Abstract
The recent revolution in sequencing technology has helped to reveal a large transcriptome of long non-coding RNAs (lncRNAs). A major challenge in the years to come is to determine what biological functions, if any, they serve. Although the purpose of these transcripts is largely unknown at present, existing examples suggest that lncRNAs play roles in a wide variety of biological processes. Exemplary cases are lncRNAs within the X-inactivation center. Indeed, lncRNAs dominate control of random X-chromosome inactivation (XCI). The RNA-based regulatory mechanisms of XCI include recruitment of chromatin modifiers, formation of RNA-based subnuclear compartments, and regulation of transcription by antisense transcription. XCI and lncRNAs now also appear to be very relevant in the development and progression of cancer. This perspective focuses on new insights into lncRNA-dependent regulation of XCI, which we believe serve as paradigms for understanding lncRNA function more generally.
INTRODUCTION: LncRNAs AND X-INACTIVATION
Chromosome-based sex determination systems create an imbalance in the dosage of X-linked genes between the two sexes. As a result of the X-Y system of sex determination, female mammals must correct for its double dosage of X-linked gene expression by transcriptionally silencing the majority of genes on one X-chromosome in a process known as X-chromosome inactivation (XCI).1 This process is driven by a series of long non-coding RNAs (lncRNAs) (Table 1). Because of the involvement of many lncRNAs and distinct mechanisms of action, XCI is an excellent model system for studying lncRNA function.
Table 1.
Summary of lncRNAs and proposed interacting protein partners for X-inactivation
| RNA | Function | cis- or trans-acting | Known protein Interactors |
|---|---|---|---|
| Xist | Required for initiation of X- inactivation6. | cis, can in some cases act in trans at autosomal Xist transgenes27 | PRC217, YY127, hnRNP-U66, ASF69 |
| Tsix | Represses Xist expression by silencing the Xist promoter37; 38; 40; 41, also required for X- chromosome pairing, counting the number of X- inactivation centers, and mutually exclusive allelic choice 14; 101; 102 | cis9 | Dnmt3a37; 38 |
| RepA | Independent transcript from Xist5′ end, helps activate Xist17 | cis17 | PRC217 |
| Jpx | Activator of Xist transcription; counting of X- chromosomes 16; 78 | trans (mild cis preference)16 | CTCF78 |
| Ftx | Potential activator of Xist expression18 | unknown18 | unknown |
One of the first lncRNAs identified was Xist2; 3; 4; 5, a 17-kb transcript expressed from the X-inactivation center (XIC) solely from the inactive X (Xi).2; 3 Xist deletions prevent X-inactivation in cis6; 7 and forced Xist expression is sufficient to induce chromosome-wide silencing.8; 9; 10 Xist RNA coats the Xi11, and the spreading of Xist RNA along one X-chromosome in cis initiates chromosome-wide silencing. Whereas Xist expression designates the Xi, Tsix expression demarcates the active X (Xa), also in cis.9; 12; 13; 14; 15 Tsix is antisense to Xist and serves as potent antagonist of Xist expression. It is therefore an excellent example of a natural antisense transcript that represses its sense partner. Noncoding genes have also been implicated in activating Xist, including Jpx, RepA, and Ftx.16; 17; 18 In this review, we will summarize recent advances in the control of XCI by lncRNAs and conclude with new insight into how one lncRNA (Xist) influences the development of cancer, a disease for which a role of the X-chromosome has long been suspected.
XIST RNA: THE RECRUITER OF SILENCING COMPLEXES
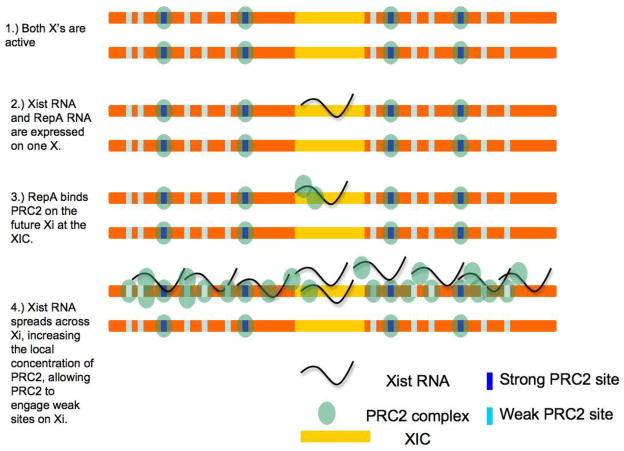
Xist RNA is one of the first examples of an RNA that recruits a chromatin- modifying complex to specific sites (Fig. 1). Polycomb repressive complex 2 (PRC2) is attracted to the X-chromosome by Xist RNA through a repeated motif at the 5′ end of the RNA, known as “Repeat A”.17 The Repeat A motif directly interacts with EZH2, the catalytic subunit of PRC2, both in vivo and in vitro. PRC2 in turn decorates the X-chromosome and silences it as it trimethylates histone H3 at lysine 27 (H3K27me3).19; 20; 21; 22 Along the X, PRC2 first binds ~150 “strong sites”, which have canonical features of known PRC2 binding sites, including a CpG-rich content and presence of bivalent domains.23 From the strong sites, PRC2 migrates laterally and locally, giving rise to thousands of non-canonical domains which may represent sites of dynamic spreading along the X chromatin.23 H3K27me3 density also spreads out from the strong sites, and H3K27me3 occupancy is anti-correlated with LINE density23; 24, an intriguing finding given a long-standing hypothesis that LINE elements serve as “booster elements” that help X-inactivation spread across the whole chromosome25. When expressed ectopically from autosomal transgenes, Xist RNA also recruits PRC2 and silences genes located in cis19; 26; 27, demonstrating that Xist RNA is both necessary and sufficient to recruit PRC2 and inactivates genes on a multi-megabase scale.
Figure 1.
Model for Xist RNA-mediated recruitment of PRC2.
While it is clear that Xist RNA spreads PRC2 to targets on the X-chromosome, mechanisms that localize Xist RNA itself are just beginning to emerge. Localization begins with loading of Xist RNA at a “nucleation center” located within exon 1 of the Xist locus.27 The transcription factor, YY1, is required for Xist RNA loading onto the nucleation site. Knocking down YY1 or mutating its three binding sites within the nucleation center eliminates Xist loading and furthermore blocks formation of the prominent cloud of Xist RNA seen in RNA fluorescence in situ hybridization (FISH) experiments. Xist RNA directly binds YY1 in vivo and in vitro, and YY1 in turn directly contacts three YY1-binding sites near “Repeat F” within Xist exon 1.27 Thus, YY1 is a “bivalent” protein (capable of binding both RNA and DNA) and acts as “bridge” between Xist DNA and RNA. In this way, YY1 tethers the Xist-PRC2 complex to the nucleation site and positions the complex to spread throughout the rest of the chromosome. It is not currently known how Xist RNA binds to other sites along the Xi. These results suggest that recruitment of PRC2 in mammals may involve not only an lncRNA but also an adaptor protein such as YY1. YY1 is a homolog of the Drosophila gene pleiohomeiotic (PHO)28, which has been shown to recruit PRC2 to polycomb response elements in Drosophila.29 Some data suggest that YY1 can interact with several components of the mammalian PRC2 30 but may not be a core component.
Expressing an Xist transgene carrying mutations in the YY1 binding sites led to the very surprising discovery that Xist RNA can act in trans.27 In such situations, transgenic Xist RNA could not bind the nucleation site in cis (on the transgene), but was observed to diffuse to the nucleation site of the Xi and then spread along the Xi. Furthermore, when the transgene is not mutated, Xist RNA produced from the Xi could migrate to the transgene nucleation site and spread along the autosome. This observation led to questions about how the Xa does not engage in binding of Xist. Further analysis indicated that YY1 binds only the nucleation site on Xi (not on Xa).27 Thus, the cis-limited action of Xist RNA occurs in the normal developmental context likely because of developmental programming which blocks YY1 nucleation sites at all but the Xi. Xist is a cis-acting RNA only to the extent that sites in trans are prevented from binding.
It is now clear that RNA-mediated recruitment of PRC2 is not unique to the inactive X-chromosome, as a myriad of transcripts associate with PRC231; 32; 33; 34; 35. The observation that PRC2 interacts with thousands of transcripts raises interesting questions regarding whether the function of RNA in PRC2 biology may extend beyond the role of targeting PRC2, given that the PRC2 “transcriptome” contains RNAs that are not strictly cis-limited. Some lncRNAs may not be directly involved in PRC2 recruitment, but instead modulate PRC2’s methyltransferase activity or its interactions with accessory proteins. Many other chromatin modifiers have been shown to interact with RNA, such as Dnmt3a, G9a, PRC1, MLL-WDR5 and LSD1-CoREST (reviewed in 36). Further understanding the interaction of the Xist-PRC2 interaction may shed light on the functions of interactions between chromatin modifiers and lncRNAs. It would also be of interest to learn whether YY1 functions as adaptor for PRC2-lncRNA complexes elsewhere in the genome.
TSIX: THE ANTISENSE REGULATOR
Tsix is another long, non-coding transcript that plays a key role in X-inactivation. A considerable amount of genetic analysis has been carried out on Tsix towards understanding the antisense mechanism of action. Tsix controls Xist expression in cis by modulating the chromatin structure and DNA methylation status of the Xist promoter.37; 38 Anti-sense transcription extending through the Xist promoter is required to silence Xist in cis.39; 40; 41 It is possible that Tsix acts as a functional RNA and recruits repressors such as the de novo methyltransferase, Dnmt3a37; 38, or titrates activators away from the Xist promoter. The act of antisense transcription through the Xist promoter could also induce a chromatin state that is refractory for sense transcription; alternatively, it could disrupt RNA polymerase function in the sense direction. Definitive experiments to test these hypotheses must separate transcription of the antisense RNA from the action of the antisense transcript.
Tsix’s mechanism of action may extend to other antisense transcripts. There are at least several hundred sense-antisense pairs within mammalian genomes42; 43; 44; 45, many of which are arranged in a structurally similar manner as the Xist-Tsix pair.44 In these cases, the antisense transcript might similarly regulate expression of the sense transcript with which it overlaps.46; 47; 48 In several well-studied examples within imprinted loci, allele-specific expression patterns have been proposed to be controlled by expression of an antisense transcript. For example, Air silences the paternal Igf2r cluster49; 50; 51 and Kcnq1ot1 silences the paternal Kcnq1 cluster.52; 53 Both antisense transcripts are implicated in binding of repressive chromatin factors, such as G9a and PRC2.50; 53 There are likely many other examples of sense-antisense transcription modules that operate like the Xist-Tsix pair. Thus, uncovering the molecular mechanisms that underpin Tsix-mediated regulation of Xist may have broad applicability for understanding the role of antisense transcription.
RNA’S RELATIONSHIP TO LARGE-SCALE CHROMOSOME INTERACTIONS
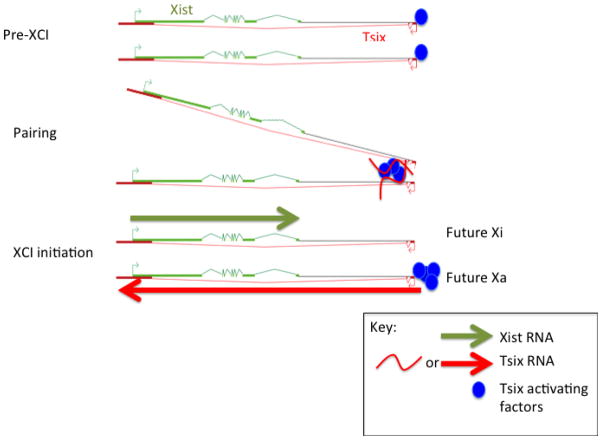
In addition to its role as an inhibitor of Xist expression on the active X, Tsix plays a role in interchromosomal contacts hypothesized to be crucial for X-chromosome choice and for properly demarcating only one active X. Before Xist is upregulated, the two Xic’s of the female cell transiently come into close contact with each other in the nucleus (Figure 2).54; 55; 56; 57 This transient “pairing” of the two Xic’s may allow a redistribution of transcriptional activators between the two alleles56; 58; 59; 60, resulting in asymmetric binding of activators to one X-chromosome and thereby enabling expression of Tsix RNA only on one chromosome. Tsix and its enhancer, Xite, are necessary for pairing56 and are also each sufficient to induce pairing when integrated onto an autosome.57 Interestingly, inhibition of transcription with actinomycin D prevents the formation of new pairing complexes but has little effect on the half-life of paired complexes already formed, suggesting that RNA may be required to attract two X-chromosome to each other but not to keep them paired.57 Whether transcripts emanating from Tsix and Xite are required is currently not known, though the two loci are clearly necessary and sufficient to induce pairing.
Figure 2. Model for XIC pairing before XCI onset.
The two X-chromosomes are epigenetically identical and euchromatic in the pre-XCI stage. The two Xs are brought together by Tsix and Xite (pairing) during cell differentiation to enable cross-talk and mutually exclusive choice of Xa and Xi. Because it is thermodynamically favorable to do so, hypothetical transcription factors, potentially OCT4 and CTCF (blue circles), that were previously randomly distributed between the two Tsix/Xite alleles stochastically shift to one X, which would then become future Xa. This shift results in monoallelic Tsix expression and differential chromatin modifications within the Xist region, which lead to repression of Xist on Xa and upregulation of Xist on Xi.
It seems likely that RNAs may generally participate in long-range chromosome interactions. There are several pieces of evidence implicating RNA as a structural component that determine higher order structures. RNA has long been known to known to co-fractionate with chromatin in eukaryotic nuclear extracts.61; 62; 63; 64 The nuclear matrix consists of a network of ribonucleoprotein particles (reviewed in 65), and it has been suggested that Xist RNA is a component of the nuclear matrix, interacting with factors such as hnRNP-U/SAF-A and ASF.66; 67; 68; 69 Recent experiments more directly implicate a relationship between non-coding RNA and chromatin architecture. A newly-discovered class of RNAs that appear to activate nearby genes in cis called “Activating RNAs” (a-RNA)70 are required for 3D contacts between enhancers and promoters of nearby genes regulated by a-RNAs.71 These RNAs are hypothesized to be predominately cis-acting non-coding transcripts that function like enhancers in enhancer assays. However, unlike classical enhancers, a-RNA activity can be blocked by siRNAs targeting the non-coding transcripts. Chromatin conformation assays revealed looping interactions between several a-RNAs and their target gene’s promoters. These looping interactions were disrupted by knockdown of the a-RNA. Another lncRNA that appears to function through looping interactions is HOTTIP, a putative activator that operates through a change in chromatin conformation in the Hox-a cluster.72 Additionally, two chromatin conformation studies73; 74 suggested that Xi assumes a more “randomized” organization relative to Xa and autosomes, thereby indirectly implicating Xist RNA in generating the less ordered configuration. Indeed, an Xi-specific deletion of Xist partially restored long-range contacts on Xi suggesting that Xist acts to disrupt long-range chromatin contacts73. These may be only a handful of examples of an entire class of lncRNAs involved large-scale chromatin interactions.
POSITIVE LncRNA REGULATORS OF XCI
Some lncRNAs of the Xic appear to be transcriptional activators. They include RepA, Jpx, and Ftx, all proposed to be activators of Xist (Table 1). The molecular mechanisms that cause these RNAs to induce Xist are currently unknown, though recent studies have suggested several intriguing possibilities.
RepA RNA is transcribed from an independent transcription unit within exon 1 of Xist and consists of a repeated motif (Repeat A) that directly binds and targets PRC2 to the Xist promoter.17 RepA is believed to induce Xist expression by increasing H3K27 trimethylation of the Xist promoter via its recruitment of PRC2.17 As PRC2 is a repressive complex, this may seem to be a counterintuitive way to activate Xist transcription. However, Xist may prefer a heterochromatic environment. Indeed, Xist remains active in the repressive context of the inactive X, it may actually be induced by repressive signals. This intriguing possibility has to be tested more directly to determine whether PRC2 recruitment to the Xist promoter actually induces Xist expression. Consistent with the idea, a mouse deletion of RepA results in loss of Xist induction.75
Jpx’s activating influence is supported by genetic analysis. When female cells aredeleted for Jpx – even on just one allele – the cells can no longer induce Xist expression 16. Jpx does not apparently function as an enhancer, as standard enhancer assays failed to reveal any activating influence on the Xist promoter. Furthermore, its ability to activate Xist in trans (when Jpx is placed in an autosomal context) also argues against an enhancer mechanism.16 Thus, Jpx’s action is distinct from enhancer-associated RNAs.70; 76; 77 Because a post-transcriptional knockdown of Jpx phenocopies a Jpx knockout, the transcript itself (not just the Jpx gene or transcription) must be the activating force. The latest work indicates that Jpx RNA is part of the X-chromosome counting mechanism and activates Xist by titrating away a repressive autosomal factor, CTCF, that normally binds and blocks the Xist promoter.78
Jpx could act in other ways as well. Jpx is trans-acting, but has a mild cis-preference.16 Consistent with a cis-preference, chromosome conformation analysis suggests that the Jpx locus makes contacts with the Xist promoter following the onset of differentiation and XCI.79 It is possible that Jpx RNA mediates the formation of these chromatin contacts, as has been shown for several other a-RNAs.71 Another possibility is that the chromatin contacts form independently of Jpx RNA, and juxtaposing nascent Jpx transcripts to the Xist promoter allows Jpx to recruit activators or titrate repressors from the Xist promoter. Studying these activating RNAs may provide several important model systems for evaluating the functions of mechanisms of recently discovered a-RNAs70 and enhancer RNAs.76; 77
XIST AND CANCER
Much work has centered on the role of Xist in initiating X-inactivation in the early developing embryo. However, it is now clear that Xist has important roles in later development and in adults as well, long after the establishment of XCI. In mouse embryonic fibroblasts 80, a conditional deletion of Xist from the Xi leads to partial X-reactivation. Since XCI silences several hundred genes, some of which are oncogenes, improper Xist expression could potentially be a mechanism underlying tumorigenesis.81; 82 Early experiments suggested a tantalizing connection between X-linked gene dosage and cancer. Loss of XCI and downregulation of XIST expression are commonly observed in basal-like cancer, BRCA1-null triple negative breast cancer83; 84; 85; 86; 87; 88; 89, and ovarian cancer lines.90; 91 Loss of XIST is most commonly caused by X isodisomy, where Xi is lost and Xa is amplified. Reactivation of Xi may be an alternative mechanism leading to loss of XIST and overedxpression of the X.85; 87; 90; 91 These observations suggest a correlation between X-chromosome dysfunction and cancer. The link between XCI and cancer was recently determined to be causal, with the finding that deleting Xist in the blood compartment leads to a highly aggressive myeloproliferative neoplasm and myelodysplastic syndrome (mixed MPN-MDS) with 100% penetrance and lethality in mice.92 This result clearly demonstrates that loss of Xist RNA and overexpression of the X in adult tissues can lead to cancer.
As Xist RNA promotes the initiation or progression of cancer, it may be reasonable to propose reactivation of Xist as a therapeutic strategy in cancer. It is known that ectopic Xist expression in adult mice can lead to ectopic gene silencing in cis and cell death in the immature precursors of the immune system.93 Therapeutic strategies may include small molecules. Indeed, there is precedence when one looks to Angelman Syndrome, a congential disorder caused by maternal deletion or mutation of the imprinted Ube3a allele. Ube3a is known to be controlled by a long antisense transcript from the Snrpn locus.94; 95; 96; 97 Topoisomerase inhibitors cause loss of imprinting of the silent paternal Ube3a allele by downregulating the antisense transcript in neurons in vitro and in mice, which may provide a strategy for rescuing the genetic defect that causes Angelman Syndrome.98 In tumors, Xist expression might also be reactivated by small molecules, offering a novel therapeutic approach that would target epigenetically functional lncRNAs.
CONCLUSIONS
LncRNAs have received a great deal of attention over the past few years as regulators of gene expression.24; 99; 100 Thousands of new transcripts have been identified in the mammalian genome, with estimates upwards of 200,000.5 A key challenge is to determine which transcripts are functional and how these transcripts regulate physiological processes. RNAs of the Xic may provide a framework towards understanding how those in the rest of the genome operate. Within the Xic, both cis- (e.g., Tsix) and trans-acting (e.g., Jpx) lncRNAs can be found. These X-linked lncRNAs can also be either activating (Jpx) or repressive (Xist). They interact with transcription factors (e.g., YY1, CTCF) and chromatin modifying complexes (e.g., PRC2, Dnmt3a). The Xic also provides a model by which to study sense-antisense pairs and the role of RNA in large-scale chromatin architecture. It has furthermore become clear that lncRNAs of the Xic are crucial not only during early embryogenesis when dosage compensation takes place, but also throughout adult life. Basic homeostatic functions of the lncRNAs have been revealed when deleting one member (Xist) results in a lethal disease (cancer). X-inactivation and other allelic phenomena have often proven to be fertile grounds for uncovering unexpected biology.94 The very first lncRNAs were discovered by applying classical genetic techniques to find genes that control X-inactivation and imprinting. In the future, emerging genomic and nano-scale technologies will likely provide many new surprising insights into how the noncoding genome contributes to normal physiology and disease.
chromosome inactivation provides a model to study lncRNA
The lncRNAs can act in cis or trans; activate or repress
Xist RNA recruits Polycomb complexes to the X-chromosome
Tsix is a model antisense RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lyon M. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2.Brown C, Ballabio A, Rupert J, Lafreniere R, Grompe M, Tonlorenzi R, Willard H. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–82. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 3.Brown C, Lafreniere R, Powers V, Sebastio G, Ballabio A, Pettigrew A, Ledbetter D, Levy E, Craig I, Willard H. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature. 1991;349:82–86. doi: 10.1038/349082a0. [DOI] [PubMed] [Google Scholar]
- 4.Brockdorff N, Ashworth A, Kay G, McCabe V, Norris D, Cooper P, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–541. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 5.Borsani G, Tonlorenzi R, Simmler M, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, Willard H, Avner P, Ballabio A. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 6.Penny G, Kay G, Sheardown S, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–138. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 7.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–66. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 8.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Molecular cell. 2000;5:695–1400. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 9.Stavropoulos N, Lu N, Lee J. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10232–10237. doi: 10.1073/pnas.171243598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luikenhuis S, Wutz A, Jaenisch R. Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol Cell Biol. 2001;21:8512–20. doi: 10.1128/MCB.21.24.8512-8520.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemson C, McNeil J, Willard H, Lawrence J. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. The Journal of cell biology. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Davidow L, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nature genetics. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 13.Lee J, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- 14.Lee J. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103:17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 15.Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128:1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- 16.Tian D, Sun S, Lee J. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–793. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J, Sun B, Erwin J, Song JJ, Lee J. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chureau C, Chantalat S, Romito A, Galvani Al, Duret L, Avner P, Rougeulle C. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Human molecular genetics. 2011;20:705–718. doi: 10.1093/hmg/ddq516. [DOI] [PubMed] [Google Scholar]
- 19.Plath K, Fang J, Mlynarczyk-Evans S, Cao R, Worringer K, Wang H, de la Cruz C, Otte A, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 20.Mak W, Baxter J, Silva J, Newall A, Otte A, Brockdorff N. Mitotically stable association of polycomb group proteins eed and enx1 with the inactive x chromosome in trophoblast stem cells. Current biology: CB. 2002;12:1016–1020. doi: 10.1016/s0960-9822(02)00892-8. [DOI] [PubMed] [Google Scholar]
- 21.Silva J, Mak W, Zvetkova I, Appanah R, Nesterova T, Webster Z, Peters A, Jenuwein T, Otte A, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Developmental cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Mager J, Chen Y, Schneider E, Cross J, Nagy A, Magnuson T. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nature genetics. 2001;28:371–375. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- 23.Pinter S, Sadreyev R, Yildirim E, Jeon Y, Ohsumi T, Borowsky M, Lee J. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome research. 2012;22:1864–1876. doi: 10.1101/gr.133751.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calabrese J, Sun W, Song L, Mugford J, Williams L, Yee D, Starmer J, Mieczkowski P, Crawford G, Magnuson T. Site-specific silencing of regulatory elements as a mechanism of x inactivation. Cell. 2012;151:951–963. doi: 10.1016/j.cell.2012.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyon M. The Lyon and the LINE hypothesis. Seminars in cell & developmental biology. 2003;14:313–318. doi: 10.1016/j.semcdb.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS biology. 2004;2 doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeon Y, Lee J. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–152. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown J, Mucci D, Whiteley M, Dirksen M, Kassis J. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Molecular cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Brown J, Cao R, Zhang Y, Kassis J, Jones R. Hierarchical recruitment of polycomb group silencing complexes. Molecular cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Paylor S, Magnuson T, Schumacher A. Juxtaposed Polycomb complexes co-regulate vertebral identity. Development. 2006;133:4957–4968. doi: 10.1242/dev.02677. [DOI] [PubMed] [Google Scholar]
- 31.Khalil A, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein B, van Oudenaarden A, Regev A, Lander E, Rinn J. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667–11739. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J, Ohsumi T, Kung J, Ogawa Y, Grau D, Sarma K, Song J, Kingston R, Borowsky M, Lee J. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Molecular cell. 2010;40:939–992. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanhere A, Viiri K, Araujo C, Rasaiyaah J, Bouwman R, Whyte W, Pereira C, Brookes E, Walker K, Bell G, Pombo A, Fisher A, Young R, Jenner R. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Molecular cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinn J, Kertesz M, Wang J, Squazzo S, Xu X, Brugmann S, Goodnough L, Helms J, Farnham P, Segal E, Chang H. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1334. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rinn J, Chang H. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Developmental cell. 2005;9:159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 38.Sun B, Deaton A, Lee J. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Molecular cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 39.Shibata S, Lee J. Tsix transcription- versus RNA-based mechanisms in Xist repression and epigenetic choice. Current biology: CB. 2004;14:1747–1754. doi: 10.1016/j.cub.2004.09.053. [DOI] [PubMed] [Google Scholar]
- 40.Sado T, Hoki Y, Sasaki H. Tsix defective in splicing is competent to establish Xist silencing. Development. 2006;133:4925–4931. doi: 10.1242/dev.02670. [DOI] [PubMed] [Google Scholar]
- 41.Ohhata T, Hoki Y, Sasaki H, Sado T. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development. 2008;135:227–235. doi: 10.1242/dev.008490. [DOI] [PubMed] [Google Scholar]
- 42.Fahey M, Moore T, Higgins D. Overlapping antisense transcription in the human genome. Comparative and functional genomics. 2002;3:244–253. doi: 10.1002/cfg.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehner B, Williams G, Campbell R, Sanderson C. Antisense transcripts in the human genome. Trends in genetics. 2002;18:63–65. doi: 10.1016/s0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- 44.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap C, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang K, Hallinan J, Mattick J, Hume D, Lipovich L, Batalov S, Engstrom P, Mizuno Y, Faghihi M, Sandelin A, Chalk A, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C, Group RGER, Genome Science G, Consortium F. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 45.Yelin R, Dahary D, Sorek R, Levanon E, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, Nemzer S, Pinner E, Walach S, Bernstein J, Savitsky K, Rotman G. Widespread occurrence of antisense transcription in the human genome. Nature biotechnology. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 46.Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y, Group RG, Members GSL. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome research. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO reports. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magistri M, Faghihi M, St Laurent G, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends in genetics. 2012;28:389–396. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sleutels F, Zwart R, Barlow D. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 50.Nagano T, Mitchell J, Sanz L, Pauler F, Ferguson-Smith A, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 51.Latos P, Pauler F, Koerner M, Senergin H, Hudson Q, Stocsits R, Allhoff W, Stricker S, Klement R, Warczok K, Aumayr K, Pasierbek P, Barlow D. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 52.Fitzpatrick G, Soloway P, Higgins M. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nature genetics. 2002;32:426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- 53.Mancini-Dinardo D, Steele S, Levorse J, Ingram R, Tilghman S. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes & development. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bacher C, Guggiari Ml, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nature cell biology. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 55.Masui O, Bonnet I, Le Baccon P, Brito I, Pollex T, Murphy N, Hupe P, Barillot E, Belmont A, Heard E. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell. 2011;145:447–458. doi: 10.1016/j.cell.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu N, Tsai CL, Lee J. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 57.Xu N, Donohoe M, Silva S, Lee J. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nature genetics. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- 58.Nicodemi M, Prisco A. Self-assembly and DNA binding of the blocking factor in X Chromosome Inactivation. PLoS Computational Biology. 2005;3:2135–2142. doi: 10.1371/journal.pcbi.0030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nicodemi M, Prisco A. Symmetry-breaking model for X-chromosome inactivation. Physical review letters. 2007;98:108104. doi: 10.1103/PhysRevLett.98.108104. [DOI] [PubMed] [Google Scholar]
- 60.Scialdone A, Nicodemi M. Mechanics and dynamics of X-chromosome pairing at X inactivation. PLoS computational biology. 2008;4 doi: 10.1371/journal.pcbi.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mondal T, Rasmussen M, Pandey G, Isaksson A, Kanduri C. Characterization of the RNA content of chromatin. Genome research. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodriguez-Campos A, Azorin F. RNA is an integral component of chromatin that contributes to its structural organization. PloS one. 2007;2 doi: 10.1371/journal.pone.0001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paul J, Duerksen J. Chromatin-associated RNA content of heterochromatin and euchromatin. Molecular and cellular biochemistry. 1975;9:9–16. doi: 10.1007/BF01731728. [DOI] [PubMed] [Google Scholar]
- 64.Huang R, Bonner J. Histone-bound RNA, a component of native nucleohistone. Proceedings of the National Academy of Sciences of the United States of America. 1965;54:960–967. doi: 10.1073/pnas.54.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nickerson J. Experimental observations of a nuclear matrix. Journal of cell science. 2001;114:463–474. doi: 10.1242/jcs.114.3.463. [DOI] [PubMed] [Google Scholar]
- 66.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Developmental cell. 2010;19:469–545. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 67.Nakagawa S, Prasanth K. eXIST with matrix-associated proteins. Trends in cell biology. 2011;21:321–327. doi: 10.1016/j.tcb.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tattermusch A, Brockdorff N. A scaffold for X chromosome inactivation. Human genetics. 2011;130:247–300. doi: 10.1007/s00439-011-1027-4. [DOI] [PubMed] [Google Scholar]
- 69.Royce-Tolland M, Andersen A, Koyfman H, Talbot D, Wutz A, Tonks I, Kay G, Panning B. The A-repeat links ASF/SF2-dependent Xist RNA processing with random choice during X inactivation. Nature structural & molecular biology. 2010;17:948–954. doi: 10.1038/nsmb.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Orom U, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lai F, Orom U, Cesaroni M, Beringer M, Taatjes D, Blobel G, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang K, Yang Y, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie B, Protacio A, Flynn R, Gupta R, Wysocka J, Lei M, Dekker J, Helms J, Chang H. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Splinter E, de Wit E, Nora E, Klous P, van de Werken H, Zhu Y, Kaaij L, van Ijcken W, Gribnau J, Heard E, de Laat W. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes & development. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nora E, Lajoie B, Schulz E, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum N, Meisig J, Sedat J, Gribnau J, Barillot E, Bluthgen N, Dekker J, Heard E. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoki Y, Kimura N, Kanbayashi M, Amakawa Y, Ohhata T, Sasaki H, Sado T. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development. 2009;136:139–146. doi: 10.1242/dev.026427. [DOI] [PubMed] [Google Scholar]
- 76.Kim TK, Hemberg M, Gray J, Costa A, Bear D, Wu J, Harmin D, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley P, Kreiman G, Greenberg M. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi B, Muller H, Ragoussis J, Wei C-L, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS biology. 2010:8. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun S, del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsai CL, Rowntree R, Cohen D, Lee J. Higher order chromatin structure at the X-inactivation center via looping DNA. Developmental biology. 2008;319:416–425. doi: 10.1016/j.ydbio.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang LF, Huynh K, Lee J. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–1399. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 81.Liu Y, Wang L, Zheng P. X-linked tumor suppressors: perplexing inheritance, a unique therapeutic opportunity. Trends in genetics. 2010;26:260–265. doi: 10.1016/j.tig.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spatz A, Borg C, Feunteun J. X-chromosome genetics and human cancer. Nature reviews Cancer. 2004;4:617–629. doi: 10.1038/nrc1413. [DOI] [PubMed] [Google Scholar]
- 83.Ganesan S, Silver D, Greenberg R, Avni D, Drapkin R, Miron A, Mok S, Randrianarison V, Brodie S, Salstrom J, Rasmussen T, Klimke A, Marrese C, Marahrens Y, Deng C, Feunteun J, Livingston D. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell. 2002;111:393–405. doi: 10.1016/s0092-8674(02)01052-8. [DOI] [PubMed] [Google Scholar]
- 84.Pageau G, Hall L, Lawrence J. BRCA1 does not paint the inactive X to localize XIST RNA but may contribute to broad changes in cancer that impact XIST and Xi heterochromatin. Journal of cellular biochemistry. 2007;100:835–850. doi: 10.1002/jcb.21188. [DOI] [PubMed] [Google Scholar]
- 85.Richardson A, Wang Z, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart J, Livingston D, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 86.Silver D, Dimitrov S, Feunteun J, Gelman R, Drapkin R, Lu S, Shestakova E, Velmurugan S, Denunzio N, Dragomir S, Mar J, Liu X, Rottenberg S, Jonkers J, Ganesan S, Livingston D. Further evidence for BRCA1 communication with the inactive X chromosome. Cell. 2007;128:991–1002. doi: 10.1016/j.cell.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 87.Sirchia S, Ramoscelli L, Grati F, Barbera F, Coradini D, Rossella F, Porta G, Lesma E, Ruggeri A, Radice P, Simoni G, Miozzo M. Loss of the inactive X chromosome and replication of the active X in BRCA1-defective and wild-type breast cancer cells. Cancer research. 2005;65:2139–2146. doi: 10.1158/0008-5472.CAN-04-3465. [DOI] [PubMed] [Google Scholar]
- 88.Sirchia S, Tabano S, Monti L, Recalcati M, Gariboldi M, Grati F, Porta G, Finelli P, Radice P, Miozzo M. Misbehaviour of XIST RNA in breast cancer cells. PloS one. 2009:4. doi: 10.1371/journal.pone.0005559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vincent-Salomon A, Ganem-Elbaz C, Manie E, Raynal V, Sastre-Garau X, Stoppa-Lyonnet D, Stern MH, Heard E. X inactive-specific transcript RNA coating and genetic instability of the X chromosome in BRCA1 breast tumors. Cancer research. 2007;67:5134–5140. doi: 10.1158/0008-5472.CAN-07-0465. [DOI] [PubMed] [Google Scholar]
- 90.Benoit MH, Hudson T, Maire G, Squire J, Arcand S, Provencher D, Mes-Masson AM, Tonin P. Global analysis of chromosome X gene expression in primary cultures of normal ovarian surface epithelial cells and epithelial ovarian cancer cell lines. International journal of oncology. 2007;30:5–17. [PubMed] [Google Scholar]
- 91.Kawakami T, Zhang C, Taniguchi T, Kim C, Okada Y, Sugihara H, Hattori T, Reeve A, Ogawa O, Okamoto K. Characterization of loss-of-inactive X in Klinefelter syndrome and female-derived cancer cells. Oncogene. 2004;23:6163–6169. doi: 10.1038/sj.onc.1207808. [DOI] [PubMed] [Google Scholar]
- 92.Yildirim E, Kirby J, Brown D, Mercier F, Sadreyev R, Scadden D, Lee J. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Molecular and cellular biology. 2006;26:7167–7177. doi: 10.1128/MCB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee J, Bartolomei M. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 95.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nature genetics. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 96.Matsuura T, Sutcliffe J, Fang P, Galjaard R, Jiang Y, Benton C, Rommens J, Beaudet A. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nature genetics. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 97.Sutcliffe J, Jiang Y, Galijaard R, Matsuura T, Fang P, Kubota T, Christian S, Bressler J, Cattanach B, Ledbetter D, Beaudet A. The E6-Ap ubiquitin-protein ligase (UBE3A) gene is localized within a narrowed Angelman syndrome critical region. Genome research. 1997;7:368–377. doi: 10.1101/gr.7.4.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang HS, Allen J, Mabb A, King I, Miriyala J, Taylor-Blake B, Sciaky N, Dutton J, Lee HM, Chen X, Jin J, Bridges A, Zylka M, Roth B, Philpot B. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481:185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rinn J, Chang H. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–211. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, Rozowsky JS, Gerstein MB, Wahlestedt C, Hayashizaki Y, Carninci P, Gingeras TR, Mattick JS. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee J. Regulation of X-chromosome counting by Tsix and Xite sequences. Science. 2005;309:768–771. doi: 10.1126/science.1113673. [DOI] [PubMed] [Google Scholar]
- 102.Vigneau S, Augui S, Navarro P, Avner P, Clerc P. An essential role for the DXPas34 tandem repeat and Tsix transcription in the counting process of X chromosome inactivation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7390–7395. doi: 10.1073/pnas.0602381103. [DOI] [PMC free article] [PubMed] [Google Scholar]