Summary
Melanoma incidence continues to rise at an alarming rate while effective systemic therapies remain very limited. Microphthalmia-associated transcription factor (MITF) is required for development of melanocytes and is an amplified oncogene in a fraction of human melanomas. MITF also plays an oncogenic role in human clear cell sarcomas, which typically exhibit melanoma-like features. Although pharmacologic suppression of MITF is of potential interest in a variety of clinical settings, it is not known to contain intrinsic catalytic activity capable of direct small molecule inhibition. An alternative drug-targeting strategy is to identify and interfere with lineage-restricted mechanisms required for its expression. Here, we report that multiple HDAC-inhibitor drugs potently suppress MITF expression in melanocytes, melanoma and clear cell sarcoma cells. Although HDAC inhibitors may affect numerous cellular targets, we observed suppression of skin pigmentation by topical drug application as well as evidence of anti-melanoma efficacy in vitro and in mouse xenografts. Consequently, HDAC inhibitor drugs are candidates to play therapeutic roles in targeting conditions affecting the melanocyte lineage.
Keywords: HDAC inhibitor, MITF, melanoma, clear cell sarcoma, pigmentation
Introduction
Microphthalmia-associated transcription factor (MITF) is a basic-helix/loop/helix-leucine-zipper transcription factor which plays an essential lineage-specific role during melanocyte development (Hodgkinson et al., 1993; Hughes et al., 1993; Steingrimsson et al., 1994). The MITF gene has a complex promoter organization, with at least nine distinct isoforms sharing exons 2 to 9. Some of these isoforms exhibit tissue-specific restriction such as M-MITF, which is expressed in melanocytes and melanoma but not most other lineages (Hershey et al., 2005; Udono et al., 2000). The melanocyte-specific promoter is located most proximal to the common downstream exons and is known as the M-MITF promoter (Fuse et al., 1996). As a transcription factor, MITF regulates multiple genes related to melanin synthesis, control of apoptosis, and cell cycle progression (Goding et al., 2000; Levy et al., 2006; Shibahara et al., 2001). Appropriate regulation of MITF is required for cell growth/survival in melanocytes and recent work has implicated MITF as a melanoma oncogene due to its amplification in ~20% of human metastatic melanomas (Garraway et al; 2005). In addition, human Clear Cell Sarcomas typically contain a chromosomal translocation which produces the EWS-ATF1 fusion protein (Brown et al., 1995; Fujimura et al., 1996; Lessnick et al., 1995). This chimeric oncoprotein directly triggers dysregulated expression of MITF, which in turn appears to play a vital role within this tumor (Davis et al., 2006). Thus, MITF is a potentially attractive molecular target for melanoma and clear cell sarcoma therapy. However being transcription factor lacking ligand dependency, it is largely considered a difficult drug target by current approaches.
In contrast, histone deacetylase inhibitors (HDACi) have been shown to exhibit anti-cancer effects, and suberoylanilide hydroxamic acid (SAHA) has been FDA approved for treatment of cutaneous T cell lymphoma (Kelly et al., 2003, 2005a,b; O’Connor et al., 2006). Cancer-related consequences of HDACi drugs include growth arrest and apoptosis as well as repression of angiogenesis (Liu et al., 2006). An effect of HDACi drugs on growth arrest and apoptosis has been shown in melanoma cells, but the mechanism has been poorly understood (Boyle et al., 2005). Recently, HDACi-treated embryos were observed to exhibit disrupted pigmentation in zebrafish and Xenopus (Gurvich et al., 2005), suggesting that HDACi might have effects on melanocyte-lineage specific factor(s).
In this study, we provide evidence that multiple HDAC inhibitor drugs, including sodium butyrate (NaB), Trichostatin A (TSA), SAHA, and LBH589, repress M-MITF expression in melanoma and clear cell sarcoma cells. Although HDACi drugs have numerous effects, we observe this lineage-selective effect in melanocytes, which is associated with transcriptional downregulation of the M-MITF promoter. The resulting data suggest that HDACi drugs may antagonize MITF, with consequent therapeutic applications either alone or perhaps in combination with other MITF suppressing strategies, to control skin pigmentation and/or survival of MITF-dependent cancers.
Results and Discussion
HDACi drugs repress M-MITF levels in melanoma and clear cell sarcoma cell lines
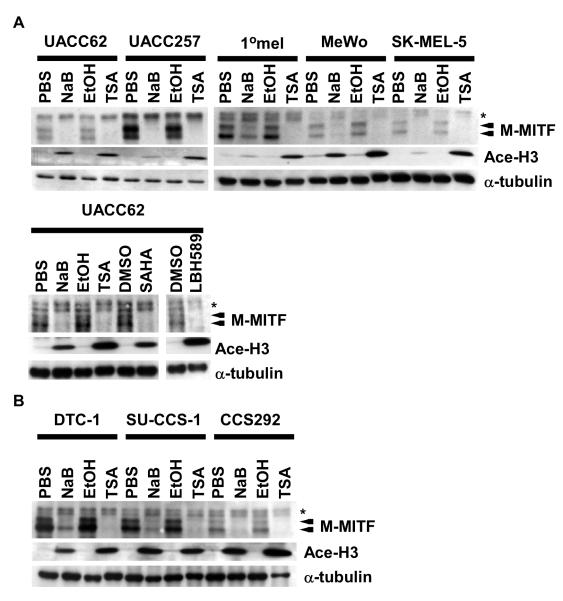
MITF is a lineage survival factor as well as amplified oncogene in ~20% of metastatic melanomas. Due to its vital role in the melanocyte lineage as well as in Clear Cell Sarcoma (Davis et al., 2006), MITF is an attractive molecular target for melanoma and clear cell sarcoma therapy. In searching for small molecules which affect MITF expression, we wanted to explore if HDACi drugs could exhibit a role as MITF antagonists given the observed disruption of pigmentation in zebrafish and Xenopus (Gurvich et al., 2005). The melanocyte-specific isoform of MITF (M-MITF) migrates as a doublet (Fig 1A and 1B, arrows) due to serine 73 phosphorylation by MAPK (Hemesath et al., 1998), whereas higher migrating MITF isoforms (asterisk) reflect non-melanocyte-specific forms of MITF derived from distinct promoters (Hershey et al., 2005; Udono et al., 2000). We subjected several established melanoma and clear cell sarcoma cell lines to HDACi treatments and observed reproducibly suppressed expression of the M-MITF doublet protein species, but not the non-melanocytic isoforms (in some cases even being associated with observable slight upregulation) (Fig 1A, 1B, and data not shown). The acetylation status of histone H3 by HDACi in human melanoma cells correlated with activity of the HDACi drugs. Other HDACi drugs, suberoylanilide hydroxamic acid (SAHA) and LBH589 (George et al., 2005), also repressed M-MITF expression in melanoma cells (Fig 1A). We detected diminished RNA levels of the M-MITF isoform in UACC62 melanoma cells with NaB and TSA (Fig. 2A and data not shown), suggesting that the decreased M-MITF protein levels result from reduced mRNA levels. To further examine whether stability of the MITF mRNA may contribute to diminished protein levels following HDACi exposure, MITF mRNA decay kinetics were tested following actinomycin D treatment, in the presence or absence of HDACi. No significant difference was observed in the rate of MITF mRNA decay by this assay (Suppl. Fig. 1A), suggesting that HDACi exposure is unlikely to affect MITF expression via altered RNA stability.
Figure 1. HDACi drugs repress M-MITF and SOX10 levels in melanoma and clear cell sarcoma cell lines.
(A) Western blotting analysis of whole-cell lysates prepared from UACC62, UACC257, MeWo, or SK-MEL-5 human melanoma cells and 1°melanocytes treated with PBS, NaB, EtOH, or TSA for 24 h (Upper panel) or from UACC62 human melanoma cells treated with PBS, NaB, EtOH, TSA, DMSO, SAHA, or LBH589 for 24 h (lower panel). Lysates were immunoblotted with antibodies against MITF, acetylated histone 3 (Ace-H3), or α-tubulin. The arrows show the bands of M-MITF protein. Asterisk indicates A-MITF protein. (B) Whole-cell lysates were prepared from DTC-1, SU-CCS-1, or CCS292 human clear cell sarcoma cells treated with PBS, NaB, EtOH, or TSA for 24 h. Other conditions are represented as indicated in Fig. 1A.
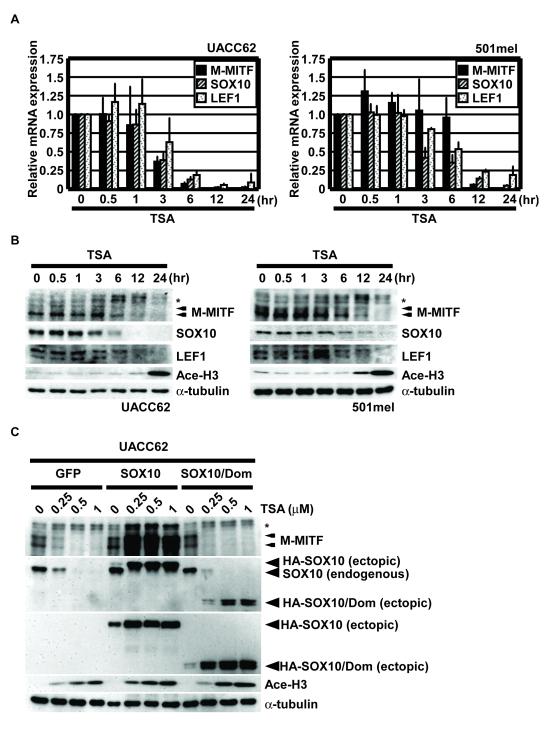
Figure 2. TSA represses SOX10 and LEF1 expression.
(A) Quantitative reverse transcription PCR analysis of total RNA isolated from UACC62 and 501mel treated with 1 μM TSA for the indicated times. M-MITF (filled bars), SOX10 (shadow bars) and LEF1 (dotted bars) mRNA levels were normalized to glyceraldehyde-3-phosphate dehydrogenase and performed in triplicate. Data are mean ± S.D. of at least three independent experiments. (B) Western blotting analysis of whole-cell lysates prepared from UACC62 and 501mel treated with 1 μM of TSA for the indicated times. Lysates were immunoblotted with antibodies against MITF, SOX10, LEF1, Ace-H3, or α-tubulin. (C) Western blotting analysis of whole-cell lysates prepared from UACC62 overexpressing GFP, SOX10, or SOX10-mut cells. Arrowheads show the ectopic expressing SOX10 or SOX10/Dom. Other conditions are represented as indicated in Fig. 1A.
Since HDACi drugs led to M-MITF specific mRNA downregulation and SOX10 and LEF1 are key upstream transcriptional regulators of the M-MITF promoter, we also examined the expression of these upstream regulators after the same treatments. Diminished protein and mRNA levels of SOX10 and LEF1 were observed in a time-dependent manner (Fig. 2A and 2B) which correlated closely with, or slightly preceded MITF’s decreases. Moreover, SOX10 overexpression was able to significantly rescue M-MITF expression and growth upon HDACi treatment in UACC62/SOX10 cells but not in UACC62/GFP (control) cells or UACC62/SOX10/Dom cells (Fig. 2C and Suppl. Fig. 1B). Collectively these data suggest that the potent suppression of M-MITF expression by HDACi drugs likely occurs via downregulation of transcription of the MITF gene and might be mediated by suppression of the upstream factors SOX10 or LEF1.
Skin-lightening effects of topical HDACi treatment
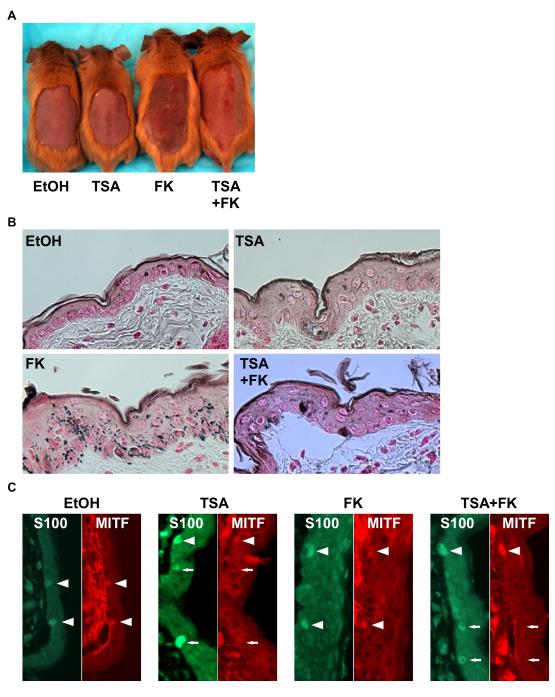
Multiple human pathologic hyperpigmentation conditions exist, including melasma, aging spots (lentigo senilis), post-inflammatory hyperpigmentation, and others. Aside from MITF’s requirement for melanocyte viability, it also plays a central role in control of pigmentation. Multiple pathways control expression and activity of MITF, including the action of α-Melanocyte Stimulating Hormone (eg in sun/UV induced pigmentation) where it is thought to activate expression of multiple enzymes in the pigment synthesis cascade and related factors involved in melanin transport/secretion (Levy et al., 2006). Moreover, mutants of MITF exist which alter pigment color, without absence of melanocytes (Steingrímsson et al., 1996) suggesting that suppression of MITF might potentially offer a topical therapeutic strategy for treatment of hyperpigmentation, if a dose were utilized which affects pigmentation without loss of melanocyte viability. To test this possibility, we used a mouse model of forskolin-induced hyperpigmentation (D’Orazio et al., 2006). The congenic C57BL/6 K14-SCF Mc1re/e mouse strain contains epidermal melanocytes due to the keratin 14 (K14) promoter driving expression of stem cell factor (Scf; also known as Kitl) (“humanized skin model”) (Kunisada et al., 1998). Mc1re/e encodes a frameshift mutation in the melanocortin receptor gene, which produces the redhair/fairskin phenotype, and which is susceptible to forkolin-induced hyperpigmentation (D’Orazio et al., 2006). As shown in Fig. 3A, dorsal pigmentation induced by topical treatment with forskolin was potently repressed by topical treatment with the HDAC inhibitor TSA. Using Fontana-Masson staining, which detects melanin in tissue sections, substantial suppression of skin pigmentation was observed in the TSA treated skin (Fig. 3B). Direct assessment of MITF expression in skin upon TSA treatment was carried out by co-staining with a monoclonal antibody against MITF as well as staining for S100-antigen (melanocytic marker) since MITF and S100-antigen are co-expressed in epidermal melanocytes (Fig. 3C; EtOH and FK). Treatment of mice with topical HDACi drug produced a significant decrease in MITF-staining, whereas S100 appeared unaffected by TSA treatment (Fig. 3C; TSA and TSA+FK). Since M-MITF appears to be the most abundantly expressed MITF isoform in melanocytes (Fig. 1A), decreasing levels of MITF-staining in S100-antigen positive cells likely reflected M-MITF reduction although the MITF antibody could detect other MITF isoforms as well. Importantly using this topical treatment regimen, melanocyte viability did not appear to be affected because the ratio of melanocytes to keratinocytes in basal layer (number. of keratinocytes/number of melanoctytes/field) among each group did not significantly change (EtOH; 5.49 ± 1.03, TSA; 6.47±1.07, FK; 5.81 ± 1.97, FK+TSA; 5.67±0.87). These data demonstrate that HDACi could repress MITF expression as well as pigmentation within skin of topically treated mice under conditions in which melanocyte viability was not compromised.
Figure 3. TSA represses the FK-induced pigmentation in the skin of mice.
(A) K14-SCF Mc1re/e mice were treated with vehicle (70 % Ethanol/ 30 % polyethylene glycol), crude forskolin, 0.7 mM trichostatin A for 2-weeks. After shaving the hair, photograph was taken. (B) Fontana-Masson staining was performed from paraffin section. Eumelanin was detected by Fontana-Masson staining. (C) MITF and S100 staining were performed from paraffin section. Cy3 (red) or FITC (green) was conjugated to secondary antibody.
Melanoma growth suppression in vitro and in vivo
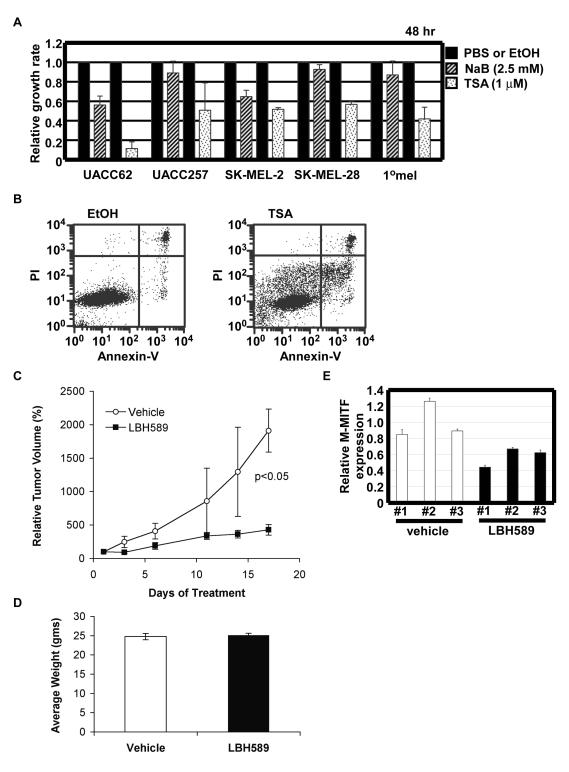
Depletion of MITF induces cell cycle arrest or loss of survival (Carreira et al. 2006; Garraway et al., 2005; Widlund et al., 2002) likely via a variety of mechanisms including induction of p27kip1 or alterations of other cell cycle or apoptotic factors in specific settings. In order to examine if HDAC inhibitors (HDACi) influence melanoma cell growth or survival, WST-1 assays were carried out using human melanoma cells treated with HDACi drugs sodium butyrate (NaB) or Trichostatin A (TSA) (Fig. 4A) Both NaB (2.5 mM) and TSA (1 μM) repressed cell growth at 48 h in UACC62 melanoma cells and UACC257 melanoma cells. The same growth inhibition was observed in other tested melanoma cell lines (data not shown). We also detected apoptosis (propidium iodide (PI) negative/Annexin V positive cells) in UACC62 cells treated with TSA (12.2%) as compared to vehicle (1.7%) by FACS analysis (Fig. 4B).
Figure 4. HDACi drugs repress melanoma growth in vitro and in vivo.
(A) UACC62, UACC257, and primary human melanocytes (1°mel) were treated with HDAC inhibitors, sodium butylate (NaB) (shadow bars) and trichostatin A (TSA) (dotted bars) for 48 h. Relative growth rate of NaB- or TSA-treated cells was normalized by PBS- or ethanol (EtOH) -treated cells (filled bars), respectively. Data are mean ± S.D. of at least three independent experiments. (B) UACC62 human melanoma cells were treated with TSA for 24 h. The cells were stained by Annexin-V and propidium iodide. (C) UACC62 human melanoma cells were injected in nude mice. In cohorts of mice with established tumors (volume 100-200 mm3), animals were treated with either LBH589 or vehicle via intraperitoneal injection. Data are represented as mean ± SEM, with n=9 per group. (D) Animals weights were determined every 2 days, and no significant differences were observed between the treatment groups. Data after 12 days of treatment are shown as mean ± SEM. (E) Quantitative reverse transcription PCR analysis of total RNA isolated from injected UACC62 treated with LBH589 or vehicle. Human M-MITF mRNA levels were normalized to glyceraldehyde-3-phosphate dehydrogenase and performed in triplicate. Relative M-MITF expression is shown as the ratio to the average obtained from vehicle samples. Data are mean ± S.D. of each experiment.
To determine whether HDAC inhibition would attenuate melanoma growth in vivo, we assessed the effects of systemic administration of LBH589 on the growth of UACC62 xenograft tumors. Mice with established tumors were treated with either vehicle or LBH589 at a dose of 10 mg/kg administered daily via intraperitoneal injection. Treatment with LBH589 significantly (p<0.05) suppressed tumor growth, such that the average tumor volume of LBH589-treated animals was 22% of that of vehicle-treated animals after 17 days of treatment (Fig. 4C). There was no appreciable difference in body weights between the two treatment groups throughput the study (Fig. 4D), demonstrating that treatment with LBH589 was well tolerated and not overtly toxic. To determine whether LBH589 treatment altered MITF expression, RNA was isolated from tumors and isoform-specific quantitative RT-PCR was used to determine M-MITF expression. Tumors from animals treated with LBH589 had reduced abundance of M-MITF mRNA compared to vehicle-treated tumors (Fig. 4E, p<0.05).
In this study, we provide evidence that the HDACi drugs sodium butylate (NaB), Trichostatin A (TSA), SAHA, and LBH589 repress MITF expression in melanoma and clear cell sarcoma cells. Moreover, the repression of MITF with HDACi was also observed in vivo using a humanized mouse skin model in which HDACi topical treatment inhibited forskolin-induced pigmentation without loss of melanocyte viability. In addition systemic HDACi treatment exhibited significant growth suppression in a human melanoma xenograft model. The mechanism of MITF suppression by HDACi’s is incompletely understood, but appears likely to involve suppression of one or several required upstream transcriptional regulators of MITF. The mechanism through which HDACi’s affect their expression awaits further analysis.
Collectively these data identify HDACi drugs as small molecule inhibitors of MITF expression, which may be worthy of clinical study in a variety of settings associated with MITF over-activity, including aberrant pigmentation or melanocytic neoplasia. As MITF-targeted small molecule targeting strategies are identified, it is plausible that combinations of such agents (including HDACi’s) may hold therapeutic promise for systemic treatment of melanoma.
Materials and Methods
Cell cultures
UACC62, UACC257, SK-MEL-2, and SK-MEL-28 human melanoma cells were cultured in RPMI-1640 medium (Mediatech Inc.) containing 10% fetal bovine serum (FBS) and penicillin/streptomycin/L-glutamine. MeWo and SK-MEL-5 human melanoma cells were cultured in DMEM medium (Mediatech Inc.) containing 10% FBS and penicillin/streptomycin/L-glutamine. 501mel human melanoma cells were cultured in Ham’s F-10 medium (Mediatech Inc.) containing 10% FBS and penicillin/streptomycin/L-glutamine. Primary human melanocytes between passages 2 and 5 were derived from neonatal foreskins and grown in TIVA media (Ham’s F12 (Mediatech Inc.), 7% fetal bovine serum, penicillin/streptomycin/glutamine(Invitrogen), 1 × 10−4 M 3-isobutyl-1-methyl xanthine (IBMX; Sigma), 50 ng ml−1 12-O-tetradecanoyl phorbol-13-acetate (TPA; Sigma), 1 μM Na3VO4, and 1 × 10−3 M N6,2′-O-dibutyryladenosine 3:5-cyclic monophosphate (dbcAMP; Sigma)). Clear cell sarcoma cell lines, DTC-1, SU-CCS-1, and CCS292, were cultured in DMEM medium containing 15% FBS and penicillin/streptomycin/L-glutamine. The stable UACC62 transformed cells containing pLNCX-GFP, pLNCX-SOX10 (full-length SOX10), and pLNCX-SOX10/Dom (truncated SOX10) were selected with G418 (500 μg/ml) (Mediatech, Inc), and referred to as UACC62/GFP (control), UACC62/SOX10, UACC62/SOX10/Dom. These stable transformed lines were grown in RPMI-1640 medium containing 10% FBS, 500 μg/ml G418, and penicillin/streptomycin/L-glutamine. The cells were treated with sodium butyrate (NaB) (2.5 mM) (Sigma-Aldrich), Trichostatin A (TSA) (1 μM) (Sigma-Aldrich), suberoylanilide hydroxamic acid (SAHA) (10 μM) (BIOMOL), or LBH589 (10 μM) (Novartis) for various times as indicated.
Western blotting analysis
Whole cell extracts (10 μg / lane) were prepared using the method of Schreiber, E. et al. (Schreiber et al., 1989) and then subjected to western blotting analysis using anti-MITF monoclonal antibody (Weilbaecher et al., 1998), anti-acetyled histone 3 (Ace-H3) polyclonal antibody (Upstate Biotechnology), anti-SOX10 polyclonal antibody (Santa Cruz Biotechnology), anti-LEF1 antibody (REMB6) (Exalpha Biologicals), anti-HA monoclonal antibody (Roche) or anti-α-tubulin monoclonal antibody (Sigma).
Real-time RT-PCR
The total RNAs were prepared by using Trizol (Invitrogen) from UACC62 and 501mel human melanoma cells with TSA for each indicated time. Real-time RT-PCR was performed by using iQ SYBR Green supermix (Bio-Rad) from cDNAs. The primers used were 5′-cat tgt tat gct gga aat gct aga a-3′ (sense) and 5′-ggc ttg ctg tat gtg gta ctt gg-3′(antisense) for M-MITF, 5′-gct gct gaa cga aag tga ca-3′ (sense) and 5′-gcc tgg gct ggt act tgt ag-3′ (antisense) for SOX10, and 5′-tca tcc cga aga gga agg cga ttt-3′ (sense) and 5′-tcc tga gag gtt tgt gct tgt ctg-3′ (antisense) for LEF1. All reactions were run in triplicate on an iCycler instrument (Bio-Rad), and MITF-M message levels were normalized to glyceraldehyde-3-phosphate dehydrogenase expression.
In vivo de-pigmentation assay
C57BL/6JJ Mc1re/e mice were crossed with K14-Scf transgenic mice (D’Orazio et al., 2006). Depilated animals were exposed to topical forskolin, TSA, or vehicle control. Skin biopsies were harvested and processed for Fontana–Masson (eumelanin) staining and immunostaining by S100 antibody (DakoCytomation), or MITF antibody (23).
In vitro growth assays
Cell metabolic activity was assessed by Cell Proliferation Reagent WST-1 (Roche Diagnostics) according to the manufacturer’s instruction. Briefly, the UACC62, UACC257, SK-MEL-2, SK-MEL-28, and Primary human melanocytes (1 × 103 cells/well) were cultured for 24 h after plating in 96-well dishes and then the HDAC inhibitors, NaB (2.5 mM) or TSA (1 μM), were added to each well. After 48 h, the WST-1 solution was added. The cells were incubated for an additional 1 h with the WST-1 solution. Finally, the absorbance of 460 nm was measured using a microplate reader. Absorbance of cells treated with HDAC inhibitors was normarized to cells treated with the appropriate control, either PBS or ethanol vehicle.
Apoptosis analysis
UACC62 cells were incubated with TSA (1μM) or EtOH for 24 hr. After incubation, the cells were collected and washed by PBS twice. The cells were resuspended in Annexin V Binding buffer (10mM Hepes, 140mM NaCl, and 2.5mM CaCl2, pH7.4). The suspensions were incubated with Annexin V Alexa647 (Invitrogen) conjugates and propidium iodide (PI).
In vivo melanoma growth assay
UACC62 xenografts were generated by subcutaneous injection of 5 × 106 cells into NCr/Nude mice. After 1 week of growth, mice with measurable tumors were segregated into groups that were treated with either LBH589 10 mg/kg IP daily or vehicle control (n=9 per group). Tumors were measured with calipers, and the tumor volume was calculated as 0.5 × length × width2. For each animal, relative tumor volume over the treatment course was determined by normalizing to the tumor volume at the start of treatment (Day 1 of treatment). Statistical significance between the two treatment arms was determined by two-tailed Student’s t-test. All animal studies were performed under the auspices of protocols approved by the Dana-Farber Cancer Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge Dr. Ruth Halaban (Yale Univ) for 501mel cells, members of the Fisher lab for useful discussions, and Yashaswi Shrestha for assistance with early portions of the project. This work was supported by a grant from NIH. DEF is Distinguished Clinical Scholar of the Doris Duke Charitable Foundation.
Footnotes
Disclosures: DEF discloses a consulting relationship with Novartis Pharmaceuticals and Magen BioSciences. AK discloses a consulting relationship with Novartis Pharmaceuticals.
References
- Boyle GM, Martyn AC, Parsons PG. Histone deacetylase inhibitors and malignant melanoma. Pigment Cell Res. 2005;18:160–166. doi: 10.1111/j.1600-0749.2005.00228.x. [DOI] [PubMed] [Google Scholar]
- Brown AD, Lopez-Terrada D, Denny C, Lee KA. Promoters containing ATF-binding sites are de-regulated in cells that express the EWS/ATF1 oncogene. Oncogene. 1995;10:1749–1756. [PubMed] [Google Scholar]
- Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IJ, Kim JJ, Ozsolak F, Widlund HR, Rozenblatt-Rosen O, Granter SR, Du J, Fletcher JA, Denny CT, Lessnick SL, et al. Oncogenic MITF dysregulation in clear cell sarcoma: defining the MiT family of human cancers. Cancer Cell. 2006;9:473–484. doi: 10.1016/j.ccr.2006.04.021. [DOI] [PubMed] [Google Scholar]
- D’Orazio JA, Nobuhisa T, Cui R, Arya M, Spry M, Wakamatsu K, Igras V, Kunisada T, Granter SR, Nishimura EK, et al. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- Fujimura Y, Ohno T, Siddique H, Lee L, Rao VN, Reddy ES. The EWS-ATF-1 gene involved in malignant melanoma of soft parts with t(12;22) chromosome translocation, encodes a constitutive transcriptional activator. Oncogene. 1996;12:159–167. [PubMed] [Google Scholar]
- Fuse N, Yasumoto K, Suzuki H, Takahashi K, Shibahara S. Identification of a melanocyte-type promoter of the microphthalmia-associated transcription factor gene. Biochem. Biophys. Res. Commun. 1996;219:702–707. doi: 10.1006/bbrc.1996.0298. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- George P, Bali P, Annavarapu S, Scuto A, Fiskus W, Guo F, Sigua C, Sondarva G, Moscinski L, Atadja P, et al. Combination of the histone deacetylase inhibitor LBH589 and the hsp90 inhibitor 17-AAG is highly active against human CML-BC cells and AML cells with activating mutation of FLT-3. Blood. 2005;15:1768–1776. doi: 10.1182/blood-2004-09-3413. [DOI] [PubMed] [Google Scholar]
- Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes and Dev. 2000;14:1712–1728. [PubMed] [Google Scholar]
- Gurvich N, Berman MG, Wittner BS, Gentleman RC, Klein PS, Green JB. Association of valproate-induced teratogenesis with histone deacetylase inhibition in vivo. FASEB J. 2005;19:1166–1168. doi: 10.1096/fj.04-3425fje. [DOI] [PubMed] [Google Scholar]
- Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- Hershey CL, Fisher DE. Genomic analysis of the Microphthalmia locus and identification of the MITF-J/Mitf-J isoform. Gene. 2005;347:73–82. doi: 10.1016/j.gene.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Hughes MJ, Lingrel JB, Krakowsky JM, Anderson KP. A helix-loop-helix transcription factor-like gene is located at the mi locus. J. Biol. Chem. 1993;268:20687–20690. [PubMed] [Google Scholar]
- Kelly WK, Richon VM, O’Connor O, Curley T, MacGregor-Curtelli B, Tong W, Klang M, Schwartz L, Richardson S, Rosa E, et al. Kelly WK, Richon VM, O’Connor O, et al. Phase I clinical trial of histone deacetylase inhibitor: suberoylanilide hydroxamic acid administered intravenously. Clin. Cancer Res. 2003;9:3578–3588. [PubMed] [Google Scholar]
- Kelly WK, Marks PA. Drug insight: Histone deacetylase inhibitors--development of the new targeted anticancer agent suberoylanilide hydroxamic acid. Nat. Clin. Pract. Oncol. 2005;2:150–157. doi: 10.1038/ncponc0106. [DOI] [PubMed] [Google Scholar]
- Kelly WK, O’Connor OA, Krug LM, Chiao JH, Heaney M, Curley T, MacGregore-Cortelli B, Tong W, Secrist JP, Schwartz L, et al. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J. Clin. Oncol. 2005;23:3923–3931. doi: 10.1200/JCO.2005.14.167. J. Clin. Oncol. 23, 3923-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisada T, Lu SZ, Yoshida H, Nishikawa S, Nishikawa S, Mizoguchi M, Hayashi S, Tyrrell L, Williams DA, Wang X, et al. Murine cutaneous mastocytosis and epidermal melanocytosis induced by keratinocyte expression of transgenic stem cell factor. J. Exp. Med. 1998;187:1565–1573. doi: 10.1084/jem.187.10.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessnick SL, Braun BS, Denny CT, May WA. Multiple domains mediate transformation by the Ewing’s sarcoma EWS/FLI-1 fusion gene. Oncogene. 1995;10:423–431. [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Liu T, Kuljaca S, Tee A, Marshall GM. Histone deacetylase inhibitors: multifunctional anticancer agents. Cancer treatment reviews. 32:157–165. doi: 10.1016/j.ctrv.2005.12.006. [DOI] [PubMed] [Google Scholar]
- O’Connor OA, Heaney ML, Schwartz L, Richardson S, Willim R, MacGregor-Cortelli B, Curly T, Moskowitz C, Portlock C, Horwitz S, et al. Clinical experience with intravenous and oral formulations of the novel histone deacetylase inhibitor suberoylanilide hydroxamic acid in patients with advanced hematologic malignancies. J. Clin. Oncol. 2006;24:166–173. doi: 10.1200/JCO.2005.01.9679. [DOI] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Müller MM, Schaffner W. Rapid detection of octamer binding proteins with ’mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara S, Takeda K, Yasumoto K, Udono T, Watanabe K, Saito H, Takahashi K. Microphthalmia-associated transcription factor (MITF): multiplicity in structure, function, and regulation. J. Investig. Dermatol. Proc. 2001;6:99–104. doi: 10.1046/j.0022-202x.2001.00010.x. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Moore KJ, Lamoreux ML, Ferre-D’Amare AR, Burley SK, Zimring DC, Skow LC, Hodgkinson CA, Arnheiter H, Copeland NG, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat. Genet. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- Steingrímsson E, Nii A, Fisher DE, Ferré-D’Amaré AR, McCormick RJ, Russell LB, Burley SK, Ward JM, Jenkins NA, Copeland NG. The semidominant Mi(b) mutation identifies a role for the HLH domain in DNA binding in addition to its role in protein dimerization. EMBO J. 1996;15:6280–6289. [PMC free article] [PubMed] [Google Scholar]
- Udono T, Yasumoto K, Takeda K, Amae S, Watanabe K, Saito H, Fuse N, Tachibana M, Takahashi K, Tamai M, et al. Structural organization of the human microphthalmia-associated transcription factor gene containing four alternative promoters. Biochim. Biophys. Acta. 2000;1491:205–219. doi: 10.1016/s0167-4781(00)00051-8. [DOI] [PubMed] [Google Scholar]
- Weilbaecher KN, Hershey CL, Takemoto CM, Horstmann MA, Hemesath TJ, Tashjian AH, Fisher DE. Age-resolving osteopetrosis: a rat model implicating microphthalmia and the related transcription factor TFE3. J. Exp. Med. 1998;187:775–785. doi: 10.1084/jem.187.5.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund HR, Horstmann MA, Price ER, Cui J, Lessnick SL, Wu M, He X, Fisher DE. Beta-catenin-induced melanoma growth requires the downstream target Microphthalmia-associated transcription factor. J Cell Biol. 2002;158:1079–1087. doi: 10.1083/jcb.200202049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.