Abstract
Together, the hypothalamus, pituitary and gonads direct the development and regulation of reproductive function in mammals. Gonadotropin-releasing hormone (GnRH) expression is limited to ∼800 neurons that originate in the olfactory placode then migrate to the hypothalamus. Coordination of the hypothalamic-pituitary-gonadal (HPG) axis is dependent upon correct neuronal migration of GnRH neurons into the hypothalamus followed by proper synthesis and pulsatile secretion of GnRH. Defects in any one of these processes causes infertility. Otx2, the vertebrate homologue of Drosophila orthodenticle, is a transcription factor that has been shown to be critical for normal brain and eye development and is expressed in both the developing GnRH neurons and the pituitary, suggesting that this gene may play a critical role in development of the HPG axis. As Otx2-null mice are embryonic lethal, we have analyzed the reproductive capacity of heterozygous Otx2 mice to determine the contribution of Otx2 gene dosage to normal HPG axis function. Our data reveal that correct dosage of Otx2 is critical for normal fertility as loss of one allele of Otx2 leads to a discernible reproductive phenotype in male mice due to disruption of the migration of GnRH neurons during development.
1. Introduction
The hypothalamic-pituitary-gonadal (HPG) axis is fundamental to the endocrine control of reproduction in mammals. Dysfunction at any level of the axis leads to pathophysiologic disorders such as infertility, polycystic ovarian syndrome, and hypogonadotropic hypogonadism. Gonadotropin-releasing hormone (GnRH) is secreted in a pulsatile pattern from a small, yet critical, population of neurons within the hypothalamus to regulate the synthesis and secretion of luteinizing hormone (LH) and follicle-stimulating hormone (FSH) from gonadotrope cells within the anterior pituitary. LH and FSH are then secreted into the bloodstream where they travel to their target organs, the gonads, to regulate spermatogenesis in males and folliculogenesis and ovulation in females.
Otx2, the vertebrate homologue of Drosophila orthodenticle, is a transcription factor that has been shown to be critical for normal brain and eye development [1-4]. During embryogenesis, Otx2 is expressed in both the developing GnRH neurons [5] and presumptive pituitary at e12.5 [2] suggesting that this gene may play a critical role in development of the HPG axis, a hypothesis supported by the identification of several heterozygous OTX2 loss-of-function mutations in patients with combined pituitary hormone deficiency [6-8]. Several germline and conditional knockout mice have been generated which have emphasized a role for Otx2 in head formation, postnatal survival and growth [1,9-11]. However, as Otx2-null mice are embryonic lethal, due to a failure to develop the forebrain, midbrain and anterior hindbrain, analysis of the development and maintenance of the HPG axis in these mice has not been possible. Recently, Diaczok et al. established that deletion of Otx2, specifically from GnRH neurons, results in hypogonadotropic hypogonadism in mice adding in vivo data to previously published reports demonstrating the important role Otx2 plays as a transcriptional regulator of GnRH expression [12-14].
In this paper, we have analyzed the reproductive capacity of heterozygous Otx2 mice to determine the contribution of Otx2 gene dosage to normal HPG axis function. We report that male Otx2 heterozygotes display compromised fertility and demonstrate that, while loss of Otx2 does not affect expression of pituitary gonadotropin genes, correct gene dosage of Otx2 is critical for normal development of the GnRH neurons and expression of GnRH in adult, male mice.
2. Materials and Methods
2.1 Mouse breeding and genotyping
Mouse colonies were maintained in agreement with protocols approved by the Institutional Animal Care and Use Committee at the University of California, San Diego. All animals were housed under a 12 h light-dark cycle and provided with food and water ad libitum. Otx2 Flox mice were generated as previously described [9] and were a kind gift from Dr. Siew-Lan Ang (MRC NIMR, London, UK). Mice heterozygous for the Otx2 allele were generated by crossing Otx2 flox mice to ZP3-Cre mice [15] to create a germ-line recombination for the deletion of Otx2. All mice were on a C57 Black6 background. Surprisingly, females were either not born or did not survive to weaning age, so studies were of male heterozygote mice only. Embryos were generated through timed-breeding with adult females with embryonic day (e) 0.5 being noon of the day the vaginal plug was detected. PCR was used to genotype the offspring for the Otx2 allele (See Table 1 for details of primer sequences).
Table 1. Quantitative RT-PCR and genotyping primer sequences.
| Otx2 Flox Genotyping Forward | 5′ GCACTGAAAATCAACTTGCC 3′ |
| Otx2 Flox Genotyping Reverse | 5′ AGGCTAAAAGACCCTGGTC 3′ |
|
| |
| Otx2 KO Genotyping Forward | 5′ TGTAGGGACTCTTGCGACCT 3′ |
| Otx2 KO Genotyping Reverse | 5′ GGGCTGAGTCTGACCACTTC 3′ |
|
| |
| Q-RT-PCR GapDH Forward | 5′ TGCACCACCAACTGCTTAG 3′ |
| Q-RT-PCRGapDH Reverse | 5′ GGATGCAGGGATGATGTTC 3′ |
|
| |
| Q-RT-PCR LHβ Forward | 5′ CTGTCAACGCAACT 3′ |
| Q-RT-PCR LHβ Reverse | 5′ ACAGGAGGCAAAGC 3′ |
|
| |
| Q-RT-PCR FSHβ Forward | 5′ GCCGTTTCTGCATAAGC 3′ |
| Q-RT-PCR FSHβ Reverse | 5′ CAATCTTACGGTCTCGTATACC 3′ |
|
| |
| Q-RT-PCR αGSU Forward | 5′ CGAGGTAATAATCTTTGGAAC 3′ |
| Q-RT-PCR αGSU Reverse | 5′ GTCATTCTGGTCATGCTGTCC 3′ |
|
| |
| Q-RT-PCR GnRHR Forward | 5′ GCCCCTTGCTGTACAAAGC 3′ |
| Q-RT-PCR GnRHR Reverse | 5′ CCGTCTGCTAGGTAGATCATCC 3′ |
|
| |
| Q-RT-PCR GnRH Forward | 5′ TGCTGACTGTGTGTTTGGAAGGCT 3′ |
| Q-RT-PCR GnRH Reverse | 5′ TTTGATCCACCTCCTTGCGACTCA 3′ |
2.2 Fertility assessments and hormone measurements
At 8 weeks of age, male mice were housed singly with a wild-type, 8-week-old, female, C57BL/6J mouse. The numbers of litters born and the number of pups per litter were recorded over a period of 180 days. For serum hormone analysis, mice were sacrificed by overdose of 5% Avertin, and blood was collected by cardiac puncture. Serum was separated by centrifugation and stored at −20°C before radioimmunoassay (RIA) analysis at the Center for Research in Reproduction Ligand Assay and Analysis Core at the University of Virginia.
2.3 Embryo collection
Plugged females were euthanized, and embryos were harvested at e13.5 and e17.5. A small amount of the tail was removed from each embryo and used to extract DNA for determination of the genotypes of the embryos. Whole embryos (e13.5) or embryo heads (e17.5) were fixed in 10% acetic acid, 30% formaldehyde, 60% ethanol, overnight at 4°C, and dehydrated in 70% EtOH prior to embedding in paraffin. Sagittal sections (10 um) were floated onto SuperFrost Plus slides (Fisher) and dried overnight at 37°C. Approximately 120 to 200 sections were processed and stained per embryo, depending on the developmental stage analyzed.
2.4 GnRH Immunohistochemistry
Immunohistochemistry was performed as previously described [16]. The primary antibody used was anti-GnRH antibody (Affinity BioReagents PA1-121; 1:1000 dilution). Biotinylated goat-anti-rabbit IgG (Vector Laboratories, 1:300 dilution) was used as a secondary antibody and GnRH peptide was then visualized using the Vectastain ABC elite kit and VIP peroxidase kit (Vector Labs). Sections from embryos were counterstained using methyl green (Vector Labs). Every section was visualized at 40× magnification using a Nikon Eclipse E800 microscope with a Nikon DS Fi1 camera and using NIS elements imaging software and the numbers of GnRH neurons present in each section, and their position along the migratory path (nasal, cribriform plate or brain) recorded. AII slides were blinded before counting so the genotype was not known.
2.5 Quantitative RT-PCR
Hypothalami and pituitaries were dissected from 3-6 month-old male mice, snap frozen, and stored at −80°C until processed. RNA was extracted using Trizol (Invitrogen) according to manufacturer's instructions and reverse transcribed using First-Strand cDNA Synthesis Kit (GE Healthcare) according to the manufacturer's instructions. Q-RT-PCR was performed as previously described [12]. For primer sequences, see Table 1.
2.6 Gonadal histology
Testes were dissected and weighed from animals of each of the three genotypes. Testes were fixed for 8 hours in Bouins fixative (Sigma) at room temperature. Gonads were paraffin embedded, serially sectioned at 10 μm and stained with hematoxylin and eosin (H&E, Sigma).
2.7 Cell culture and transient transfections for luciferase reporter assays
Cell lines used were GT1-7 and LβT2. GT1-7 cells represent a fully differentiated GnRH neuron that secretes high levels of GnRH in a pulsatile manner [17] and were used to assess the effect of Otx2 on GnRH promoter activity. LβT2 cells represent a mature gonadotrope cell that expresses both luteinizing and follicle stimulating hormone [18] and were used to assess the effect of Otx2 on gonadotropin promoter activity. All cells were cultured in DMEM (Mediatech) containing 10% fetal calf serum (Gemini Bio-Products), and 1% penicillin/streptomycin (Invitrogen) in a humidified 5% CO2 incubator at 37°C. Cells were seeded into 24-well plates and incubated overnight at 37°C before being transiently transfected using FuGENE reagent (Roche Applied Science). Luciferase reporters were pGL3-1800-αGSU (−1.8 kb of the human alpha GSU regulatory region), pGL3-1800-LH (−1800 bp of the mouse LH-Beta regulatory region), pGL3-398-FSH (−398 bp of the mouse FSH-Beta regulatory region, pGL3-1200-GnRHr (−1200 bp of the mouse GnRH receptor regulatory region) and pGL3-5kb-GnRH (5 kb of the rat GnRH regulatory region). Cells were transfected with 200 ng of expression plasmid (pSG5-Otx2), 400 ng of luciferase-reporter plasmid and 100 ng of the internal-control TK -109 bp promoter on β-galactosidase. Cells were harvested after 48 h, lysed then assayed for luciferase and β-galactosidase as previously described [19]. Luciferase values were divided by β-galactosidase values to control for transfection efficiency. All experiments were performed in triplicate and repeated a minimum of three times.
2.8 Statistical Analysis
Raw data were analyzed by Student's t-test or One-way ANOVA using the statistical package GraphPad Prism. Significant differences were designated as p < 0.05.
3. Results
3.1 Otx2 heterozygote males display abnormal fertility
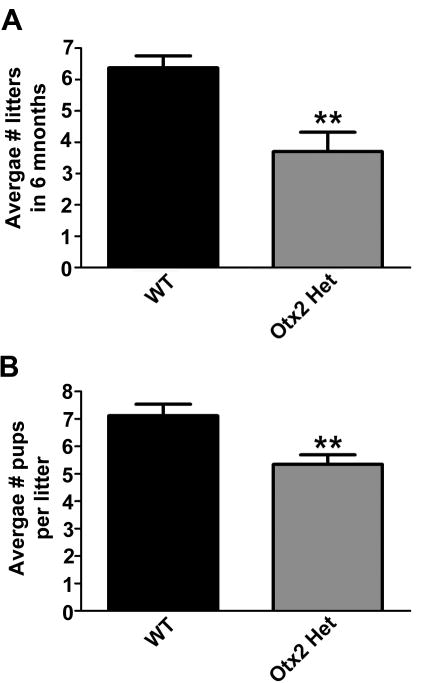
Continuous mating studies were performed to assess fertility of male Otx2 heterozygous mice. Eight-week-old males were housed individually with a wild-type, eight-week-old, C57Black6 female. The number of litters per month and the number of pups born per litter were measured. A continuous 6-month fertility assessment determined that Otx2 heterozygotes displayed significantly reduced fertility during the 180-day assessment period (Figure 1A). The average number of pups per litter was also significantly decreased (Figure 1B), however, this was to be expected given that Otx2het mice can also have development abnormalities resulting in perinatal lethality [20].
Figure 1. Global loss of one Otx2 allele results in reduced fertility in males.
(A) Average number of litters sired in 6 months. (B) Average number of pups per litter. Results shown are average ± SEM. Student's t-tests were performed on wild-type (WT) vs. Otx2 Het and established statistical significance as ** p<0.01.
3.2 Loss of Otx2 expression results in decreased LH serum levels
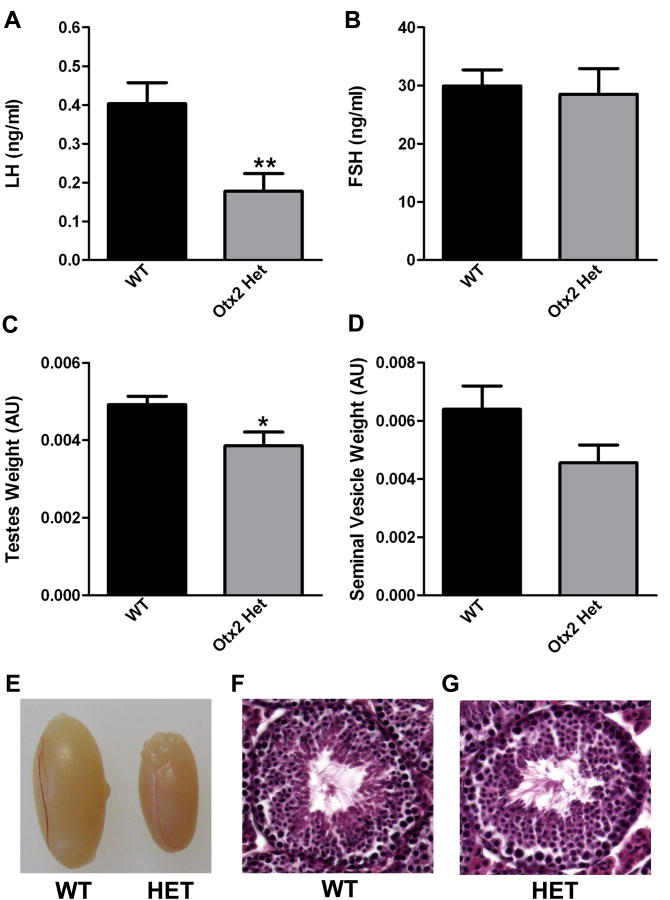
Changes in gonadotropin hormone release [follicle-stimulating hormone (FSH) and luteinizing hormone (LH)] were measured in WT and Otx2 heterozygotes by radioimmunoassay (RIA). While LH levels were significantly reduced in Otx2 heterozygotes (Figure 2A), FSH levels were comparable between the mice (Figure 2B). Comparison of testes and seminal vesicle weights (Figures 2C and 2D) revealed no significant differences in seminal vesicle (SV) weight between the two genotypes; however, testes weight was significantly reduced in the Otx2 heterozygotes (Figure 2C and 2E). Histological examination of testes from both groups of mice revealed no striking differences in either architecture or spermatogenesis (Figure 2E-G).
Figure 2. Loss of Otx2 expression results in decreased LH serum levels.
(A) Average serum LH levels of 4-8 month old male mice. (B) Average serum FSH levels of 4-8 month old male mice. (C) Average testes weight (normalized to body weight) of 4-8 month old males. (D) Average seminal vesicle weight (normalized to body weight) of 4-8 month old males. Results shown are average ± SEM. Student's t-tests were performed on WT vs. Otx2 Het and established statistical significance as * p<0.05 and ** p<0.01. (E) Representative image demonstrating comparable size of testes dissected from wild-type (WT) or Otx2 heterozygous (HET) mice. (F) and (G) H&E stained testes sections from 12 week old wild-type (WT, panel F) and heterozygous (HET, panel G) mice. No significant abnormalities were observed. Magnification is 40×.
3.3 Overexpression of Otx2 has no effect on gonadotropin transcription
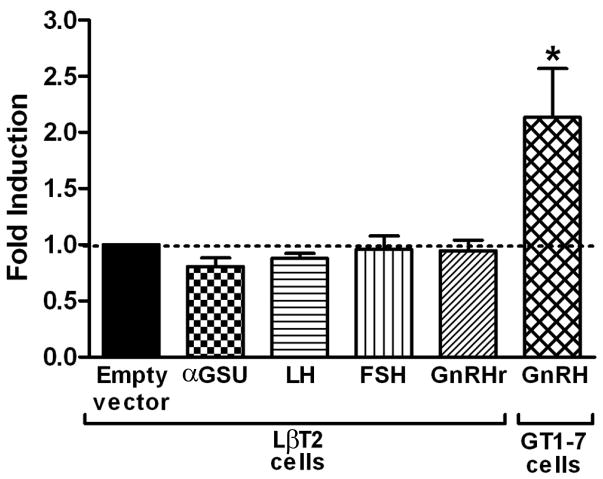
Given that Otx2 heterozygotes display compromised fertility, and Otx2 is expressed in both the developing pituitary and hypothalamus [2,5], we investigated the role of Otx2 in transcriptional control of genes known to be critical for normal reproductive function. While a positive control using the GnRH promoter in GT1-7 cells (mouse GnRH neuronal cell line) showed significantly increased activity in response to over-expression of Otx2, there was no significant effect of over-expressing Otx2 on the activity of any of the gonadotropin luciferase reporter genes in LBT2 cells (mouse gonadotrope cell line) indicating that the decreased fertility observed in Otx2 heterozygotes was unlikely to be due to a primary effect on pituitary gonadotropin hormone gene transcription (Figure 3).
Figure 3. Overexpression of Otx2 in LβT2 gonadotrope cells does not regulate gonadotrope-specific genes.
Effect of over-expression of Otx2 on various luciferase reporters in LβT2 and GT1-7 cells. Fold-induction compared to ‘empty’ vector alone (dashed line) is indicated, corrected for β-galactosidase, which was used as an internal control. All experiments were performed in triplicate and repeated three times. Results shown are average ± SEM. One-way ANOVA with post hoc Tukey test established statistical significance as ‘*’, p<0.05.
3.4 Loss of Otx2 expression results in decreased GnRH mRNA levels in adult hypothalamus
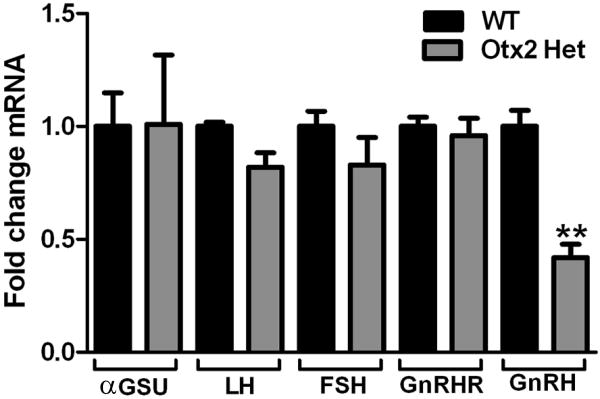
Given that loss of Otx2 resulted in a significant decrease in serum LH levels (Figure 2), it was important to determine the levels of gonadotropin and GnRH gene expression in these mice. Q-RT-PCR analysis of pituitaries confirmed transient transfection data and revealed no effect of decreased Otx2 expression on gonadotropin gene expression. In contrast, Q-RT-PCR analysis of hypothalami revealed that loss of one Otx2 allele results in a 71% decrease in GnRH mRNA levels (Figure 4).
Figure 4. Loss of Otx2 expression results in decreased GnRH mRNA expression.
Quantitative RT-PCR analysis of LH, FSH, and GnRH mRNA in 4- to 8-month-old WT and Otx2 heterozygous mice. Results are expressed as arbitrary units (AU) of GnRH mRNA levels normalized against GapDH mRNA levels and are the mean of three separate experiments performed in triplicate. Students t-tests were performed and established statistical significance as * p<0.05.
3.5 Otx2 expression is required for GnRH neuronal development and migration
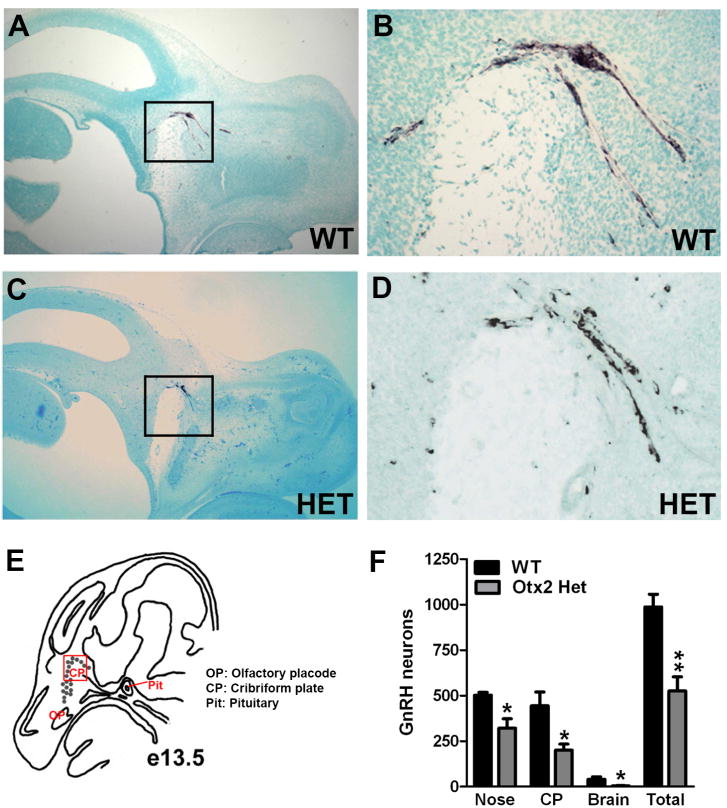
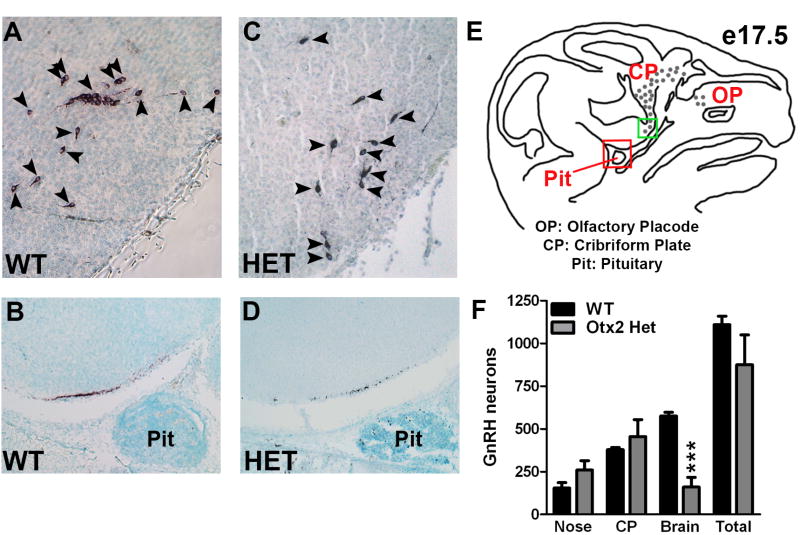
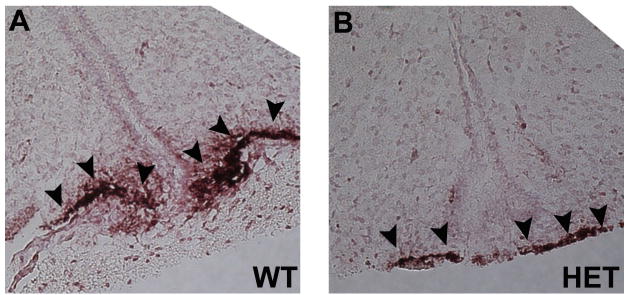
To analyze the role of Otx2 in the development and migration of GnRH neurons, GnRH immunohistochemistry was performed on sagittal sections of Otx2het embryos and littermates. By e13.5, the full complement of GnRH neurons should be located predominantly within the nasal and cribriform plate regions [21]. As expected, at e13.5, all wild-type embryos had numbers of GnRH positive cells consistent with previous findings [16,22,23] (987 ± 69, Figure 5). In contrast, Otx2 heterozygous embryos had significantly reduced numbers of GnRH neurons (525 ± 78, Figure 5). To determine the location of these GnRH neurons along the migratory pathway, the route from the olfactory placode, to the hypothalamus, was divided into three areas: nasal, cribriform plate (CP) and brain and the specific location of the GnRH neurons along this pathway recorded. Otx2 heterozygous mice had significantly reduced numbers of GnRH-positive cells at all stages of the migratory pathway when compared to wild-type littermates (Figure 5F). These results suggest that Otx2 expression is required for the birth and initiation of migration of GnRH neurons towards the hypothalamus. To determine whether the decrease in GnRH neuron number persisted during development, we analyzed mice at e17.5. Interestingly, at this later stage of development, Otx2 heterozygous mice showed similar total numbers of GnRH neurons to wild-type littermates (WT 1110 ± 50 vs. Het 875 ± 175, Figure 6). While the numbers of GnRH neurons in the nasal region and crossing the cribriform plate were similar in all mice at e17.5, the number of GnRH-positive neurons that had successfully crossed the cribriform plate and arrived within the basal forebrain was significantly lower in Otx2 heterozygous mice (Figure 6). No mis-migration of GnRH neurons was observed in any of the embryos analyzed. In addition, the intensity of the GnRH staining in neuronal terminals at the median eminence was also substantially reduced in both Otx2 heterozygous embryos at e17.5 (Figure 6B and D) and 3-month-old mice (Figure 7A and 7B)
Figure 5. Loss of Otx2 expression disrupts GnRH neuronal migration at e13.5.
Immunohistochemical staining for GnRH on sagittal sections (X40) of e13.5 WT (A and B) and Otx2 Het (C and D) embryos. The black boxes in A and C indicate the area of the section that has been enlarged (X200) in B and D respectively. (E) Diagram of e13.5 mouse head anatomy in a sagittal section showing the location of migrating GnRH neurons (grey dots). The red box indicates the area shown in B and D. (F) Average numbers of GnRH neurons at e13.5. Student's t-tests were performed on WT vs. Otx2 Het and established statistical significance as * p<0.05 and ** p<0.01.
Figure 6. Loss of Otx2 expression results in delayed progress of GnRH neurons along the migratory route at e17.5.
Immunohistochemical staining for GnRH on sagittal sections (X200) of e17.5 WT (A and B) and Otx2 Het (C and D) embryos. Panels A and C show GnRH neurons within the basal forebrain. Black arrowheads indicate GnRH neurons. Panels B and D show targeting of GnRH neurons at the median eminence. (E) Diagram of e17.5 mouse head anatomy in a sagittal section showing the location of migrating GnRH neurons (grey dots). The green box indicates the area shown in A and C. The red box indicates the area shown in B and D. (F) Average total number of GnRH neurons at e17.5. (G) Average numbers of GnRH neurons counted in the nasal, cribriform plate (CP) or brain regions at e17.5. Student's t-tests were performed on WT vs. Otx2 Het and established statistical significance as *** p<0.001.
Figure 7. Loss of Otx2 expression results in decreased targeting of GnRH axons to the median eminence in 3 month old mice.
Immunohistochemical staining for GnRH on sagittal sections (X200) of 3 month old WT (A) and Otx2 Het (B) brains. Black arrowheads indicate targeting of GnRH neuron axons to the median eminence.
4. Discussion
Correct expression and secretion of GnRH from the hypothalamus, and the subsequent transcription and secretion of gonadotropin hormones (LH and FSH) from the pituitary, is critical for achieving normal reproductive function. Understanding the in vivo contribution of genes that control the synthesis and release of GnRH, LH and FSH is therefore vital to understanding fertility. In humans, Otx2 mutations/deletions have been associated with craniofacial abnormalities [24,25],abnormal gonadotropin secretion [6,7,26-28] and short stature [6,7,27]. Although there is a wealth of information regarding the molecular regulation of GnRH expression by Otx2 [12-14], it is hard to establish whether the low LH, FSH and sex steroids seen in Otx2 heterozygous patients [6,7,29] are due to a hypothalamic (GnRH neuronal) or pituitary (gonadotrope) defect. Therefore we assessed the reproductive effects of the loss of one allele of Otx2 in male mice. Our data reveal that decreased expression of Otx2 leads to a discernible reproductive phenotype in male mice and that the primary site of Otx2 action along the HPG axis is the hypothalamus.
We observed that Otx2 heterozygotes sired significantly fewer litters than WT littermates during a 6-month assessment period (Figure 1) indicating that correct dosage and expression of Otx2 is critical for normal reproductive function. Further analysis determined that the decreased fertility was likely due to a significant fall in LH serum levels (Figure 2) because of a substantial decrease in GnRH expression (Figure 4). Pituitary gonadotropin gene expression was normal in all mice analyzed (Figure 4). Given that during embryogenesis GnRH neurons must travel along a migratory pathway from the olfactory placode and arrive within the hypothalamus prior to birth, any delay to this migration can have serious consequences to the final number of functional GnRH neurons within the hypothalamus, and therefore levels of GnRH expression. Detailed analysis of GnRH neuronal development and migration in Otx2 heterozygotes revealed that decrease in Otx2 gene dosage significantly delays the progress of GnRH neurons along the migratory pathway and GnRH axon targeting to the median eminence (Figures 5,6 and 7).
Our findings suggest that, in males at least, the idiopathic hypogonadotrophic hypogonadism (IHH) seen in patients with Otx2 mutations cannot solely be attributed to abnormal pituitary development and that these patients may display abnormal GnRH neuronal migration that could be a significant contributing factor to their infertility. Further investigations are required to determine whether the short stature seen in a high proportion of OTX2 patients is due to hypothalamic abnormalities, pituitary abnormalities, or a combination of both. Although, as Otx2 has not been reported to be expressed in growth hormone releasing hormone neurons, it seems likely that the growth hormone deficiency seen in these patients is a pituitary phenotype.
Unfortunately, we were unable to generate meaningful numbers of female Otx2 heterozygotes (beyond the embryonic stage) to establish whether a loss of one functional copy of Otx2 would have an impact on the female reproductive axis. No reports of gender bias with regards to Otx2 gene dosage have been reported in other mouse studies, however, most of this research focused on the role of Otx2 during development. Certainly, where OTX2 mutations in humans are concerned, equal numbers of affected males and females are seen so this phenomenon may perhaps represent something specific to our mouse line/strain. However, because the female reproductive axis is traditionally thought to be much more sensitive to disruption than that of the male, we would hypothesize that, should they survive to puberty, female Otx2 heterozygotes would also display reduced fertility.
Highlights.
Correct dosage of Otx2 is critical for normal fertility in male mice
Loss of one allele of Otx2 significantly decreases hypothalamic GnRH expression
Decrease is likely due to abnormal migration of GnRH neurons during development
Acknowledgments
We thank Hanne Hoffmann for critical reading of the manuscript and members of the Mellon laboratory for helpful discussion and support throughout this work. DNA sequencing was performed by the DNA-sequencing shared resource, UCSD Cancer Center, which is funded in part by NCI Cancer Support Grant P30 CA023100. Serum hormone assays were performed by The University of Virginia Ligand Assay Core Laboratory, which is supported through National Institute of Child Health and Human Development Grant U54 HD028934. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, et al. Forebrain and midbrain regions are deleted in OTX2-/- mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- 2.Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D'Apice R, et al. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm of the gastrulating mouse embryo. EMBO J. 1993;12:2735–2774. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frantz GD, Weimann JM, Levin ME, McConnell SK. Otxl and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci. 1994;14:5725–5740. doi: 10.1523/JNEUROSCI.14-10-05725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puelles E, Annino A, Tuorto F, Usiello A, Acampora D, et al. Otx2 regulates the extent, identity and fate of neuronal progenitor domains in the ventral midbrain. Development. 2004;131:2037–2048. doi: 10.1242/dev.01107. [DOI] [PubMed] [Google Scholar]
- 5.Mallamaci A, DiBlas E, Briata P, Boncinelli E, Corte G. OTX2 homeoprotein in the developing central nervous system and migratory cells of the olfactory area. Mech Devel. 1996;58:165–178. doi: 10.1016/s0925-4773(96)00571-0. [DOI] [PubMed] [Google Scholar]
- 6.Dateki S, Kosaka K, Hasegawa K, Tanaka H, Azuma N, et al. Heterozygous orthodenticle homeobox 2 mutations are associated with variable pituitary phenotype. J Clin Endocrinol Metab. 2010;95:756–764. doi: 10.1210/jc.2009-1334. [DOI] [PubMed] [Google Scholar]
- 7.Dateki S, Fukami M, Sato N, Muroya K, Adachi M, et al. OTX2 mutation in a patient with anophthalmia, short stature, and partial growth hormone deficiency: functional studies using the IRBP, HESX1, and POU1F1 promoters. J Clin Endocrinol Metab. 2008;93:3697–3702. doi: 10.1210/jc.2008-0720. [DOI] [PubMed] [Google Scholar]
- 8.Diaczok D, Romero C, Zunich J, Marshall I, Radovick S. A novel dominant negative mutation of OTX2 associated with combined pituitary hormone deficiency. J Clin Endocrinol Metab. 2008;93:4351–4359. doi: 10.1210/jc.2008-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, et al. A targeted mouse OTX2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996;122:243–252. doi: 10.1242/dev.122.1.243. [DOI] [PubMed] [Google Scholar]
- 10.Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse OTX2 functions in the formation and patterning of rostral head. Genes & Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 11.Fossat N, Chatelain G, Brun G, Lamonerie T. Temporal and spatial delineation of mouse Otx2 functions by conditional self-knockout. EMBO Rep. 2006;7:824–830. doi: 10.1038/sj.embor.7400751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larder R, Mellon PL. Otx2 induction of the gonadotropin-releasing hormone promoter is modulated by direct interactions with Grg co-repressors. J Biol Chem. 2009;284:16966–16978. doi: 10.1074/jbc.M109.002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HH, Wolfe A, Cohen RN, Eames SC, Johnson AL, et al. In vivo identification of a 107 bp promoter element mediating neuron-specific expression of mouse GnRH. Mol Endocrinol. 2007;21:457–471. doi: 10.1210/me.2005-0216. [DOI] [PubMed] [Google Scholar]
- 14.Kelley CG, Lavorgna G, Clark ME, Boncinelli E, Mellon PL. The Otx2 homeoprotein regulates expression from the gonadotropin-releasing hormone proximal promoter. Mol Endocrinol. 2000;14:1246–1256. doi: 10.1210/mend.14.8.0509. [DOI] [PubMed] [Google Scholar]
- 15.Lewandoski M, Wassarman KM, Martin GR. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol. 1997;7:148–151. doi: 10.1016/s0960-9822(06)00059-5. [DOI] [PubMed] [Google Scholar]
- 16.Larder R, Clark DD, Miller NL, Mellon PL. Hypothalamic dysregulation and infertility in mice lacking the homeodomain protein Six6. J Neurosci. 2011;31:426–438. doi: 10.1523/JNEUROSCI.1688-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wetsel WC, Valenca MM, Merchenthaler I, Liposits Z, Lopez FJ, et al. Intrinsic pulsatile secretory activity of immortalized luteinizing hormone-releasing hormone-secreting neurons. Proc Natl Acad Sci U S A. 1992;89:4149–4153. doi: 10.1073/pnas.89.9.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alarid ET, Windle JJ, Whyte DB, Mellon PL. Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development. 1996;122:3319–3329. doi: 10.1242/dev.122.10.3319. [DOI] [PubMed] [Google Scholar]
- 19.McGillivray SM, Bailey JS, Ramezani R, Kirkwood BJ, Mellon PL. Mouse GnRH receptor gene expression is mediated by the LHX3 homeodomain protein. Endocrinology. 2005;146:2180–2185. doi: 10.1210/en.2004-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makiyama Y, Shoji S, Mizusawa H. Hydrocephalus in the Otx2+/− mutant mouse. Exp Neurol. 1997;148:215–221. doi: 10.1006/exnr.1997.6638. [DOI] [PubMed] [Google Scholar]
- 21.Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- 22.Givens ML, Rave-Harel N, Goonewardena VD, Kurotani R, Berdy SE, et al. Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J Biol Chem. 2005;280:19156–19165. doi: 10.1074/jbc.M502004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller NL, Wevrick R, Mellon PL. Necdin, a Prader-Willi syndrome candidate gene, regulates gonadotropin-releasing hormone neurons during development. Hum Mol Genet. 2009;18:248–260. doi: 10.1093/hmg/ddn344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ragge NK, Brown AG, Poloschek CM, Lorenz B, Henderson RA, et al. Heterozygous mutations of OTX2 cause severe ocular malformations. Am J Hum Genet. 2005;76:1008–1022. doi: 10.1086/430721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson RA, Williamson K, Cumming S, Clarke MP, Lynch SA, et al. Inherited PAX6, NF1 and OTX2 mutations in a child with microphthalmia and aniridia. Eur J Hum Genet. 2007;15:898–901. doi: 10.1038/sj.ejhg.5201826. [DOI] [PubMed] [Google Scholar]
- 26.Nolen LD, Amor D, Haywood A, St Heaps L, Willcock C, et al. Deletion at 14q22-23 indicates a contiguous gene syndrome comprising anophthalmia, pituitary hypoplasia, and ear anomalies. Am J Med Genet A. 2006;140:1711–1718. doi: 10.1002/ajmg.a.31335. [DOI] [PubMed] [Google Scholar]
- 27.Elliott J, Maltby EL, Reynolds B. A case of deletion 14(q22.1-->q22.3) associated with anophthalmia and pituitary abnormalities. J Med Genet. 1993;30:251–252. doi: 10.1136/jmg.30.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaczok D, DiVall S, Matsuo I, Wondisford FE, Wolfe AM, et al. Deletion of Otx2 in GnRH neurons results in a mouse model of hypogonadotropic hypogonadism. Mol Endocrinol. 2011;25:833–846. doi: 10.1210/me.2010-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyatt Bakrania P, Bunyan DJ, Osborne RJ, Crolla JA, et al. Novel heterozygous OTX2 mutations and whole gene deletions in anophthalmia, microphthalmia and coloboma. Hum Mutat. 2008;29:E278–283. doi: 10.1002/humu.20869. [DOI] [PubMed] [Google Scholar]