Summary
Human African sleeping sickness (HAT) is caused by the parasitic protozoan Trypanosoma brucei. Polyamine biosynthesis is an important drug target in the treatment of HAT. Previously we showed that trypanosomatid S-adenosylmethionine decarboxylase (AdoMetDC), a key enzyme for biosynthesis of the polyamine spermidine, is activated by heterodimer formation with an inactive paralog termed prozyme. Furthermore, prozyme protein levels were regulated in response reduced AdoMetDC activity. Herein we show that T. brucei encodes three prozyme transcripts. The 3’UTRs of these transcripts were mapped and chloramphenicol acetyltransferase (CAT) reporter constructs were used to identify a 1.2 kb region that contained a 3’UTR prozyme regulatory element sufficient to up regulate CAT protein levels (but not RNA) upon AdoMetDC inhibition, supporting the hypothesis that prozyme expression is regulated translationally. To gain insight into trans-acting factors, genetic rescue of AdoMetDC RNAi knockdown lines with human AdoMetDC was performed leading to rescue of the cell growth block, and restoration of prozyme protein to wild-type levels. Polyamine and AdoMet metabolite analysis showed that prozyme protein levels were inversely proportional to intracellular levels of decarboxylated AdoMet (dcAdoMet). These data suggest that prozyme translation may be regulated by dcAdoMet, a metabolite not previously identified to play a regulatory role.
Keywords: Trypanosoma brucei, polyamines, spermidine, gene regulation
Introduction
Human African trypanosomiasis (HAT) is a fatal vector borne disease of sub-Saharan Africa with up to 70 million people at risk in disease transmission areas (Stuart et al., 2008, Kennedy, 2008, Brun et al., 2010). HAT is caused by the parasitic protozoan Trypanosoma brucei leading to a disabling neurological disease, which if untreated leads to coma and eventually death in most patients. The recent introduction of nifurtimox/eflornithine combination therapy has provided a simplified treatment option that is safer and more efficacious than prior therapies, however, significant limitations still hinder treatment (Burri, 2010, Jacobs et al., 2011, Barrett, 2010). Eflornithine, also known as α-difluoromethylornithine (DFMO), exerts its anti-trypanosomal activity by inhibiting polyamine biosynthesis. The proven clinical value of targeting this pathway has led to interest in understanding the role and function of polyamines in T. brucei (Jacobs et al., 2011).
Polyamines are ubiquitous small organic cations that are essential for eukaryotic cell growth and have demonstrated roles in regulating transcription, translation, chromatin structure and ion channel function (Pegg, 2009a, Pegg & Casero, 2011). Additionally, spermidine is required for the formation of the hypusine modification of eIF5A, which is an essential translation factor in eukaryotes (Park et al., 2010). Because polyamines are essential for proliferation, enzymes involved in their biosynthesis and metabolism have been targeted for the treatment of proliferative diseases, including cancer and protozoan pathogens (Pegg, 2009a, Pegg & Casero, 2011, Jacobs et al., 2011). The only clinically approved systemic application of polyamine biosynthetic inhibitors is DFMO for the treatment of HAT.
DFMO is a mechanism-based inhibitor of ornithine decarboxylase (ODC), which catalyzes the formation of putrescine, the first committed step in the biosynthesis of polyamines (Jacobs et al., 2011) (Scheme 1). In the subsequent steps the polyamine spermidine is synthesized from putrescine and decarboxylated S-adenosylmethionine (dcAdoMet), which is the product of the reaction catalyzed by pyruvoyl-dependent S-adenosylmethionine decarboxylase (AdoMetDC). Uniquely in the trypanosomatids, spermidine is then conjugated to two molecules of glutathione to form trypanothione, the cofactor required to maintain reduced cellular thiol pools (Krauth-Siegel & Comini, 2008). The polyamine and trypanothione biosynthetic enzymes, including ODC, AdoMetDC and spermidine synthase (SpdSyn), have been shown to be essential in T. brucei by genetic methods (e.g. gene knockdown by interfering RNA (RNAi) or gene knockout) (Willert & Phillips, 2012). In addition to DFMO, inhibitors of AdoMetDC (Bacchi et al., 2009, Barker et al., 2009, Brun et al., 1996, Bacchi et al., 1996, Tekwani et al., 1992, Bacchi et al., 1992, Bitonti et al., 1990) and trypanothione synthetase (TrypSyn) (Wyllie et al., 2009, Spinks et al., 2012) have been reported to have anti-trypanosomal activity.
Scheme 1. Polyamine metabolic pathway in T. brucei.
The functional subunit structure of AdoMetDC is novel in the trpanosomatid parasites (Willert & Phillips, 2012). In mammals AdoMetDC is a (α2β2) homodimer and the pyruvoyl cofactor is generated in an autocatalytic reaction that cleaves the peptide backbone to form the α and β chains with the pyruvate cofactor at the N-terminus of the α chain (Pegg, 2009b). In the trypanosomatids, AdoMetDC is a heterodimer formed from two paralogous gene products, one that retains limited catalytic activity, and the other, an inactive homolog termed prozyme that has a regulatory role (Willert et al., 2007, Velez et al., 2013, Willert & Phillips, 2009). Heterodimer formation between the active AdoMetDC subunit and prozyme leads to a 102- to 103-fold stimulation of activity of the T. brucei and T. cruzi enzymes.
Polyamine biosynthesis is tightly regulated in mammals, yeast and plants through transcriptional, translational and post-translational mechanisms that influence the levels of ODC and AdoMetDC (Pegg & Casero, 2011, Pegg, 2009a, Pegg, 2009b, Casero & Pegg, 2009, Persson, 2009, Nowotarski et al., 2011, Kahana, 2009, Ivanov et al., 2010). Analogous mechanisms have not been found in the trypanosomatids (Willert & Phillips, 2012), possibly due to the atypical mechanism of mRNA biogenesis in these parasites. Gene expression in T. brucei occurs via the formation of polycistronic messages that are processed by coupled trans-splicing/polyadenylation to generate the mature capped messages, and thus transcriptional regulation is uncommon (Clayton & Shapira, 2007, Ouellette & Papadopoulou, 2009, Gunzl, 2010, Kramer & Carrington, 2011, Kramer, 2012). The 5’UTRs are short and do not typically contain elements for regulatory control. Gene expression is instead controlled by the 3’UTR, and the major mechanisms for regulation are mRNA stability, translation and protein stability.
Recently we uncovered evidence that in T. brucei the polyamine biosynthetic pathway is regulated by a novel mechanism involving the AdoMetDC activator subunit prozyme. We found that either knockdown of AdoMetDC by RNAi, or treatment of bloodstream form (BSF) parasites with a potent irreversible AdoMetDC inhibitor MDL 73811, led to significant upregulation of prozyme protein levels (Willert & Phillips, 2008). Protein turnover and mRNA expression were unaltered suggesting that prozyme translation was regulated. Unlike in other eukaryotic cells where spermidine is the regulatory signal that controls expression of polyamine biosynthetic enzymes, spermidine did not appear to be involved in the regulation of T. brucei prozyme. Knockdown of ODC or SpdSyn by RNAi led to similar decreases in spermidine levels as found with AdoMetDC knockdown, however knockdown of these other biosynthetic enzymes did not lead to increased prozyme expression (Xiao et al., 2009). Additionally, while spermidine could rescue the growth defect observed upon knockdown of AdoMetDC, prozyme levels remained elevated under these conditions. Thus the mechanistic basis for the effects of knockdown or inhibition of AdoMetDC on prozyme translation remained enigmatic.
To further study the mechanisms that regulate prozyme expression we have evaluated the prozyme mRNA 3’ untranslated region (UTR) in order to identify potential AdoMetDC inhibitor responsive regulatory elements. We mapped and sequenced the 3’UTR, then characterized the expression of CAT reporter constructs containing complete or partial 3’UTR sequences. Three distinct messages encoded by the prozyme gene were identified, and we identified sequences within the 3’UTR that activate CAT protein expression in the presence of the AdoMetDC inhibitor MDL 73811. These studies provide additional evidence that prozyme expression is regulated by translational control. To gain insight into potential trans-acting factors, genetic rescue of the AdoMetDC RNAi knockdown lines was performed. We observed a correlation between dcAdoMet levels and prozyme expression, suggesting that dcAdoMet may regulate prozyme translation in T. brucei. Decarboxylated AdoMet has not previously been reported to have a regulatory role in protein expression, suggesting that prozyme translation is regulated by a novel mechanism.
Results
Prozyme regulation occurs in both mammalian and insect cell stages
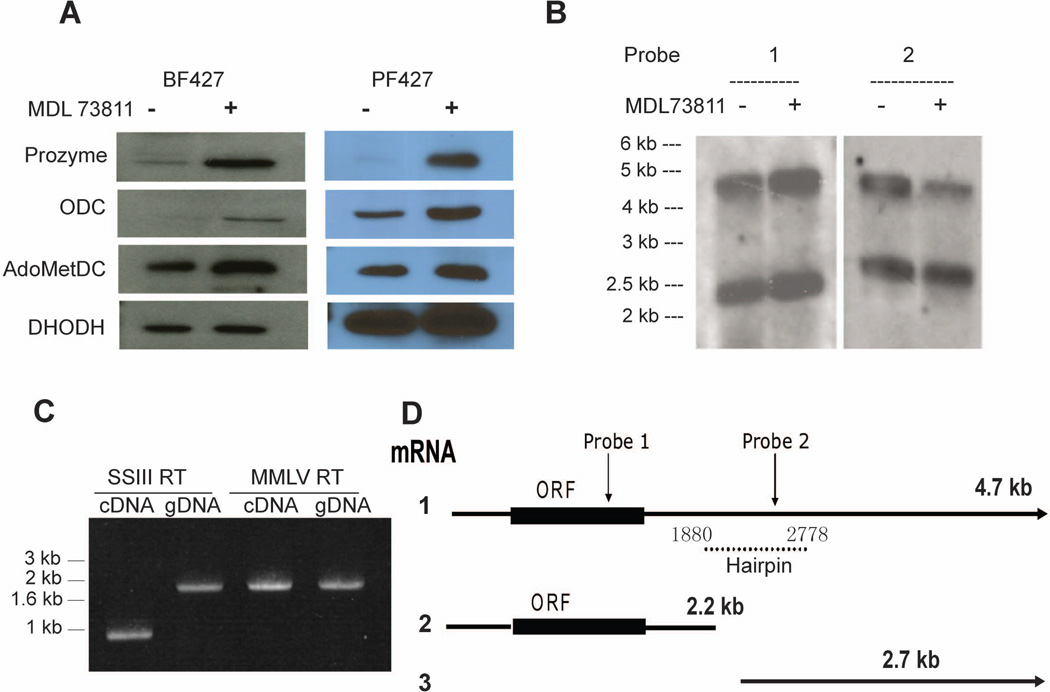
In our prior studies we demonstrated that treatment of T. brucei bloodstream form (BSF) cells with MDL 73811 led to the induction of prozyme, and to a lesser extent of ODC (Willert & Phillips, 2008). Here we reproduce this result and show that substantial upregulation of prozyme, and to some extent ODC, is observed in both the BSF and the insect-stage procyclic form (PF) cells after 48 h of MDL 73811 treatment (Figure 1A). The response in PF cells was even stronger than that observed for BSF cells. These results demonstrate that the regulatory control mechanisms for prozyme gene expression are not stage-specific.
Figure 1. Structure of the prozyme 3’UTR and the effects of AdoMetDC inhibitors on prozyme mRNAs.
(A) The effects of chemical inhibition of AdoMetDC on expression of polyamine biosynthetic enzymes. BSF and PF form T. brucei 427 cells were treated with MDL 73811 (75 nM). Cells were harvested after 48 h incubation and analyzed by western blot. DHODH is included as a loading control. (B) Northern blot analysis of BSF mRNA from T. brucei 427 cells in the presence or in the absence of MDL 73811 with two different probes (probe primers see Table 3S). (C) PCR amplification of prozyme 3’UTR using genomic BSF DNA (gDNA) or cDNA generated with either SuperScript III (SSIII) or MMLV RT. Primer set 2 (Table S3) was used for PCR amplification. (D) Schematic of the prozyme 3’UTRs as mapped by RACE (rapid amplification of cDNA ends). The location of the probes used in northern analysis (part B above) are shown.
Mapping of the prozyme 3’UTR
Northern analysis of BSF cells using an RNA probe containing sequence from the prozyme open reading frame (ORF) (probe 1, nts 451–972) demonstrated the presence of both 4.7 kb and 2.2 kb messenger RNAs (mRNAs) (Figure 1B, panel 1). Our prior northern analysis had only uncovered the 2.2 kb mRNA (Willert & Phillips, 2008). The different outcome can be ascribed to the fact that in the current study the transfer conditions were run to ensure efficient transfer of larger RNAs from the gel to the membrane. Both mRNAs were observed in the presence or absence of MDL 73811, and as was observed previously for the smaller mRNA, drug treatment did not lead to any significant changes in abundance of either transcript.
We previously reported the sequence of the 70-nucleotide (nt) prozyme 5’UTR (Willert et al., 2007). As most gene expression in T. brucei is controlled by elements in the 3’UTR, we undertook mapping of the prozyme 3’UTR. Mapping was performed by Rapid Amplification of cDNA ends (RACE) on mRNA isolated from BSF 427 cells using a combination of oligo dT and gene specific primers (Figures S1 and S2). The polyadenylation (poly(A)) addition site of the 4.7 kb mRNA could be unambiguously assigned to nt 4699 in the prozyme gene (Figure S2). A poly(Y) tract, which is the typical signal to initiate a coupled splicing and polyadenylation event in T. brucei, is located 21 nts downstream of the poly(A) site at nt 4720 in the prozyme gene (Figure S1). This distance is atypically short (Siegel et al., 2005, Nilsson et al., 2010, Kolev et al., 2010). The splice site acceptor AG used for splicing of the downstream AdoMetDC mRNA (Willert & Phillips, 2008) is 58 nt downstream of this tract (Figure S2B). Mapping of the poly(A) site of the 2.2 kb mRNA was complicated by the presence of internal poly(A) tracts in the cDNA at nts 2004 and 3324 that led to generation of RACE products. Polyadenylation at nt 3324 would not generate either of the observed mRNAs, suggesting this product was initiated from the internal poly(A) tract. However polyadenylation at nt 2004 is consistent with the 2.2 kb mRNA size, supporting assignment of the poly(A) site for the 2.2 kb mRNA to the region near nt 2004. Furthermore, no other RACE products were identified based on oligo dT priming that could lead to the generation of the 2.2 kb mRNA. The size of the 4.7 kb mRNA 3’UTR (3651 nt) that we mapped is in good agreement with the 3’UTR length (3617 nt) reported from the genome wide RNA sequencing studies (Siegel et al., 2010, Kolev et al., 2010), though the exact poly(A) site does not precisely match. An RNA ending at nt 2004 was also observed in that study. Our sequence of the full length 3’UTR (3651 nt) is 98% identical to that obtained in the study mapping mRNA in procyclic 427 cells. The differences likely reflect lab strain differences in the currently propagated isolates.
In order to better define the end of the 2.2 kb mRNA we performed additional northern analysis using a probe 3’ to the coding region designed to begin downstream of the likely poly(A) site in the 2.2 kb mRNA (probe 2; position nts 2229 – 2778) (Figures 1B, 1D and S2). Probe 2 hybridized to the 4.7 kb larger mRNA but not the 2.2 kb mRNA showing that this mRNA indeed ends upstream of nt 2229. This analysis revealed the presence of yet a third message of 2.7 kb that did not contain the prozyme ORF, but which began within the 3’UTR, and would be predicted to end near the stop site of the 4.7 kb mRNA. The 2.7 kb mRNA does not contain an ORF greater than 96 amino acids in the sense direction and is not predicted to encode a protein. The data are consistent with a poly(A) splice site for the 2.2 kb mRNA at position 2004 (identified by the RACE analysis), and the coupled 5’ splice site for the 2.7 kb mRNA occurring at one of three possible down stream splice sites (nts 2091, 2121 or 2180). A requisite poly(Y) track is present between these sites at nt 2054.
Identification of secondary structure elements in the 3’UTR
During the sequencing of the clones generated by RACE we identified some clones that lacked 899 bases from nts 1879 – 2278 in the 3’UTR (Figures S1 and S2). These clones were obtained with reverse strand primers ending at nt 2838 and 2928 but not with oligo dT primers. Additional PCR analysis showed that the 899 nts were missing only when cDNA (generated with Superscript reverse transcriptase (RT)), but not genomic DNA, was used as the template (Figure 1C). We noted that 14 nts on either side of the junction (5’ of nt 1879 and 3’ of nt 2278) are exactly complementary in sequence (Figure S1), suggesting that the region on either side of the missing 899 nt would form a tight hairpin structure in the mRNA that might account for the excision of this sequence by the Superscript RT reaction. RT excisions of this type have been reported for regions of strong secondary structure and they were found to be dependent on the type of RT used for the reaction (Houseley & Tollervey, 2010). To test if this was the case for the 899 nt prozyme sequence we regenerated cDNA using Moloney murine leukemia virus (MMLV) RT. In this reaction both cDNA and genomic DNA yielded the full length PCR fragment without deletion of the 899 bases (Figure 1C). These data support the presence of a secondary structure region at nts 1879 – 2278. Furthermore, the alternative explanation, the presence of an intron was unlikely to account for the observation as the junction sites (CT/CA) at the excision points were not typical of the major splice sites (GT/AG) expected at an intron boundary (Nilsson et al., 2010).
As a final study to rule out the possibility of an intron we performed RNase protection using a 208 nt probe that spanned across the deleted 899 nt sequence (114 nt upstream of the splice site and 94 nt downstream of the splice site). The 208 nt fragment was protected when RNA was isolated from cells transfected with a positive control plasmid (clone 4b in Figure 2), which lacked the 899 nt sequence, however when RNA was isolated from wild-type T. brucei 427 cells or cells transfected with a negative control construct (clone 4a in Figure 2), the full length probe was not protected (Figure S3). Thus the RNase protection study shows that an intron is not present at nts 1879–2278.
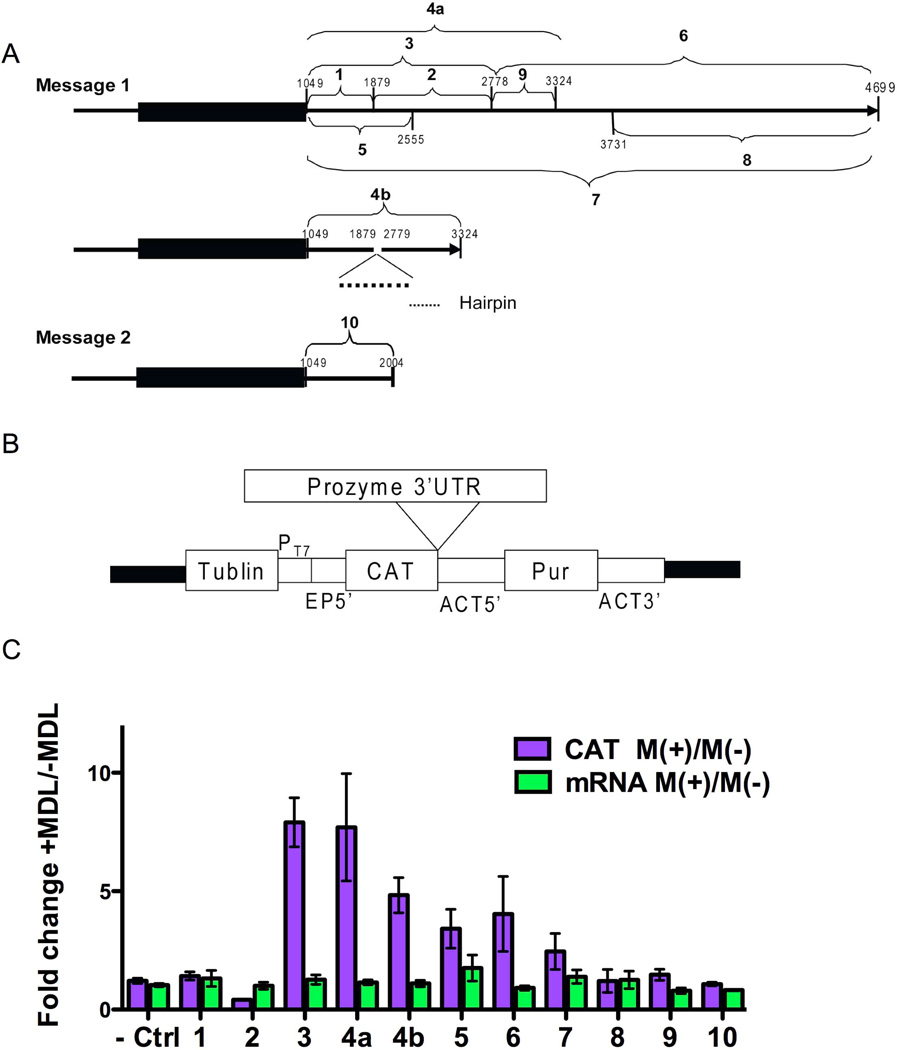
Figure 2. The effect of AdoMetDC inhibition on CAT expression levels from prozyme 3’UTR reporter constructs.
(A) Schematic of the prozyme 3’UTR fragments that were cloned into pHD1437 reporter construct vector. Number (from 1 to 10) indicates the fragment number and subsequent construct number. Nucleotide positions are displayed. The start and end points of each fragment are shown in Table 4S. The gene is numbered with position 1 representing the first gene specific base after the splice leader in the 5’UTR (Figures S1 and S2) (B) Structure of the reporter gene vector pHD1437: the black line of the structure of pHD1437 shows the plasmid backbone. Other components include a region from the tubulin locus (Tublin), a T7 polymerase promoter (PT7), a 5’-splice site and UTR from EP1 gene (EP5’), a CAT reporter gene (CAT), 5’UTR from the actin locus (ACT5’), a puromycin selectable marker gene (Pur), and the actin 3’UTR (ACT 3’). (C) CAT reporter plasmids containing different prozyme 3’UTR fragments were transiently transfected into BSF 90–13 cells. The empty pHD1437 vector was transfected as the control (Ctrl). Cells were grown for 3 h before the addition of MDL 73811 (75 nM), and then incubated for additional 16 h before harvesting. The CAT ELISA assay was used to determine the level of CAT enzyme activity (CAT), and mRNA levels were quantitated by qPCR with primers from table 3S (mRNA). The fold change was calculated as M(+)/M(−). Errors represent the standard error of the mean for n=3–4 independent biological replicates.
Mapping regulatory elements in the prozyme 3’UTR with CAT reporter constructs
In order to determine if the prozyme 3’UTR contained regulatory elements that were responsive to MDL 73811 we cloned the full length UTR from the 4.7 kb (clone 7) and from the 2.2 kb (clone 10) mRNA into the pHD1437 construct (Colasante et al., 2007, Robles & Clayton, 2008) (Figure 2A), which contains a CAT reporter gene under the control of the T7 promoter, and a EP1 5’ splice site (Figure 2B). The 3’UTR was supplied from the prozyme gene. In addition to the full-length UTRs we also cloned a number of smaller fragments from the 4.7 kb 3’UTR into this vector in an attempt to narrow down any regulatory control elements to smaller regions of the UTR. In order to understand the potential role of the secondary structure elements discovered above we also studied clone 4b, which lacked the 899 nt sequence. The plasmid constructs (10 in total) were transiently transfected into BSF 90-13 cells and MDL 73811 was added 3 h after transfection, followed by harvesting for CAT expression analysis 16 h later.
Treatment of the cells harboring the CAT expression constructs with MDL 73811 led to a significant and reproducible 5 – 9-fold average induction of CAT protein expression for two of the nine constructs (clones 3 and 4a) with both showing similar activity (Clone 4a activation average = 9-fold, range 5 – 14-fold and clone 3 activation average = 8-fold, range 6 – 10-fold) (Figure 2C and Table S5). The full length 3’UTR (clone 7) from the 4.7 kb mRNA was only modestly activated by drug treatment (2.5-fold), suggesting the presence of negative regulatory elements on this longer message that suppressed the response elements observed in the shorter fragments. Clone 3 represented the shortest sequence that provided a full MDL 73811 response, suggesting that the prozyme regulatory elements are located between nt 1049–2278. Notably this sequence encompasses the full 956 nts of the 2.2 kb 3’UTR, and all of the hairpin secondary structure region. Attempts to narrow down the response element to a shorter fragment however led to inactivation of the element as clones 1, 2 and 9 showed no activation of CAT expression in the presence of MDL 73811 and behaved identically to the negative empty vector control while clones 4b, 5 and 6, which contain parts of the sequence in 4a showed activation of CAT protein expression in the presence of MDL 73811, but at a reduced level. The construct containing the full-length UTR from the shorter 2.2 kb message (clone 10) was not activated by MDL 73811 treatment suggesting that expression of the shorter mRNA may be constitutive, while expression of the larger one is regulated. Thus these data show that the 4.7 kb prozyme mRNA 3’UTR contains elements that are responsive to the inhibition of AdoMetDC by MDL 73811, leading to increased expression of CAT protein from the transfected reporter construct. In contrast, CAT mRNA levels were not changed by MDL 73811 treatment for any of the tested clones (Figure 3 and Table S5). Thus, as was observed for the effects on prozyme expression when cells were treated with MDL 73811 (Willert & Phillips, 2008), the effect of the compound is confined to changes in CAT protein expression levels, providing additional support that prozyme expression is regulated at the translational level.
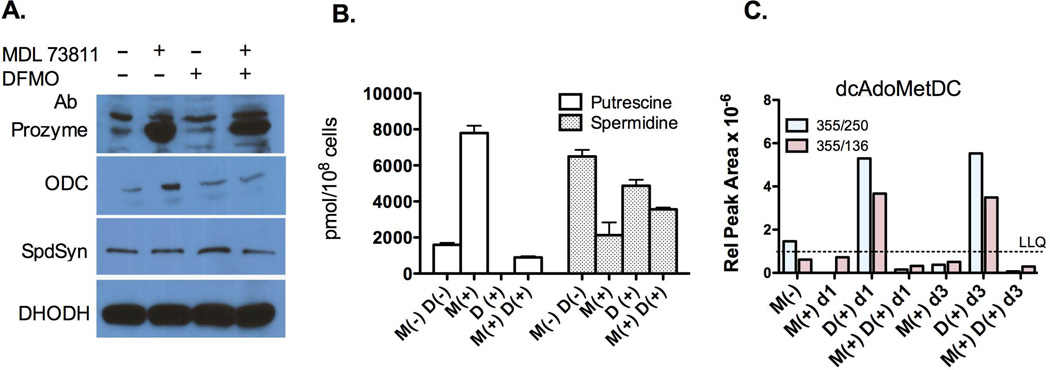
Figure 3. The effects of chemical inhibitors of AdoMetDC and/or ODC on the expression of polyamine biosynthetic enzymes and intracelluar polyamine and dcAdoMetDC pools.
(A) Western blot analysis of polyamine biosynthetic enzymes (40 µg total protein/lane). BSF 427 cells were grown in the absence (−) or presence (+) of MDL 73811 (M) (75 nM) and/or DFMO (D) (12.5 µM) for three days before harvesting. DHODH is shown as a loading control. (B) HPLC analysis of polyamine biosynthesis pathway metabolites for n=4 replicates. Cells were treated as in A. (C) LC-MS/MS analysis of dcAdoMet pools followed by two separate ion peaks (represented by the blue and red bars). Cells were treated as in A except that incubation with drugs was for either 1 (d1) or 3 (d3) days. Data are displayed as the relative peak area. The lower limit of quantitation (LLQ) for dcAdoMet was a relative peak area of 10−6.
The effects of putrescine levels on prozyme expression in BSF T. brucei cells
While reduced spermidine levels are unlikely to be involved in regulating prozyme expression, changes in putrescine levels were potentially implicated. Putrescine increased 10-fold after induction of AdoMetDC RNAi, or in the presence of MDL 73811, while it was decreased or unaffected by the knockdown of other enzymes in the pathway (e.g. ODC and SpdSyn) (Xiao et al., 2009, Willert & Phillips, 2008). In order to determine whether elevated putrescine levels have a role in the induction of prozyme expression we treated cells with a combination of the ODC inhibitor DFMO (12.5 µM) and with MDL 73811 (75 nM) for 3 d. We reasoned that DFMO would reduce putrescine pools by inhibiting biosynthesis and thereby might reverse the increases observed upon inhibition of AdoMetDC with MDL 73811. Prozyme expression was induced by treatment with MDL 73811 but not by DFMO, and prozyme protein levels remained high in cells treated simultaneously with both inhibitors (Figure 3A). Treatment of cells with MDL 73811 led to an increase in putrescine levels (5-fold), while treatment with DFMO alone led to a near 100% reduction in putrescine pools (Figure 3B). Treatment with DFMO and MDL 73811 in combination had a balancing effect, and putresciene levels were found to be ~50% of wild-type. Spermidine levels were decreased in the presence of both inhibitors, but the reduction was modest (3–4-fold in the presence of MDL 73811 and only 1.3-fold in the presence of DFMO). As prozyme protein levels were increased by both MDL 73811 alone, or by the combination treatment with MDL 73811 and DFMO, these data suggest putrescine levels are not involved in regulating prozyme protein expression.
The intracellular levels of AdoMet, dcAdoMet and methylthioadenosine (MTA) were monitored by LC-MS/MS. Neither the AdoMet nor MTA levels were significantly affected by treatment with either drug (Figure S4A). In contrast, treatment with DFMO led to an increase in dcAdoMet levels, whereas in cells treated with MDL 73811, either alone or in combination with DFMO, dcAdoMet levels were below the level of detection (~1 × 10−6 relative peak area for the more abundant fragment ion) (Figure 3C). Thus, the data suggested the possibility that reduced levels of dcAdoMet could play a role in inducing prozyme expression levels, while elevated levels correlate with repressed or base-line expression.
Complementation of the T. brucei BSF AdoMetDC RNAi cell line with human AdoMetDC (hAdoMetDC)
To obtain additional mechanistic insight into trans-acting factors that contribute to the induction of prozyme expression we wanted to explore two potential hypotheses in greater detail. The first was the possibility that AdoMetDC could function as a RNA binding protein to regulate translation of the prozyme mRNA. Precedence for this hypothesis comes from the study of dihydrofolate reductase, which has been shown to regulate its own translation in both human and Plasmodium cells by binding its own mRNA and thereby inhibiting translation (Chu et al., 1993, Ercikan-Abali et al., 1997, Hsieh et al., 2009, Zhang & Rathod, 2002). Secondly, we sought to explore the possibility that prozyme expression is induced in response to depletion of dcAdoMet pools. While there is no precedence for dcAdoMet as a regulatory molecule, riboswitch-mediated regulation by AdoMet has been described in bacteria (Wang & Breaker, 2008, Batey, 2011).
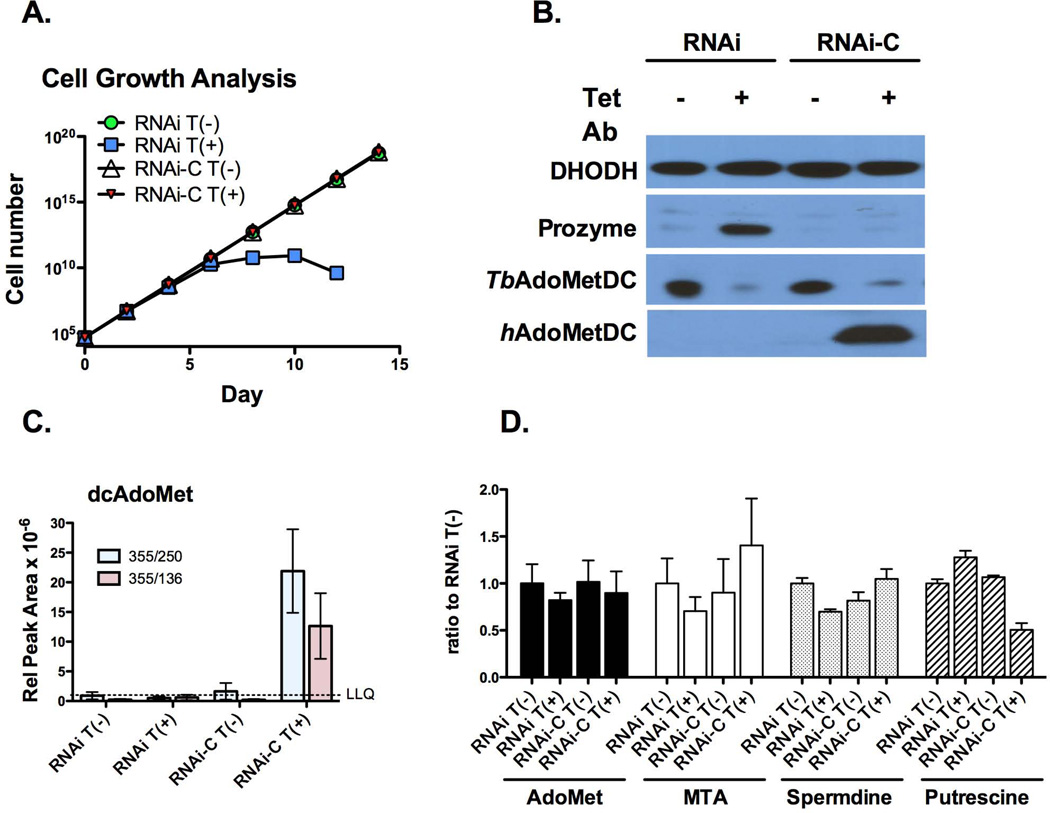
To test these hypotheses we transfected the BSF AdoMetDC RNAi cell line (RNAi) with a Tet-inducible expression construct for hAdoMetDC to generate the complemented RNAi line (RNAi-C). In this line the addition of Tet should lead to the simultaneous knockdown of T. brucei AdoMetDC and to the expression of hAdoMetDC. We reasoned that if T. brucei AdoMetDC functions as an RNA binding protein, hAdoMetDC would be unlikely to replace this function because it would be unlikely to have evolved to bind T. brucei specific RNA sequences. However, if dcAdoMet levels are the important factor, than hAdoMetDC should restore both normal growth and wild-type prozyme levels to the cell. The AdoMetDC RNAi cell line and the hAdoMetDC RNAi-C line were grown in the presence and absence of Tet and the cell growth rate was followed over 15 days (Figure 4A). Levels of AdoMetDC and prozyme were monitored by western blot on day 3 post Tet induction (Figures 4B). As previously observed in the AdoMetDC RNAi line (Willert & Phillips, 2008), the addition of Tet led to the induction of the RNAi response, to cell growth arrest, to the loss of the AdoMetDC protein and to a substantial increase in prozyme protein levels. For the RNAi-C line, addition of Tet led to simultaneous knockdown of T. brucei AdoMetDC and to expression of hAdoMetDC. Under these conditions cells grew at the same rate as the control cells and prozyme protein levels were similar to control levels.
Figure 4. Expression of hAdoMetDC rescues the effects of AdoMetDC knockdown by RNAi.
(A) Representative growth curve analysis comparing AdoMetDC RNAi cells (RNAi) with the same cell line transfected with the human AdoMetDC expression construct (RNAi-C). Tet (1 µg/ml) was added on day 0 resulting in the co-induction of AdoMetDC RNAi and human AdoMet expression. Cell numbers were below detection levels for the RNAi induced cells after day 14. Total cell numbers were calculated as cell density × the total dilution factor and are plotted versus time in days after Tet induction. (B) Representative western analysis. Cells were harvested 3 d after Tet induction and analyzed by western using antibodies to prozyme, T. brucei AdoMetDC and human AdoMetDC. DHODH is shown as the loading control (40 µg total protein/lane). (C) LC-MS/MS analysis of intracellular dcAdoMet levels. Cells were harvested as in B. Data are displayed as the relative peak area. Data represent the mean of n=3 independent biological replicates. Two separate ion fragments were quantitated for each condition (red and blue bars). The LLQ for dcAdoMet was a relative peak area of 10−6(D) LC-MS/MS analysis of intracellular AdoMet, MTA, spermidine and putrescine levels. Cells were harvested as in B. Data represent the ratio to the uninduced AdoMetDC RNAi line and are shown for the highest abundance ion peak of the two measured. Raw data are displayed in Figure S4. Errors represent the standard error of the mean for n=3 independent biological replicates.
Polyamine (spermidine and putrescine) and AdoMet (AdoMet, dcAdoMet and MTA) pools were also evaluated on day 3 post Tet induction using LC-MS/MS (Figure 4C, D and Figure S4B). The dcAdoMet levels in the uninduced cell lines (-Tet) and in the AdoMetDC RNAi line +Tet were at or below the limit of detection, not allowing a clear distinction between these conditions (Figure 4C). However in the RNA-C line, when hAdoMetDC was expressed (+Tet), dcAdoMet levels increased at least 20-fold in comparison to the no Tet control. There were no significant differences in the AdoMet or MTA pools under any of the tested conditions (Figure 4D). A modest 30% decrease in spermidine was observed in the Tet-induced AdoMetDC RNAi line, whereas wild-type levels were restored in the complemented cells. A 50% reduction in putrescine was observed in the RNA-C line in the presence of Tet, but putrescine was otherwise similar to the no Tet controls under the remaining conditions. The modest effects of the AdoMetDC knockdown on the spermidine pools are consistent with previous results showing spermidine levels did not drop below 50% until day 4 after Tet induction of the AdoMetDC RNAi, whereas prozyme induction occurred within the first 12 h (Willert & Phillips, 2008). In parallel, the metabolite levels were also determined for BSF 427 cells plus and minus MDL 73811 (Figure S4B), and the results were similar to those shown in Figures 3 and S4A. Thus the data show a clear correlation between prozyme expression and dcAdoMet levels: prozyme levels were low when dcAdoMet levels were high.
The effects of exogenous dcAdoMet
To test if dcAdoMet added exogenously to the cell cultures could be used to manipulate intracellular levels of dcAdoMet and regulate prozyme expression levels we synthesized the compound enzymatically and purified it using column chromatography. To determine if exogenous dcAdoMet could provide functional rescue of the intracellular dcAdoMet pools we tested the effects on growth of cells that were either chemically or genetically depleted of AdoMetDC activity. We found that dcAdoMet (50 µM) added to the cultures was unable to rescue the cell growth arrest caused by either treatment with MDL 73811 or by Tet induction of the RNAi response in the AdoMetDC RNAi cell line (Figure S5). Concentrations above 100 µM dcAdoMet began to show toxicity to the cells preventing testing of higher concentrations. These data show that exogenously added dcAdoMet is unable to replace the function of intracellular dcAdoMet pools and thus an independent conformation of the effects of dcAdoMet on prozyme expression within the parasite was not possible. These results are in contrast to what we observed when exogenous spermidine was added to the AdoMetDC RNAi line, as spermidine was able to rescue growth arrest demonstrating that the AdoMetDC RNAi effect was on target (Willert & Phillips, 2008). Further proof that the AdoMetDC RNAi was target-specific was obtained by the finding that hAdoMetDC was able to rescue the cell growth affects of RNAi induced depletion of TbAdoMetDC (as discussed above and shown in Figure 4). Thus the results suggest that dcAdoMet is either not transported into the parasite or that it is unstable in culture. Decarboxylated AdoMet has been reported to be highly labile in basic pH conditions (Zappia et al., 1977).
Discussion
Polyamine metabolism is highly regulated in eukaryotic cells (Pegg & Casero, 2011, Pegg, 2009a, Pegg, 2009b, Casero & Pegg, 2009, Persson, 2009, Nowotarski et al., 2011), including in trypanosomatids where AdoMetDC is regulated by a novel mechanism requiring heterodimer formation with a catalytically-dead paralog (prozyme) to form the active enzyme (Willert et al., 2007). Our prior data suggested that prozyme protein expression was regulated translationally in response to loss of AdoMetDC (Willert & Phillips, 2008). In extending these studies we sought to provide additional support for the hypothesis that prozyme expression is regulated at the translational level and we additionally focused on obtaining mechanistic insight into both cis- and trans-acting factors that contribute to the control of prozyme expression.
Gene expression in T. brucei is typically controlled post-transcriptionally and sequence elements that modulate mRNA stability and control translation are usually found in the 3’UTR (Clayton & Shapira, 2007, Ouellette & Papadopoulou, 2009, Gunzl, 2010, Kramer & Carrington, 2011, Kramer, 2012) (Horn, 2008). We identified three separate mRNAs that were transcribed from the prozyme gene including two that contained the prozyme ORF (4.7 and 2.2 kb) and one that contained only the 3’ end of the 3’UTR (2.7 kb). The 2.2 kb and 2.7 kb mRNAs appear to be generated by an alternative splicing of the full length 3’UTR found in the 4.7 kb mRNA. However, despite the existence of multiple prozyme mRNAs, neither alternative splicing nor changes in mRNA levels were found to control prozyme protein levels as the mRNAs were found in equal abundance before or after the addition of the AdoMetDC inhibitor MDL 73811 to the cells. The role of the non-coding 2.7 kb mRNA remains unclear, though it is most likely simply a by-product of mRNA processing, as was previously demonstrated for procyclin mRNAs 1 (Vassella et al., 1994, Hug et al., 1994).
In order to determine if elements in the 3’UTR regulate prozyme protein expression we used CAT reporter constructs and deletion analysis to identify regulatory control elements. Several fragments of the 3’UTR displayed regulated CAT protein expression in the presence of MDL 73811 while CAT mRNA levels remained unchanged, thus recapitulating the effects observed for prozyme expression from the endogenous locus. These results provide additional support for the hypothesis that regulation of prozyme protein expression occurs at the translational level. The key regulatory response elements in the mRNA 3’UTR appear to be localized to the first 1.2 kb of the 3’UTR, but attempts to further narrow down the functional region by deletion analysis led to reduced activity. A region of strong secondary structure was identified at nts 1879 – 2778, based on excision of this sequence during RT reactions. This region of the sequence is contained within the prozyme regulatory element. The full length 3’UTR from the 4.7 kb mRNA was less responsive to the addition of MDL 73811 than smaller regions of the UTR, though expression of the CAT reporter was increased 2.5-fold in the construct containing this sequence. These data suggest that additional elements in the mRNA 5’UTR or the coding sequence function together with the prozyme regulatory elements in the 3’UTR in order to fully manifest increased prozyme expression from the full length mRNAs.
To identify potential trans-acting factors that contribute to the regulation of prozyme translation we addressed the question of whether or not T. brucei AdoMetDC might regulate translation by binding to the prozyme mRNA. As precedence for this hypothesis dihydrofolate reductase has been shown to be an RNA binding protein that regulates its own translation in both human and Plasmodium cells (Chu et al., 1993, Ercikan-Abali et al., 1997, Hsieh et al., 2009, Zhang & Rathod, 2002). To determine if this mechanism could operate in the control of prozyme expression we rescued the T. brucei AdoMetDC RNAi cell line with human AdoMetDC, which provided a source of dcAdoMet in the background of low T. brucei AdoMetDC levels. Expression of hAdoMetDC fully rescued the effects of AdoMetDC knockdown by RNAi. Since hAdoMetDC would be unlikely to have evolved to bind to specific elements in the T. brucei AdoMetDC RNA, our results show that a T. brucei AdoMetDC RNA binding mechanism for controlling prozyme expression is unlikely to contribute to the observed regulation.
Finally we further explored the possibility that small molecule metabolites from the pathway were involved in the regulation of prozyme translation. Spermidine has been shown to be the key regulator of polyamine biosynthesis in mammals and yeast (Pegg & Casero, 2011, Pegg, 2009a, Pegg, 2009b, Casero & Pegg, 2009, Persson, 2009, Nowotarski et al., 2011). However, we showed previously that spermidine levels do not correlate with changes in prozyme protein expression (Xiao et al., 2009, Willert & Phillips, 2008). We again observed that spermidine levels are only modestly decreased (~30%) by knockdown of AdoMetDC under conditions that led to prozyme upregulation, including a different time course for these events. We noted previously a trend for putrescine levels to be increased under conditions that led to prozyme induction (e.g. when AdoMetDC activity was reduced), however, in these current studies we showed that by treating cells with both MDL 73811 and DFMO in combination, we were able to restore putrescine levels to near wild-type levels demonstrating elevated putrescine does not appear to play a role in inducing prozyme expression.
In contrast, dcAdoMet levels inversely correlated with prozyme protein levels, providing the first evidence that a metabolite may function to regulate prozyme protein expression. These studies suggested that prozyme translation may be feedback controlled by the product of the AdoMetDC reaction, thus in this model when dcAdoMet levels are low additional prozyme would be expressed to increase the catalytic efficiency of AdoMetDC. In our previous studies we showed that during normal cell growth prozyme levels were present at limiting levels relative to AdoMetDC, providing the potential for the increased prozyme protein to lead to higher AdoMetDC activity (Willert & Phillips, 2008). Under normal growth conditions dcAdoMet represents <0.5% of the total AdoMetDC pool in BSF T. brucei (Xiao et al., 2009), which is similar to the findings in mammalian cells (Pegg et al., 2011). This suggests that small fluctuations in its synthetic or degradation rate could lead to rapid and significant changes in steady-state levels, and thus provide a sensitive mechanism to regulate gene expression. The finding that exogenously added dcAdoMet could not replace the function of intracellular dcAdoMet prevented a an independent conformation of this hypothesis and leaves open the possibility that other metabolites restored by the expression of human AdoMetDC could still play a role in the regulation.
Control of gene expression by dcAdoMet would represent a novel mechanism of translational regulation. There are no reports implicating dcAdoMet as a regulatory molecule in eukaryotic or prokaryotic cells. However, both AdoMet and S-adenosylhomocysteine sensing riboswitches have been reported in bacteria (Wang & Breaker, 2008, Bastet et al., 2011, Heppell et al., 2011) and recently genomic approaches have uncovered evidence that some riboswitches may be located in the 3’UTR (Weinberg et al., 2010). The strong secondary structure region we identified in the mRNA (nt 1879 – 2278), including the hairpin region, is likely to play a role in prozyme regulation and could form a key structural element in a potential riboswitch or other regulatory structure.
The metabolite dcAdoMet represents a potentially novel trans-activating factor involved in the control of prozyme translation. Only a handful of trans-acting factors involved in regulation of mRNA stability and protein translation have been described in T. brucei (Kramer & Carrington, 2011). These examples include, the CCCH-class Zn RNA binding proteins (TbZFP1, TbZFP2 and TbZFP3) involved in regulation of procyclin isoform expression in procyclic cells (Walrad et al., 2009, Paterou et al., 2006), the RNA binding protein PUF9 which stabilizes transcripts during S-phase (Archer et al., 2009) and TbRBP6, a RNA-binding protein that regulates differentiation into the infective metacyclic forms (Kolev et al., 2012, Walrad et al., 2009). Both glucose depletion and cis-aconitate addition can trigger differentiation of BSF cells. The RNA binding protein RBP10 has been implicated in the control of expression of BSF specific proteins and in the response of cells to cis-aconitate during differentiation (Wurst et al., 2012). The stumpy-specific surface transporter PAD1 is differentially expressed between slender and stumpy forms, and expression is regulated by 3’UTR elements that are derepressed in the presence of stumpy induction factor or cAMP (MacGregor & Matthews, 2012).
In T. brucei while there are many examples where regulation of mRNA stability is used to control gene expression, translational control has been less well explored. However, in addition to our example of prozyme translational regulation, a number of other genes also appear to be regulated translationally. Genome-wide analysis comparing proteome changes to transcriptome changes during development from blood stage to insect stage identified many examples of likely regulation at the translational or posttranslational level (Gunasekera et al., 2012). Furthermore specific examples include the RNA-binding protein TbZFP3 that regulates EP1 surface expression at the level of translation (Walrad et al., 2009), RBP10 that represses translation of some mRNAs needed for PF gene expression (Wurst et al., 2012), and developmental regulation of the Nrk protein kinase at the translational level (Gale et al., 1994).
In conclusion we have demonstrated that elements in the 3’UTR of the prozyme gene are able to modulate the expression levels of the prozyme protein, thus identifying cis-acting elements that control prozyme gene expression and showing that this regulation is likely to be translational. We further showed that prozyme protein levels are low when dcAdoMet levels are high, whereas prozyme protein levels are induced under conditions that would lead to reduced dcAdoMet levels. Thus dcAdoMet is a likely trans-acting factor controlling translation of the prozyme gene, which would represent the first example of dcAdoMet acting in a regulatory role. It remains to be determined if a novel RNA binding protein mediates this regulation, if dcAdoMet binds directly to the mRNA or if other factors are involved.
Experimental Procedures
Gene accession numbers for genes and proteins described in the study
Gene accession numbers have been taken from http://www.tritryp.org and are as follows: AdoMetDC, Tb927.6.4460/Tb927.6.4410; prozyme, Tb927.6.4470; ODC, Tb11.01.5300; SpdSyn, Tb09.v1.0380; TrypSyn, Tb927.2.4370, trypanothione reductase, Tb10.406.0520 and from GenBank, hAdoMetDC, NP_001625.2.
Trypanosome cultures
BSF trypanosomes, including wild-type 427 cells, 90-13 cells, and AdoMetDC RNAi lines were cultured in HMI-9 medium with 10% chicken serum at 37°C under 5% CO2 (Willert & Phillips, 2008). Chicken serum is used in place of fetal calf serum (FCS) when polyamines may be added to the culture for analysis because it prevents toxicity due to generation of reactive species by polyamine oxidases that are present in FCS but not chicken serum. Cells were maintained in mid-log phase (105-106 cells/ml) and selected in medium containing the appropriate antibiotics (2.5 µg/ml of G418, 2.5 µg/ml of hygromycin, and 2.5 µg/ml of phleomycin). For experiments with MDL 73811 (75 nM) or DFMO (12.5 µM) cells were treated for 1 or 3 d where concentrations represent the approximate ED50 for growth inhibition using the available compound stocks. For experiments with dcAdoMet, 50 µM was added freshly to the culture either every 2 days for experiments with the AdoMetDC RNAi line and or every day for experiments with MDL 73811. Cell densities were determined by counting with a hemocytometer (Fisher). PF trypanosomes 427 were grown in SDM-79 media with 10% FCS at 28 °C (Brun & Schonenberger, 1979). Cells were grown to mid-log phase (105–106 cells/ml) and diluted 1:100 about every 4 days into fresh media. Total cell numbers were calculated as the product of cell density × the total dilution factor × culture volume. Cells were scored as dead when no live parasites were observed in the culture flask by microscopy after monitoring for 2 – 4 days.
Preparation of total RNA and mRNA
Cells (~1 × 108 ) were centrifuged at 3000 × rpm and cell pellets were lysed in TRIzol (Invitrogen) after vortexing. Total RNA was extracted with RNeasy mini kit (Qiagen). To obtain mRNA, total RNA was purified with Dynabeads mRNA purification kit (Invitrogen).
Preparation of genomic DNA
For the extraction of genomic DNA, 1 × 107 of cells were centrifuged at 3000 × rpm, washed twice in 20 ml phosphate buffered saline (PBS) pH 7.4, and resuspended in 0.5 ml of lysis buffer (100 mM Tris pH 8.0, 5 mM EDTA pH 8.0, 200 mM NaCl, 0.2% SDS). Proteinase K (Sigma) was added to a final concentration of 0.2 mg/ml, and the suspension was incubated at 55°C overnight with shaking. After sequential extraction with phenol-chloroform, the DNA was precipitated by the addition 0.3 M sodium acetate and 2.5 volumes of ethanol. The pellet was washed with 70% ethanol, air dried, and resuspended in distilled water.
Prozyme 3’UTR mapping
3’- RACE was performed with a GeneRacer kit with SuperScript III TOPO TA cloning kit (Invitrogen) using RNA isolated from T. brucei BSF 427 cells as template. cDNA was synthesized using oligo dT primers and PCR products were generated from the cDNA using prozyme gene specific primers (Table S1) and/or a GeneRacer 3’ primer homologous to the oligo dT primer. PCR products were cloned into a pCR 2.1-TOPO vector (Invitrogen) and sequenced in their entirety in both directions at the McDermott sequencing core (UT Southwestern). Sequenced clones are displayed in Figure 1S. Additional cDNA synthesis was also performed with Moloney murine leukemia virus (MMLV) RT (Invitrogen) as a replacement to SuperScript III RT. Touchdown PCR methodology was used for mapping as described with modification (Yue et al., 2010). Reactions contained PCR buffer (60 mM Tris-SO4, 18 mM NH4SO4), 2.5 mM MgSO4, 0.2 mM each dNTP, T. brucei cDNA (100 ng), 1.5 U of Platinum® Taq DNA Polymerase High Fidelity (Invitrogen) (total volume 0.05 ml). The touchdown PCR protocol consisted of two phases: phase 1 included an initial step of 94°C for 2 m, followed by 5- cycles of 94°C for 30 s, 72°C for 3 m, and 5-cycles of 94°C for 30 s, 70°C for 3 m. The subsequent 3 cycles were at 94°C for 30 s, with annealing temperatures of 68°C, 66°C and 72°C respectively, for 3 m each. Phase 2 consisted of 22 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 3 m, followed by a final 15 m extension cycle at 68°C.
Generation of Prozyme 3’UTR plasmid reporter constructs
The 3’UTR of the prozyme mRNA, and fragments therein (9 in total), were cloned into pHD1437 reporter construct vector (Colasante et al., 2007, Robles & Clayton, 2008) which allows the expression of the CAT gene to serve as a reporter for the strength of the regulatory sequences that are cloned 3’ to the gene. Fragments were amplified by PCR (except fragment 4a, 4b and 7) and cloned into pHD1437 immediately after the CAT ORF. Cloning was performed after digestion of both the vector and the PCR fragments with Bam HI and Mlu I, followed by subsequent purification of the fragments from agarose gels using high pure PCR product purification kit (Roche). Forward strand primers contained a Bam H1 site while reverse strand primers contained the Mlu I site. To generate construct 4a, construct 3 was digested with Mlu I and Sal I and ligated to the PCR product containing fragment 9MS generated with primers that contained Mlu I and Sal I restriction sites, respectively; construct 4b was made similarly by annealing fragment 9MS to construct 1 after digestion of both with Mlu I and Sal I; and construct 7 was made by annealing fragment 6MS to construct 3 again digested with Mlu I and Sal I. See Table S2 for primer pairs used to generate the described clones and Table S4 for a list of the base pair regions included in each construct.
Transient transfection of cell lines with CAT reporter constructs
To monitor CAT activity cultures of BSF 90-13 cells were transiently transfected with the prozyme 3’UTR plasmid constructs as described (Wirtz et al., 1994). Briefly, BSF 90-13 cells (3 × 107) were electroporated with 15 µg of circular plasmid constructs by a single pulse on the AMAXA Nucleofector II (Lonza). AdoMetDC inhibitor MDL 73811 (75 nM) or a vehicle control were added 3 h after transfection and cells were allowed to grow for 16 h before harvesting. Cells were collected by centrifugation at 3000 × rpm (Sorvall legend RT). For CAT ELISA assays cells were washed twice with 20 ml of PBS and resuspended in lysis buffer from the CAT Elisa assay kit (Roche). Supernatants were collected after centrifugation at 3000 × rpm (10 min) for subsequent CAT activity assays. Cells to be used for the isolation of RNA for quantitative real-time PCR analysis were lysed in Trizol, and the RNA was extracted with RNase mini kit (Qiagen) according to the manufacturer’s protocol.
CAT activity assays
CAT activity was monitored using the CAT ELISA assay kit (Roche), a colorimetric enzyme immunoassay that monitors CAT protein levels. Absorbance was monitored at 405 nm using a 96-well plate reader (PowerWave X, Bio-Tek instrument, Inc). CAT protein levels were quantitated using standard curves prepared using CAT protein standards (included within the ELISA kit). CAT level in each lysate were verified to be in the linear range of quantitation by diluting lysates over a 1000-fold range.
Generation of the AdoMetDC RNAi-hAdoMetDC cell line
hAdoMetDC (Gene ID: NP_001625.2) was amplified by PCR (primers in Table 3S) using a previously reported E. coli expression vector as template (Clyne et al., 2002), and cloned into a modified pLEW100 vector containing a resistance selectable marker for blasticidin in place of phleomycin (Willert & Phillips, 2008). The construct was transfected into the previously generated AdoMetDC RNAi line derived from 90-13 BSF cells, which contains a 620 base pair (bp) fragment of the AdoMetDC gene cloned into a stem loop hairpin under the control of a tetracycline (Tet) regulated promoter with a phleomycin resistance selection marker(Willert & Phillips, 2008). Transfection was performed as described above for the CAT reporter constructs except stable cell lines were generated by selection with blasticidin to drive plasmid integration into the rRNA locus. The coexpression of hAdoMetDC and the double stranded RNA (RNAi) targeting the T. brucei AdoMetDC message was induced by the addition of Tet (1 µg/ml) added into the culture every 24 h; blasticidin and phleomycin were added to the culture to maintain the AdoMetDC RNAi and hAdoMetDC expression constructs.
Northern blotting analysis
All reagents were purchased from Applied Biosystems unless otherwise indicated. Template DNA for RNA probes was generated by PCR from genomic DNA isolated from BSF 427 cells using primers shown in Table S3. The antisense strand primer contained the T7 polymerase minimal promoter to allow direct transcription to generate the probe. RNA probes were synthesized from the PCR products using MAXIscript T7 kit (Invitrogen), and subjected to purification with RNase mini kit (Qiagen). Purified RNA probes were labeled with psoralen-biotin using BrightStar Psoralen-biotin kit (Invitrogen). T. brucei mRNA (isolated as describe above) was denatured with formaldehyde, and separated on 1% denaturing agarose gels (2 µg/lane). The gel was soaked in 0.05 M NaOH/1.5 M NaCl for 30 m with gentle shaking, rinsed with DEPC-treated water twice, then soaked in 10X SSC buffer (1.5 M NaCl and 0.15 M sodium citrate) for 40 m before blotting onto a positively charged nylon membrane (BrightStar-Plus) (Invitrogen) to ensure efficient transfer of larger mRNAs. Blots were cut into equal sections for analysis. Blots were hybridized overnight in ULTRAhyb buffer containing 0.1 nM psoralen-biotin labeled RNA (Yue et al., 2010) probe at 68°C, washed and visualized by BrightStar® BioDetect nonisotopic detection kit after subsequent exposure to film. Primers used to generate the various probes (Figures 1 and S2) for Northern analysis are shown in Table S3.
RNase protection assay
A non-coding strand RNA probe was synthesized spanning 94 bases of sequence 5’ to the region of secondary structure in the prozyme cDNA and 114 bases down stream of the 3’ region of secondary structure, while eliminating the intervening sequence (Table S3 and Figure S3). A 41-base tag was included on the 5’ end of the probe that contained the 20 nucleotide T7 polymerase site to allow transcription and 21 bases of random sequence to distinguish the undigested and digested probes. The probe DNA was synthesized by Genscript and cloned into the Bam HI/Hind III sites of pUC57. The Bam HI/Hind III fragment was excised from this vector, gel purified and used directly in the reaction to synthesize the RNA probe in the presence of alpha-[32P]UTP and T7 RNA polymerase (Invitrogen). Total RNA was extracted from BSF 427 cells and from BSF 90-13 cells transiently transfected with construct 4a or 4b to serve as controls. Gel purified RNA probe and DNase-treated RNA were mixed in 30 µl of Hybe Buffer (40 mM Pipes pH 6.4; 1 mM EDTA; 0.4 M NaCl; 80% di-formamide), and incubated at 45°C overnight. After adding 300 µl RNase digestion buffer (300 mM NaCl; 10 mM Tris-HCl, pH 7.5; 5 mM EDTA), 1.5 ug of RNaseA (Life techonology) and 45 units of RNaseT1 (Life technology), digestion of single-strand RNAs was performed by incubation at 30°C for 30 min. The digestion was stopped by adding 10 µl of 20% SDS and 10 µl of 20 mg/ml proteinase K and further incubation at 37°C for 15 min. The RNAs were treated with phenol/chloroform, ethanol precipitated in the presence of 25 µg of carrier yeast tRNA, and resuspended in 10 µl of urea gel-loading buffer (8M urea, 0.15% bromophenol blue, 0.15% xylene cyanol, 2.5 mM EDTA, pH 8.0, 1x TBE buffer (890 mM Tris, 890 mM boric acid, 20 mM EDTA). Protected RNA fragments were fractionated in 6% urea page gel. After exposure to x-ray film, gels were analyzed with a PhosphoImaging system.
Western blot analysis
Protein levels were evaluated by western blot analysis as described (Xiao et al., 2009). Briefly, cells (1 × 108) were harvested, washed twice in 20 ml phosphate buffered saline PBS pH 7.4, resuspended in lysis buffer (50 mM HEPES pH 8.0, 100 mM NaCl, 5 mM 2-mercaptoethanol, 2 mM phenylmethylsulfonyl fluoride (PMSF), 1 µg/ml leupeptin, 2 µg/ml antipain, 10 µg/ml benzamidine, 1 µg/ml pepstatin, and 1 µg/ml chymostatin) and subjected to three freeze/thaw cycles. The supernatant was collected by centrifugation at 3000 × rpm for 10 m. Total protein concentration was determined using the protein assay dye reagent from Bio-Rad Labs. Cell lysates in SDS-sample buffer were boiled for 5 m, total protein (40 ug) was separated by 16% of SDS-PAGE, and transferred to a polyvinylidene difluoride (PVDF) membrane (Hybond-P, Life techonology). Membranes were probed with rabbit polyclonal antibody raised against T. brucei ODC, T. brucei AdoMetDC, T. brucei Prozyme, and T. brucei dihydroorotate dehydrogenase (DHODH), using antibody dilutions and conditions as previously described (Willert & Phillips, 2008, Arakaki et al., 2008), followed by visualization with ECL Western blotting detection reagents (Amersham Biosciences/GE Healthcare). hAdoMetDC antibody was a generous gift from David Feith and was used as described (Wallick et al., 2005).
Quantitative real-time PCR analysis
Gene expression was measured by quantitative real-time PCR as described (Kalidas et al., 2011). Total RNA was treated with DNAse I (Invitrogen) to remove genomic DNA and reverse-transcribed using SuperScript® III First-Strand Synthesis System (Invitrogen). First-strand cDNA was used as a template for subsequent real time PCR amplification with the iTaq SYBR Green Supermix With ROX kit (Bio-Rad) and CFX 96 Real-time System (Bio-Rad). Primers are listed in Table S3. Reactions contained 0.1 µM primer, 30 ng of first strand cDNA, and 2x ready-to-use reaction mix (0.4 mM dATPs, 0.4 mM dCTPs, 0.4 mM dGTPs, 50 unit/ml iTaq DNA polymerase, 6 mM Mg2+, SYBR® Green I, 1 µM ROX reference dye) in a final volume of 20 µl. PCR cycling conditions were as follows: 95 °C for 3 m, followed by 45 cycles of 95 °C for 10 s and 55 °C for 30 s. Gene expression levels were quantified by the comparative ΔΔCT method (Winer et al., 1999) by using telomerase reverse transcriptase (TERT) as the internal standard for normalization. TERT was previously shown to be an optimal control transcript (Brenndorfer & Boshart, 2010). The ΔCT value was determined by subtracting the CT value of each sample from that of TERT in the corresponding sample. The ΔΔCT values were then calculated by subtracting the highest mean ΔCT value as an arbitrary constant from all other ΔCT values. Relative gene expression levels were calculated using the equation of 2−ΔΔCt. To control for the presence of contaminating genomic DNA a no-reverse transcriptase control was included in all runs and the signal generated by primer dimers was determined through no-template controls. Melting curve analysis (65–95 °C) was performed and confirmed no visible nonspecific amplification of any PCR products from genomic DNA or primer dimers.
Quantification of intracellular polyamines by HPLC
Quantitation of intracellular polyamines by HPLC was performed as previously described (Willert & Phillips, 2008, Xiao et al., 2009). T. brucei cells (1×107 ) were collected and washed twice in 20 ml PBS pH 7.4, resuspended in lysis buffer (100 mM MOPS buffer pH 8.0, 50 mM NaCl, 2 mM EDTA, and 20 mM MgCl2), and subjected to three freeze/thaw cycles. Protein was precipitated with trichloroacetic acid (9.2% final concentration), and the supernatants were collected by centrifugation (13500 rpm for 3 m). Aliquots (5 µl) were labeled with AccQ-tag reagent (Waters) followed by HPLC analysis (System Gold Nouveau, Beckman Instruments) on an AccQ-TAG column (3.9 × 150 mm) (Waters) using Buffer A (450 mM NaAcetate, 17 mM triethylamine, 0.01% w/v NaN3, pH 4.95) and Buffer B (60% acetonitrile, and 0.01% acetone) with a modified gradient (0–20 % solvent B over 4 m, 20–50% solvent B over 45 m, 50–100% solvent B over 5 m). Fluorescent peaks were detected with λEx at 250 nm and λEm at 395 nm. Polyamines were quantitated by the peak areas relative to commercial standards.
Quantitation of polyamines and AdoMet metabolites by LC-MS/MS
Polyamine metabolites (Spermidine, AdoMet, dcAdoMet, and MTA) were also quantitated by LC-MS/MS as previously described (Tu et al., 2007). Briefly, cells were lysed with cold lysis buffer (50% methanol, 0.1% formic acid), and subjected to ten freeze/thaw cycles. The supernatant was collected by centrifugation, vacuum dried and re-suspended in formic acid buffer (0.1% in HPLC grade water) before filtering through a 0.2 µm spin column (Grace Davison). A Shimadzu Prominence LC20/SIL-20AC HPLC coupled to a ABSCIEX 3200 QTRAP triple quadrupole mass spectrometer was used for LC-MS/MS quantification of metabolites. Chromatographic separation of metabolites was performed on a C18-based column with polar embedded groups (Synergi Fusion, 150 × 2.0 mm 4 µ, Phenomenex). for detection of sulfur-containing metabolites in positive mode, a 0.1% formic acid /methanol gradient was used based on previously published methods (Tu et al., 2007). Infusion quantitative optimization was performed to acquire optimal product ion mass for each metabolite. Multiple reaction monitoring (MRM) was used to detect and quantitate metabolites. The two most abundant daughter ions were used when possible and metabolite concentrations were normalized to total ion content. For dcAdoMet a relative peak area of 1 ×10−6 for the more abundant ion represents the lower limit of quantitation. Daughter ion pairs were as follows: AdoMet (ion 1, 355/250; ion 2, 399/136), dcAdoMet (ion 1, 355/250; ion 2, 355, 136), MTA (ion 1, 298/136; ion 2, 298/119), putrescine (ion 1, 89/72; ion 2, 89/71) and spermidine (ion 1, 146/72; ion 2, 146/84).
Synthesis of dcAdoMet
Enzymatic synthesis and purification of dcAdoMet was performed as described (Zappia et al., 1977). Briefly, AdoMet (8 mM) (Affymetrix) was incubated with recombinant T. brucei AdoMetDC/Prozyme (1 µM; expressed and purified as described (Velez et al., 2013)) for 3 h in 5 mL 300 mM HEPES buffer, pH 7.7, 50 mM NaCl, 5 mM DTT. The reaction was stopped with 2.5 mL 1.5 M HClO4. After centrifugation at 17,000 g for 15 m, 3 M KHCO3 was added to adjust the pH to 4.5. The solution was cleared by centrifugation at 3,500 g for 15 min, the supernatant was boiled for 1 h, cooled and then applied to Dowex 50 W resin (8 % cross-linked, dry mesh 200–400, from Sigma) packed in a 4 × 2.5 cm column (20 mL bed volume) and prepared by washing with 1 M NaCl until neutral pH followed by water until conductivity reached zero. Conductivity and UV at 254 nM were monitored on an AKTAPurifier UPC-900 monitor. After sample was applied, the column was washed with 0.1 M NaCl, followed by 3 M HCL to elute homoserine and methylthioadenosine, products of acid hydrolysis of AdoMet, and then dcAdoMet was eluted with 6 M HCl. The solution was evaporated and the pellet re-dissolved in water. The presence of dcAdoMet was confirmed by LC-MS on an Agilent 6430 triple Quadrupole LC/MS system and purity was determined to be >90%. Levels were quantified assuming an extinction coefficient of 1.53E4 M−1cm−1 at 260 nm.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants (R01AI34432 and R37AI034432) (to MAP) and the Welch Foundation grant I-1257 (to MAP). MAP holds the Beatrice and Miguel Elias Distinguished Chair in Biomedical Science and the Carolyn R. Bacon Professorship in Medical Science and Education. We thank Dr. Christine Clayton for providing the pHD1437 reporter construct, for helpful discussions and critical reading of the manuscript, Dr. Nick Conrad for helpful advice on the RNase protection assay and Dr. Anthony J. Michael for helpful discussions.
Abbreviations
- HAT
human African trypanosomiasis
- AdoMetDC
S-adenosylmethionine decarboxylase
- AdoMetDC/prozyme
the purified complex between AdoMetDC and prozyme
- hAdoMetDC
human AdoMetDC
- ODC
ornithine decarboxylase
- AdoMet
(S-adenosylmethionine)
- dcAdoMet
decarboxylated AdoMet
- SpdSyn
spermidine synthase
- TrypSyn
trypanothione synthase
- CAT
chloramphenicol acetyl transferase
- RT
reverse transcriptase
- DHODH
dihydroorotate dehydrogenase
- DFMO
α-difluromethylornithine (also known as eflornithine)
- (BSF)
T. brucei bloodstream form
- (PF)
T. brucei procyclic form
- (RACE)
Rapid Amplification of cDNA ends
- (poly(A))
polyadenylation
- (poly(Y))
pyrimidine track
- (nt)
nucleotide
- (bp)
base-pair
- (ORF)
open reading frame
- RT
reverse transcriptase
Footnotes
The authors declare no conflict of interest.
Supplemental Tables and Figures
Figure S1. Cloned 3’UTR RACE products used to sequence the prozyme 3’UTR.
Figure S2. Sequence of the prozyme gene
Figure S3. RNase protection assay of T. brucei RNA extracted from BSF 427 cells.
Figure S4. LC-MS/MS analysis of samples described in Figures 4 and 5
Figure S5. T. brucei growth curves in the presence of dcAdoMet for both MDL 73811-treated cells and the AdoMetDC RNAi line.
Table S1. PCR primers used for RACE analysis and gene sequencing
Table S2. Cloning primers used to generate CAT reporter constructs
Table S3. Primers used for PCR, real time PCR and synthesis of RNA probes
Table S4. Description of the 3’UTR fragments used to generate CAT reporter constructs
Table S5. Summary table of the effects of MDL 73811 on CAT enzyme and mRNA levels
References
- Arakaki TL, Buckner FS, Gillespie JR, Malmquist NA, Phillips MA, Kalyuzhniy O, Luft JR, Detitta GT, Verlinde CL, Van Voorhis WC, Hol WG, Merritt EA. Characterization of Trypanosoma brucei dihydroorotate dehydrogenase as a possible drug target; structural, kinetic and RNAi studies. Mol Microbiol. 2008;68:37–50. doi: 10.1111/j.1365-2958.2008.06131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Luu VD, de Queiroz RA, Brems S, Clayton C. Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle. PLoS Pathog. 2009;5:e1000565. doi: 10.1371/journal.ppat.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchi CJ, Barker RH, Jr, Rodriguez A, Hirth B, Rattendi D, Yarlett N, Hendrick CL, Sybertz E. Trypanocidal activity of 8-methyl-5'-{[(Z)-4-aminobut-2-enyl]-(methylamino)}adenosine (Genz-644131), an adenosylmethionine decarboxylase inhibitor. Antimicrob Agents Chemother. 2009;53:3269–3272. doi: 10.1128/AAC.00076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchi CJ, Brun R, Croft SL, Alicea K, Buhler Y. In vivo trypanocidal activities of new S-adenosylmethionine decarboxylase inhibitors. Antimicrob Agents Chemother. 1996;40:1448–1453. doi: 10.1128/aac.40.6.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchi CJ, Nathan HC, Yarlett N, Goldberg B, McCann PP, Bitonti AJ, Sjoerdsma A. Cure of murine Trypanosoma brucei rhodesiense infections with an S-adenosylmethionine decarboxylase inhibitor. Antimicrob Agents Chemother. 1992;36:2736–2740. doi: 10.1128/aac.36.12.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker RH, Jr, Liu H, Hirth B, Celatka CA, Fitzpatrick R, Xiang Y, Willert EK, Phillips MA, Kaiser M, Bacchi CJ, Rodriguez A, Yarlett N, Klinger JD, Sybertz E. Novel S-adenosylmethionine decarboxylase inhibitors for the treatment of human African trypanosomiasis. Antimicrob Agents Chemother. 2009;53:2052–2058. doi: 10.1128/AAC.01674-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett MP. Potential new drugs for human African trypanosomiasis: some progress at last. Curr Opin Infect Dis. 2010;23:603–608. doi: 10.1097/QCO.0b013e32833f9fd0. [DOI] [PubMed] [Google Scholar]
- Bastet L, Dube A, Masse E, Lafontaine DA. New insights into riboswitch regulation mechanisms. Mol Microbiol. 2011;80:1148–1154. doi: 10.1111/j.1365-2958.2011.07654.x. [DOI] [PubMed] [Google Scholar]
- Batey RT. Recognition of S-adenosylmethionine by riboswitches. Wiley Interdiscip Rev RNA. 2011;2:299–311. doi: 10.1002/wrna.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitonti AJ, Byers TL, Bush TL, Casara PJ, Bacchi CJ, Clarkson AB, Jr, McCann PP, Sjoerdsma A. Cure of Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense Infections in Mice with an Irreversible Inhibitor of S-Adenosylmethionine Decarboxylase. Antimicrob Agents Chemother. 1990;34:1485–1490. doi: 10.1128/aac.34.8.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenndorfer M, Boshart M. Selection of reference genes for mRNA quantification in Trypanosoma brucei. Mol Biochem Parasitol. 2010;172:52–55. doi: 10.1016/j.molbiopara.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Brun R, Blum J, Chappuis F, Burri C. Human African trypanosomiasis. Lancet. 2010;375:148–159. doi: 10.1016/S0140-6736(09)60829-1. [DOI] [PubMed] [Google Scholar]
- Brun R, Buhler Y, Sandmeier U, Kaminsky R, Bacchi CJ, Rattendi D, Lane S, Croft S, Snowdon D, Yardley V, Caravatti G, Frei J, Stanek J, Mett H. In vitro trypanocidal activities of new S-adenosylmethionine decarboxylase inhibitors. J Med Chem. 1996;40:1442–1447. doi: 10.1128/aac.40.6.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R, Schonenberger M. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- Burri C. Chemotherapy against human African trypanosomiasis: is there a road to success? Parasitology. 2010;137:1987–1994. doi: 10.1017/S0031182010001137. [DOI] [PubMed] [Google Scholar]
- Casero RA, Pegg AE. Polyamine catabolism and disease. Biochem J. 2009;421:323–338. doi: 10.1042/BJ20090598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu E, Takimoto CH, Voeller D, Grem JL, Allegra CJ. Specific binding of human dihydrofolate reductase protein to dihydrofolate reductase messenger RNA in vitro. Biochemistry. 1993;32:4756–4760. doi: 10.1021/bi00069a009. [DOI] [PubMed] [Google Scholar]
- Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Clyne T, Kinch L, Phillips M. Putrescine activation of Trypanosoma cruzi S-adenosylmethionine decarboxylase. Biochemistry. 2002;41:13207–13261. doi: 10.1021/bi026541d. [DOI] [PubMed] [Google Scholar]
- Colasante C, Robles A, Li CH, Schwede A, Benz C, Voncken F, Guilbride DL, Clayton C. Regulated expression of glycosomal phosphoglycerate kinase in Trypanosoma brucei. Mol Biochem Parasitol. 2007;151:193–204. doi: 10.1016/j.molbiopara.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Ercikan-Abali E, Banerjee D, Waltham M, Skacel N, Scotto K, Bertino J. Dihydrofolate reductase protein inhibits its own translation by binding to dihydrofolate reductase mRNA sequences within the coding region. Biochemistry. 1997;36:12317–12322. doi: 10.1021/bi971026e. [DOI] [PubMed] [Google Scholar]
- Gale MJ, Carter V, Parsons M. Translational control mediates the developmental regulation of the Trypanosoma brucei Nrk protein kinase. J. Biol. Chem. 1994:31659–31665. [PubMed] [Google Scholar]
- Gunasekera K, Wuthrich D, Braga-Lagache S, Heller M, Ochsenreiter T. Proteome remodelling during development from blood to insect-form Trypanosoma brucei quantified by SILAC and mass spectrometry. BMC Genomics. 2012;13:556. doi: 10.1186/1471-2164-13-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunzl A. The pre-mRNA splicing machinery of trypanosomes: complex or simplified? Eukaryot Cell. 2010;9:1159–1170. doi: 10.1128/EC.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppell B, Blouin S, Dussault AM, Mulhbacher J, Ennifar E, Penedo JC, Lafontaine DA. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol. 2011;7:384–392. doi: 10.1038/nchembio.563. [DOI] [PubMed] [Google Scholar]
- Horn D. Codon usage suggests that translational selection has a major impact on protein expression in trypanosomatids. BMC Genomics. 2008;9:2. doi: 10.1186/1471-2164-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS One. 2010;5:e12271. doi: 10.1371/journal.pone.0012271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YC, Skacel NE, Bansal N, Scotto KW, Banerjee D, Bertino JR, Abali EE. Species-specific differences in translational regulation of dihydrofolate reductase. Mol Pharmacol. 2009;76:723–733. doi: 10.1124/mol.109.055772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug M, Hotz HR, Hartmann C, Clayton C. Hierarchies of RNA-processing signals in a trypanosome surface antigen mRNA precursor. Mol Cell Biol. 1994;14:7428–7435. doi: 10.1128/mcb.14.11.7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Atkins JF, Michael AJ. A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res. 2010;38:353–359. doi: 10.1093/nar/gkp1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs RT, Nare B, Phillips MA. State of the art in African trypanosome drug discovery. Curr Top Med Chem. 2011;11:1255–1274. doi: 10.2174/156802611795429167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana C. Regulation of cellular polyamine levels and cellular proliferation by antizyme and antizyme inhibitor. Essays Biochem. 2009;46:47–61. doi: 10.1042/bse0460004. [DOI] [PubMed] [Google Scholar]
- Kalidas S, Li Q, Phillips MA. A Gateway((R)) compatible vector for gene silencing in bloodstream form Trypanosoma brucei. Mol Biochem Parasitol. 2011 doi: 10.1016/j.molbiopara.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PG. The continuing problem of human African trypanosomiasis (sleeping sickness) Annals Neurol. 2008;64:116–126. doi: 10.1002/ana.21429. [DOI] [PubMed] [Google Scholar]
- Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6:e1001090. doi: 10.1371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Ramey-Butler K, Cross GA, Ullu E, Tschudi C. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 2012;338:1352–1353. doi: 10.1126/science.1229641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S. Developmental regulation of gene expression in the absence of transcriptional control: the case of kinetoplastids. Mol Biochem Parasitol. 2012;181:61–72. doi: 10.1016/j.molbiopara.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Kramer S, Carrington M. Trans-acting proteins regulating mRNA maturation, stability and translation in trypanosomatids. Trends Parasitol. 2011;27:23–30. doi: 10.1016/j.pt.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauth-Siegel RL, Comini MA. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochim Biophys Acta. 2008;1780:1236–1248. doi: 10.1016/j.bbagen.2008.03.006. [DOI] [PubMed] [Google Scholar]
- MacGregor P, Matthews KR. Identification of the regulatory elements controlling the transmission stage-specific gene expression of PAD1 in Trypanosoma brucei. Nucleic Acids Res. 2012;40:7705–7717. doi: 10.1093/nar/gks533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D, Gunasekera K, Mani J, Osteras M, Farinelli L, Baerlocher L, Roditi I, Ochsenreiter T. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 2010;6:e1001037. doi: 10.1371/journal.ppat.1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotarski SL, Origanti S, Shantz LM. Posttranscriptional regulation of ornithine decarboxylase. Methods Mol Biol. 2011;720:279–292. doi: 10.1007/978-1-61779-034-8_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette M, Papadopoulou B. Coordinated gene expression by post-transcriptional regulons in African trypanosomes. J Biol. 2009;8:100. doi: 10.1186/jbiol203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Nishimura K, Zanelli CF, Valentini SR. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterou A, Walrad P, Craddy P, Fenn K, Matthews K. Identification and stage-specific association with the translational apparatus of TbZFP3, a CCCH protein that promotes trypanosome life-cycle development. J Biol Chem. 2006;281:39002–39013. doi: 10.1074/jbc.M604280200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE. Mammalian polyamine metabolism and function. IUBMB Life. 2009a;61:880–894. doi: 10.1002/iub.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE. S-Adenosylmethionine decarboxylase. Essays Biochem. 2009b;46:25–45. doi: 10.1042/bse0460003. [DOI] [PubMed] [Google Scholar]
- Pegg AE, Casero RA., Jr Current status of the polyamine research field. Methods Mol Biol. 2011;720:3–35. doi: 10.1007/978-1-61779-034-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE, Wang X, Schwartz CE, McCloskey DE. Spermine synthase activity affects the content of decarboxylated S-adenosylmethionine. Biochem J. 2011;433:139–144. doi: 10.1042/BJ20101228. [DOI] [PubMed] [Google Scholar]
- Persson L. Polyamine homoeostasis. Essays Biochem. 2009;46:11–24. doi: 10.1042/bse0460002. [DOI] [PubMed] [Google Scholar]
- Robles A, Clayton C. Regulation of an amino acid transporter mRNA in Trypanosoma brucei. Mol Biochem Parasitol. 2008;157:102–106. doi: 10.1016/j.molbiopara.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010;38:4946–4957. doi: 10.1093/nar/gkq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Tan KS, Cross GA. Systematic study of sequence motifs for RNA trans splicing in Trypanosoma brucei. Mol Cell Biol. 2005;25:9586–9594. doi: 10.1128/MCB.25.21.9586-9594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinks D, Torrie LS, Thompson S, Harrison JR, Frearson JA, Read KD, Fairlamb AH, Wyatt PG, Gilbert IH. Design, synthesis and biological evaluation of Trypanosoma brucei trypanothione synthetase inhibitors. ChemMedChem. 2012;7:95–106. doi: 10.1002/cmdc.201100420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K, Brun R, Croft S, Fairlamb A, Gurtler RE, McKerrow J, Reed S, Tarleton R. Kinetoplastids: related protozoan pathogens, different diseases. J Clin Invest. 2008;118:1301–1310. doi: 10.1172/JCI33945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekwani BL, Bacchi CJ, Secrist JA, Pegg AE. Irreversible inhibition of S-adenosylmethionine decarboxylase of Trypanosoma brucei brucei by S-adenosylmethionine analogs. Biochem Pharmacol. 1992;44:905–911. doi: 10.1016/0006-2952(92)90122-y. [DOI] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci USA. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassella E, Braun R, Roditi I. Control of polyadenylation and alternative splicing of transcripts from adjacent genes in a procyclin expression site: a dual role for polypyrimidine tracts in trypanosomes? Nucleic Acids Res. 1994;22:1359–1364. doi: 10.1093/nar/22.8.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez N, Brautigam CA, Phillips MA. Trypanosoma brucei S-adenosylmethionine decarboxylase N-terminus is essential for allosteric activation by the regulatory subunit prozyme. J Biol Chem. 2013;288:5232–5240. doi: 10.1074/jbc.M112.442475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallick CJ, Gamper I, Thorne M, Feith DJ, Takasaki KY, Wilson SM, Seki JA, Pegg AE, Byus CV, Bachmann AS. Key role for p27Kip1, retinoblastoma protein Rb, and MYCN in polyamine inhibitor-induced G1 cell cycle arrest in MYCN-amplified human neuroblastoma cells. Oncogene. 2005;24:5606–5618. doi: 10.1038/sj.onc.1208808. [DOI] [PubMed] [Google Scholar]
- Walrad P, Paterou A, Acosta-Serrano A, Matthews KR. Differential trypanosome surface coat regulation by a CCCH protein that co-associates with procyclin mRNA cis-elements. PLoS Pathog. 2009;5:e1000317. doi: 10.1371/journal.ppat.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Breaker RR. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem Cell Biol. 2008;86:157–168. doi: 10.1139/O08-008. [DOI] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11:R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert E, Phillips MA. Regulation and function of polyamines in African trypanosomes. Trends Parasitol. 2012;28:66–72. doi: 10.1016/j.pt.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Willert EK, Fitzpatrick R, Phillips MA. Allosteric regulation of an essential trypanosome polyamine biosynthetic enzyme by a catalytically dead homolog. Proc Natl Acad Sci USA. 2007;104:8275–8280. doi: 10.1073/pnas.0701111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert EK, Phillips MA. Regulated expression of an essential allosteric activator of polyamine biosynthesis in African trypanosomes. PLoS Pathog. 2008;4:e1000183. doi: 10.1371/journal.ppat.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert EK, Phillips MA. Cross-species activation of trypanosome S-adenosylmethionine decarboxylase by the regulatory subunit prozyme. Mol Biochem Parasitol. 2009;168:1–6. doi: 10.1016/j.molbiopara.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Hartmann C, Clayton C. Gene expression mediated by bacteriophage T3 and T7 RNA polymerases in transgenic trypanosomes. Nucleic Acids Res. 1994;22:3887–3894. doi: 10.1093/nar/22.19.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst M, Seliger B, Jha BA, Klein C, Queiroz R, Clayton C. Expression of the RNA recognition motif protein RBP10 promotes a bloodstream-form transcript pattern in Trypanosoma brucei. Mol Microbiol. 2012;83:1048–1063. doi: 10.1111/j.1365-2958.2012.07988.x. [DOI] [PubMed] [Google Scholar]
- Wyllie S, Oza SL, Patterson S, Spinks D, Thompson S, Fairlamb AH. Dissecting the essentiality of the bifunctional trypanothione synthetase-amidase in Trypanosoma brucei using chemical and genetic methods. Mol Microbiol. 2009;74:529–540. doi: 10.1111/j.1365-2958.2009.06761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, McCloskey DE, Phillips MA. RNA interference-mediated silencing of ornithine decarboxylase and spermidine synthase genes in Trypanosoma brucei provides insight into regulation of polyamine biosynthesis. Eukaryot Cell. 2009;8:747–755. doi: 10.1128/EC.00047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X, Schwartz JC, Chu Y, Younger ST, Gagnon KT, Elbashir S, Janowski BA, Corey DR. Transcriptional regulation by small RNAs at sequences downstream from 3' gene termini. Nat Chem Biol. 2010;6:621–629. doi: 10.1038/nchembio.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappia V, Galletti P, Oliva A, de Santis A. New methods for preparation and analysis of S-adenosyl-(5')-3-methylthiopropylamine. Anal Biochem. 1977;79:535–543. doi: 10.1016/0003-2697(77)90427-4. [DOI] [PubMed] [Google Scholar]
- Zhang K, Rathod P. Divergent regulation of dihydrofolate reductase between malaria parasite and human host. Science. 2002;296:545–547. doi: 10.1126/science.1068274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.