Abstract
The synthetic homotetrameric ββα (BBAT1) protein possesses a stable quaternary structure with a ββα fold. Because of its small size (a total of 84 residues), the homotetramer is an excellent model system with which to study the self-assembly and protein-protein interactions. We find from replica exchange molecular dynamics simulations with the coarse-grain UNRES force field that the folding and association pathway consists of three well-separated steps, where that association to a tetramer precedes and facilitates folding of the four chains. At room temperature the tetramer exists in an ensemble of diverse structures. The crystal structure becomes energetically favored only when the molecule is put in a dense and crystal-like environment. The observed picture of folding promoted by association may mirror the mechanism according to which intrinsically unfolded proteins assume their functional structure.
Keywords: Protein association, protein folding, molecular dynamics
1 Introduction
Self-assembly of proteins1,2 and protein complexes3 are key processes in the biochemistry of cells.4 Knowledge of these processes is crucial for the understanding of disease pathways and for insight into the working of drugs at the level of cells. Take as an example the multi drug exporter AcrB, a 1053 residue protein that needs to assemble into a homotrimer in order to be functional.5 This suggests that the assembly of the trimer is a potential target for developing new drugs in the fight against multi-drug resistant bacteria and, for this reason, various groups are studying the mechanism of the association of AcrB.6 However, probing such association processes in experiments can be a challenge.
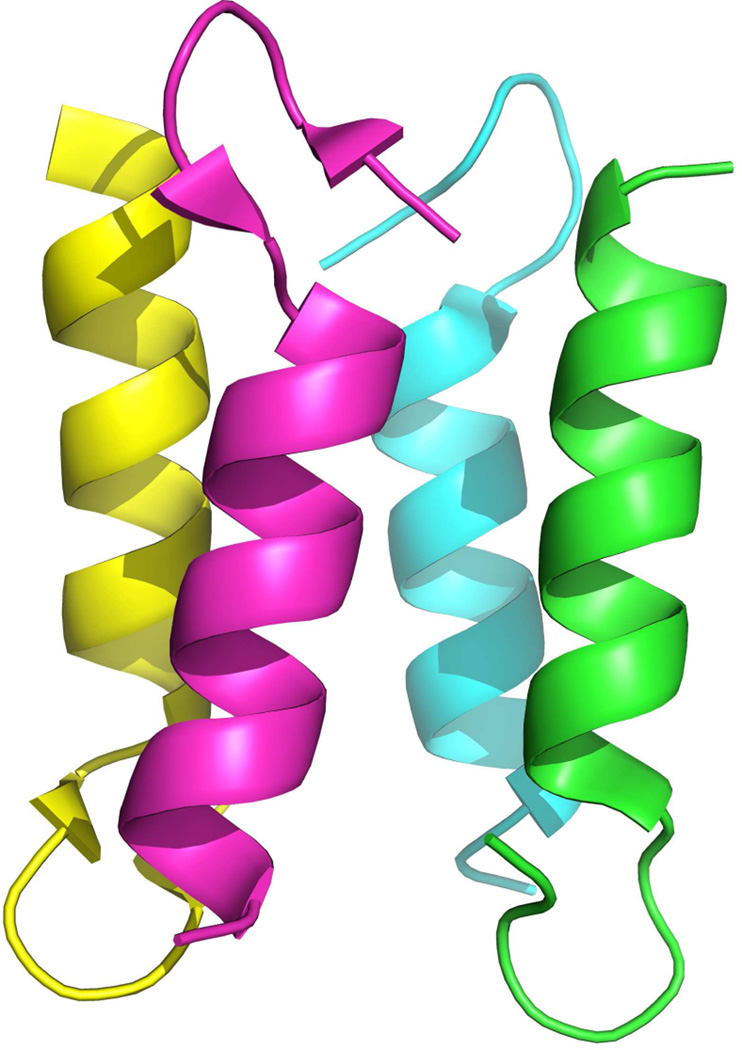
Computer simulations can complement experiments,7 but are not yet at a point that they enable one to study protein association of proteins such as AcrB with more than 1000 residues. Hence, there is a need for model systems for which one can study the mechanism of folding and association of functional protein assemblies in a systematic way. A prime candidate is the homotetrameric ββα motif (BBAT1)8 of figure 1, one of the simplest examples for self-assembly of protein complexes. It is build from a synthetic 21-residue mini-protein with a ββα-fold9,10 that has been derived from a closely related protein with similar fold and size, BBA5. The aim of this paper is to explore the interplay between the folding of monomers and association to the tetramer.
Fig. 1.
Crystal structure of a homologue of BBAT1 (PDB: 1SN9). The pictures was prepared with the PyMOL software.36
While BBAT1 is one of the smallest protein complexes, with a total of 84 residues it still poses an enormous computational challenge. This is because all-atom molecular dynamics simulations are limited to small systems and time scales. Even for very small (50 amino-acid residue) systems and with the use of special purpose computers one is restricted to time scales of 200 – 500µs.11 Overcoming these sampling difficulties requires a combination of enhanced sampling techniques12 and coarse-grained models.13
In the present paper, we use multiplexed replica exchange molecular dynamics simulations with the UNRES force field14–17 to determine the folding and association pathway of BBAT1 in solution. We study for this simple system the coupling between folding and conformational changes of the monomers and their assembly into a tetramer. Association of the monomers precedes their folding, and distinct intermediates are observed before the BBAT1 assumes its final fold. The folding of the individual chains follows the same mechanism as for isolated monomers but the various transitions along the folding pathway are facilitated by the tetramer environment. Tetramers with various different symmetries but equal energies are observed in our simulations, and the crystal structure becomes energetically favored only when the molecule is put in a crystal-like environment. We conclude with discussing the implication of the observed mechanism of association modulated folding.
2 Methods
2.1 UNRES force field
Our simulations rely on the UNRES force field,14–16 in which a polypeptide chain is represented by a sequence of α-carbon atoms with united side chains attached, and peptide groups positioned halfway between two consecutive α-carbons. The effective energy function is defined as the free energy of the chain constrained to a given coarse-grained conformation plus the surrounding solvent. This potential of mean force of the virtual-bond chain contains temperature dependent pre factors and its explicit form can be found in ref.17 The version of the UNRES force field calibrated with the 1GAB protein15 was used. The force field was supplemented with the new torsional, double-torsional, correlation, and side-chain rotamer terms that have recently been introduced to treat proteins with D-amino-acid residues, such as the D-proline in BBAT1. Additionally, the parameters of the side chain of the benzyloxy diaminopropionic acid [Dap(Bz)] residue of BBAT1 have been adopted from the phenylalanine residue except that the dimensions are increased and the mass changed to that of Dap(Bz).
2.2 Replica exchange simulation parameters and first-time-passage optimization
In the Replica Exchange Molecular Dynamics (REMD) method,18–21 multiple simulations are carried out simultaneously on a ladder of increasing temperatures. After every m steps, an exchange of replicas between two neighboring temperatures is attempted, and the exchange is accepted or rejected according to a Metropolis criterion that accounts for the temperature dependence of the UNRES energy function. The multiplexed variant of the REMD by Rhee and Pande22 differs from the original method in that several trajectories run at a given temperature.
The efficiency of REMD depends on the flow of replicas through temperature space, from the target temperature (usually the lowest temperature) to the highest temperature (chosen such that all relevant energy barriers can be crossed) and back. We use the techniques described in Refs.23 to derive a distribution of temperatures that maximizes this flow.
2.3 Analysis methods
For analysis of of our data we use the weighted histogram analysis method15,24 to evaluate thermodynamic quantities at any temperature. Configurations are clustered using Ward’s minimum variance method.25 For calculation of RMSD, we account for the permutational symmetry (4 chains with same sequence) by defining RMSD(X) = min𝒫{RMSD[𝒫(ABCD)]} where X denotes a given conformation of the tetramer and 𝒫 denotes a permutation of the four chains (A, B, C, and D).
2.4 Technical details
The starting structures of the tetramer were constructed from four extended monomer chains, with all virtual-bond-dihedral angles γ set at 180°. Each chain was rotated by a random angle about the z axis, followed by rotation by a random angle about the y axis and then by a rotation by a random angle about the x axis. The chains were subsequently translated by random vectors whose lengths were within the interval , where l is the length of the extended chain. If the distance between the geometric centers of any two chains was greater than the length of the extended chain, the structure was discarded and another attempt was made. A different starting structure was generated for each trajectory.
Version 3.0 of UNRES (available at www.unres.pl) with multiplexed replica extension16 of the coarse-grained molecular dynamics algorithm for UNRES14 was employed with modifications for D-proline and Dap(BZ) as described above. The Berendsen thermostat26 was used to control temperature, with angular momentum reset every 1000 steps and maximum acceleration change set at 4. The equations of motions were integrated by using the adaptive multiple time step (A-MTS) algorithm27 as implemented in the UNRES program.
The simulations were performed at flow-distribution optimized23 set of 36 temperatures with 4 copies of each temperature. This optimized distribution was obtained in four iterations from a distribution that was equally spaced in the inverse temperatures. The final temperatures are: 250, 256, 262, 268, 274, 278, 281, 284, 287, 290, 293, 297, 301, 305, 309, 313, 317, 321, 325, 329, 333, 338, 344, 351, 359, 368, 379, 391, 405, 421, 439, 450, 485, 520, 560 and 600 K. Even with this optimized temperature distribution the simulation required about 400 ns (around 80 million steps with a step time frame of 4.89 fs) to converge. The convergence was tested by comparing the specific heat curve of 3) computed from consecutive time windows, each with the length of 4 million time steps for each replica. Once the form of the curve did no longer change, the convergence was assumed to have been reached. The simulation were carried out in two modes. The first series of simulations were confined, for which the maximum distance between chains (defined as the distance from the C-terminal Cα atom of a given chain to the N-terminal Cα atom of a next chain) with no energy penalty was 30 Å. In the free simulations the maximal distance between chains with no energy penalty was set to 60 Å. Therefore, the two series of simulations mimic different peptide concentration.
For the simulations of BBAT1 in a crystal-like environment, we used representative structures of the most common clusters, obtained in our previous confined and free simulations, as starting conformations of the folded tetramers. Each of these structures was copied four times and positioned on a square of edge length 30 Å. Simulations were run at 32 temperatures with 2 replicas at each temperature.
3 Results and Discussion
In order to gain insight into the mechanism of folding and association of BBAT1, the folding of an isolated monomer is studied first and compared with that of the closely related BBA5 peptide. In 2, the specific heat is displayed as a function of temperature. The curve has two distinct peaks. The peak at a higher temperature Tcu ≈ 305 K corresponds to the minimum of the end-to-end distance, while the secondary peak at a lower temperature Tf ≈ 270 K marks a rapid decrease in average root-mean-square deviation (rmsd) to the structure that BBAT1 chains take in the tetramer crystal. Assuming a correspondence between thermal ordering and folding pathways, this suggests that the isolated BBAT1 monomer folds in a two-step process. The first step is the collapse into a compact, bend-like form, where the C- and N- termini are closer together than in the folded structure. The helix at the C-terminus is only partially folded, but the bending creates already a hydrophobic core. The transition from a coil configuration into this pre-folded intermediate is enthalpy driven and leads to the pronounced peak in the specific heat. Lowering the temperature further leads to an increase of the end-to-end distance, i.e. a stretching of the monomer that is due to further growth of the helix and the formation of the β-turn. Hence, the second step in the folding of isolated BBAT1 monomers is the formation of secondary structure. The associated transition leads to configurations that are close to ones BBAT1 chains take in the crystal structure. This transition happens at a temperature Tf ≈ 270 K that corresponds to the secondary peak in the specific heat. Here we define the Tf by the condition that on average 50 % of all native contacts are formed. This folding process, where secondary structure forms after an initial collapse, differs from that of the closely related BBA5 where the secondary structure has to be completed before the peptide can form the hydrophobic contacts that stabilize its tertiary structure.28,29
In the next step, we compare this folding pathway with that of BBAT1 chains in the tetramer. The specific heat is shown in 3 for both the “free” protein and (in the inset) for the protein confined to a box. Both curves are similar indicating that the energetics of the folding and association mechanism does not change by confining BBAT1. With lowering the temperature, the specific heat increases; with the growth accelerating strongly for temperatures lower than T ≈ 350 K. Assuming again an equivalence of thermal and temporal ordering, the form of the specific heat curve together with figure 5 suggests that the process of folding and association of BBAT1 starts with the association of the four chains to a tetramer. This is illustrated in Figure 4, in which representative structures at a high temperature (T = 600 K; A), a moderate temperature but still above the second heat-capacity peak (T = 372 K; B) and, finally, at a temperature below the second heat-capacity peak (T = 315 K; C) are presented. While configurations with four separated monomers dominate at high temperatures, their frequency decreases with lowering the temperature, and they are replaced by dimers, trimers, and, finally, tetramers. Here a monomer is defined as a chain that does not form contacts involving residues Ile4, Phe9,Leu13,16,17 with those of other chains, a dimer as two chains that have such contacts between themselves but not with other chains, etc.
Fig. 5.
Fraction of monomers (red), dimers (green), trimers (blue) and tetramers (black) as function of temperature.
Fig. 4.
Representative structures of the BBAT1 tetramer obtained at (A) T = 600 K (above the second heat-capacity peak), (B) at T = 372 K (between the first and the second heat-capacity peak), and (C) at T = 315 K (below the first heat-capacity peak).
A rapid decrease of the fraction of monomers (around T ≈ 450 K) and even more steep increase of the fraction of tetramers is observed at Ta ≈ 390 K. This temperature corresponds to a shoulder in the specific heat; however, a steep increase of the specific heat only starts after about 90 % of all configurations are tetramers, i.e. the association process does not seem to be driven by energy. Dimers and trimers are found with frequency of more than 10 % only at temperatures above T ≈ 350 K. This indicates that dimers and trimers are less stable than tetramers.
During the association stage (i.e. for Ta ≈ 390 K) the monomers are not folded. This can be seen from 6 where the frequency of folded monomers (defined as having RMSD less than 4 Å) is displayed as a function of temperature. The folding transition for the monomers, is with Tfm ≈ 290 K much lower than the association temperature Ta. On the other hand, the most important inter-chain contacts, such involving residues Ile4, Phe9,Leu13,16,17 that mark the association of the chains, start forming above T ≈ 350 K and before intra-chains are folded. The curves for the free and the confined system have again the same form.
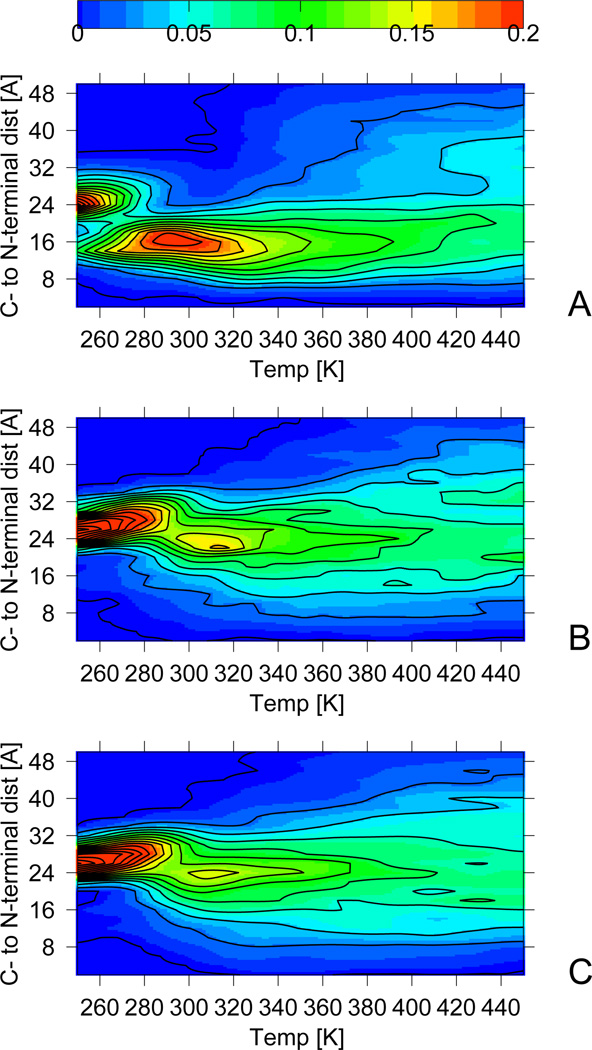
As for isolated monomers, the next step is the collapse of the constituting chains into a compact “curly” state consisting of randomly associated compact monomers with partially formed helical structure at the C-terminus and a hydrophobic core formed by the bending of the monomer chains. This process starts when the frequency of tetramer configurations is about 90 %, indicating that the tetramer environment is important to facilitate this step. This can be seen from 7 where we plot the frequency of configurations as function of temperature and distance between the N- and the C-terminus in a monomer. The increase of that quantity corresponds to the one of the specific heat and accelerates for temperatures below T ≈ 350 K. The maximum around Tcu ≈ 320 K in 3 corresponds to the secondary peak in the specific heat capacity curve of the free system and to the shoulder of the confined system indicating that this state is a stable intermediate. Confining BBAT1 does not change the folding mechanism. The collapse transition into the curly state is similar to that in the folding of isolated monomers and leads to similar structures. However, the transition temperature Tcu is increased by about 20 K and the peak in the specific heat capacity is less pronounced than for isolated monomers. For BBAT1 confined to a box, the peak is even reduced to a shoulder. Hence, the formation of the “curly” state in the tetramer system requires crossing of much smaller barriers than in isolated monomers. This observation suggests that, in the tetramer environment, the monomers prefer compact structures stabilizing each other due to hydrophobic contacts between the chains.30,31 These contacts involving residues Ile4, Phe9,Leu13,16,17 form before the chains are folded, at temperatures above T ≈ 350 K.
We did not find an experimental heat capacity profile of the system; however, the temperature of the second heat-capacity peak (Tcu ≈ 320 K) is in good agreement with the melting temperature derived from circular-dichroism (CD) measurements (Tm = 313 K).10 The later indicates that parts of secondary structure are form, but does not imply that the protein is fully folded. As the heat-capacity maximum at Tcu ≈ 320 K corresponds to the creation of compact state with partially formed secondary structure, we conjecture that the experimentally measured melting temperature can be interpreted also as a transition between states with unfolded monomers and such with partially formed helical structure (as observed in the curly state).
7 shows that the probability of structures for which the chains have larger end-to-end distance increases with further lowering of temperature. This indicates that the third stage of the folding of BBAT1 involves the stretching of the BBAT1 monomers into their folded structures. As for isolated monomers, the stretching results from further growth of the helix and creating a β-turn through forming contacts between Tyr2 and Tyr7 or Phe9. Hence, as for isolated monomers, the BBAT1 chains in the tetramer environment form secondary structure after an initial collapse transition. At the temperature Tf = 295 K, about 50 % of the monomers possess native-like structure. This transition corresponds to the primary heat capacity peak (3) and is driven by forming thermodynamically stable α-helices and β-turns, as in isolated monomers. The folding temperature Tf is again raised by approximately 20 K in the tetramer system and, unlike for the isolated monomer, this transition leads to the primary peak in specific heat here. We conjecture that this is because in the tetramer environment the collapse of chains starts from configurations that are more confined than the isolated-monomer conformers. Hence, the change in energy through the collapse transition is less pronounced in the tetramer environment than for the isolated monomer. However, the frequency distributions of 7 indicate that the folding transition, resulting from secondary structure formation, is also less pronounced in tetramer systems than in the isolated monomers, with lower free energy barriers. It is worth-while to note that, even at the lowest simulation temperatures, the β-turn is not observed for all configurations; indicating that this structure is flexible, and likely only well formed in the crystal (see the discussion below).32
Since in the replica exchange molecular dynamics, a replica performs a random walk in temperature space, it is not possible, in the strict sense, to extract the correct folding pathways (as observed at a constant temperature) from a replica trajectory. However, if the temperature variation is small over the length of the folding event, the trajectories will be valid approximations of the correct folding dynamics. A visual analysis of such pseudo trajectories reveals an order of events that corresponds closely to the thermal ordering described above.
The folding and association pathway starts with the arrangement of four unfolded monomers into a tetramer. Depending on trajectory, either an association of monomers to dimers that then form tetramers or a stepwise growths of monomers to dimers, trimers and, finally, tetramers is observed. The final tetramer environment seems to facilitate the folding of the individual monomers, but the folding processes of the four chains are not synchronized. The folding trajectory often (but not always) starts with the formation of a loop and subsequent partial formation of the C-terminal α-helix in the “curly” state. The propagation of this helix leads to the elongation of the structure and to the increase of end-to-end distance, and it finishes after β-turn formation. The sequence of events is the same as for isolated monomers; however, as the thermal analysis above shows, the monomers stabilize each other in the tetramer environment. This stabilizing force may be due to the confinement of the monomers as indicated by the inset of 6 where the frequency of folded tetramers is plotted as function of temperature for both free and confined simulations. It is also consistent with the experimentally observed concentration dependence of folded BBAT1 complexes in Ref.33
The folding and association pathway described above leads at low temperatures to an ensemble of tetrameric structures, but not necessarily to the crystal structure. A cluster analysis of the folded configurations shows that the tetramers consist of two pairs of chains shifted toward each other by a small and variable angle. In the crystal-structure, this angle is zero and the structure has the C4s symmetry. The energy differences between tetramer structures differing in this angle is of the order of 0.5 kcal/mol and is comparable to thermal fluctuations. This observation suggests that the solution structure of BBAT1 is more flexible than than that in the crystal. This finding is consistent with that of a recent work32 that demonstrated the influence of crowding on the folding process and structure. Configurations with crystal-like fold, i.e. with the C4s symmetry (8A) where the angle between the two pairs close to zero, appear with a ≈ 64 % frequency in the simulation of the confined system, but only with a ≈ 51 % frequency in the “free” simulation. This can also be seen in the inset of 6 where the frequency of folded and associated BBAT1 tetramers is plotted against temperature. The confined and the “free” simulations differ strongly in the frequency of correctly assembled BBAT1 tetramers.The difference becomes larger as the monomers are formed completely. In general, structures in the free environment are more diverse than in the confined one, with the difference resulting from the variation of the angle between the two pairs of chains in the tetramer. For instance the perpendicular alignment is observed in 8C observed with a 49 % frequency. The same residues that create the hydrophobic core in the crystal structure, (Phe9,Leu13,16,17), now serve as an anchor for the perpendicular structure. The bulk of interactions that stabilize BBAT1 tetramer are the interactions between the helices. However, both in simulations and in the experimental structures interactions are also observed between the N-terminal beta-hairpin segments. It has been suggested8 that the loop takes important part in the association process. This is consistent with our observation that Ile4 takes part in the association of the tetramer. In particular the interactions between the N-terminal hairpins seem to be holding the chains in parallel orientation.
It is worthwhile to note that even our simulations of a confined environment, the cluster with the second highest frequency (≈ 35 %) has a C2s symmetry (8B) and, therefore, differs largely from the crystal structure. This observation suggests that the crystal structure of BBAT1 does not arise because of a crowding effect but emerges because of interaction with the neighboring molecules in the protein crystal. In order to test this assumption, systems of four BBAT1 tetramers set on a square have been simulated. This procedure enables us to take into account, in a simplistic way, the effect of the crystal environment on the structure of BBAT1. The starting configurations of the four BBAT1 molecules are either such with C4s symmetry (i.e. crystal-like structures), such with C2s symmetry, or such where the two pairs are almost perpendicular to each other.
In the simulation of the system constructed from four BBAT1 molecules with the C4s symmetry, all tetramers are stable and do not dissolve. However, they rearrange their position: assuming that the z-axis runs along the monomer helices, the centers of mass of the four units are all in the xy-plane in the starting configuration, but move later to the xz-plane. On the other hand, the BBAT1 molecules with the C2s symmetry do change their structure into perpendicular form or even into the crystal structure with the C4s symmetry. When the simulation started from the perpendicular structure the molecules either remained perpendicular or the tetramer changed its symmetry to the C4s but never into a configuration with C2s symmetry. In all of the above simulations modeling the crystal environment, the BBAT1 molecules arranged as in the crystal structure have a 4 kcal/mol lower energy on average than those forming tetramers with the C2s symmetry, and a 3 kcal/mol lower energy than such where the tetramer is built out of two perpendicular dimeric units. This observation indicates that it is not mere crowding but the interaction with other BBAT1 tetramers in the crystal that favors the crystal structure of BBAT1. It should also be noted that the β-turn is almost always formed, indicating again that the formation of this turn results from crystal packing. Moreover, no higher oligomers than tetramers were formed during the simulations. This observation suggests that the tetramer is the most stable oligomer.
4 Conclusions
The assembly of protein complexes is a process with important implications in material and life sciences. Yet it is only poorly understood and difficult to tackle experimentally. Computational investigations can complement experiments but their usefulness is limited by the large size of these systems. For this reason, in the present study, we focus on a model system, BBAT1, which is one of the smallest and simplest protein complexes. It is therefore an ideal test case to study the mechanism of self assembly of protein structures. We find that this process is of surprising complexity, even for such a simple model system. Our simulations indicate that, for BBAT1, association into a tetramer precedes the folding of the individual four chains. The association of the four chains into a tetramer creates hydrophobic contacts between the chains that facilitates their folding and stabilize the complex already at an early stage.30,31
The influence of the tetramer environment does not change the form of the folding pathway of individual chains. Both the isolated BBAT1 monomers and chains in the tetramer form a bend-like structure in a number of distinct steps, starting with a closer end-to-end distance than that in native state and only partially formed secondary structure. A comparison with the experimentally measured heat capacity curves suggests that the measured melting temperatures are not the folding temperatures but instead characterize the formation of these “pre-folded” states. In a second step, the helices in the BBAT1 chains elongate increasing their end-to-end distance. The folding of the individual chains finishes with the formation of the C-terminal β-turn. In solution, this process leads to an ensemble of structures where the four chains in the tetramer arrange themselves in pairs that form an angle between each other. The various structures differ little in their energy. As a consequence, only in a small percentage of configurations is this angle zero (as it is in the crystal structure). The crystal-structure tetramer becomes energetically more favorable in a crystal-like environment. This is not a crowding effect but results from the interaction with other molecules in the protein crystal. Hence, the structures of protein assemblies in crowded environments such as cells may not only differ from those in a diluted solution32 but also from the crystal structure.
It is interesting to observe that BBAT1 does not form in a hierarchical process where elementary units form first before assembling into a complex structure, but that rather the association to a tetramer creates an environment which then encourages the folding of individual chains. The results of our present study also suggest that folding promoted by association may also be the mechanism by that intrinsically unfolded proteins assume their functional structure.34 More studies of different systems are, however, required to corroborate this hypothesis. Promoting the folding by encouraging the formation for hydrophobic contacts may also be an important mechanism here, and a deeper knowledge of how association of proteins influences their structure is likely important for drug design and understanding of protein interaction networks and their role in diseases. In future we want to study the process of association-mediated folding in more detail by investigating various mutations of the key residues Phe9 and Leu13,16,17 that either stabilize or destabilize the association to a tetramer.
Although the helix-helix contacts seem to be dominant in determining the stability of the tetramer, the interaction involving the hydrophobic Ile4 residue from the N-terminal β-hairpin appear also to be important for correct antiparallel alignment of the helices. It would, therefore, be worthwhile to determine the influence of the N-terminal segment on the structure and stability of the tetramer. The kinetics of the folding and association process also needs to be addressed and we are now developing techniques to extract the kinetic information from multiplexed replica exchange molecular dynamics simulations with the UNRES force field.
Fig. 2.
Specific heat (red), end-to-end distance (black), and rmsd (green) as function of temperature for an isolated monomer. The solid lines represent data obtained from weighted histogram analysis, and error bars are the standard errors.
Fig. 3.
Specific heat as a function of temperature for the BBAT1 tetramer. The inset displays the same quantity for the confined molecule.
Fig. 6.
Percentage of chains whose rmsd to the corresponding chains in the crystal structure is within 4 Å. The red and blue lines represent data from confined and free simulations, respectively. Green and black lines represents the percentage that at least one of the four chains is within the cutoff. The Inset displays the frequency of configurations where the tetramer is within 8 Å to the crystal structure.
Fig. 7.
Distribution of chain end-to-end distance as a function of temperature. Isolated monomers (top), confined (middle) and free (bottom) tetrameric BBAT1.
Fig. 8.
Cartoon representation of the most frequent tetramers at T = 268K. The figures were prepared using PyMOL36
Acknowledgments
This work was supported, in part, by research grant GM62838 of the National Institutes of Health (USA) and grants UMO-2011/01/N/ST4/01772 and DS/8372-4-0138-12 from the National Science Center of Poland. Calculations were carried out using (a) the supercomputer resources at the Informatics Center of the Metropolitan Academic Network (IC MAN) in Gdańsk, (b) the 624-processor Beowulf cluster at the Baker Laboratory of Chemistry, Cornell University, (c) our 184-processor Beowulf cluster at the Faculty of Chemistry, University of Gdańsk, and (d) the SOONER cluster at the University of Oklahoma.
References
- 1.Coté S, Laghaei R, Derreumaux P, Mousseau NJ. Phys. Chem. B. 2012;116:4034–4055. doi: 10.1021/jp2126366. [DOI] [PubMed] [Google Scholar]
- 2.Deane R, Singh I, Sagare AP, Bell RD, Ross NT, LaRue B, Love R, Perry S, Paquette N, Deane RJ, Thiyagarajan M, Zarcone T, Fritz G, Friedman AE, Miller BL, Zlokovic BV. J. Clin. Invest. 2012;122:1377–1392. doi: 10.1172/JCI58642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinesh, Goswami A, Surech P, Thirunavukkarasu C, Wieergräber O, Kumar M. Bioinformation. 2011;7:21–28. [PMC free article] [PubMed] [Google Scholar]
- 4.Sinitskiy AV, Saunders MG, Voth GA. J. Phys. Chem. B. 2012;116:8363–8374. doi: 10.1021/jp2108895. [DOI] [PubMed] [Google Scholar]
- 5.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 6.Lu W, Zhong M, Wei YJ. Mol. Biol. 2011;411:264–274. doi: 10.1016/j.jmb.2011.05.042. [DOI] [PubMed] [Google Scholar]
- 7.Deng M, Sun F, Chen T. Pac. Symp. Biocomput. 2003;8:140–151. [PubMed] [Google Scholar]
- 8.Ali MH, Peisach E, Allen KN, Imperiali B. Proc. Natl. Acad. Sci. USA. 2004;101:12183–12188. doi: 10.1073/pnas.0401245101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mezo AR, Cheng RP, Imperiali BJ. Am. Chem. Soc. 2001;123:3885–3891. doi: 10.1021/ja004292f. PMID: 11457138. [DOI] [PubMed] [Google Scholar]
- 10.McDonnell KA, Imperiali BJ. Am. Chem. Soc. 2002;124:428–433. doi: 10.1021/ja016991d. PMID: 11792213. [DOI] [PubMed] [Google Scholar]
- 11.Lindorff-Larsen K, Trbovic N, Maragakis P, Piana S, Shaw DE. J. Am. Chem. Soc. 2012;134:3787–3791. doi: 10.1021/ja209931w. [DOI] [PubMed] [Google Scholar]
- 12.Zimmermann O, Hansmann UH. Biochim. Biophys. Acta, Proteins Proteomics. 2008;1784:252–258. doi: 10.1016/j.bbapap.2007.10.010. Inhibitors of Protein Kinases (5th International Conference, IPK-2007) and Workshop Session on Molecular Design and Simulation Methods Warsaw, Poland June 23–27, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Sanbonmatsu KY, Voth GA. J. Am. Chem. Soc. 2011;133:16828–16838. doi: 10.1021/ja2028487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liwo A, Khalili M, Scheraga HA. Proc. Natl. Acad. Sci. U.S. A. 2005;102:2362–2367. doi: 10.1073/pnas.0408885102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liwo A, Khalili M, Czaplewski C, Kalinowski S, Ołdziej S, Wachucik K, Scheraga HJ. Phys. Chem. B. 2007;111:260–285. doi: 10.1021/jp065380a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czaplewski C, Kalinowski S, Liwo A, Scheraga HA. J. Chem. Theor. Comput. 2009;5:627–640. doi: 10.1021/ct800397z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen H, Liwo A, Scheraga HA. J. Phys. Chem. B. 2009;113:8738–8744. doi: 10.1021/jp901788q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geyer CJ, Thompson EA. J. Am. Stat. Assoc. 1995;90:909–920. [Google Scholar]
- 19.Hukushima K, Nemoto K. Journal of the Physical Society of Japan. 1996;65:1604–1608. [Google Scholar]
- 20.Hansmann UHE, Okamoto Y. Phys. Rev. E. 1996;54:5863–5865. doi: 10.1103/physreve.54.5863. [DOI] [PubMed] [Google Scholar]
- 21.Sugita Y, Okamoto Y. Phys. Rev. Lett. 2000;329:261–270. [Google Scholar]
- 22.Rhee YM, Pande VS. Biophys. J. 2003;84:775–786. doi: 10.1016/S0006-3495(03)74897-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadler W, Meinke JH, Hansmann UH. E. Phys. Rev. E. 2008;78:061905. doi: 10.1103/PhysRevE.78.061905. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 25.Murtagh F. Multidimensional clustering algorithms. Vienna: Physica-Verlag; 1985. [Google Scholar]
- 26.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 27.Rakowski F, Grochowski P, Lesyng B, Liwo A, Scheraga HA. J. Chem. Phys. 2006;125:204107. doi: 10.1063/1.2399526. [DOI] [PubMed] [Google Scholar]
- 28.Mohanty S, Hansmann UH. Biophys. J. 2006;91:3573–3578. doi: 10.1529/biophysj.106.087668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim E, Jang S, Pak YJ. Chem. Phys. 2009;131:195102. doi: 10.1063/1.3266510. [DOI] [PubMed] [Google Scholar]
- 30.Dunker A, et al. J. Mol. Graphics Modell. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 31.Vajda S, Wheng Z, Rosenfeld R, DeLisi C. Biochemistry. 1994;33:13977–13988. doi: 10.1021/bi00251a004. PMID: 7947806. [DOI] [PubMed] [Google Scholar]
- 32.Feig M, Sugita YJ. Phys.Chem. B. 2012;116:599–605. doi: 10.1021/jp209302e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mezo AR, Ottesen JJ, Imperiali BJ. Am. Chem. Soc. 2001;123:1002–1003. doi: 10.1021/ja0038981. PMID: 11456646. [DOI] [PubMed] [Google Scholar]
- 34.Drescher M, Huber M, Subramaniam V. ChemBioChem. 2012;13:761–768. doi: 10.1002/cbic.201200059. [DOI] [PubMed] [Google Scholar]
- 35.Ali MH, Taylor CM, Grigoryan G, Allen KN, Imperiali B, Keating AE. Structure. 2005;13:225–234. doi: 10.1016/j.str.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 36.PyMOL(TM) Molecular Graphics System, Version 1.2r2. Palo Alto, CA: DeLano Scientific LLC; 2009. [Google Scholar]