Abstract
Estrogen receptor (ER)-negative tumors represent 20–30% of all breast cancers, with a higher proportion occurring in younger women and women of African ancestry1. The etiology2 and clinical behavior3 of ER-negative tumors are different from those of tumors expressing ER (ER positive), including differences in genetic predisposition4. To identify susceptibility loci specific to ER-negative disease, we combined in a meta-analysis 3 genome-wide association studies of 4,193 ER-negative breast cancer cases and 35,194 controls with a series of 40 follow-up studies (6,514 cases and 41,455 controls), genotyped using a custom Illumina array, iCOGS, developed by the Collaborative Oncological Gene-environment Study (COGS). SNPs at four loci, 1q32.1 (MDM4, P = 2.1 × 10−12 and LGR6, P = 1.4 × 10−8), 2p24.1 (P = 4.6 × 10−8) and 16q12.2 (FTO, P = 4.0 × 10−8), were associated with ER-negative but not ER-positive breast cancer (P > 0.05). These findings provide further evidence for distinct etiological pathways associated with invasive ER-positive and ER-negative breast cancers.
ER-negative tumors are associated with a worse short-term prognosis3 and have weaker associations with reproductive risk factors2 than ER-positive tumors. There are also important differences in genetic susceptibility to these two types of tumors. BRCA1 mutations predispose primarily to ER-negative disease, whereas most known common susceptibility loci for breast cancer show stronger associations with ER-positive than with ER-negative tumors4. Exceptions are three loci tagged by rs10069690 on chromosome 5p15 (ref. 5) (TERT-CLPTM1L), rs8170 at 19p13 (ref. 6) (BABAM1, also known as MERIT40) and rs2284378 at 20q11 (ref. 7), which predispose primarily to ER-negative tumors, and loci at 6q25 (ref. ref. 8) that confer higher risk for ER-negative than for ER-positive tumors. With the aim of identifying susceptibility loci specific for invasive ER-negative disease, we analyzed three genome-wide association studies (GWAS) in populations of European ancestry and followed-up promising signals from each GWAS in the Breast Cancer Association Consortium (BCAC).
The 3 GWAS included a total of 4,193 ER-negative breast cancer cases and 35,194 controls of European ancestry drawn from 23 studies participating in the National Cancer Institute Breast and Prostate Cancer Cohort Consortium (BPC3), the Triple-Negative Breast Cancer Consortium (TNBCC) and the Combined BCAC ER-negative GWAS (C-BCAC) (Online Methods and Supplementary Table 1).We selected 13,276 SNPs on the basis of rank P values from the 3 GWAS, and these were genotyped in an independent set of 6,514 ER-negative cases and 41,455 controls of European ancestry from 40 BCAC studies forming part of the COGS Project (Online Methods and Supplementary Table 1). Samples were genotyped using the iCOGS custom Illumina Infinium array that included a total of 211,155 SNPs selected in collaboration with other cancer consortia (Online Methods). We performed a fixed-effects meta-analysis of odds ratio (OR) estimates from the GWAS and follow-up studies (quantile- quantile plot shown in Supplementary Fig. 1) and identified four loci newly associated with ER-negative disease at P < 5 × 10−8 (Fig. 1 and Table 1; cluster plots shown in Supplementary Fig. 2).
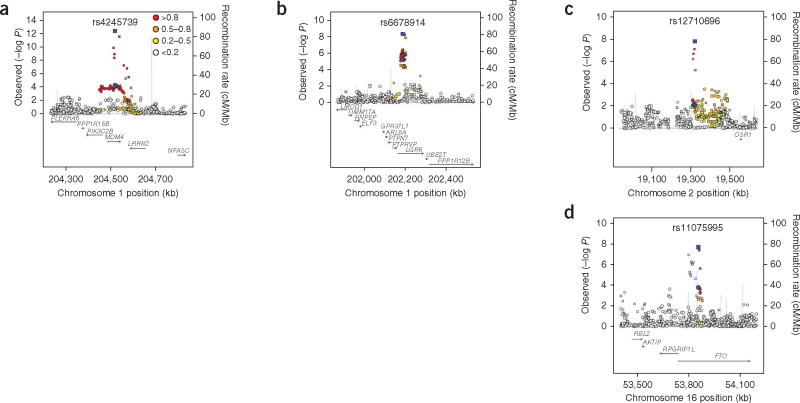
Figure 1.
Association and recombination plots. (a–d) Results are shown for the 1q32.1 (rs4245739; MDM4) (a), 1q32.1 (rs6678914; LGR6) (b), 2p24.1 (rs12710696) (c) and 16q12.2 (rs11075995; FTO) (d) loci in populations of European ancestry. Data from ER-negative breast cancer GWAS are plotted as circles; LD between each SNP and the top SNP (blue) is indicated by the color of the symbol. Estimates from the combined analysis of GWAS and BCAC replication data are plotted as squares, with the top SNP shown in blue. Recombination rates, plotted in light blue, are based on the HapMap CEU samples (Utah residents of Northern and Western European ancestry), and genomic coordinates are based on GRCh37 of the human genome.
Table 1.
Associations of SNPs and ER-negative breast cancer risk in populations of European ancestry
| SNP | Cytoband | Gene | Positiona | Stage | T/Ib | Studies | Cases | Controls | RAF | OR (95% CI) | P | Phet | I2 study het. (%)c |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs4245739 | 1q32.1 | MDM4 | 202785465 | GWAS | |||||||||
| BPC3 | I | 7 | 2,069 | 25,385 | 0.27 | 1.07 (0.97–1.17) | 0.177 | ||||||
| TNBCC | I | 11 | 1,562 | 3,399 | 1.20 (1.08–1.32) | 4.6 × 10-4 | |||||||
| C-BCAC | I | 5 | 562 | 6,410 | 0.28 | 1.17 (1.02–1.35) | 0.024 | ||||||
| Follow-up | |||||||||||||
| BCAC/iCOGS | T | 40 | 6,512 | 41,451 | 0.26 | 1.13 (1.09–1.18) | 8.5 × 10-9 | ||||||
| Meta-analysis | 63 | 10,705 | 76,645 | 0.26 | 1.14 (1.10–1.18) | 2.1 × 10-12 | 0.413 | 3.2 | |||||
| GWAs | |||||||||||||
| rs6678914 | 1q32.1 | LGR6 | 20045399 | BPC3 | I/T | 7 | 2,069 | 25,385 | 0.59 | 1.12 (1.03–1.22) | 0.007 | ||
| TNBCC | T | 11 | 1,562 | 3,399 | 0.59 | 1.16 (1.05–1.27) | 0.003 | ||||||
| C-BCAC | I/T | 5 | 562 | 6,410 | 0.59 | 1.15 (1.01–1.30) | 0.032 | ||||||
| Follow-up | |||||||||||||
| BCAC/iCOGS | T | 40 | 6,514 | 41,452 | 0.59 | 1.08 (1.04–1.12) | 1.8 × 10−4 | ||||||
| Meta-analysis | 63 | 10,707 | 76,646 | 0.59 | 1.1 (1.06–1.13) | 1.4 × 10-8 | 0.481 | 0.0 | |||||
| rs12710696 | 2p24.1 | Non-genic | 19184284 | GWAs | |||||||||
| BPC3 | I | 7 | 2,069 | 25,385 | 0.37 | 1.05 (0.96–1.14) | 0.304 | ||||||
| TNBCC | I | 11 | 1,562 | 3,399 | 1.17 (1.06–1.29) | 0.001 | |||||||
| C-BCAC | I | 5 | 562 | 6,410 | 0.37 | 1.00 (0.88–1.14) | 0.947 | ||||||
| Follow-up | |||||||||||||
| BCAC/iCOGS | T | 40 | 6,512 | 41,453 | 0.36 | 1.10 (1.06–1.15) | 1.4 × 10-6 | ||||||
| Meta-analysis | 63 | 10,705 | 76,647 | 0.36 | 1.1(1.06–1.13) | 4.6 × 10-8 | 0.464 | 0.0 | |||||
| rs11075995 | 16q12.2 | KIAA1752-FTO | 52412792 | GWAs | |||||||||
| BPC3 | I | 7 | 2,069 | 25,385 | 0.24 | 1.15 (1.04–1.28) | 0.008 | ||||||
| TNBCC | I | 11 | 1,562 | 3,399 | 1.15 (1.03–1.28) | 0.010 | |||||||
| C-BCAC | I | 5 | 562 | 6,410 | 0.24 | 1.09 (0.92–1.28) | 0.328 | ||||||
| Follow-up | |||||||||||||
| BCAC/iCOGS | T | 40 | 6,513 | 41,453 | 0.24 | 1.10 (1.05–1.15) | 4.2 × 10-5 | ||||||
| Meta-analysis | 63 | 10,706 | 76,647 | 0.24 | 1.11 (1.07–1.15) | 4.0 × 10-8 | 0.079 | 24.3 | |||||
Results are shown for the SNPs showing the strongest association in four loci reaching association P < 5 × 10−8 in meta-analyses of GWAS and follow-up data. RAF, risk allele frequency; freq., frequency.
NCBI Build 36.
Imputed (I) and typed (T) SNPs: rs6678914 was typed in one BPC3 study (WGHS), three C-BCAC studies (ABCFS, SASBCAC, UK2) and all TNBCC studies and imputed in allother GWAS studies.
Result of Q test for heterogeneity of estimated ORs.
Two independently associated loci were located on chromosome 1q32.1 and were tagged by two uncorrelated (r2 < 0.001) markers (from reference sequence NCBI Build 36): rs4245739 (P = 2.1 × 10−12, OR = 1.14, 95% confidence interval (CI) = 1.10–1.18) and rs6678914 (P = 1.4 × 10−8, OR = 1.10, 95% CI = 1.06–1.13). Conditional analyses of the two SNPs in BCAC follow-up data showed comparable estimates, indicating that these are two distinct signals (Supplementary Table 2). The other two loci were located at 2p24.1 (rs12710696,P = 4.6 × 10−8, OR = 1.10, 95% CI = 1.06–1.13) and 16q12.2 (rs11075995, P = 4.0 × 10−8, OR = 1.11, 95% CI = 1.07–1.15). For each region, there was little evidence for heterogeneity of effect by study (Table 1 and Supplementary Fig. 3a–d), and genotype-specific risks for rs4245739, rs6678914 and rs12710696 were consistent with a log-additive model. For rs11075995, departure from a log-additive model was significant (P = 0.039), and genotype-specific estimates suggested a recessive effect (Supplementary Table 3).
The strength of the association for each SNP differed significantly by ER status, and none of the SNPs showed significant associations with ER-positive disease in the analysis of 25,227 ER-positive cases and 41,455 controls of European ancestry in BCAC (Supplementary Tables 4 and 5). Notably, we observed no significant differences in ORs for ER-negative tumors with and without the triple-negative phenotype (defined as ER-negative, progesterone receptor (PR)-negative and HER2-negative) for rs6678914 (1q32.1, LGR6), rs12710696 (2p24.1) and rs11075995 (16q12.2). However, rs4245739 (1q32.1, MDM4) seemed to be specific to triple-negative tumors (case-only heterogeneity P value (Phet) by triple-negative status = 0.005;Supplementary Table 5).
None of the four SNPs showed significant (P < 0.05) associations in studies of Asian ancestry in BCAC, and only the 16q12.2 (FTO) variant was associated at P = 0.05 in combined analyses of studies of African-American ancestry in BCAC and the African-American Breast Cancer Consortium5 (AABC; Supplementary Table 6). However, estimates for Asian and African-American populations were not significantly different from those in Europeans (P > 0.05), and larger studies in these populations are needed to determine whether risk associations exist. None of the markers were significantly associated with increasing age at the onset of ER-negative disease in the BCAC follow-up data (Ptrend ≥ 0.314), although there were some differences in age-specific estimates (Supplementary Table 7). Furthermore, OR estimates were not significantly different for women with and without a family history of any breast cancer in at least one first-degree relative, and risk alleles were not over-represented in cases with a positive family history (Supplementary Table 8).
rs4245739 (1q32.1) is located in the 3’ region of the MDM4 oncogene. MDM4 is a repressor of TP53 and TP73 transcription and is important for cell cycle regulation and apoptosis. rs4245739 resides in a linkage disequilibrium (LD) block of approximately 230 kb (Supplementary Fig. 4a) that also contains the tRNALys transcript and the genes PIK3C2B and LRRN2 (Supplementary Fig. 5a). MDM4, tRNALys and PIK3C2B but not LRRN2 are expressed in normal breast epithelium, breast cancer cell lines and breast tumors9–11.There are no nonsynonymous SNPs correlated with rs4245739 in the 1000 Genomes Project populations of European ancestry (r2 > 0.10); however, correlated SNPs are located in the promoter region of PIK3C2B (rs3014606, r2 = 0.94 and rs2926534, r2 = 0.94) and in the tRNALys transcript (rs11240753, r2 = 0.78 and rs4951389, r2 = 0.78). Variants in the MDM4 locus correlated with rs4245739 have also been associated with breast cancer in BRCA1 mutation carriers who have predominantly ER-negative tumors12. Thus, this region seems to be specifically associated with ER-negative disease and not with overall breast cancer risk, as suggested by a previous, smaller candidate gene study13. To our knowledge, no studies before the COGS collaboration have evaluated rs4245739 in relation to the risk of ER-negative disease.
rs6678914 on chromosome 1q32.1 is located in intron 1 of the LGR6 gene (Supplementary Fig. 4b). LGR6 and several other genes in this region, including UBE2T and PTPN7, are expressed in breast tumors9.A correlated SNP (rs12032424, r2 = 0.96) is located in a putative enhancer region in the same intron of LGR6 in normal breast epithelial cells, although not in the triple-negative breast cancer cell line MDA-MB-231 (Supplementary Fig. 5b). The rs6678914 SNP is not correlated with nonsynonymous SNPs in LGR6 (r2 > 0.10 in 1000 Genomes Project populations of European ancestry).
The SNP rs12710696 on chromosome 2p24.1 is located in an intergenic region, more than 200 kb from the nearest gene (OSR1) (Supplementary Fig. 4c). It is possible that the allele marked by rs12710696 could influence a set of active enhancers, as the region contains multiple overlapping chromatin marks in normal breast epithelial cells and the MDA-MB-231 triple-negative breast cancer cell line (Supplementary Fig. 5c).
The signal found on chromosome 16q12.2 is located in the fat mass– and obesity-associated gene FTO (Supplementary Fig. 4d). This signal is tagged by rs11075995, located in a ~40-kb LD block in intron 1 of FTO, within an enhancer region that appears to be active in both normal and triple-negative breast cancer cells (Supplementary Fig. 5d). rs11075995 is located ~40 kb distal to a region in intron 1 that contains multiple SNPs associated with obesity in the Genetic Investigation of ANthropometric Traits (GIANT) Consortium14,15, as well as a SNP associated with overall breast cancer risk (rs17817449)8. rs11075995 is not correlated with any of the previously reported SNPs associated with obesity at genome-wide significant levels in GIANT or with rs17817449 (P = 3.7 × 10−60, based on 123,864 subjects in GIANT; ref. 15). However, rs11075995 is associated with body mass index (BMI), both in GIANT (P = 1.51 × 10−6, based on 121,427 subjects) and our control population (P = 2.8 × 10−5, based on 20,952 controls in iCOGS; data not shown). Analyses adjusting and stratifying by BMI on the basis of 3,071 ER-negative cases and 20,130 controls from 19 studies genotyped on the iCOGS array indicated that the association between rs11075995 and ER-negative disease is not explained or modified by our measure of BMI (BMI-adjusted OR = 1.16, 95% CI = 1.09–1.24, P = 1.1 × 10−5; Pinteraction = 0.912; data not shown). Furthermore, conditional analyses indicated that the ER-negative disease–specific signal (rs11075995) is independent of rs17817449 (Supplementary Table 2). This finding adds to the increasing evidence of distinct signals at the same locus for different subtypes of cancers occurring at the same site, including, for example, 5p15.33 (TERT-CLPTM1L)16 and 14q24.1 (RAD51B, also known as RAD51L1)8 in breast cancer and 5p15.33 (TERT-CLPTM1L)16 and HNF1B17 in ovarian cancer. Detailed fine mapping of known and newly identified breast cancer–associated regions will be required to determine whether additional subtype-specific signals exist in these regions.
In an attempt to investigate the likely genes responsible in the observed risk associations, we examined associations between SNPs with available genotype (rs4245739, rs12710696 and rs6678914) and RNA expression in data from 382 primary breast tumors, including 81 ER-negative samples in The Cancer Genome Atlas (TCGA) database. None of the associations were significant after Bonferroni adjustment for multiple comparisons, whether considering only the immediately neighboring genes or all genes within a 1-Mb window of the lead SNP (data not shown).
To provide a comprehensive analysis of common genetic loci for ER-negative breast cancer, we also evaluated associations between 67 known loci for overall breast cancer risk (26 previously reported and 41 newly identified8) and ER-negative disease. On the basis of our meta-analysis of 10,707 ER-negative cases and 76,649 controls, 7 regions influenced risk of ER-negative disease at P < 5 × 10−8: 1p36.22 (PEX14), 5p15 (TERT-CLPTM1L), 2 independent loci at 6q25.1 (ESR1), 12p11.22 (PTHLH), 16q12.1 (TOX3) and 19p13.1 (BABAM1) (Supplementary Table 9). Only seven loci identified so far, the four reported here and the three previously reported located at 5p15 (ref. 5), 19p13.1 (ref. 6) and 20q11 (ref. 7), are specific to ER-negative disease.
In summary, our analyses provide further evidence for distinct etiological pathways for invasive ER-positive and ER-negative breast cancers. Fine mapping and functional studies of the susceptibility loci for ER-negative disease should provide important insights into the biological mechanisms of ER-negative breast cancer, potentially leading to the identification of new targets for therapy and prevention of this aggressive form of breast cancer.
ONLINE METHODS
ER-negative breast cancer GWAS
Three GWAS of ER-negative breast cancer were conducted in populations of European ancestry by National Cancer Institute (NCI) BPC3 (refs. 7,18), TNBCC5,6 and C-BCAC.
ER-negative status for BPC3 and C-BCAC cases was determined from review of medical records or state cancer registry information. TNBCC focused on triple-negative cases, defined as individuals with ER-negative, PR-negative and HER2-negative breast cancer using data from medical records5,6. The BPC3 GWAS included 2,188 ER-negative cases and 26,477 controls from 8 studies (CPSII, EPIC, MEC, NHS, NHSII, PLCO, PBCS and WGHS), geno-typed using different versions of Illumina SNP arrays7,18. A total of 1,718 triple-negative cases from 11 studies (ABCTB, BBCC, DFCI, FCCC, GENICA, HEBCS, MARIE, MCBCS, MCCS, POSH and SBCS) were genotyped for the TNBCC GWAS using Illumina SNP arrays5. Data for TNBCC controls (N = 3,670) were obtained from a Finnish study (HEBCS) and publicly available controls of European ancestry from the United States (CGEMS), Germany (KORA), Australia (QIMR) and the UK (Wellcome Trust Case Control Consortium 2, WTCCC2) genotyped using Illumina arrays5. Samples from the four latter studies are not counted in the total number of TNBCC studies because they only provided controls for other studies. C-BCAC performed a meta-analysis of 9 GWAS that included data on 10,052 breast cancer cases and 12,575 controls8. Five studies (ABCFS, MARIE, HEBCS, SASBAC and UK2) provided data on ER status from medical records or cancer registries and contributed data on 702 ER-negative cases and 7,713 controls of European ancestry. All C-BCAC studies were genotyped with versions of Illumina arrays. Control data for C-BCAC were obtained from individual studies or publicly available data.
Standard genotyping quality control procedures were performed for each GWAS as previously described5,7,8. Estimated per-allele log(OR) and standard error were calculated for each SNP using unconditional logistic regression on allele counts (dosages), as implemented in ProbABEL19. Analyses were adjusted by study, country of origin or principal components as previously described5,7,8. Analyses assumed a log-additive genetic model, and P values were based on the 1-degree-of-freedom Wald test. Quantile-quantile plots from each GWAS showed no substantial evidence for cryptic population substructure or differential genotype calling between cases and controls. The estimated inflation factor (λ) was 1.02 for BPC3 (ref. 7), 1.04 for TNBCC6 and 0.98 for C-BCAC (Supplementary Fig. 1).
SNPs were selected for the iCOGS custom genotyping array separately by each participating group (see details in Michailidou et al.8). BPC3 nominated independent SNPs with a 1-degree-of-freedom log-additive trend test P < 0.02 or with P < 0.02 for one of several auxiliary tests, including tests for dominant or recessive effects of the minor allele and case-only tests comparing PR-positive to PR-negative tumors. SNPs from C-BCAC were selected on the basis of the 1-degree-of-freedom trend test for ER-negative disease. TNBCC nominated SNPs on the basis of log-additive trend test P < 0.01. Subsequent analyses that combined OR estimates across GWAS and follow-up samples only included SNPs that had been directly genotyped on the iCOGS array and had passed genotyping quality control. SNPs successfully genotyped on iCOGS but not included on the chips used for the GWAS were imputed within each GWAS before combining results with iCOGS data. Imputation was performed within each study and genotyping array using the HapMap Phase 2 CEU reference panel and MACH software package v1.0. SNPs with low imputation quality (r2 < 0.3) or minor allele frequency (MAF) < 1% were excluded.
iCOGS genotyping
Samples for follow-up analyses were drawn from 50 studies participating in BCAC (40 from populations of predominantly European ancestry (including CTS, DEMOKRITOS, NBCS, NBHS, OSUCCG, RPCI and SKKDKFZS from TNBCC), 9 of Asian ancestry and 1 of African-American ancestry) with information on ER status. Most breast cancer cases in BCAC studies have not been tested for BRCA1 mutations; however, the frequency of mutations in the studied populations is expected to be low. Samples were genotyped as part of the COGS Project using a custom Illumina Infinium array (iCOGS) at four genotyping centers (Supplementary Table 1). The most common source of data for ER, PR and HER2 status was medical records, followed by immunohistochemistry performed on tumor tissue microarrays (TMAs) or whole-section tumor slides. Breast cancer cases in the BCAC follow-up with missing data on ER status and cases from one study (PBCS) that included only ER-positive cases are excluded from this report. Studies were required to provide ~2% of samples in duplicate.
The iCOGS chip included a total of 211,155 SNPs selected in collaboration with other consortia of BRCA1 and BRCA2 mutation carriers (CIMBA), ovarian cancer (OCAC) and prostate cancer (PRACTICAL). Genotype calling and quality control analyses were conducted by a single analysis center at the University of Cambridge 8. A total of 13,276 SNPs proposed by the combined ER-negative GWAS yielded high-quality genotype data (5,738 from BPC3, 4,628 from TNBCC and 2,910 from C-BCAC).
Statistical analysis
After quality control exclusions8, BCAC follow-up data were analyzed using the Genotype Library and Utilities (GLU) package to estimate per-allele ORs and standard errors for each SNP using unconditional logistic regression. Analyses were stratified by ancestry (European, Asian or African). For samples of European ancestry, BCAC follow-up analyses were adjusted for seven principal components (the first six plus an additional component to reduce inflation for the LMBC study).
GWAS and BCAC follow-up results were combined using inverse variance–weighted fixed-effects meta-analysis, as implemented in METAL20. Forest plots showing study-specific estimates and fixed-effects meta-analysis for SNPs showing genome-wide significance were drawn using the command metan in STATA v.12. Samples that overlapped among the three GWAS and the BCAC follow-up were identified by concordance of genotypes and removed from either the GWAS or follow-up data set before this analysis so that each data set contributing to the meta-analysis was independent of the others (see Supplementary Table 1 for the counts of case and control included in the analyses after removing overlapping samples). Heterogeneity by study was evaluated using the Q statistic.
Analyses in this report focused first on the 13,276 SNPs proposed by the ER-negative breast cancer GWAS. For SNPs showing evidence of association with ER-negative breast cancer at P < 1 × 10−6, we also evaluated correlated SNPs in the rest of COGS and reported on the most significant SNP in the region. For the regions that reached genome-wide statistical significance (P < 5 × 10−8), we performed additional analyses examining heterogeneity in the associated effect by tumor type and subject characteristics using the most significant SNP in the region. The associations between these SNPs and ER-positive breast cancer were assessed using 25,227 ER-positive cases of European ancestry in BCAC who had been genotyped as part of the COGS Project. Differences in the strength of the associations with ER-positive and ER-negative breast cancers were assessed using case-only analyses (Supplementary Table 5). Stratum-specific estimates of per-allele OR by categories of age and family history of disease were obtained from logistic regression models (Supplementary Tables 6 and 7), and differences in ORs across strata were tested using an ordinal-product interaction term.
We also assessed associations between the most significant markers and ER-negative breast cancer in Asian and African-American populations. The Asian-ancestry analyses included 1,547 ER-negative cases and 6,624 controls in 9 studies from BCAC. The African-American analyses included 91 ER- negative cases and 252 controls in 1 BCAC study and 988 ER-negative cases and 2,745 controls in 9 studies from AABC5 (Supplementary Table 1). Both the Asian-ancestry and African-American analyses adjusted for the first two principal components of genetic variation, calculated separately in each ancestry group. Differences by ancestry were tested by a χ2 test comparing summary ORs across the three ancestry groups.
Bioinformatics
In an attempt to identify functionality in regions of interest, we used the open-source R/Bioconductor package FunciSNP version 0.1.14 (Functional Integration of SNPs)21 (S.K.R., S.G. Coetzee, H. Noushmehr, C. Yan, J.M. Kim et al., unpublished data), which systematically integrates 1000 Genomes Project SNP data (June 2011 data release) with chromatin features of interest. For each of the four newly associated ER-negative breast cancer markers we analyzed all SNPs within a 1-Mb window that were in LD (r2 > 0.5) with the index marker (according to the 1000 Genome Project CEU panel). We assessed whether these SNPs colocalized with 13 different chromatin features that capture open chromatin regions and enhancers across the genome, using data generated by next-generation sequencing technologies. Information on open chromatin states (H3K9ac and H3K14ac), nucleosome-depleted regions (DNase I and FAIRE), enhancers (H3K4me1) and active/engaged enhancers (H3K27ac) was either generated by the Coetzee Laboratory (S.K.R. et al., unpublished data) or harvested from the Encyclopedia of DNA Elements (ENCODE) Project. All chromatin features were identified in normal human mammary epithelial cells (HMECs) and triple-negative breast cancer cells (MDA-MB-231). We used the UCSC Genome Browser (see URLs) with potentially functional SNPs identified using FunciSNP and chromatin features tracks to generate images (Supplementary Fig. 5).
Ethics
All women in participating studies provided written consent for the research, and approval for the study was obtained from the local ethical review board relevant to each institution. Collection of blood samples and clinical data from subjects was performed in accordance with local guidelines and regulations.
Supplementary Material
Acknowledgments
The authors wish to thank all the individuals who took part in these studies and all the researchers, clinicians and administrative staff who have enabled this work to be carried out. We are very grateful to Illumina, in particular J. Stone, S. McBean, J. Hadlington, A. Mustafa and K. Cook, for their help with designing the array. BCAC is funded by Cancer Research UK (C1287/A10118 and C1287/A12014) and by the European Community’s Seventh Framework Programme under grant agreement 223175 (HEALTH-F2-2009-223175) (COGS). Meetings of BCAC have been funded by the European Union European Cooperation in Science and Technology (COST) programme (BM0606). BPC3 is funded by US National Cancer Institute cooperative agreements U01-CA98233, U01-CA98710, U01-CA98216 and U01-CA98758 and the Intramural Research Program of the US National Institutes of Health (NIH)/National Cancer Institute, Division of Cancer Epidemiology and Genetics. TNBCC is supported by Mayo Clinic Breast Cancer Study (MCBCS) (US NIH grants CA122340 and a Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201)), grants from the Komen Foundation for the Cure and the Breast Cancer Research Foundation. Genotyping on the iCOGS array was funded by the European Union (HEALTH-F2-2009-223175), Cancer Research UK (C1287/A10710), US NIH grant CA122340, the Komen Foundation for the Cure, the Breast Cancer Research Foundation, the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer program (J. Simiard and D.E.) and Ministry of Economic Development, Innovation and Export Trade of Quebec grant PSR-SIIRI-701 (J. Simiard, D.E. and P.H.). J. Simiard holds the Canada Research Chair in Oncogenetics. Combination of the GWAS data was supported in part by US NIH Cancer Post-Cancer GWAS initiative grant U19 CA 148065-01 (DRIVE, part of the GAME-ON initiative) and Breakthrough Breast Cancer Research.
Footnotes
URLs. BCAC,http://www.srl.cam.ac.uk/consortia/bcac/index.html; CIMBA, http://www.srl.cam.ac.uk/consortia/cimba/index.html/; COGS, http://www.cogseu.org/; GIANT, http://www.broadinstitute. org/collaboration/giant/index.php/GIANT_consortium; OCAC,http://www.srl.cam.ac.uk/consortia/ocac/index.html; PRACTICAL, http://www.srl.cam.ac.uk/consortia/practical/index.html; TCGA, http://www.cancergenome.nih.gov/; 1000 Genomes Project, http://www.1000genomes.org/; GLU (Genotype Library and Utilities), http://code.google.com/p/glu-genetics/; UCSC Genome Browser, http://genome.ucsc.edu/.
Accession codes. Reference sequences for the human genome of the regions containing the following genes are available at NCBI under the indicated accessions: LRRN2, NC_000001.10; UBE2T, NC_000001.10; PTPN7, NC_000001.10; PEX14, NC_000001.10; LGR6, NC_000001.10; MDM4, NC_000001.10; TP73, NC_000001.10; PIK3C2B, NC_000001.10; OSR1, NC_000002.11; TERT, NC_000005.9; CLPTM1L, NC_000005.9; ESR1, NC_000006.11; PTHLH, NC_000012.11; RAD51B, NC_000014.8; FTO, NC_000016.9; TOX3, NC_000016.9; HNF1B, NC_000017.10; BRCA1, NC_000017.10; TP53, NC_000017.10; MERIT40, NC_000019.9.
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
M.G.-C., F.J.C., S.L., K. Michailidou, M.K.S., P.D.P.P., C.V., D.F.E., C.A.H. and P. Kraft formed the writing group and drafted the manuscript. M.G.-C. coordinated the writing group. F.J.C., S.L., K. Michaildou, D.F.E., C.A.H. and P. Kraft performed statistical analyses of GWAS data. M.G.-C. and M.N.B. performed statistical analyses of BCAC follow-up studies and meta-analyses. P. Kraft coordinated the BPC3 GWAS, and M.G.-C., E.R., H.S.F., L.L.M., J.E.B., W.C.W., D.J.H. and S.J.C.led individual studies in the BPC3 scan. F.J.C. and C.V. coordinated the TNBCC GWAS, and D.E., P. Miron, P.A.F., J.C.-C., J.C., A.A., H.N., H. Brauch and G.G.G. led individual studies in the TNBCC scan. D.E. coordinated the C-BCAC GWAS, and H.N., J.L.H., J.C.-C. and P.H. led individual studies in the C-BCAC scan. D.F.E. conceived and coordinated the synthesis of the iCOGS array and led BCAC. P.H. coordinated COGS, and J.B. led the BCAC genotyping working group. A.G.-N., G.P., M.R.A., D.V., F.B., D.C.T. and F.J.C. coordinated genotyping of the iCOGS array. M.G.-C., P.D.P.P. and M.K.S. led the pathology working group in BCAC. M.E.S. was the lead pathologist in BCAC. W.J.H. performed automated scoring of tissue microarrays. A.M.D. and G.C.-T. led the quality control working group.J.D. and N.O. provided bioinformatics support. S.K.R. and G.A.C. performed FunciSNP bioinformatics analyses. M.K.B. and Q. Wang provided data management support for BCAC. G.G., A.A., A. Broeks, A.B.E., A.C., U.H., A.-S.D., A.G.U., A.H., A.H.W., A.I., the ABCTB Investigators, A.J.-V., A.J., A.K.G., R.W., A. Lindblom, A. Lophatananon, A.M.D., A.M.M., A.M.W.v.d.O., A.R., A. Swerdlow, A. Schneeweiss, B.B., B.E.H., B.G.N., B.M.-M., B.P., C.B., C.B.A., C.-Y.C., C.C., C.D.B., C.-N.H., C.H.M.v.D., C.H.Y., C.J., C.M., C.M.S., C.O., C.R., C.-Y.S., C. Sohn, C. Stegmaier, C.-C.T., C.T., C.W.C., D.C., D.C.T., D.F.-J., D.G., D.I.C., D.J.P., D.J.S., D.K., D.L., D.O.S., D.S., D.T., D.V.D.B., E.D., C.V., E.J.R., E.J.S., E.M., E.M.J., E.V.B., E.W., F.A., FBCS, F.C.-C., F.C., F.H., F.L., F.M., F.R., F.S., G.A.C., G.C.-T., G.K.C., G.S., G.W.M., H.A.-C., H.C., H.F., H. Ito, H. Iwata, H. Müller, H. Miao, H.M.-H., H.P., H.T., H.W., I.d.S.S., I.K., I.L.A., I.T., J.A.K., J.D.F., J.E.O., J.I.A.P., J.J.H., J. Long, J. Lubinski, J. Liu, J. Lissowska, J.L.R.-G., J.M.H., J.P., J. Stone, J. Simard, J.W., J.-C.Y., K. Aittomäki, K. Aaltonen, K.C., K.D., K.J., K.-T.K., K.L., K. Muir, K. Matsuo, K.P., K.S., K.S.C., L. Bernard, L. Baglietto, L. Bernstein, L. Beckmann, L.D., L.G., L.J.V.V., L.N.K., L.S., M.B., M.C.S., M.D., M.F.P., M.G.S., M. Jones, M. Johansson, M.J.H., M.J.K., M.K., M.K.B., M.L., M.M.G., M.P.L., M. Shrubsole, M. Shah, M.W.B., M.W.R.R., N.A.M.T., N.D., N.G.M., N.J., N.M., N.N.A., N.R., N.S., N.V.B., O.F., P.G., P.H., P.H.P., P. Kerbrat, P.L.-P., P.L., P. Menénde, P.N., P.P., P.R., P. Siriwanarangsan, P. Sharma, P.-E.W., Q.C., Q. Wang, Q. Waisfisz, R.B., R.G.Z., R.H., R.K., R.K.S., R.L.M., R.M.M., R.N.H., R.P., R.A.E.M.T., R. Tumino, R. Travis, S.A.I., S.E.B., S.E.H., S.F.N., S.G., S.H.T., S.K., S.K.R., S.L.D.-H., S.M., S.M.J., S. Nickels, S. Nyante, S.P.B., S. Sangrajrang, S.S.-B., S. Slager, S.S.C., T.A.M., T.B., T.D., T.H., T.T., V.A., V. Kristensen, V. Kataja, V.-M.K., W.B., W.L., W.R.D., W.T., X.-O.S., X.W., Y.F., Y.-T.G., Y.-D.K. and Y.Y. contributed to GWAS and/or BCAC follow-up studies. M.G.-C., F.J.C., S.L., K. Michailidou, M.K.S., P.D.P.P., C.V., D.F.E., C.A.H., P. Kraft, M.N.B., E.R., H.S.F., L.L.M., J.E.B., W.C.W., D.J.H., S.J.C., D.E., P.A.F., J.C.-C., J.C., A. Broeks, H.N., H. Brauch, H. Brenner, G.P., G.G.G., J.L.H., P. Miron, J.B., A.G.-N., M.R.A., D.V., F.B., M.E.S., W.J.H., G.G., A.A., A. Beck, A.B.E., A.C., U.H., A.-S.D., A.G.U., A.H., A.H.W., A.I., the ABCTB Investigators, A.J.-V., A.J., A.K.G., R.W., A. Lindblom, A. Lophatananon, A.M.D., A.M.M., A.M.W.v.d.O., A.R., A. Swerdlow, A. Schneeweiss, B.B., B.E.H., B.G.N., B.M.-M., B.P., C.B., C.B.A., C.-Y.C., C.C., C.D.B., C.-N.H., C.H.M.v.D., C.H.Y., C.J., C.M., C.M.S., C.O., C.R., C.-Y.S., C. Sohn, C. Stegmaier, C.-C.T., C.T., C.W.C., D.C., D.C.T., D.F.-J., D.G., D.I.C., D.J.P., D.J.S., D.K., D.L., D.O.S., D.S., D.T., D.V.D.B., E.D., C.V., E.J.R., E.J.S., E.M., E.M.J., E.V.B., E.W., F.A., FBCS, F.C.-C., F.C., F.H., F.L., F.M., F.R., F.S., G.A.C., G.C.-T., G.K.C., G.S., G.W.M., H.A.-C., H.C., H.F., H. Ito, H. Iwata, H. Müller, H. Miao, H.M.-H., H.P., H.T., H.W., I.d.S.S., I.K., I.L.A., I.T., J.A.K., J.D., J.D.F., J.E.O., J.I.A.P., J.J.H., J. Long, J. Lubinski, J. Liu, J. Lissowska, J.L.R.-G., J.M.H., J.P., J. Stone, J. Simard, J.W., J.-C.Y., K. Aittomäki, K. Aaltonen, K.C., K.D., K.J., K.-T.K., K.L., K. Muir, K. Matsuo, K.P., K.S., K.S.C., L. Bernard, L. Baglietto, L. Bernstein, L. Beckmann, L.D., L.G., L.J.V.V., L.N.K., L.S., M.B., M.C.S., M.D., M.F.P., M.G.S., M.H., M. Jones, M. Johansson, M.J.H., M.J.K., M.K., M.K.B., M.L., M.M.G., M.P.L., M. Shrubsole, M. Shah, M.W.B., M.W.R.R., N.A.M.T., N.D., N.G.M., N.J., N.M., N.N.A., N.O., N.R., N.S., N.V.B., O.F., P.G., P.H., P.H.P., P. Kerbrat, P.L.-P., P.L., P. Menénde, P.N., P.P., P.R., P. Siriwanarangsan, P. Sharma, P.-E.W., Q.C., Q. Wang, Q. Waisfisz, R.B., R.G.Z., R.H., R.K., R.K.S., R.L.M., R.M.M., R.N.H., R.P., R.A.E.M.T., R. Tumino, R. Travis, S.A.I., S.E.B., S.E.H., S.F.N., S.G., S.H.T., S.K., S.K.R., S.L.D.-H., S.M., S.M.J., S. Nickels, S. Nyante, S.P.B., S. Sangrajrang, S.S.-B., S. Slager, S.S.C., T.A.M., T.B., T.D., T.H., T.T., V.A., V. Kristensen, V. Kataja, V.-M.K., W.B., W.L., W.R.D., W.T., X.-O.S., X.W., Y.F., Y.-T.G., Y.-D.K. A. Mannermaa, A. Meindl, W.Z., P.D., M.S.G. and Y.Y. provided critical review of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Chu KC, Anderson WF. Rates for breast cancer characteristics by estrogen and progesterone receptor status in the major racial/ethnic groups. Breast Cancer Res Treat. 2002;74:199–211. doi: 10.1023/a:1016361932220. [DOI] [PubMed] [Google Scholar]
- 2.Yang XR, et al. Associations of breast cancer risk factors with tumor subtypes:a pooled analysis from the Breast Cancer Association Consortium studies. J Natl Cancer Inst. 2011;103:250–263. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blows FM, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mavaddat N, Antoniou AC, Easton DF, Garcia-Closas M. Genetic susceptibility to breast cancer. Mol Oncol. 2010;4:174–191. doi: 10.1016/j.molonc.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haiman CA, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor–negative breast cancer. Nat Genet. 2011;43:1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevens KN, et al. 19p13.1 is a triple negative–specific breast cancer susceptibility locus. Cancer Res. 2012;72:1795–1803. doi: 10.1158/0008-5472.CAN-11-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siddiq A, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21:5373–5384. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michailidou K, et al. Large-scale genotyping identifies 41 new breast cancer susceptibility loci . Nat Genet. 2013 Mar 27; doi: 10.1038/ng.2563. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turashvili G, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55. doi: 10.1186/1471-2407-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham K, et al. Gene expression in histologically normal epithelium from breast cancer patients and from cancer-free prophylactic mastectomy patients shares a similar profile. Br. J Cancer. 2010;102:1284–1293. doi: 10.1038/sj.bjc.6605576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Yan C. A small-molecule p53 activator induces apoptosis through inhibiting MDMX expression in breast cancer cells. Neoplasia. 2011;13:611–619. doi: 10.1593/neo.11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couch FJ, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atwal GS, et al. Altered tumor formation and evolutionary selection of genetic variants in the human MDM4 oncogene. Proc Natl Acad Sci USA. 2009;106:10236–10241. doi: 10.1073/pnas.0901298106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bojesen SE, et al. Multiple independent TERT variants associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013 Mar 27; doi: 10.1038/ng.2566. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen H, et al. Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat Comm. 2013 Mar 27; doi: 10.1038/ncomms2629. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coetzee SG, Rhie SK, Berman BP, Coetzee GA, Noushmehr H. FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res. 2012;40:e139. doi: 10.1093/nar/gks542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.