Summary
Embryonic cells transcriptionally repress the expression of endogenous and exogenous retroelements. Trim28, a key player in this silencing, is known to act in a large DNA-bound complex, but the other components of the complex are not fully characterized. Here we show that the zinc finger protein Yin Yang 1 (YY1) is one such component. YY1 binds to the LTR region of both exogenous and endogenous retroviruses (ERVs). Deletion of the YY1 binding site from the retroviral genome leads to major loss of silencing in embryonic cells, and a coordinated loss of repressive epigenetic marks from the proviral chromatin. Depletion of YY1 protein results in marked upregulation of expression of exogenous viruses and of selected ERVs. Finally, we report an embryonic cell-specific interaction between YY1 and Trim28. Our results suggest a major role for YY1 in the silencing of both exogenous and endogenous retroviruses in embryonic cells.
Introduction
Retroviruses integrate their genome into the host DNA as an obligate part of their life cycle, and by infection of germline cells have made permanent contributions to the genome of their hosts (Stoye, 2012). Transcription of the integrated proviruses results in assembly and release of virions, chronic infections that initiate leukemias, and potential genomic instability. Endogenous and exogenous retroviruses are strongly silenced in embryonic stem (ES) and embryonic carcinoma (EC) cells by both chromatin and DNA methylation (Katz et al., 2007; Rowe and Trono, 2011; Smith et al., 2012), even though ES cells are globally transcriptionally hyperactive and express large regions of the genome at low levels due to open chromatin structure (Meshorer and Misteli, 2006). The mechanisms for initiation of this silencing process, which is essential to protect genome integrity, remain poorly characterized.
Moloney murine leukemia virus (MMLV) is transcriptionally silenced in mouse embryonic cells (Barklis et al., 1986; Teich et al., 1977) through recruitment of chromatin modifiers to the proviral DNA by the universal silencing protein, Trim28/Kap-1/Tif1b (Wolf and Goff, 2007). Trim28 is recruited to the provirus by a specific zinc finger DNA binding protein, ZFP809, which recognizes and directly binds a short DNA sequence, the so-called proline primer binding site (PBS) (Wolf and Goff, 2009; Wolf and Goff, 2007). Trim28 in turn recruits many factors involved in transcriptional silencing and heterochromatin formation, several of which are essential for ERVs silencing. These include the histone H3K9 methyltransferases ESET (Matsui et al., 2010a) and G9a (Leung et al., 2011) and the heterochromatin associated protein HP1 (Sripathy et al., 2006), involved in the heterochromatinization and repression of class I and II ERVs and some exogenous retroviruses; the polycomb group (PcG) I and II proteins, responsible for histone H3K27trimethylation of these ERV classes (Casa and Gabellini, 2012; Leeb et al., 2010); and KDM1a/LSD1, which mediates H3K4 demethylation of class III ERVs (Macfarlan et al., 2011). Most of the retroviral vectors and ERVs used in these studies utilize alternative PBS sequences that are not recognized by the PBS dependent silencing machinery but they are still subject to transcriptional repression through PBS-independent mechanisms (Cherry et al., 2000; Rowe and Trono, 2011; Schlesinger and Goff, 2012), suggesting that another DNA binding protein may be able to tether the complex to other sites on the DNA. One candidate for such a protein is Yin Yang 1 (YY1), a DNA/RNA-binding zinc finger protein, ubiquitously expressed in all tissues and highly conserved in vertebrates from Xenopus laevis to humans (Shi et al., 1997). It is able to activate or repress gene expression in different cellular contexts and interacts with a wide variety of regulatory proteins (Atchison et al., 2011). YY1 also has important functions in regulation of viral sequences; for example, YY1 can directly and indirectly bind and repress HIV-1 (Coull et al., 2000; He and Margolis, 2002). YY1 was also shown to bind the Negative Control Region (NCR) in the U3 region of the MMLV LTR (Flanagan et al., 1992; Flanagan et al., 1989), the intracisternal A-type particle (IAP) genome (Satyamoorthy et al., 1993), and many other viruses (Hyde-DeRuyscher et al., 1995). Interestingly, the transcription factor Rex1, a member of the YY1 family, was recently suggested to be involved in silencing ERVs (Guallar et al., 2012).
Here we show that YY1 plays a major role in repression of endogenous and exogenous retroviral DNAs in mouse ES cells. YY1 binds the LTR of MMLV as well as different ERVs in vitro and in vivo. Deletion of the YY1 binding site in a retroviral genome results in an embryonic-cell specific relief of repression by up to 4-6 fold relative to wt virus. YY1 knockdown (KD) in EC cells also relieves the repression of incoming MMLV (up to 24 fold change). Chromatin immunoprecipitation (ChIP) analyses of YY1 KD cells with Trim28 antibody show lower enrichment of Trim28 in the LTR of the YY1 KD line relative to the wild type, suggesting that the assembly of a silencing complex containing Trim28 is at least in part dependent on YY1. Moreover, we show that YY1 binds Trim28, but that this binding is specific to the ES cell stage and is not detected 8 days or more after onset of differentiation or in differentiated cell lines, even though both proteins are present. The findings highlight the interaction between YY1 and Trim28 as a novel site of regulation of transcriptional silencing by ES cells.
Results
YY1 is required for MMLV silencing in embryonic cells
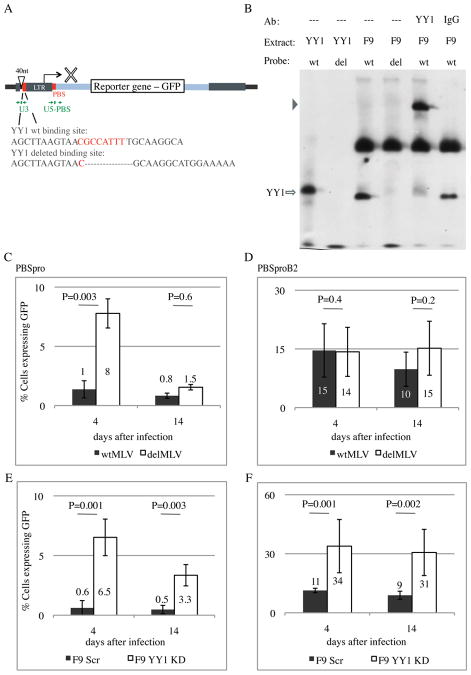
To test the possibility that YY1 plays a role in retroviral silencing in embryonic cells, we directly assayed expression of reporter constructs. Four MMLV-based vector genomes expressing a GFP reporter were generated and packaged into virus particles (Fig 1A). The vectors contained either wild-type virus regulatory sequences, or a deletion in the U3 YY1 binding site (delMLV), or a mutation in the PBS sequence (PBSproB2), or mutations in both (Fig S1A). YY1 was shown to bind to the wild-type LTR and not to the deleted sequence by electrophoretic mobility shift assay (EMSA) performed with YY1 recombinant protein or nuclear cell extracts (Fig 1B). Adding YY1 antibody, but not control IgG, to the nuclear extract resulted in a supershift of the band, proving YY1 to be present in the bound complex. EMSA with nuclear extracts of differentiated NIH3T3 or of F9 embryonic carcinoma (EC) cells after knockdown of ZFP809 showed similar results (Fig S1B), demonstrating that the in vitro YY1 DNA binding activity is not dependent on cell line or on PBS targeting machinery.
Figure 1. Functional assays demonstrate the importance of YY1 for retroviral silencing in F9 EC cells.
(A) Map of proviral DNA with the YY1 binding site and the PBS indicated in red. 40 random nucleotides (40nt) were added 5′ to the regulatory region (Ooi et al. 2010) to enable detection of proviral DNA via a specific PCR product. Taq-man qPCR primers and probe for the U3 and U5-PBS regions are indicated in green (see Table S1 for sequences). Oligonucleotides used for detection of YY1 DNA-binding activity by EMSA are shown. (B) Electrophoretic mobility shift assays (EMSAs) using 250 ng recombinant human YY1 protein (YY1) or F9 nuclear extract (F9). Oligonucleotide probes were as indicated: wt, wild-type U3 YY1 binding site probe; del, deleted YY1 binding site probe. Arrow indicates specific YY1-DNA complex. Arrowhead indicates YY1 supershift formed after pre-incubation with YY1 Ab, but not IgG Ab. A background band of unknown identity is apparent in all nuclear extracts lanes (see also Fig S1B). (C) Flow analysis of GFP-positive cells at 4 and 14 days after infection by wt MLV and YY1 deleted binding site MLV (delMLV) with PBSpro and PBSproB2 (D) in F9 EC cells. Averages ± standard errors of mean (SEM) from 3 independent experiments are shown. For ES-E14 cells results see Fig S1E, F. (E) Same assay on F9 Scrambled KD pool cells and on F9 YY1 KD pool cells (see Fig S1G, H for KD verification) with PBSpro and PBSproB2 (F) in F9 EC cells. Averages ± SEM from 3 independent experiments are shown. Statistical significance, p value, is determined by Student's t test.
To test for expression of the viral genomes in EC cells, we infected F9 cells with viruses containing the four different constructs and assessed GFP reporter expression by flow analysis. Infections were performed at multiplicities <1, and DNA copy numbers of expressing and nonexpressing cell populations were measured and found to be comparable (Figure S1C). Wild-type virus was rapidly and efficiently silenced, and mutation of the PBSpro caused a dramatic loss of that silencing as expected (Barklis et al., 1986). Deletion of the YY1 binding site alone also resulted in significant release from silencing as compared to the wild type at early times after infection (Fig 1C). This change was temporary, however, and 14 days after infection both viruses were almost completely silenced. Virus with mutations of both U3 YY1 binding site and PBSpro were indistinguishable from virus with the PBSpro mutation alone and showed no significant silencing (Fig 1D). These results suggest that the YY1 binding site acts in concert with the wild-type PBSpro element, and is important for the rapid onset of the wt virus silencing but is not important at later times after infection. The deletion of the YY1 binding site had no effect on virus infection and expression in differentiated NIH3T3 cells (Fig S1D), indicating that the release from silencing was specific to embryonic cells. Infections of authentic ES cells with these viruses gave similar results to F9 cells (Fig S1E, F), indicating that the results were not limited to EC cells.
To test for the importance of the YY1 protein itself in silencing, F9 lines were generated in which YY1 expression was knocked down by shRNAs (Fig. S1G, H). Infection of the YY1 knockdown (YY1 KD) and control scrambled (Scr) shRNA F9 lines with the GFP expressing vectors gave different results from those seen with the YY1 binding site mutants. Here we detected a major loss of silencing in the KD cells at early times as before, but in this case the loss of silencing continued up to four weeks after infection. The effect was seen for both wt PBSpro and PBSproB2 containing viruses with a somewhat larger effect for the PBSpro virus (Fig 1E,F). No change in expression of other silencing genes – Trim28 and ZFP809 – was observed (Fig S1G). While this experiment confirms the previous finding that YY1 serves as a negative regulator of viral expression (Flanagan et al., 1992), the distinctive effects of mutation in the YY1 binding site and of YY1 knockdown indicate a dual phase mechanism of action. At first, YY1 binding to its binding site, together with ZFP809 binding to the PBS, results in a rapid onset of proviral silencing, prevented either by loss of the YY1 binding site or YY1 KD. However, YY1 has a second, PBS-independent role in silencing, as indicated by the superior and longer-lasting release from silencing seen in the YY1 KD cells.
YY1 binding to the LTR has different requirements at different times after infection
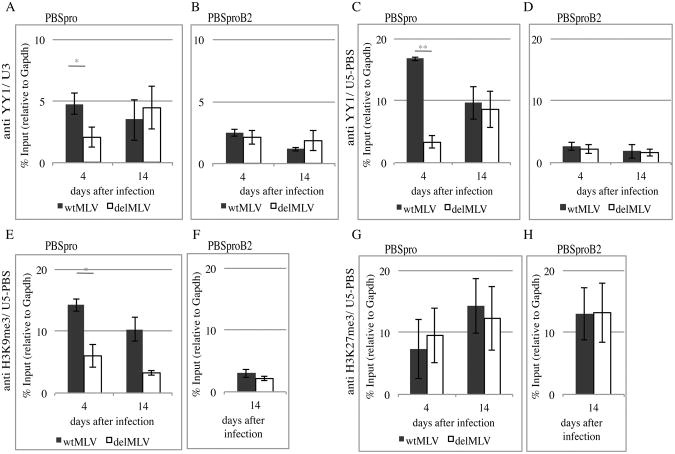
To monitor the binding of YY1 to the proviral DNA in vivo, Chromatin Immunoprecipitation (ChIP) experiments with YY1 antibody were performed. After crosslinking, DNA was immunoprecipitated and quantitative PCR was performed with primers that amplified various sequences along the LTR of the newly introduced viruses (Fig 1A). The levels were normalized to the signals in the input DNA fraction and then to the housekeeping Gapdh gene. Enrichment of YY1 on positive control genes was observed for each ChIP experiment (Fig S2A-C), and nonspecific IgG antibody always gave background values. Control ChIP experiments showed that Rex1, an ES cell transcription factor and YY1 family member also involved in ERV silencing (Guallar et al., 2012) was not enriched on the MMLV LTR (Fig S2D). The results showed that YY1 was consistently enriched near its U3 binding site in wt virus at early times after infection, but not in virus lacking this U3 site (delMLV) (Fig 2A,B). Surprisingly, YY1 was even more highly enriched at the U5-PBS region, 500 bp away from the YY1 binding site (Fig 2C,D). This enrichment at the PBS region was not observed in the delMLV virus. This remote enrichment could be a result of either spreading of YY1 along the viral DNA or a result of protein-protein interactions in the YY1-containing complex that bring the two DNA sequences close together. More surprisingly, the enrichment of YY1 on its own binding site and on the PBS was not seen in the PBSproB2 viruses (Fig. 2B,D). This loss of YY1 enrichment on the PBSProB2 virus (Table S2) was statistically significant. Thus, the binding of YY1 to the proviral DNA at early times after infection required both the wild-type PBS as well as the wild-type YY1 binding site.
Figure 2. YY1 enrichment throughout the viral promoter is dynamic, and associated with H3K9me3 histone modification.
(A, B) ChIP–based measurement of YY1 at the viral U3 region in cells infected with wt or delMLV containing either wt PBSpro or mutant PBSproB2 at 4 and 14 days after infection. (C, D) YY1 enrichment at the U5-PBS site in cells infected with wt or delMLV containing PBSpro or PBSproB2 at 4 and 14 days after infection. (E, F) ChIP assay for H3K9me3 at the U5-PBS region of proviruses in cells infected with wt or delMLV containing PBSpro at 4 and 14 days after infection, or 14 days after infection of cells with PBSproB2. (G, H) Same essay for H3K27me3 on PBSpro or PBSproB2. All graphs show the mean enrichment ± SEM from 3 independent experiments relative to the total input samples and normalized to the signal of negative control (Gapdh). Student's t test was used for statistical analysis; one asterisk denotes P value < 0.05, two denote P < 0.01. A control with IgG antibody (Ab) gave background enrichment (not shown).
At later times, 14 days after infection, YY1 was bound to the LTR at both sites on both the wild-type and the delMLV viruses. Thus, the later enrichment on the LTR occurs even without the YY1 binding site, correlating with the slower silencing seen with the delMLV reporter. This later binding was not observed in the PBSproB2 mutants (Fig. 2B). Thus, the binding of YY1 is dependent on the wild-type PBSpro at all times, but is only YY1 binding site-specific early after integration.
Taken together, these ChIP results are concordant with the reporter expression data, indicating a strong correlative link between YY1 binding and viral silencing. Wild-type virus at early times after infection is highly enriched for YY1 and strongly silenced; deletion of the YY1 binding site or YY1 KD reduces binding and silencing. In addition, the fact that YY1 does not bind the PBSproB2 viruses under any circumstances or at any time after infection explains the lack of effect on expression caused by the YY1 binding site deletion in the context of the PBSproB2 mutation.
We next probed the mechanism of YY1 silencing further by performing ChIP analysis for histone modifications on chromatin of the different proviruses. Our previous studies showed that the proviruses in embryonic cells were enriched for two silencing histone tail trimethylation modifications, H3K9me3 and H3K27me3, and that the silencing correlated well with the H3K9me3 mark and not with theH3K27me3 modification (Schlesinger and Goff, 2012). In agreement with that finding, our results here showed that depletion of the early YY1 binding through mutations of the U3 binding site or the PBS correlated with loss of H3K9me3 enrichment from the LTR (Fig 2E, G). The loss of H3K9me3 at early times was continued to late times for virus lacking the U3 binding site, even though after 14 days YY1 binding on the provirus was restored. Thus, the establishment of the mark was dependent on the early YY1 binding to the U3 site. In contrast to the findings with H3K9 modification, the H3K27me3 enrichment was similar in all times and on the different proviruses. All the ChIP numerical enrichment values are provided in Table S2.
Trim28 and YY1 interaction
To probe the interaction of YY1 with the silencing complex on the provirus, we examined protein-protein interactions with Trim28, the main regulator of retroviral silencing in embryonic cell. We immunoprecipitated YY1 from F9 and NIH3T3 nuclear cell extracts and tested for bound Trim28 by Western blot. The endogenous YY1 and Trim28 proteins indeed interacted strongly in F9 cells (Fig 3A) and E14 ES cells (Fig S3A). Although both proteins are present, almost no Trim28 was bound to YY1 in the differentiated NIH3T3 cell line (Fig 3A, see also Fig S3A,B,C), in Balb3T3, or E14 ES cell differentiated for 8 d (Fig S3A). These results indicate that the interaction of these proteins is regulated, and may reflect a mechanism for the specificity of retroviral silencing in embryonic cells. The embryonic cell-specific interaction is not dependent on the presence of ZFP809, as can be seen from the CoIP performed on ZFP809-KD cells (Fig S3B). In addition, treating the nuclear extracts with RNAse and DNAse before IP had no effect on the result (Fig S3C), suggesting that RNA or DNA bridges do not mediate the binding. A quantification of the enrichment values of all CoIPs as determined using LiCOR and fold difference between F9 and NIH3T3 cells are provided in Figure S3D.
Figure 3. Endogenous YY1 interacts with Trim28 and mediates deposition of chromatin and DNA methylation marks.
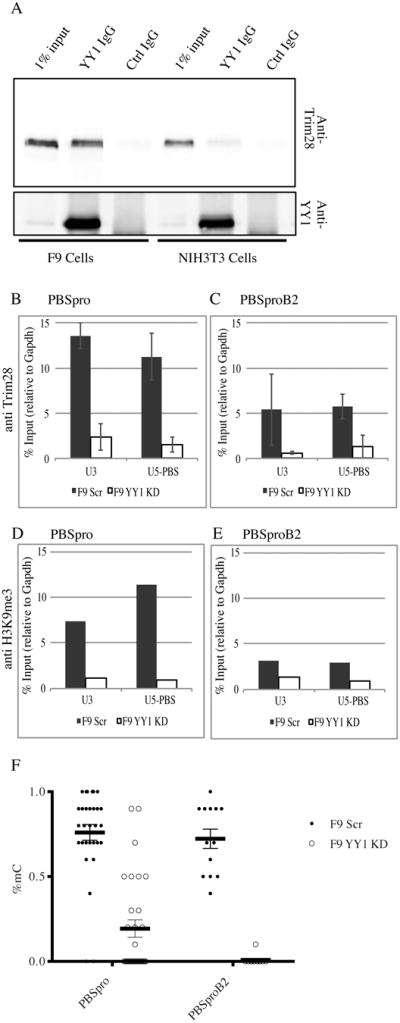
(A) Anti-YY1 immunoprecipitates from F9 and NIH3T3 nuclear extracts were analyzed by Western blot analysis with anti-Trim28 and anti-YY1 antibodies as indicated. Total proteins from 1% of the input to the immunoprecipitates are shown for comparison (for quantification see Fig S3D). (B, C) ChIP–based measurement of Trim28 at the viral U3 and U5-PBS regions on F9 YY1 KD cells and control F9 scrambled KD cells infected with MLV vectors containing either wt PBSpro or mutant PBSproB2 at 4 days after infection. The graphs show the mean enrichment ± SEM from 4 independent experiments relative to the total input samples and normalized to the signal of negative control (Gapdh). (D, E) ChIP assay for H3K9me3 on PBSpro or PBSproB2 vectors at 4 days after infection. All graphs show enrichment values relative to the total input samples and normalized to the signal of negative control (Gapdh). One experiment out of two is shown. (F) Bisulfite sequencing analysis of the 5′LTR of the infecting virus was performed on F9 YY1 KD pool and control F9 scrambled KD pool; percentages of methylated CpGs are shown for 10 to 20 cloned DNA molecules per cell and infection type (see Fig S4 for PBSpro data). Statistical significance determined using the HolmSidak method, with alpha=5.000%.
Chromatin and DNA modifications require YY1
To test whether Trim28 binding to the provirus in vivo is dependent on YY1, we carried out ChIP experiments on the F9 YY1 KD and Scr control lines with Trim28 and H3K9me3 antibodies. Both Trim28 and the H3K9me3 mark were highly enriched on the LTR of the wt PBSpro virus (at both the U3 and U5-PBS regions) in wild-type F9 cells, and these were both dramatically depleted in the YY1 KD cells (Fig 3B, D). Thus, YY1 was critically important in targeting the silencing machinery to the LTR. Trim28 was enriched at lower levels on the LTR of the PBSproB2 virus at both regions of wild-type F9 cells, and surprisingly, this enrichment was also critically dependent on YY1 expression (Fig. 3C). Enrichments on positive and negative control genes were seen as expected (Fig. S3E). The H3K9me3 mark was not enriched on the PBSproB2 virus (Fig. 3E), as seen previously (Fig. 2F).
Examination of the methylation status of the proviral DNA in wild-type cells revealed that there was no methylation at early times after infection but extensive methylation after 14 days (Fig 3F). However, proviral CpG methylation was decreased in the YY1KD cells two weeks (Fig 3F, and Fig S4A-E) and even four weeks after infection (data not shown), consistent with the long lasting effect of the YY1 KD on viral expression. Examination of the DNA methylation status of the Oct4 locus revealed a normal unmethylated status of undifferentiated cells (Fig S4F). These results suggest that YY1 is needed for the localization of Trim28, the marking of histones, and DNA methylation of proviral DNAs mediated by a stably DNA-bound silencing complex.
ERV silencing mediated by YY1
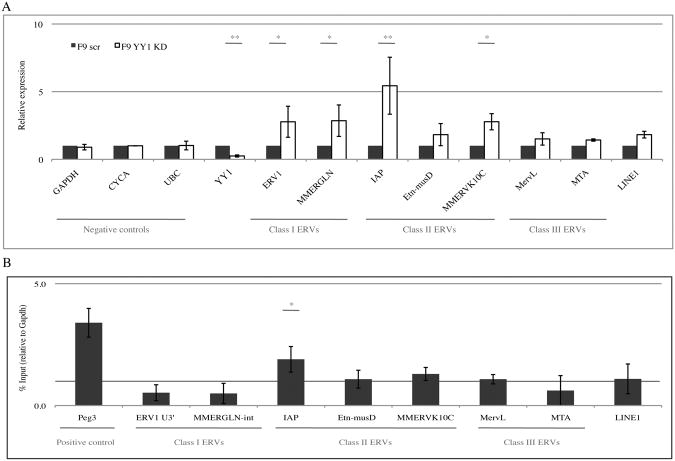
Endogenous retroviruses (ERVs) are members of the long terminal repeat (LTR)-containing transposable elements, constituting about 10% of the mouse genome. The ERVs can be divided into three classes based on sequence phylogeny: the class I (eMLVs), class II (intracisternal A-type particles or IAPs) and class III (MERVL). The expression of these elements is tightly regulated during embryonic development. Trim28 is fundamental for ERV silencing (Matsui et al. 2010; Rowe et al. 2010). This silencing is PBS-independent, and the sequence targeted by the silencing complex lies within the LTR (Rowe et al., 2010a). To determine if YY1 also plays a role in ERV silencing, we tested for changes in their expression levels in the YY1 KD cell line as compared to a Scrambled KD control line. RNA levels of 2-3 members of each of the ERV classes were analyzed by RT-qPCR and compared to three negative control genes (Fig 4A). ERV classes I and II were generally up regulated in the YY1 KD cells, while no significant change was seen in class III or in control genes. These results indicate an important role for YY1 in selected ERV silencing in embryonic cells. To test if this is attributable to YY1 binding to the ERVs LTRs we performed ChIP as described above, using previously described ERV-specific primers (Fig 4B). The results show significant enrichment of YY1 only for the IAPs, correlated with their expression being the most strongly up regulated by YY1 KD. Interestingly, IAPs are also the ERV group that showed the highest up regulation by Trim28 deletion in mouse ES cells and in early embryos (Rowe et al., 2010a). We found that Trim28 enrichment on the IAPs was lost in YY1 KD cells (Fig. S3E). Thus, we conclude that, together with Trim28, YY1 likely plays a role in the complex responsible for silencing ERVs, especially the class II IAPs. Rex1, another member of the YY1 family that was shown to bind and regulate ERVs, regulate primarily class III (Guallar at el., 2012), suggesting that these family members may each regulate a specific subset of ERVs.
Figure 4. YY1 Is required for silencing of some endogenous retroviruses in pluripotent cells.
(A) Quantitative RT-PCR expression analysis of different ERVs in YY1 KD cells relative to control scrambled KD cells. Levels of RNAs in F9 Scrambled KD cells were set to 1. Depletion of YY1 mRNA in F9 YY1 KD cells, and negative control genes are shown. The minus RT control values were below detection. The values are averages of three or more independent experiments ± SEM. One asterisk denotes P value < 0.05, two denote P < 0.01. (B) YY1 ChIP with primers specific for 7 different transposable elements, mainly ERVs. Positive control gene (Peg3) gave the expected results. Graphs show the mean enrichment ± SEM from 3 independent experiments relative to the total input samples and normalized to the signal of negative control (Gapdh).
Discussion
Embryonic cells suppress the expression of incoming and endogenous retroviruses. Here we propose that a Trim28-YY1-LTR complex is a key component of this silencing machinery. Mutating or knocking down each of these components results in decrease of viral restriction in embryonic cells. Manipulating the various components, however, leads to distinctive effects on the course of silencing. For example, deletion of the U3 YY1 binding site from the MMLV LTR had different effects than KD of YY1. At the initiation of silencing immediately after infection, eliminating the binding site results in no YY1 binding, the absence of histone H3K9me3 from the provirus, and impaired silencing. The U3 YY1 binding site is only significant in the initial recruitment of Trim28 by YY1 and ZFP809, and is not needed at later times. In contrast, reduction of the YY1 protein levels through RNAi knockdown results in a global, robust and more long-lasting effect on all retroviral vectors and at all times. Thus, we suggest that YY1 has a bimodal role in silencing: first, YY1 binds to the U3 binding site and together with ZFP809, recruits the silencing complex which mediates the chromatin modifications responsible for the PBSpro-dependent silencing. This immediate mode of action is probably important only for the silencing of an incoming MMLV infection. Second, the YY1 protein is part of the silencing complex bound to the LTR, acting independently of its U3 binding site, at all times after MMLV infection, and also acting on selected ERV families. This second mode seems to involve YY1 binding to the provirus but not only to its canonical binding site as has been shown in other settings (Alexandrov et al., 2012; Arvey et al., 2012). This mode of action of YY1 may also be operating in the silencing by ES cells of other promoters that lack obvious YY1 binding sites.
The requirement for both the U3 and PBSpro sites for full silencing, and for tethering of Trim28 and YY1 to both sites, suggests that the silencing complex is bound to DNA by two proteins: by ZFP809 binding to the PBSpro and by YY1 binding to U3. Trim28, in the form of a homotrimer, uses its RING-B box-coiled coil (RBCC) domain to bind to KRAB domain zinc finger proteins such as ZFP809 (Peng et al., 2000) and induce a long-range repression effect, through the spread of heterochromatin (Groner et al., 2010) and DNA methylation (Quenneville et al., 2012). Trim28 binding to many promoters, however, is RBCC independent, and is mediated by contacts with DNA binding proteins that, like YY1, do not have KRAB domains (Iyengar et al., 2011). The Trim28-ZFP809-YY1 complex could thus bind both U3 and PBS DNA sequences simultaneously, necessitating the formation of a large DNA loop between the sites. The loss of the YY1 from DNA with mutation of either site suggests that the high-affinity binding of the complex requires the simultaneous cooperative interaction with both sites.
As previously seen for retrovirus silencing (Karimi et al., 2011; Schlesinger and Goff, 2012), the early YY1 binding to viral DNA correlates with the H3K9me3 mark, but not the H3K27me3 mark. Mutation of the U3 YY1 binding site blocked both silencing and the associated H3K9me3 mark at early times, and H3K9 trimethylation was not established later, even when the later silencing was established. This observation suggests that at later times the silencing is mediated by other means, such as DNA methylation.
In YY1 KD cells, proviruses remained stably depleted of both histone and DNA methylation marks, correlating with stable expression of the reporter gene. This effect on DNA methylation is similar to the loss of methylation of endogenous and exogenous proviruses seen in Trim28 and ESET KO ES cells (Rowe et al., 2013). Thus YY1 may well be also involved in recruiting Trim28 and ESET and imposing the de novo DNA methylation in this setting. YY1 has been implicated in the DNA methylation associated with control of imprinted genes during early development (Kim et al., 2009), but its major role in controlling DNA methylation of exogenous retroviral DNAs was not previously known.
In YY1 KD cells, we observed upregulation of the IAPs and to a lesser degree other class I and II ERVs, the same ERV classes upregulated in Trim28 and ESET KO cells (Matsui et al., 2010b; Rowe et al., 2010b). These observations further reinforce the shared role and mode of action of these proteins in silencing exogenous and endogenous viruses. Many other cellular genes may also be affected by YY1 KD, and the changes in the levels of these gene products may indirectly mediate many of the changes in silencing that we observe.
The fact that YY1 and Trim28 are both general transcription regulators but achieve cell-type specificity is noteworthy. Both are extremely versatile factors that derive their versatility from combinatorial zinc finger usage (Coull et al., 2000; Iyengar and Farnham, 2011; Shi et al., 1997). YY1 can serve as transcriptional activator or repressor, and may have functions in imprinting and in immunoglobulin maturation. How YY1 protein achieves developmental, allelic, and mechanistic specificity has always aroused interest, given that the protein is ubiquitous and has a large repertoire of possible binding sites in the genome. The idea of combinatorial regulation through interaction with other cofactors gains further ground from the work presented here. We suggest that the major mechanism of action of YY1 is the tethering of Trim28 to the provirus, and that the limited developmental stage in which this interaction happens may be a key aspect of embryonic cell-specific silencing.
The biochemical basis for the cell type-specific YY1-Trim28 interaction is not known, but it could be controlled by phosphorylation, SUMOylation, or other modifications, or by bridging proteins. The phosphorylated form of Trim28 was shown to be important in the maintenance of pluripotency in ES cells (Hu et al., 2009; Seki et al., 2010). Trim28 is an E3 SUMO-protein ligase that undergoes autoSUMOylation, which is an important step for transcriptional repression (Ivanov et al., 2007). The roles that these modifications may play in the interaction of Trim28 with YY1 and in the proviral silencing, are yet unknown. Taken together, our data suggest that the interaction of Trim28 and YY1 may be important for the genetic stability of embryonic cells and thus necessary for differentiation.
Experimental Procedures
Cell culture and stable RNAi cell line production and transduction
Cells were cultured as described in Extended Experimental Procedures. RNAi knockdown was performed as in (Wolf and Goff, 2009). For shRNA sequences and detailed description see Extended Experimental Procedures, Viruses for transduction assays were prepared as described using pNCA-GFP vectors (Ooi et al., 2010). Each experiment was repeated 3 or more times.
Flow cytometry
GFP-positive cells data were acquired on an automated cell analyzer (LSR II; BD Bioscience) and analyzed with FlowJo software (Treestar). Percent of cells expressing GFP shown relative to percent NIH3T3 cells expressing GFP infected with the same virus in parallel.
Chromatin Immunoprecipitation (ChIP) was performed with Magna ChIP™ kit (Millipore) and DNA was purified using QIAquick PCR purification kit (Qiagen). Antibodies and primers are listed in Extended Experimental Procedures and Table S1.
EMSA
Nuclear extract were prepared as previously described (Wolf and Goff, 2007). Double-stranded DNA probes were end-labeled using LightShift Chemiluminescent EMSA kit (cat# 20148, Thermo Scientific). Binding reactions were performed as recommended by the manufacturer. For supershifts, antibody was added at same time as probe for 30 min. Binding reactions were analysed by electrophoresis on 10% native polyacrylamide gels.
Co-immunoprecipitations
Co-immunoprecipitations were performed after incubation of nuclear extracts (250 ug total protein) with rabbit anti-YY1 antibody or control antibody (4 ug; Santa Cruz Biotech) for 16h. Pre-washed protein A/G dynabeads were added to the lysates for 1h, and the bound proteins were eluted and analyzed by western blot. Detailed protocol and antibodies are presented in Extended Experimental Procedures.
RNA Extraction and RT-PCR protocols are presented in Extended Experimental Procedures.
Statistical analysis
Statistical evaluation was performed using Student's unpaired t test analysis of variance. Data presented as the mean ± SEM of 3 or more independent biological replicates, and p ≤ 0.05 was considered statistically significant.
Supplementary Material
Highlights.
Deletion of YY1 binding site from the MMLV LTR impairs rapid onset of silencing.
In YY1 KD cells, silencing of endogenous and exogenous retroviruses is impaired.
YY1 binding correlates with H3K9me3 histone marks but not with H3K27me3 marks.
YY1 interacts strongly with Trim28 in embryonic cells and less in differentiated cells.
Acknowledgments
This work was supported by NCI grant R37 CA 30488 and NYSTEM grant N08G-152/contract #C024329 from the New York State Department of Health. SPG is an Investigator of the Howard Hughes Medical Institute.
We thank for Prisma Lopez and Stella Chung for experimental assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandrov B, Fukuyo Y, Lange M, Horikoshi N, Gelev V, Rasmussen K, Bishop A, Usheva A. DNA breathing dynamics distinguish binding from nonbinding consensus sites for transcription factor YY1 in cells. Nucleic acids research. 2012;40:10116–10123. doi: 10.1093/nar/gks758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvey A, Agius P, Noble W, Leslie C. Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome research. 2012;22:1723–1734. doi: 10.1101/gr.127712.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison M, Basu A, Zaprazna K, Papasani M. Mechanisms of Yin Yang 1 in oncogenesis: the importance of indirect effects. Critical reviews in oncogenesis. 2011;16:143–161. doi: 10.1615/critrevoncog.v16.i3-4.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barklis E, Mulligan RC, Jaenisch R. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell. 1986;47:391–399. doi: 10.1016/0092-8674(86)90596-9. [DOI] [PubMed] [Google Scholar]
- Casa V, Gabellini D. A repetitive elements perspective in Polycomb epigenetics. Frontiers in genetics. 2012;3:199. doi: 10.3389/fgene.2012.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D, Jaenisch R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol. 2000;20:7419–7426. doi: 10.1128/mcb.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coull J, Romerio F, Sun J, Volker J, Galvin K, Davie J, Shi Y, Hansen U, Margolis D. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. Journal of virology. 2000;74:6790–6799. doi: 10.1128/jvi.74.15.6790-6799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J, Becker K, Ennist D, Gleason S, Driggers P, Levi B, Appella E, Ozato K. Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Molecular and cellular biology. 1992;12:38–44. doi: 10.1128/mcb.12.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J, Krieg A, Max E, Khan A. Negative control region at the 5′ end of murine leukemia virus long terminal repeats. Molecular and cellular biology. 1989;9:739–746. doi: 10.1128/mcb.9.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Denervaud N, Bucher P, Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS genetics. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guallar D, Pérez-Palacios R, Climent M, Martínez-Abadía I, Larraga A, Fernández-Juan M, Vallejo C, Muniesa P, Schoorlemmer J. Expression of endogenous retroviruses is negatively regulated by the pluripotency marker Rex1/Zfp42. Nucleic acids research. 2012;40:8993–9007. doi: 10.1093/nar/gks686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Margolis D. Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Molecular and cellular biology. 2002;22:2965–2973. doi: 10.1128/MCB.22.9.2965-2973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde-DeRuyscher R, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic acids research. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Farnham P. KAP1 protein: an enigmatic master regulator of the genome. The Journal of biological chemistry. 2011;286:26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Ivanov A, Jin V, Rauscher F, Farnham P. Functional analysis of KAP1 genomic recruitment. Molecular and cellular biology. 2011;31:1833–1847. doi: 10.1128/MCB.01331-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Goyal P, Maksakova I, Bilenky M, Leung D, Tang J, Shinkai Y, Mager D, Jones S, Hirst M, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell stem cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R, Jack-Scott E, Narezkina A, Palagin I, Boimel P, Kulkosky J, Nicolas E, Greger J, Skalka A. High-frequency epigenetic repression and silencing of retroviruses can be antagonized by histone deacetylase inhibitors and transcriptional activators, but uniform reactivation in cell clones is restricted by additional mechanisms. Journal of virology. 2007;81:2592–2604. doi: 10.1128/JVI.01643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kang K, Kim J. YY1's role in DNA methylation of Peg3 and Xist. Nucleic acids research. 2009;37:5656–5664. doi: 10.1093/nar/gkp613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes & development. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D, Dong K, Maksakova I, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager D, et al. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5718–5723. doi: 10.1073/pnas.1014660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T, Gifford W, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews S, Franco L, Rosenfeld M, Ren B, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes & development. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova I, Miyachi H, Kimura H, Tachibana M, Lorincz M, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010a;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010b;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Wolf D, Hartung O, Agarwal S, Daley GQ, Goff SP, Bestor TH. Dynamic instability of genomic methylation patterns in pluripotent stem cells. Epigenetics Chromatin. 2010;3:17. doi: 10.1186/1756-8935-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, Rauscher FJ., 3rd Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. Journal of molecular biology. 2000;295:1139–1162. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D. The KRAB-ZFP/KAP1 System Contributes to the Early Embryonic Establishment of Site-Specific DNA Methylation Patterns Maintained during Development. Cell reports. 2012;2:766–773. doi: 10.1016/j.celrep.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development (Cambridge, England) 2013;140:519–529. doi: 10.1242/dev.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard P, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010a;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Rowe H, Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411:273–287. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010b;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Satyamoorthy K, Park K, Atchison M, Howe C. The intracisternal A-particle upstream element interacts with transcription factor YY1 to activate transcription: pleiotropic effects of YY1 on distinct DNA promoter elements. Molecular and cellular biology. 1993;13:6621–6628. doi: 10.1128/mcb.13.11.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger S, Goff S. Silencing of proviruses in embryonic cells: efficiency, stability and chromatin modifications. EMBO reports. 2012 doi: 10.1038/embor.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Kurisaki A, Watanabe-Susaki K, Nakajima Y, Nakanishi M, Arai Y, Shiota K, Sugino H, Asashima M. TIF1beta regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10926–10931. doi: 10.1073/pnas.0907601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lee J, Galvin K. Everything you have ever wanted to know about Yin Yang 1. Biochimica et biophysica acta. 1997;1332:66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- Smith Z, Chan M, Mikkelsen T, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripathy S, Stevens J, Schultz D. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Molecular and cellular biology. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye J. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nature reviews Microbiology. 2012;10:395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- Teich N, Weiss R, Martin G, Lowy D. Virus infection of murine teratocarcinoma stem cell lines. Cell. 1977;12:973–982. doi: 10.1016/0092-8674(77)90162-3. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff S. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.