Abstract
SAMHD1 restricts human immunodeficiency virus-1 (HIV-1) infection of dendritic and other myeloid cells at an early stage in the replication cycle. SIVsm/HIV-2 lineage viruses counteract SAMHD1-mediated restriction by encoding Vpx, a virion-packaged accessory protein that targets SAMHD1 for degradation. We show that SAMHD1 restricts HIV-1 infection of monocyte-derived macrophages (MDM) by hydrolyzing the cellular deoxynucleotide triphosphates (dNTP), reducing their level to below that required for the synthesis of the viral genomic DNA. Vpx prevented the SAMHD1-mediated decrease in dNTP. The restriction was partially alleviated in MDM by the addition of exogenous deoxynucleosides. HIV-1 with a V148I mutation in reverse transcriptase that lowers its affinity for dNTP was particularly sensitive to SAMHD1-mediated restriction. Nucleotide starvation could serve as a mechanism to protect cells from infection by a wide variety of infectious agents that replicate through a DNA intermediate.
SAMHD1 was recently identified as a host factor that restricts HIV-1 replication in dendritic and myeloid cells. The HD domain of SAMHD1 has putative nucleotidase and phosphodiesterase activities and is required for restriction1-4. SAMHD1 has weak homology with the bacterial nucleotide metabolic enzyme, EF1143 from Enterococcus faecalis5 (Supplementary Fig. 1), further suggesting a role in nucleotide biochemistry. Mutations in the genes encoding SAMHD1, the exonuclease TREX1 and RNAse H2 are associated with Aicardi-Goutieres syndrome (AGS), a rare genetic inflammatory encephalopathy characterized by inappropriate immune activation and over-production of alpha-interferon3,6. SAMHD1 is expressed at low level in a variety of cell-types but at moderate levels in MDM and high levels in dendritic cells (DC)1. It is not detectably expressed in CD4+ T cells and these cells are permissive for infection. SAMHD1 has been proposed to play a role as a negative regulator of the innate immune response, analogous to TREX-1, a protein that is thought to dampen innate immune responses to cytoplasmic DNA7.
In viruses of the SIVsm/HIV-2 lineage, SAMHD1-mediated restriction is counteracted by the Vpx accessory protein1,2. Vpx is a small nuclear protein that is packaged in virions through a specific interaction with an amino acid motif in the p6 protein of the Gag precursor8,9. In the cell, Vpx is associated with a DCAF1-based E3 ubiquitin ligase. When expressed in a cell or introduced exogenously via virus-like particles (VLP), Vpx binds to SAMHD1 inducing its degradation. Curiously, HIV-1 does not encode Vpx yet is highly sensitive to SAMHD1-mediated restriction1,2,10.
The presence of an HD domain in SAMHD1 suggests a possible role for dNTPs in its antiviral activity. Cells differ in their content of dNTP depending upon cell-type, differentiation state, activation and cell cycle. In terminally differentiated, nonreplicating MDM, the dNTP concentration (20-40 nM) is about 200-fold lower than that of activated CD4+ T cells (2-4 μM)11. Lentiviruses have evolved to replicate under conditions of low dNTP by virtue of a reverse transcriptase (RT) with a low Km for dNTP11,12. Nevertheless, reverse transcription of HIV-1 proceeds slowly in MDM as compared to activated T cells13,14. Addition of deoxynucleosides (dN) to the culture medium, accelerates HIV-1 reverse transcription in infected MDM, indicating that the intracellular dNTP pool is limiting in these cells11,12,15,16. Despite an efficient RT, viruses of the HIV-2/SIVsm lineage depend upon Vpx, which has no counterpart in HIV-1, for productive infection of monocytic cells17,18.
These low levels of dNTP in MDM coupled with the homology of SAMHD1 to the Enterococcus nucleotide metabolic enzyme, EF1143, led us to hypothesize that SAMHD1 regulates the intracellular dNTP pool, influencing the efficiency of reverse transcription. Here we demonstrate that SAMHD1 presents a metabolic barrier to lentiviral reverse transcription by limiting the dNTP supply in MDM, thereby blocking virus replication and that Vpx enhances dNTP levels by reducing the amount of SAMHD1 in the cell.
RESULTS
SAMHD1 is a trinucleotide phosphohydrolase that regulates the intracellular pool of deoxynucleotide triphosphates
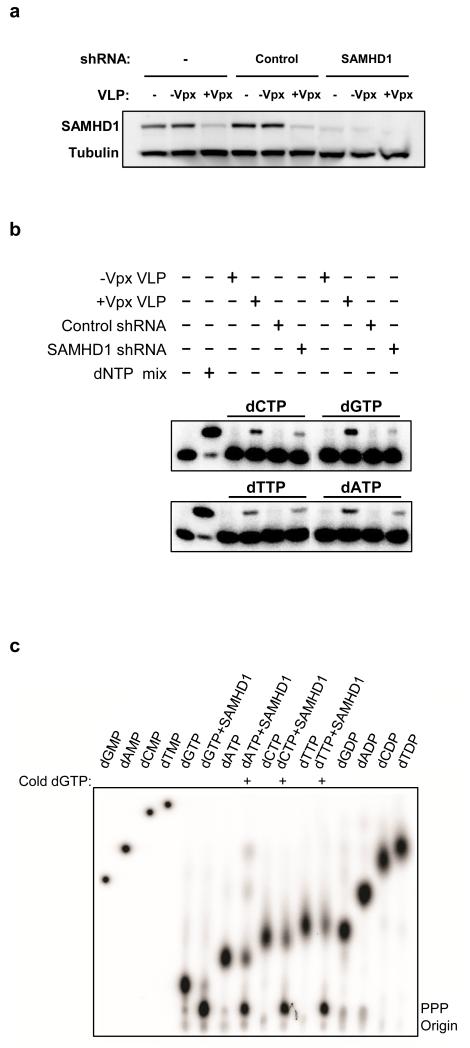
To test whether SAMHD1 controls the intracellular dNTP pool, we determined the effect of SAMHD1 knock-down on dNTP levels in Phorbol 12-myristate 13-acetate (PMA) treated THP-1 cells. PMA-treated THP-1 cells model MDM with respect to their nonpermissiveness for Δvpx SIV and their sensitivity to Vpx-containing VLP (1,19 and Supplementary Fig. 2). To quantify dNTP, we used the single nucleotide elongation assay described by Diamond et al.11 in which a 5′-32P labeled primer is extended one nucleotide by reverse transcriptase, individually, for each of the four dNTP. In THP-1 cells in which SAMHD1 was stably knocked down by transduction with shRNA (Fig. 1a), the dNTP pool was elevated compared to control cells (Fig. 1b). Pre-incubation of PMA-treated THP-1 cells with Vpx-containing VLP also increased the dNTP pool, mimicking the effect of SAMHD1 knock-down (Fig. 1b). This finding suggests that Vpx increases the dNTP supply by acting on SAMHD1.
Figure 1.
SAMHD1 knockdown and Vpx enhance the dNTP pool in PMA-treated THP-1 cells. (a) THP-1, engineered to stably express shRNA scrambled (control) or shRNA specifically targeting SAMHD1, were differentiated overnight with PMA. Where indicated, the cells were incubated for 2 h with Vpx-containing and control VLP (+/−Vpx). Cell extracts were analyzed by western blot with the indicated antibodies. (b) dNTP concentrations were determined by the single nucleotide incorporation assay described in Diamond et al 11. The experiment was performed in duplicate and one representative result is shown. (c) dNTP triphosphohydrolase activity of recombinant SAMHD1. dNTPase activity assay was performed as described in the methods section in the presence of the indicated dNTP, 1 μCi of the corresponding γ-32P-dNTP and 1 μM of wild type SAMHD1. Where indicated 200 μM of unlabeled dGTP was added. The non-labeled standards were visualized by UV shadowing and γ-32P-labeled nucleotides were visualized using a phosphorimager (Fluorescent Image Analyzer FLA3000 (Fuji).
Because of its ability to decrease intracellular levels of dNTP and in light of its homology to bacterial nucleotide hydrolase, we tested recombinant SAMHD1 for the ability to hydrolyze dNTP in vitro. We incubated recombinant SAMHD1 separately with each of the four deoxynucleotides and analyzed the reaction products by thin-layer chromatography. We found that SAMHD1 hydrolyzed dGTP, liberating inorganic triphosphate (Fig. 1c) but did not degrade the other dNTP (data not shown). However, addition of unlabeled dGTP, stimulated the hydrolase activity of SAMHD1 for the other three nucleotide triphosphates. Analysis of the cleavage products shows that the three phosphates were removed in a single step, resulting in free inorganic triphosphate (supplementary Fig. 3). During the preparation of this manuscript, Goldstone et al.20 and Powell et al.21 reported that SAMHD1 is a dGTP-activated dNTP triphosphohydrolase20,21, consistent with our findings.
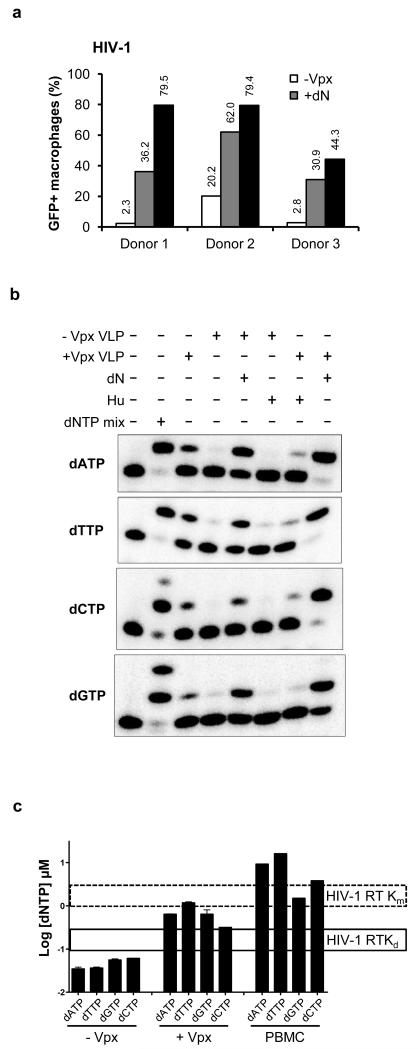
Next, we tested the role of SAMHD1 in regulating the dNTP pool in primary MDM. We reasoned that if SAMHD1 limits virus infection by decreasing the availability of dNTP, providing a source of dNTP to the MDM would relieve the restriction to HIV-1 infection. To increase the level of intracellular dNTP, we treated the cells with exogenous dN. These are taken-up and directly converted to dNTP through the salvage pathway of dNTP synthesis, increasing the intracellular dNTP pool13,22,23. In addition, if SAMHD1 serves to decrease the levels of dNTP in MDM, then inducing its degradation by the addition of Vpx-containing VLP ought to increase the dNTP levels. To determine whether dN treatment would relieve the block to HIV-1 infection, we treated MDM purified from the peripheral blood mononuclear cells (PBMC) of three healthy donors with dN or with Vpx-containing and control VLP (Fig. 2a). We found that dN treatment and Vpx-containing VLP both enhanced HIV-1 infection as compared to the untreated cells. The dN treatment was less effective than the Vpx-containing VLP at relieving the block to infection, probably because the cells still have SAMHD1 that continues to remove dNTP as they are generated in the cell. To determine the effect of the dN and Vpx treatments on each of the four dNTPs, we measured their intracellular concentrations in the single nucleotide extension assay (Fig. 2b). We found that Vpx-containing VLP and dN treatment increased the level of the four dNTPs in the MDM (Fig. 2a), increasing their levels 5 to 33-fold, depending on the nucleotide (Supplementary Table 1). To further test the role of SAMHD1 in regulating dNTP concentrations in MDM, we treated the cells with hydroxyurea (HU). HU is an inhibitor of ribonucleotide reductase (RNR), the enzyme that converts rNDP into dNDP24. If SAMHD1 reduces the levels of dNTPs in MDM, then the increase in dNTP that occurs when SAMHD1 is degraded by Vpx ought to be prevented by inactivation of RNR with HU. This would occur because even without the hydrolysis of dNTP by SAMHD1 there would be no source of dNTP production. The results showed that, consistent with this hypothesis, HU blocked the Vpx-mediated increase in the dNTP supply (Fig. 2b). The results further show that production of dNTP by RNR is required for Vpx to be effective. SAMHD1 is not expressed in activated CD4+ T cells and as a result, the cells are fully susceptible to HIV-1 and Δvpx SIV. Moreover, treatment of primary activated CD4+ cells with Vpx-containing VLP had only a slight effect on the dNTP level, demonstrating that Vpx acts in the cell by directly targeting SAMHD1 (supplementary Fig. 4). In activated cells, it is likely that the dNTPs degraded by SAMHD1 are rapidly replaced. In addition, replicating cells may regulate the activity of SAMHD1 to maintain the high levels of dNTP that are required in S phase of the cell cycle. SAMHD1, expressed in SupT1 T cells from a retroviral vector, also failed to inhibit viral infection but when RNR was inhibited by treatment with HU, the dNTP levels dropped, revealing the SAMHD1 activity in these cells (supplementary Fig. 5).
Figure 2.
Vpx increases the intracellular pool of dNTP in MDM. (a) MDM (2 × 106) from three healthy donors were pre-incubated for 4 h with Vpx-containing or control VLP or with 1.5 mM dN. The cells were infected with HIV-GFP reporter virus (MOI=1) and the GFP+ cells were quantified by flow cytometry. (b) MDM were treated with VLP, dN or 2 mM hydroxyurea. dNTP were visualized by the single nucleotide extension method. (c) The data were quantified and are plotted as a histogram. The Km and Kd values of HIV-1 RT for dNTP substrates are indicated25.
Quantification of the individual dNTP concentrations, shows that Vpx-containing VLP increased the dNTP level to about 5-fold below that of activated PBMC, a level that is above the Kd but below the Km of HIV-1 RT25 (Fig. 2c). Thus, in MDM, unlike activated T cells, the dNTP concentration is suboptimal for proviral DNA synthesis. Upon delivery of Vpx to the cells, the dNTP concentration rose to a level higher than the Kd and closer to the Km for HIV-1 RT. This finding predicts that by elevating the level of dNTP in MDM, Vpx serves to accelerate the rate of proviral DNA synthesis by RT.
The antiviral activity of SAMHD1 is mediated by a dramatic reduction of the intracellular dNTP pool
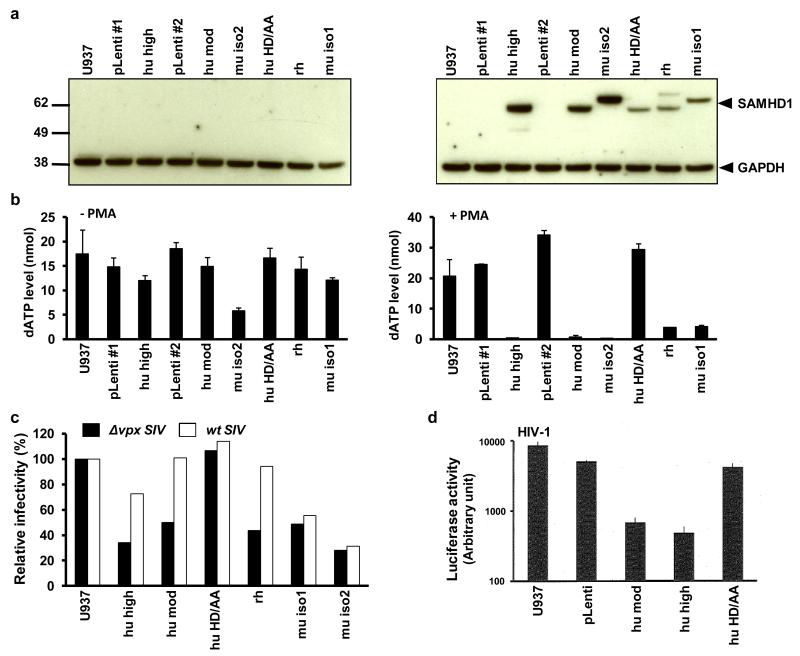
To determine whether SAMHD1 is sufficient to reduce the intracellular dNTP level, we transduced U937 cells, a monocytoid cell line that does not express endogenous SAMHD1, with lentiviral expression vectors (Fig. 3a). U937 cells are highly susceptible to HIV-1 infection. They are somewhat more susceptible than THP-1 cells that have been treated with Vpx-containing VLP or that have been stably depleted of SAMHD1 by shRNA transduction (supplementary Fig. 6a). The cell lines express human SAMHD1 at high or moderate level (hu-SAMHD1 high and mod), mouse isoform 2 and 1 (mu-iso2 and mu-iso1), rhesus macaque SAMHD1 (rh-SAMHD1) or human SAMHD1 with the consensus histidine and aspartate residues mutated to alanine (SAMHD1 HD/AA). Matched control lentiviral vector cell lines were established in parallel (pLenti-1 and pLenti-2). Analysis of the cell lines by western blot showed that without PMA-treatment, SAMHD1 was undetectable (Fig. 3a, left). Accordingly, the dNTP levels in these cells were relatively unaffected by the lentiviral vectors (Fig. 3b and supplementary Fig. 6b). In contrast, treatment of the cells with PMA resulted in robust expression of the SAMHD1 proteins (Fig. 3a, right). Expression of SAMHD1 was accompanied by a dramatic decrease in the dNTP pool (Fig. 3b and supplementary Fig. 6c). In the two cell lines that expressed wild-type hu-SAMHD1 at high and moderate amounts, the dNTP level was reduced 41- and 20-fold, respectively. In the mouse iso-2 cell line, in which SAMHD1 was expressed at the highest level, the dNTP level was reduced 53-fold. The HD/AA mutant had no effect on the dNTP level, indicating that the reduction was dependent on SAMHD1 catalytic activity. rh-SAMHD1 and mouse iso-1 which were expressed at a somewhat lower level, caused a less pronounced decrease in dNTP concentration. Interestingly, in the U937 cells not treated with PMA, a small reduction in dNTP level was detected in the cell line that expressed mouse isoform 2 SAMHD1 (Fig. 3a, left), suggesting that expression of even trace amounts of SAMHD1 were sufficient to detectably impact the dNTP level.
Figure 3.
SAMHD1 reduces the intracellular dNTP pool and is counteracted by human and rhesus but not mouse SAMHD1. U937 cells were transduced with a control lentiviral vector (pLenti), or with a vector that expressed human SAMHD1 (hu high and hu mod), human HD206-7AA SAMHD1 (hu HD/AA), rhesus macaque SAMHD1(rh) or the two mouse SAMHD1 isoforms (mu iso1 and mu iso2). Two independent control cell lines were established (pLenti#1 and pLenti#2) in parallel with human SAMHD1 cell lines that expressed at high or moderate levels (hu high and hu mod). (a) The cells were either untreated (left) or PMA-treated (right) and SAMHD1 expression level was determined by immunoblot analysis. (b) dNTP levels were quantified in the untreated and PMA-treated U937 cells. (c) The PMA-treated U937 cell lines were infected with Δvpx or wild-type SIVmac239-HSA reporter viruses. The infected cells were stained with anti-murine CD24 monoclonal antibody and the infected cells were quantified by flow cytometry. (d) The cell lines were infected with HIV-1 luciferase reporter virus and luciferase activity was determined after two days.
Analysis of the infectabilty of the PMA-treated U937 cells with single-cycle wild-type and Δvpx SIVmac239 HSA (mouse CD24) reporter virus confirmed the antiviral activity of the introduced SAMHD1 (Fig. 3c). The restriction was dependent on SAMHD1 catalytic activity as shown by the lack of antiviral effect of the HD/AA mutant. Vpx counteracted the restriction mediated by human and rhesus SAMHD1 but interestingly, mouse SAMHD1 was resistant. The finding that Vpx packaged in SIV virions can counteract SAMHD1 in the target cell is important because previous reports have all relied on Vpx-containing VLP, an artificial system in which the cell is flooded with Vpx. Analysis of the cell lines with HIV-1 reporter virus showed that the SAMHD1 proteins restricted the virus and that the activity was dependent on the catalytic activity of the enzyme (Fig. 3d).
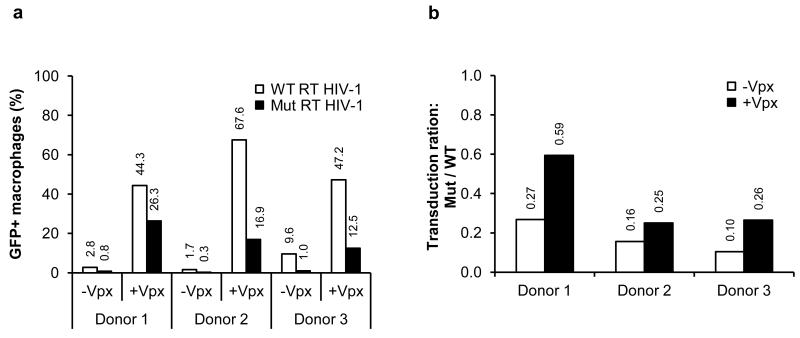
HIV-1 that carries a V148I mutation in RT, replicates in primary CD4+ T cells but not MDM. The inability of the virus to replicate in MDM is caused by an increased Km and reduced binding affinity of the mutant RT for dNTP coupled with the low level of dNTP in these cells11,26. If SAMHD1 is responsible for the low level of dNTP in MDM, we hypothesized that causing SAMHD1 degradation by the introduction of Vpx would rescue the ability of the V148I mutant HIV-1 to infect cells. To test this hypothesis, we added Vpx-containing and control VLP to MDM isolated from three donors, infected the cells with wild-type and V148I HIV-GFP and quantified the number of infected cells by flow cytometry. We found that the V148I RT mutant was less infectious than the wild-type virus and that Vpx enhanced the infectivity of the wild-type and mutant viruses (Fig. 4a). However, in the presence of Vpx, the difference in infectivity of the wild-type and mutant viruses was less pronounced (Fig. 4b). Although this difference was small, it supports the hypothesis that SAMHD1 reduces the availability of dNTP, limiting the ability of HIV-1 to productively infect MDM. SAMHD1 could be more restrictive to viruses that have a RT with reduced affinity for dNTP as that may be the case for primary viruses in vivo.
Figure 4.
Vpx rescues the infection of an HIV-1 that encodes a mutant RT with a reduced affinity for dNTP. (a) MDM were pre-incubated with Vpx-containing or control VLP and then infected with wild-type or RT V148I mutant HIV-GFP at an MOI of 1. Four days post-infection, GFP+ cells were quantified by flow cytometry. (b) The efficiency of infection in the presence and absence of Vpx is displayed as the ratio of mutant/wild-type virus. A ratio of 1 indicates equivalent infection efficiency of wild-type and RT V148I mutant HIV-1.
Providing extra-cellular nucleotides to MDM compensates for Vpx
If SAMHD1 restricts virus replication by limiting the availability of dNTP, then the block to Δvpx SIV (a virus that is unable to degrade SAMHD1) ought to be relieved by providing a source of dNTP to the cell. To test this hypothesis, MDM from three healthy donors were exposed to an increasing concentration of dN together with Vpx-containing VLP or control VLP and then infected with Δvpx SIVsm-GFP (Fig. 5). The results showed that as the dN concentration increased, the infectivity of the Δvpx SIV without Vpx approached that of the virus with Vpx. This finding further supports the hypothesis that SAMHD1 restricts lentivirus replication by reducing the intracellular pool of dNTP and that the role of Vpx is to induce the degradation of SAMHD1 to increase the intracellular pool of dNTP.
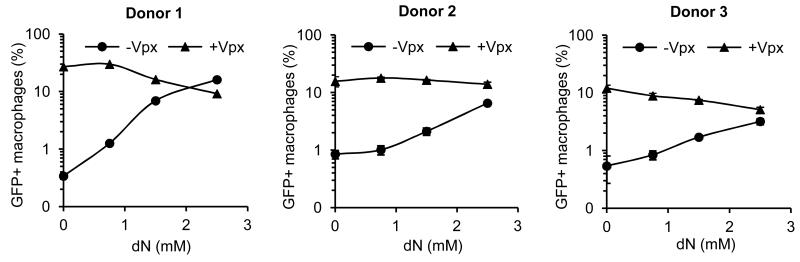
Figure 5.
The salvage pathway of dNTP synthesis partially rescues infectivity of Δvpx SIVmac in MDM. MDM were pre-incubated with Vpx-containing and control VLP and were infected with Δvpx SIVmac GFP reporter virus (MOI=1). The salvage pathway of dNTP synthesis was stimulated by addition of 0.75, 1.5 or 2.5 mM dN to the media (from 2 h before infection to 20 h post-infection) where indicated. Four days post-infection, the percent GFP+ MDM was determined by flow cytometry. Three representative donors are shown.
DISCUSSION
Taken together, our findings demonstrate that the primary mechanism by which SAMHD1 restricts lentivirus infection is to reduce the intracellular pool of dNTP. SAMHD1 activity is sufficient to reduce the level of dNTP below the Km of RT, thereby blocking the synthesis of viral reverse transcripts in the newly infected target cell. To counteract the restriction, lentiviruses of the HIV-2/SIVsm lineage have evolved Vpx, an accessory protein that induces the proteosomal degradation of SAMHD1 as a means of increasing the dNTP level to an extent that permits efficient viral DNA synthesis. Curiously, HIV-1 lacks the ability to degrade SAMHD1 and as a result, is restricted in its ability to infect MDM and DC. It has been suggested that for HIV-1, the restriction may be a strategy to avoid provoking an innate immune response in these cells27. The block to viral DNA synthesis imposed in MDM by SAMHD1 would preclude the activation of cytoplasmic nucleic acid sensors, thereby limiting the induction of inflammatory cytokines. DC have been shown to express a cryptic receptor that detects HIV-1 and activates a potent inflammatory response27. This sensor is thought to act late in the virus replication cycle and thus is not likely to be SAMHD1 itself. Nevertheless, as SAMHD1 is interferon-inducible, the sensor could serve to activate SAMHD1 expression in nearby cells.
SAMHD1 restriction is novel and differs from the restriction factors Trim5α, APOBEC3G and BST-2/tetherin by acting on a cellular factor rather than a viral component (reviewed in28). As for APOBEC3G and BST2/tetherin, SAMHD1 is counteracted by a viral accessory protein that induces its degradation. In addition to inducing the degradation of SAMHD1, Vpx has been reported to counteract the cellular cytidine deaminase APOBEC3A29. The role of Vpx in counteracting APOBEC3A is unclear, but it is possible that under conditions of low dNTP caused by SAMHD1, in which reverse transcription is slowed, APOBEC3A more effectively deaminates nascent reverse transcripts. It is noteworthy that a large homozygous deletion in the SAMHD1 gene has been associated with mitochondrial DNA deletions, as mitochondrial diseases are often the result of mutations in the genes encoding enzymes that are involved in dNTP metabolism30-32.
A distinctive feature of lentiviruses among the retrovirus family is their ability to infect nondividing cells33. Unlike gamma retroviruses such as MLV that depend on nuclear membrane dissolution occurring during mitosis, lentiviruses, following synthesis of their double-stranded DNA genome, negotiate the barrier imposed by the nuclear membrane by traversing the nuclear pore34-38. The cellular and viral determinants that allow the lentiviral preintegration complex to traverse the nuclear pore have been the subject of intensive study39. A less appreciated challenge faced by lentiviruses is the limited supply of dNTP present in nondividing and quiescent cells13,40. In myeloid cells targeted by HIV-1, the problem presented by the low level of dNTP may have been under-appreciated because of the remarkable adaptation of lentiviruses to unfavourable reverse transcription conditions by having evolved an RT with a low Km for dNTP11,12,15,16. The requirement by other retroviruses for a high level of dNTP precludes processivity of reverse transcription in quiescent cells and nondividing cells in which the dNTP level is low41,42. Our findings highlight the fact that despite their efficient RT, HIV-1 and HIV-2 remain sensitive to the low dNTP content in the cytoplasm of their natural targets.
The role of SAMHD1 in limiting the supply of dNTP provides a novel mechanism by which the host attempts to limit virus replication. To successfully replicate their genomes, DNA viruses and viruses that replicate through a DNA intermediate need access to an intracellular supply of dNTP. Viruses have evolved several strategies to ensure dNTP availability. Small DNA viruses preferentially infect mitotic cells; adenoviruses, polyomaviruses and papillomaviruses encode proteins that drive quiescent cells into S phase43; herpesviruses and poxviruses encode an RNR that converts NTP to dNTP43,44. By reducing the pool of available dNTP, SAMHD1 effectively starves the virus of a building block at the heart of its replication strategy. Lentiviruses of the HIV-2/SIV lineage counteract this host defense through Vpx. It is paradoxical that although HIV-1 is sensitive to SAMHD1-mediated restriction it lacks a Vpx gene. Whether it has another mechanism to counteract the effects of SAMHD1 on dNTP levels or whether its RT is robust enough to synthesize dNTP in the face of the low dNTP environment will require further investigation. It is noteworthy that HIV-2 reverse transcriptase appears less active than the HIV-1 enzyme45.
The advent of Vpx by lentiviruses as a means of removing SAMHD1 demonstrates an ingenious and unexpected strategy by which lentiviruses thwart cellular antiviral defenses. The ability of Vpx packaged in an incoming virion to promote SAMHD1 degradation such that dNTP concentration is increased to an extent sufficient to allow reverse transcription is remarkable. The mechanism explains how SAMHD1, which is localized in the nucleus, exerts its phenotype in the cytoplasm where the virus is reverse transcribed. These findings bring to light a new and unexpected mechanism of innate immunity to virus infection and raise the question of whether pharmacologic alteration of intracellular dNTP pools could be a new therapeutic approach to virus infection. The implications of nucleotide starvation as a host defense mechanism are potentially far reaching.
METHODS
Plasmid construction
HA-tagged SAMHD1 constructs were generated by PCR using primers containing BamH-I and Xho-I cleavage sites. The PCR products were cloned into the BamH-I and Sal-I sites of pLenti-puro lentiviral vector (Cell Biolabs). Human, rhesus and mouse SAMHD1 were amplified from cDNA generated by reverse transcription of RNA from THP-1 cells, rhesus PBMCs and mouse bone marrow-derived DCs using transcriptor reverse transcriptase (Roche).
Cells
Human monocytes were purified from buffy coats purchased from “Etablissement Français du Sang” (EFS). PBMC were prepared by ficoll density gradient separation (GE-Healthcare). Monocytes were isolated by positive selection on CD14 magnetic microbeads (Miltenyi Biotec). Purified monocytes were cultured in BD Primaria™ flasks with R10 medium (RPMI 1640 GlutaMAX™-I, 10 mM HEPES, 1 mM sodium pyruvate, 1% nonessential amino acids, 10% heat-inactivated FCS) and antibiotics. MDM were obtained from monocytes by culturing the cells for 7 to 9 days in R10 medium containing 10 ng/ml GM-CSF and 20 ng/ml M-CSF. The cells were detached using PBS containing 13 mM lidocaine and 10 mM EDTA. THP-1 cells were cultured in R10 medium. HeLa and 293T cells were cultured in Dulbecco’s Modified Eagle Medium glutaMAX™-I containing 10% FCS and antibiotics.
THP-1 cells expressing scrambled shRNA (control) or shRNA-N°4 specifically targeting SAMHD1 were already described1.
To establish U937 cells that stably expressed SAMHD1, lentiviral vector stocks were generated by calcium phosphate transfection of 293T with pLenti vectors for human, rhesus and mouse SAMHD1 and pRSV-Rev, pMDG gag/pol and VSV-G expression plasmid. U937 cells were spin infected with lentiviral vectors and the transduced cells were selected in 1μg/ml puromycin.
Immunoblot analysis
SAMHD1 stable U937 cells lines were differentiated with 30 ng/mL PMA for 20 h, lysed in RIPA buffer containing Halt Protease Inhibitor (ThermoScientific) and normalized for protein content by Bradford assay. Whole cell lysates containing 10 μg total protein were separated by SDS PAGE on a 4-12% gradient gel. The proteins were transferred to a PVDF membrane and probed with a mixture of an anti-HA Mab HA.11 (Covance) and an anti-GAPDH Mab (Ambion) and probed with goat anti-mouse horse radish peroxidase-conjugated antibody (Pierce). The proteins were visualized using Super Signal West Pico chemiluminescent substrate (ThermoScientific). SAMHD1 and alpha-tubulin expression in THP-1 cells was controlled using antibodies purchased respectively from Abcam (ab67820) and Sigma (T9026).
Viruses, VLP and infections
Viruses were produced in 293T cells co-transfected with reporter virus plasmid and VSG-G by the calcium phosphate method. Wild-type and RT V148I mutant HIV-1 GFP reporter viruses were produced by cotransfection of 293T cells with pRRLsin.eGFP and pCMVΔ8.2 or pCMVΔ8.2 V148I and VSV-G plasmid46-48. Δvpx SIVmac virus was produced by cotransfection with the pSIV3+ Δvpx and the minimal SIV genome pGAE 1.0 in which GFP expression is driven by the CMV promoter49-51. Vpx-containing and control VLP were produced by transfecting 293T cells with pSIV3+ (VLP Vpx+) or pSIV3+ Δvpx (VLP Vpx−) in the absence of a viral genome49-51. The culture supernatants were harvested and filtered 24 to 48 h post-transfection and concentrated by polyethylene glycol precipitation.
The cells were infected as previously described for DC infections49. The cells were pre-incubated with VLP for 2 h before infection. The minimal amount of VLP (Vpx+) required for a maximal helper effect towards HIV-1 infection was determined by titration. Equivalent amounts of Vpx-containing and control VLP normalized by p27 capsid protein by ELISA. After pre-incubation, the cells were infected for 2 h with a GFP reporter virus at an MOI of 1 predetermined by titration on HeLa cells. The infected cells were treated with dN which consisted of a mixture of dA (D8668), dC (D0776), dG (D0901) and dT (T1895) (Sigma Aldrich) or with 2 mM hydroxyurea. Four days post-infection, the GFP+ cells were quantified by flow cytometry.
Δenv SIVmac239-HSA and ΔenvΔvpx SIVmac239-HSA reporter viruses were produced as VSV-G pseudotypes. The viruses were titered on 293T cells by determining the number of CD24+ cells by flow cytometry. U937 cells were differentiated for 20 h in 30 ng/ml PMA and then infected at an MOI of 1 with reporter virus. The infected cells were quantified by flow cytometry. For luciferase reporter viruses, the cells were infected with 10 ng VSV-G pseudotyped NL.LucE−R−. Luciferase activity was measured after 48 h using Steadylite Luminescence Reporter Gene Assay (Perkin Elmer).
Quantification of whole-cell dNTP pool
dNTP were quantified as previously described11. Briefly, the cells were washed twice with ice-cold PBS and lysed in 60% (V/V) aqueous methanol at −20°C for 30 min. The lysates were heated 7 min at 100°C and clarified by centrifugation at 16,000 × g for 15 min at 4° C. The supernatant fraction was dried under vacuum, resuspended in ultrapure water and stored at −80°C. A 32P-5′-end labeled 18-mer primer (5′-GTCCCTGTTCGGGCGCCA-3′) was annealed to four different 19-mer templates (5′-NTGGCGCCCGAACAGGGAC-3′) with nucleotide variation at the 5′-end (primer/template 1:4 ratio). A total of 200 fM template/primer complex, 2 μl of 0.5 mM dNTP mix or cell extract, 25 mM Tris-HCl pH8.0, 2 mM DTT, 100 mM KCl, 5 mM MgCl2, and 5 μM oligo dT were used in each 20 μl reaction. The reactions were incubated at 37° C for 5 min in the presence of excess HIV-1 RT and terminated by adding 10 μl stop dye (40 mM EDTA and 99% formamide) and denatured for 5 min at 95° C. The extended and unextended products were resolved by 16% urea PAGE and analyzed on a phosphorimager. The percentage of extended primers (sum of extended and unextended) was determined with QuantityOne software. The dNTP content of a standard dilution of the samples was determined. The sample volume was then adjusted to obtain a signal within the linear range of the assay. Addition of pure dNTP to dNTP prepared from SAMHD1 expressing cells showed that the data were not influenced by the presence of inhibitors in the cellular dNTP preparation (not shown).
dNTPase activity assay
The assay was performed in 50 mM Tris-HCl pH 8.0, 50 mM KCl, 5 mM MgCl2, 0,1% Triton X-100, 200 μM of the indicated dNTP, 1 μCi of the corresponding γ-32P-dNTP and 1 μM of wild-type SAMHD1 at 37°C for 3 h. Where indicated, 200 μM of unlabeled dGTP was added to the reaction. The reaction was stopped by incubating for 5 min at 70°C. Samples were separated on polyethyleneimine cellulose thin-layer chromatography (TLC) plates (Macherey Nagel) using an 0.8 M LiCl mobile phase. After drying the TLC plates, the unlabeled standards were visualized by UV shadowing and γ-32P-labeled nucleotides were visualized using a phosphorimager (Fluorescent Image Analyzer FLA3000 (Fuji)).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the Agence Nationale de la Recherche sur le SIDA et les hépatites virales (ANRS), SIDACTION, Fondation de France, the American Foundation for AIDS Research and the National Institutes of Health (grants AI049781 and A1077401 to BK, A1067059 to NRL and F31 GM095190 to WD). Work in MB’s lab was supported by grants from the ERC (250333) and ANRS. HL was supported by ANRS; CM and DA by Paris Diderot University and the Ministère de l’Enseignement Supérieur et de la Recherche. MB and NL were supported by SIDACTION fellowships. The authors acknowledge Laurence Stouvenel and Karine Labroquère from the Cochin Flow Cytometry Facilitie. They thank Joseph Hollenbaugh and Steven Dewhurst for critical reading of the manuscript. The authors declare no competing financial interests.
Footnotes
AUTHOR CONTRIBUTIONS
HL, WD, MoB, NRL, CT, BK and FMG conceived and designed the experiments. HL, MoB, CT, BK, NRL and FMG wrote the paper. DA, HH, ECL, NB, NBLD, CM, MaB, BC and SP designed some experiments.
References
- 1.Laguette N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rice GI, et al. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmerman MD, Proudfoot M, Yakunin A, Minor W. Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5′-deoxyribonucleotidase YfbR from Escherichia coli. J Mol Biol. 2008;378:215–226. doi: 10.1016/j.jmb.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vorontsov II, et al. Characterization of the deoxynucleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex. J Biol Chem. 2011;286:33158–33166. doi: 10.1074/jbc.M111.250456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crow YJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 7.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, Conway JA, Kim J, Kappes JC. Localization of the Vpx packaging signal within the C terminus of the human immunodeficiency virus type 2 Gag precursor protein. J Virol. 1994;68:6161–6169. doi: 10.1128/jvi.68.10.6161-6169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sunseri N, O’Brien M, Bhardwaj N, Landau NR. HIV-1 modified to package SIV Vpx efficiently infects macrophages and dendritic cells. J Virol. 2011;85:6263–6274. doi: 10.1128/JVI.00346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goujon C, et al. With a little help from a friend: increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIV(MAC) Gene Ther. 2006;13:991–994. doi: 10.1038/sj.gt.3302753. [DOI] [PubMed] [Google Scholar]
- 11.Diamond TL, et al. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J Biol Chem. 2004;279:51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodside AM, Guengerich FP. Effect of the O6 substituent on misincorporation kinetics catalyzed by DNA polymerases at O(6)-methylguanine and O(6)-benzylguanine. Biochemistry. 2002;41:1027–1038. doi: 10.1021/bi011495n. [DOI] [PubMed] [Google Scholar]
- 13.O’Brien WA, et al. Kinetics of human immunodeficiency virus type 1 reverse transcription in blood mononuclear phagocytes are slowed by limitations of nucleotide precursors. J Virol. 1994;68:1258–1263. doi: 10.1128/jvi.68.2.1258-1263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collin M, Gordon S. The kinetics of human immunodeficiency virus reverse transcription are slower in primary human macrophages than in a lymphoid cell line. Virology. 1994;200:114–120. doi: 10.1006/viro.1994.1169. [DOI] [PubMed] [Google Scholar]
- 15.Furge LL, Guengerich FP. Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo- and human immunodeficiency virus-1 reverse transcriptase using steady-state and pre-steady-state kinetics. Biochemistry. 1997;36:6475–6487. doi: 10.1021/bi9627267. [DOI] [PubMed] [Google Scholar]
- 16.Ueno T, Shirasaka T, Mitsuya H. Enzymatic characterization of human immunodeficiency virus type 1 reverse transcriptase resistant to multiple 2′,3′-dideoxynucleoside 5′-triphosphates. J Biol Chem. 1995;270:23605–23611. doi: 10.1074/jbc.270.40.23605. [DOI] [PubMed] [Google Scholar]
- 17.Fletcher TM, 3rd, et al. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM) EMBO J. 1996;15:6155–6165. [PMC free article] [PubMed] [Google Scholar]
- 18.Kawamura M, Sakai H, Adachi A. Human immunodeficiency virus Vpx is required for the early phase of replication in peripheral blood mononuclear cells. Microbiol Immunol. 1994;38:871–878. doi: 10.1111/j.1348-0421.1994.tb02140.x. [DOI] [PubMed] [Google Scholar]
- 19.Goujon C, et al. Characterization of simian immunodeficiency virus SIVSM/human immunodeficiency virus type 2 Vpx function in human myeloid cells. J Virol. 2008;82:12335–12345. doi: 10.1128/JVI.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011 doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 21.Powell RD, Holland PJ, Hollis T, Perrino FW. The Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J Biol Chem. 2011 doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyerhans A, et al. Restriction and enhancement of human immunodeficiency virus type 1 replication by modulation of intracellular deoxynucleoside triphosphate pools. J Virol. 1994;68:535–540. doi: 10.1128/jvi.68.1.535-540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamburuthugoda VK, Chugh P, Kim B. Modification of human immunodeficiency virus type 1 reverse transcriptase to target cells with elevated cellular dNTP concentrations. J Biol Chem. 2006;281:13388–13395. doi: 10.1074/jbc.M600291200. [DOI] [PubMed] [Google Scholar]
- 24.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy EM, et al. Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J Biol Chem. 2010;285:39380–39391. doi: 10.1074/jbc.M110.178582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diamond TL, et al. Mechanistic understanding of an altered fidelity simian immunodeficiency virus reverse transcriptase mutation, V148I, identified in a pig-tailed macaque. J Biol Chem. 2003;278:29913–29924. doi: 10.1074/jbc.M211754200. [DOI] [PubMed] [Google Scholar]
- 27.Manel N, et al. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 2010;467:214–217. doi: 10.1038/nature09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neil S, Bieniasz P. Human immunodeficiency virus, restriction factors, and interferon. J Interferon Cytokine Res. 2009;29:569–580. doi: 10.1089/jir.2009.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger G, et al. APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog. 2011;7:e1002221. doi: 10.1371/journal.ppat.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leshinsky-Silver E, et al. A large homozygous deletion in the SAMHD1 gene causes atypical Aicardi-Goutieres syndrome associated with mtDNA deletions. Eur J Hum Genet. 2011;19:287–292. doi: 10.1038/ejhg.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bourdon A, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 32.Brahimi N, et al. The first founder DGUOK mutation associated with hepatocerebral mitochondrial DNA depletion syndrome. Mol Genet Metab. 2009;97:221–226. doi: 10.1016/j.ymgme.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Yamashita M, Emerman M. Retroviral infection of non-dividing cells: old and new perspectives. Virology. 2006;344:88–93. doi: 10.1016/j.virol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 34.Bieniasz PD, Weiss RA, McClure MO. Cell cycle dependence of foamy retrovirus infection. J Virol. 1995;69:7295–7299. doi: 10.1128/jvi.69.11.7295-7299.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis P, Hensel M, Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis PF, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roe T, Reynolds TC, Yu G, Brown PO. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinberg JB, Matthews TJ, Cullen BR, Malim MH. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J Exp Med. 1991;174:1477–1482. doi: 10.1084/jem.174.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamashita M, Emerman M. The cell cycle independence of HIV infections is not determined by known karyophilic viral elements. PLoS Pathog. 2005;1:e18. doi: 10.1371/journal.ppat.0010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Triques K, Stevenson M. Characterization of restrictions to human immunodeficiency virus type 1 infection of monocytes. J Virol. 2004;78:5523–5527. doi: 10.1128/JVI.78.10.5523-5527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chowdhury K, Kaushik N, Pandey VN, Modak MJ. Elucidation of the role of Arg 110 of murine leukemia virus reverse transcriptase in the catalytic mechanism: biochemical characterization of its mutant enzymes. Biochemistry. 1996;35:16610–16620. doi: 10.1021/bi961462l. [DOI] [PubMed] [Google Scholar]
- 42.Shi Q, Singh K, Srivastava A, Kaushik N, Modak MJ. Lysine 152 of MuLV reverse transcriptase is required for the integrity of the active site. Biochemistry. 2002;41:14831–14842. doi: 10.1021/bi0258389. [DOI] [PubMed] [Google Scholar]
- 43.Lembo D, Brune W. Tinkering with a viral ribonucleotide reductase. Trends Biochem Sci. 2009;34:25–32. doi: 10.1016/j.tibs.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Productive infection of primary macrophages with human herpesvirus 7. J Virol. 2001;75:10511–10514. doi: 10.1128/JVI.75.21.10511-10514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Post K, et al. Human immunodeficiency virus type 2 reverse transcriptase activity in model systems that mimic steps in reverse transcription. J Virol. 2003;77:7623–7634. doi: 10.1128/JVI.77.13.7623-7634.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skasko M, Kim B. Compensatory role of human immunodeficiency virus central polypurine tract sequence in kinetically disrupted reverse transcription. J Virol. 2008;82:7716–7720. doi: 10.1128/JVI.00120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mangeot PE, et al. High levels of transduction of human dendritic cells with optimized SIV vectors. Mol Ther. 2002;5:283–290. doi: 10.1006/mthe.2002.0541. [DOI] [PubMed] [Google Scholar]
- 48.Naldini L, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 49.Berger G, et al. A simple, versatile and efficient method to genetically modify human monocyte-derived dendritic cells with HIV-1-derived lentiviral vectors. Nat Protoc. 2011;6:806–816. doi: 10.1038/nprot.2011.327. [DOI] [PubMed] [Google Scholar]
- 50.Goujon C, et al. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology. 2007;4:2. doi: 10.1186/1742-4690-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gramberg T, Sunseri N, Landau NR. Evidence for an activation domain at the amino terminus of simian immunodeficiency virus Vpx. J Virol. 2010;84:1387–1396. doi: 10.1128/JVI.01437-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.