMed] [Google Scholar]
- 8.Garrity GM, Lilburn TG, Cole JR, Harrison SH, Euzeby J, Tindall BJ. Michigan State University Board of Trustees, editor. Release 7.7. 2007. Taxonomic Outline of the Bacteria and Archaea; pp. 6–31. [Google Scholar]
- 9.Huber H, Prangishvili D. The Sulfolobales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E, editors. The Prokaryotes. 3. Springer; 2006. pp. 23–50. [Google Scholar]
- 10.Huber H, Stetter KO. Thermoplasmatales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E, editors. The Prokaryotes. 3. Springer; 2006. pp. 101–112. [Google Scholar]
- 11.Plumb JJ, Haddad CM, Gibson JA, Franzmann PD. Acidianus sulfidivorans sp. nov. an extremely acidophilic, thermophilic archaeon isolated from a solfatara on Lihir Island, Papua New Guinea, and emendation of the genus description. Int J Syst Evol Microbiol. 2007;57:1418–1423. doi: 10.1099/ijs.0.64846-0. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida N, Nakasato M, Ohmura N, Ando A, Saiki H, Ishii M, Igarashi Y. Acidianus manzaensis sp. nov. a novel thermoacidophilic archaeon growing autotrophically by the oxidation of H2 with the reduction of Fe3+ Curr Microbiol. 2006;53:406–411. doi: 10.1007/s00284-006-0151-1. [DOI] [PubMed] [Google Scholar]
- 13.Kozubal M, Macur RE, Korf S, Taylor WP, Ackerman GG, Nagy A, Inskeep WP. Isolation and distribution of a novel iron-oxidizing crenarchaeon from acidic geothermal springs in Yellowstone National Park. Appl Environ Microbiol. 2008;74:942–949. doi: 10.1128/AEM.01200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh T, Yoshikawa N, Takashina T. Thermogymnomonas acidicola gen. nov., sp. nov. a novel thermoacidophilic, cell wall-less archaeon in the order Thermoplasmatales, isolated from a solfataric soil in Hakone, Japan. Int J Syst Evol Microbiol. 2007;57:2557–2561. doi: 10.1099/ijs.0.65203-0. [DOI] [PubMed] [Google Scholar]
- 15••.Reysenbach AL, Liu Y, Banta AB, Beveridge TJ, Kirshtein JD, Schouten S, Tivey MK, Von Damm KL, Voytek MA. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature. 2006;442:444–447. doi: 10.1038/nature04921. Discusses strategy used to isolate a new order of extremely thermoacidophilic organisms in a low pH deep-sea environment. [DOI] [PubMed] [Google Scholar]
- 16.Grogan DW, Carver GT, Drake JW. Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci U S A. 2001;98:7928–7933. doi: 10.1073/pnas.141113098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Frols S, Gordon PM, Panlilio MA, Duggin IG, Bell SD, Sensen CW, Schleper C. Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J Bacteriol. 2007;189:8708–8718. doi: 10.1128/JB.01016-07. Genome-wide transcriptional response analysis of Sso PH1 and Sso PH1 (SSV1) to UV irradiation to elucidate the DNA damage/repair mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gotz D, Paytubi S, Munro S, Lundgren M, Bernander R, White M. Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biology. 2007;8:R220. doi: 10.1186/gb-2007-8-10-r220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frols S, Gordon PM, Panlilio MA, Schleper C, Sensen CW. Elucidating the transcription cycle of the UV-inducible hyperthermophilic archaeal virus SSV1 by DNA microarrays. Virology. 2007;365:48–59. doi: 10.1016/j.virol.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 20.Contursi P, Jensen S, Aucelli T, Rossi M, Bartolucci S, She Q. Characterization of the Sulfolobus host-SSV2 virus interaction. Extremophiles. 2006;10:615–627. doi: 10.1007/s00792-006-0017-2. [DOI] [PubMed] [Google Scholar]
- 21.Abella M, Rodriguez S, Paytubi S, Campoy S, White MF, Barbe J. The Sulfolobus solfataricus radA paralogue sso0777 is DNA damage inducible and positively regulated by the Sta1 protein. Nucleic Acids Res. 2007;35:6788–6797. doi: 10.1093/nar/gkm782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han CJ, Park SH, Kelly RM. Acquired Thermotolerance and Stressed-Phase Growth of the Extremely Thermoacidophilic Archaeon Metallosphaera sedula in Continuous Culture. Appl Environ Microbiol. 1997;63:2391–2396. doi: 10.1128/aem.63.6.2391-2396.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trent JD, Osipiuk J, Pinkau T. Acquired thermotolerance and heat shock in the extremely thermophilic archaebacterium Sulfolobus sp. strain B12. J Bacteriol. 1990;172:1478–1484. doi: 10.1128/jb.172.3.1478-1484.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peeples TL, Kelly RM. Bioenergetic Response of the Extreme Thermoacidophile Metallosphaera sedula to Thermal and Nutritional Stresses. Appl Environ Microbiol. 1995;61:2314–2321. doi: 10.1128/aem.61.6.2314-2321.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagawa HK, Yaoi T, Brocchieri L, McMillan RA, Alton T, Trent JD. The composition, structure and stability of a group II chaperonin are temperature regulated in a hyperthermophilic archaeon. Mol Microbiol. 2003;48:143–156. doi: 10.1046/j.1365-2958.2003.03418.x. [DOI] [PubMed] [Google Scholar]
- 26.Tachdjian S, Kelly RM. Dynamic metabolic adjustments and genome plasticity are implicated in the heat shock response of the extremely thermoacidophilic archaeon Sulfolobus solfataricus. J Bacteriol. 2006;188:4553–4559. doi: 10.1128/JB.00080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clissold PM, Ponting CP. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr Biol. 2000;10:R888–890. doi: 10.1016/s0960-9822(00)00858-7. [DOI] [PubMed] [Google Scholar]
- 29.Daines DA, Wu MH, Yuan SY. VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease. J Bacteriol. 2007;189:5041–5048. doi: 10.1128/JB.00290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salzano AM, Febbraio F, Farias T, Cetrangolo GP, Nucci R, Scaloni A, Manco G. Redox stress proteins are involved in adaptation response of the hyperthermoacidophilic archaeon Sulfolobus solfataricus to nickel challenge. Microb Cell Fact. 2007;6:25. doi: 10.1186/1475-2859-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Schelert J, Drozda M, Dixit V, Dillman A, Blum P. Regulation of mercury resistance in the crenarchaeote Sulfolobus solfataricus. J Bacteriol. 2006;188:7141–7150. doi: 10.1128/JB.00558-06. Discusses the use of genetic tools in S. solfataricus to elucidate the regulation of a mercury resistance mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Remonsellez F, Orell A, Jerez CA. Copper tolerance of the thermoacidophilic archaeon Sulfolobus metallicus: possible role of polyphosphate metabolism. Microbiology. 2006;152:59–66. doi: 10.1099/mic.0.28241-0. A study of Cu efflux and correlation to polyP accumulation and enzyme activity in selected Sulfolobus species with high and low Cu tolerances. [DOI] [PubMed] [Google Scholar]
- 33.Ettema TJ, Brinkman AB, Lamers PP, Kornet NG, de Vos WM, van der Oost J. Molecular characterization of a conserved archaeal copper resistance (cop) gene cluster and its copper-responsive regulator in Sulfolobus solfataricus P2. Microbiology. 2006;152:1969–1979. doi: 10.1099/mic.0.28724-0. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Lee SB. Rare codon clusters at 5′-end influence heterologous expression of archaeal gene in Escherichia coli. Protein Expr Purif. 2006;50:49–57. doi: 10.1016/j.pep.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Koma D, Sawai T, Harayama S, Kino K. Overexpression of the genes from thermophiles in Escherichia coli by high-temperature cultivation. Appl Microbiol Biotechnol. 2006;73:172–180. doi: 10.1007/s00253-006-0448-9. [DOI] [PubMed] [Google Scholar]
- 36.Albers S-V, Driessen AJ. Conditions for gene disruption by homologus recombination of exogenous DNA into the Sulfolobus solfataricus genome. Archaea. 2007;2:145–149. doi: 10.1155/2008/948014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Worthington P, Hoang V, Perez-Pomares F, Blum P. Targeted disruption of the alpha-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol. 2003;185:482–488. doi: 10.1128/JB.185.2.482-488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin A, Yeats S, Janekovic D, Reiter WD, Aicher W, Zillig W. SAV 1, a temperate u.v. -inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius isolate B12. EMBO J. 1984;3:2165–2168. doi: 10.1002/j.1460-2075.1984.tb02107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clore AJ, Stedman KM. The SSV1 viral integrase is not essential. Virology. 2007;361:103–111. doi: 10.1016/j.virol.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Aucelli T, Contursi P, Girfoglio M, Rossi M, Cannio R. A spreadable, non-integrative and high copy number shuttle vector for Sulfolobus solfataricus based on the genetic element pSSVx from Sulfolobus islandicus. Nucleic Acids Res. 2006;34:e114. doi: 10.1093/nar/gkl615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berkner S, Grogan D, Albers SV, Lipps G. Small multicopy, non-integrative shuttle vectors based on the plasmid pRN1 for Sulfolobus acidocaldarius and Sulfolobus solfataricus, model organisms of the (cren-)archaea. Nucleic Acids Res. 2007;35:e88. doi: 10.1093/nar/gkm449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Albers SV, Jonuscheit M, Dinkelaker S, Urich T, Kletzin A, Tampe R, Driessen AJ, Schleper C. Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl Environ Microbiol. 2006;72:102–111. doi: 10.1128/AEM.72.1.102-111.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schelert J, Dixit V, Hoang V, Simbahan J, Drozda M, Blum P. Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J Bacteriol. 2004;186:427–437. doi: 10.1128/JB.186.2.427-437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du Plessis CA, Batty JD, Dew DW. Commercial Applications of Thermophile Bioleaching. In: Rawlings DE, Johnson DB, editors. Biomining. Springer-Verlag; 2007. [Google Scholar]
- 45.Mikkelsen D, Kappler U, McEwan AG, Sly LI. Archaeal diversity in two thermophilic chalcopyrite bioleaching reactors. Environ Microbiol. 2006;8:2050–2056. doi: 10.1111/j.1462-2920.2006.01115.x. [DOI] [PubMed] [Google Scholar]
- 46•.Rawlings DE, Johnson DB. The microbiology of biomining: development and optimization of mineral-oxidizing microbial consortia. Microbiology. 2007;153:315–324. doi: 10.1099/mic.0.2006/001206-0. Consortia optimization strategy considerations based on microbiology of biocatalysts, as well as chemical engineering characteristics of biomining operations. [DOI] [PubMed] [Google Scholar]
- 47.Chen L, Brugger K, Skovgaard M, Redder P, She Q, Torarinsson E, Greve B, Awayez M, Zibat A, Klenk HP, et al. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J Bacteriol. 2005;187:4992–4999. doi: 10.1128/JB.187.14.4992-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawarabayasi Y, Hino Y, Horikawa H, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, Kosugi H, Hosoyama A, et al. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7. DNA Res. 2001;8:123–140. doi: 10.1093/dnares/8.4.123. [DOI] [PubMed] [Google Scholar]
- 49.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC-Y, Clausen Ib G, Curtis BA, De Moors A, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Auernik KS, Maezato Y, Blum PH, Kelly RM. The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl Environ Microbiol. 2008;74:682–692. doi: 10.1128/AEM.02019-07. Discusses genome features relevant to biomining and genome-wide transcriptional response to the presence of ferrous iron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castresana J, Lubben M, Saraste M. New archaebacterial genes coding for redox proteins: implications for the evolution of aerobic metabolism. J Mol Biol. 1995;250:202–210. doi: 10.1006/jmbi.1995.0371. [DOI] [PubMed] [Google Scholar]
- 52.Lubben M, Arnaud S, Castresana J, Warne A, Albracht SP, Saraste M. A second terminal oxidase in Sulfolobus acidocaldarius. Eur J Biochem. 1994;224:151–159. doi: 10.1111/j.1432-1033.1994.tb20006.x. [DOI] [PubMed] [Google Scholar]
- 53.Purschke WG, Schmidt CL, Petersen A, Schafer G. The terminal quinol oxidase of the hyperthermophilic archaeon Acidianus ambivalens exhibits a novel subunit structure and gene organization. J Bacteriol. 1997;179:1344–1353. doi: 10.1128/jb.179.4.1344-1353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hiller A, Henninger T, Schafer G, Schmidt CL. New genes encoding subunits of a cytochrome bc1-analogous complex in the respiratory chain of the hyperthermoacidophilic crenarchaeon Sulfolobus acidocaldarius. J Bioenerg Biomembr. 2003;35:121–131. doi: 10.1023/a:1023742002493. [DOI] [PubMed] [Google Scholar]
- 55.Kappler U, Sly LI, McEwan AG. Respiratory gene clusters of Metallosphaera sedula - differential expression and transcriptional organization. Microbiology. 2005;151:35–43. doi: 10.1099/mic.0.27515-0. [DOI] [PubMed] [Google Scholar]
- 56••.Bathe S, Norris PR. Ferrous iron- and sulfur-induced genes in Sulfolobus metallicus. Appl Environ Microbiol. 2007;73:2491–2497. doi: 10.1128/AEM.02589-06. Identification of a new terminal oxidase cluster and ETC components involved in ferrous iron oxidation in selected Sulfolobus species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Urich T, Gomes CM, Kletzin A, Frazao C. X-ray Structure of a self-compartmentalizing sulfur cycle metalloenzyme. Science. 2006;311:996–1000. doi: 10.1126/science.1120306. Structural resolution of an intracellular sulfur oxidoreductase and implications for its mechanism of operation via creation of a microenvironment. [DOI] [PubMed] [Google Scholar]
- 58•.Kletzin A. Metabolism of inorganic sulfur compounds in Archaea. In: Garrett RA, Klenk HP, editors. Archaea: Evolution, Physiology, and Molecular Biology. Blackwell; 2007. pp. 261–276. A thorough summary of known inorganic sulfur metabolism in archaea and comparison to bacterial metabolism. [Google Scholar]
- 59••.Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–1786. doi: 10.1126/science.1149976. Outline of a new inorganic carbon fixation cycle in archaea and phylogenetic diversity of microorganisms containing key enzymes. [DOI] [PubMed] [Google Scholar]
- 60.Hugler M, Krieger RS, Jahn M, Fuchs G. Characterization of acetyl-CoA/propionyl-CoA carboxylase in Metallosphaera sedula Carboxylating enzyme in the 3-hydroxypropionate cycle for autotrophic carbon fixation. Eur J Biochem. 2003;270:736–744. doi: 10.1046/j.1432-1033.2003.03434.x. [DOI] [PubMed] [Google Scholar]
- 61.Alber B, Olinger M, Rieder A, Kockelkorn D, Jobst B, Hugler M, Fuchs G. Malonyl-coenzyme A reductase in the modified 3-hydroxypropionate cycle for autotrophic carbon fixation in archaeal Metallosphaera and Sulfolobus spp. J Bacteriol. 2006;188:8551–8559. doi: 10.1128/JB.00987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alber BE, Kung JW, Fuchs G. 3-Hydroxypropionyl-coenzyme A synthetase from Metallosphaera sedula, an enzyme involved in autotrophic CO2 fixation. J Bacteriol. 2008;190:1383–1389. doi: 10.1128/JB.01593-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Huttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- 64.Omer AD, Zago M, Chang A, Dennis PP. Probing the structure and function of an archaeal C/D-box methylation guide sRNA. RNA. 2006;12:1708–1720. doi: 10.1261/rna.31506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ciammaruconi A, Gorini S, Londei P. A bifunctional archaeal protein that is a component of 30S ribosomal subunits and interacts with C/D box small RNAs. Archaea. 2007;2:151–158. doi: 10.1155/2008/472786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lillestol RK, Redder P, Garrett RA, Brugger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sorek R, Kunin V, Hugenholtz P. CRISPR - a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2007 doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 68.Sleytr UB, Egelseer EM, Ilk N, Pum D, Schuster B. S-Layers as a basic building block in a molecular construction kit. FEBS J. 2007;274:323–334. doi: 10.1111/j.1742-4658.2006.05606.x. [DOI] [PubMed] [Google Scholar]
- 69.Engelhardt H. Are S-layers exoskeletons? The basic function of protein surface layers revisited. J Struct Biol. 2007;160:115–124. doi: 10.1016/j.jsb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 70.Engelhardt H. Mechanism of osmoprotection by archaeal S-layers: a theoretical study. J Struct Biol. 2007;160:190–199. doi: 10.1016/j.jsb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 71.Bode ML, Buddoo SR, Minnaar SH, du Plessis CA. Extraction, isolation and NMR data of the tetraether lipid calditoglycerocaldarchaeol (GDNT) from Sulfolobus metallicus harvested from a bioleaching reactor. Chem Phys Lipids. 2008 doi: 10.1016/j.chemphyslip.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 72.Huber H, Stetter KO. Desulfurococcales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E, editors. The Prokaryotes. 3. Springer; 2006. pp. 52–68. [Google Scholar]
- 73.Huber H, Huber R, Stetter KO. Thermoproteales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E, editors. The Prokaryotes. 3. Springer; 2006. pp. 10–22. [Google Scholar]


