Abstract
The asymmetric polarization of cells allows specialized functions to be performed at discrete subcellular locales. Spatiotemporal coordination of polarization between groups of cells allowed the evolution of metazoa. For instance, coordinated apical-basal polarization of epithelial and endothelial cells allows transport of nutrients and metabolites across cell barriers and tissue microenvironments. The defining feature of such tissues is the presence of a central, interconnected luminal network. Although tubular networks are present in seemingly different organ systems, such as the kidney, lung, and blood vessels, common underlying principles govern their formation. Recent studies using in vivo and in vitro models of lumen formation have shed new light on the molecular networks regulating this fundamental process. We here discuss progress in understanding common design principles underpinning de novo lumen formation and expansion.
Introduction
The essence of metazoa is the organization of cells into tissues. The most fundamental type of tissue is epithelia, which consist of a layer of polarized cells that line a surface and thus serve to divide the organism into compartments. Some epithelia cover the outside of the organism, but almost all metazoa contain internal hollow spaces or lumens, which are lined by a layer of epithelial cells. Such lumens may serve to isolate specific functions, such as digestion, or to allow the movement of fluids, gases or cells between different parts of larger animals. Some very small lumens are surrounded by a single cell, such as the terminal branches of the Drosophila trachea, but most lumens are encompassed by multiple cells [1]. The simplest overall structure of lumen-containing organs is a sphere, such as the thyroid follicle. Most typically, though, these organs are elongated into tubules, which can be unbranched (e.g. sweat gland) or branched, often ending in spherical caps, termed acini or alveoli (e.g. mammary gland or lung) [2]. Some tubules form anastomosing networks, such as the vasculature, which is lined by specialized epithelia known as endothelia. All of these networks have in common a central lumen.
Lumens form during development by remarkably diverse mechanisms, including the wrapping, folding, invagination or evagination of polarized cell sheets to generate a hollow lumen [2]. Loosely adherent mesenchymal cells can also convert into polarized epithelia, termed the mesenchymal–epithelialtransition [2], and create lumens between the cells. Several reviews of tubule formation have described the molecular control of these processes in different organs [1–7].
Certain common design principles underpin the seemingly enormous diversity of lumen and tubule formation mechanisms. In nearly all cases, lumens are lined by the apical surfaces of the limiting epithelial cells [3]. (A fascinating variation is the circulatory system of certain invertebrates, which lacks endothelial cells and in which the basal surfaces of cells line the lumen, which is initially filled with extracellular matrix (ECM) [8].) Formation of the apical surface involves the coordination of membrane trafficking machinery with the polarity complexes that define polarized plasma membrane domains [9]. Moreover, in the case of multicellular lumens, cells must coordinate the orientation of their apical surfaces to face the lumen, which requires interaction of the cell with other cells and the ECM [10].
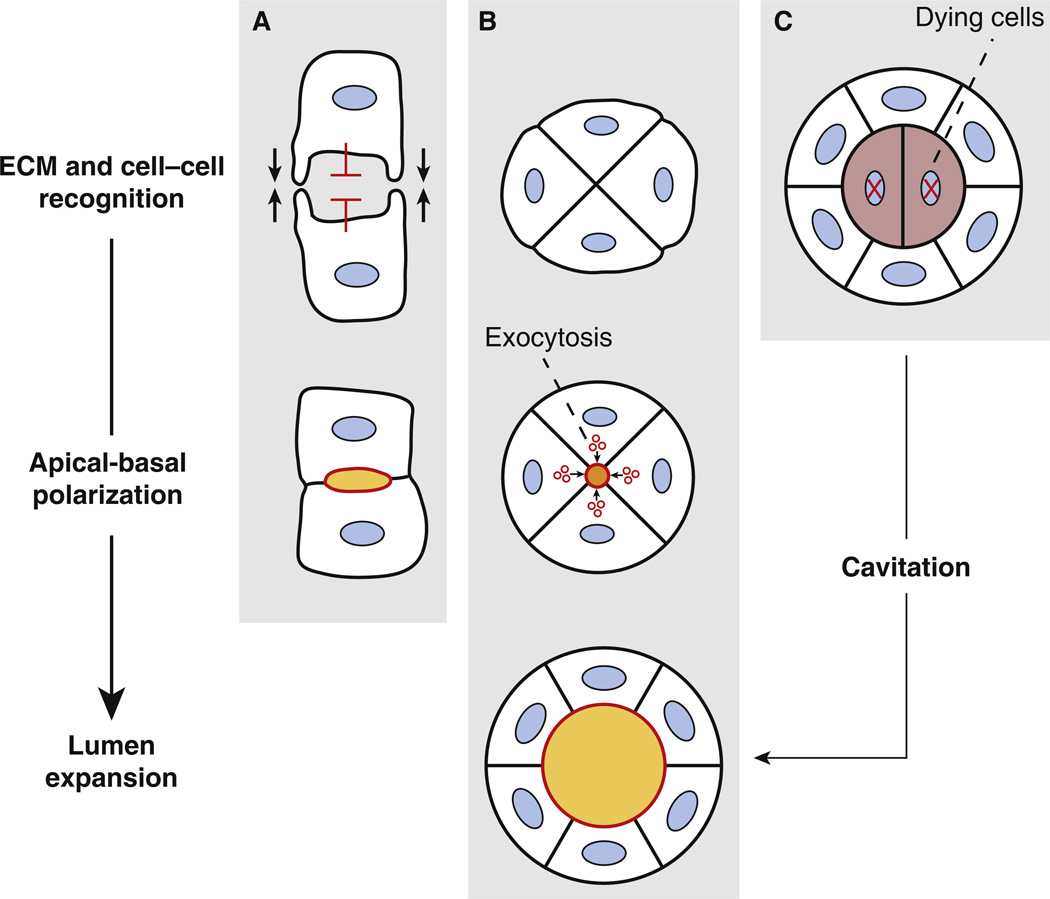
What basic design principles are required for cells to form a lumen de novo? The first principle must involve cell–matrix and cell–cell recognition — sensing one’s environment and neighbors. This is a pre-requisite for determining where to form the lumen. The second principle must involve apical-basal polarization, spatiotemporally coordinated with neighboring cells. This can happen by one of at least three principal ways: hollowing, i.e. vectorial apical membrane transport to a common point between apposing cells, generating luminal space de novo; cavitation, i.e. clearing of non-ECM-contacting inner cells from a cell cluster, such as by apoptosis, resulting in a polarized layer surrounding luminal space;or focalized contact, where adjacent cells adhere only at their lateral-most apposing edges, generating luminal space between contacts (Figure 1). A third design principle involves the expansion of the luminal space, such as by fluid and ion efflux. Here, we consider recent advances in our understanding of common design principles, across different species, tissues, and cells, of de novo lumen generation and expansion.
Figure 1. Design principles for de novo lumen formation.
Lumen formation in various contexts relies on the co-ordination of three consecutive basic design principles: extracellular matrix (ECM) and cell–cell recognition, apical-basal polarization, and lumen expansion. Molecular instructions for whether, where and how lumens will be generated are provided from integrating signals from the ECM (depicted by gray shading in all panels) and cell–cell contacts. In (A), adhesive contacts occur between neighboring cells only at discrete foci (indicated by black arrows), with non-contacting regions undergoing active repulsion (indicated by red inhibitory arrows). This leads to the formation of a luminal space between adhesions (called focalized contact), which allows apical-basal polarization. This occurs, for example, in Drosophila heart tube formation [3]. In (B), clusters of cells contacting the ECM initially adhere without a luminal space, then vesicles containing luminal components are exocytosed in a coordinated fashion to a central luminal region, generating apical-basal polarization (called hollowing). This occurs, for example, in developing mouse aorta [42] and MDCK cysts [32]. In (C), clusters of cells initially adhere without a luminal space; however, unlike (B) some cells do not contact the ECM, and thus undergo apoptosis. This results in generation of luminal space as these inner cells die (a process called cavitation). In addition, apical-basal polarization must occur. This occurs, for example, in mammary terminal end buds [5]. Thus, although de novo lumen formation occurs through seemingly different morphogenetic events, all make use of the common principles of ECM and cell–cell recognition and apical-basal polarization. Similarly, once lumens have formed, such as through these different processes, lumen expansion will occur to generate the appropriate lumen diameter. Red lines indicate apical/luminal membrane; blue ovals, nuclei; grey, ECM; maroon, dying cells.
Cell–Cell and Cell–Matrix Recognition
When non-epithelial cells coalesce to form tubular epithelia de novo, polarization must begin with a cell determining the directionality of lumenogenesis. Typically, lumens form at a shared position between neighboring cells, often perpendicular to the ECM-contacting surface, such as in kidney tubular epithelium, although other luminal positions (e.g. laterally between hepatocytes) can occur. Signals from the ECM provide one axis from which to orient lumen positioning; neighboring cells, through cell–cell contacts, provide a second axis. These combinatorial inputs provide molecular cues, and thus spatial coordinates, for generation and positioning of apical membranes.
Role of Cell–Matrix and Cell–Cell Recognition
How are signals transduced from the ECM into cells to effect polarization? Heterodimeric integrin molecules, consisting of an α- and β-integrin pair, play crucial roles in sensing ECM in a variety of cell types, with β1-integrin-containing complexes having key roles in tissue polarization [11,12]. In Drosophila tracheal terminal branches, deficiency of certain β- or α-integrins, or talin, which connects integrins to the cytoskeleton, leads to multiple lumens [13]. In mice, global loss of β1 integrin leads to embryonic lethality [14]. Tissue-specific knockouts have varying severity, although lumens are often perturbed. While the loss of β1-integrin from kidney collecting ducts does not abolish polarization or luminal network formation, kidneys were hypoplastic, and lumens were often cystic or dilated [15]. Endothelial-specific β1-integrin knockout mice are embryonic lethal, associated with luminal and branching defects [16,17]. Analysis of later blood vessel development using hypomorphic alleles revealed perturbed vessel polarity, filled lumens, and mistargeting of cell–cell junction proteins [18]. In these knockouts, the polarity protein Par3 was downregulated, and Par3 re-expression partially rescued lumen occlusion, suggesting that polarity proteins are key β1-integrin signaling targets. Indeed, in 3D cultures (Figure 2B) a signaling module involving α2β1 integrin, the adhesion proteins Jam-B/C, and the polarity proteins Par-3–Par6β–Cdc42 controls ECM remodeling by the matrix metalloprotease MT1-MMP to form endothelial tubes [19].
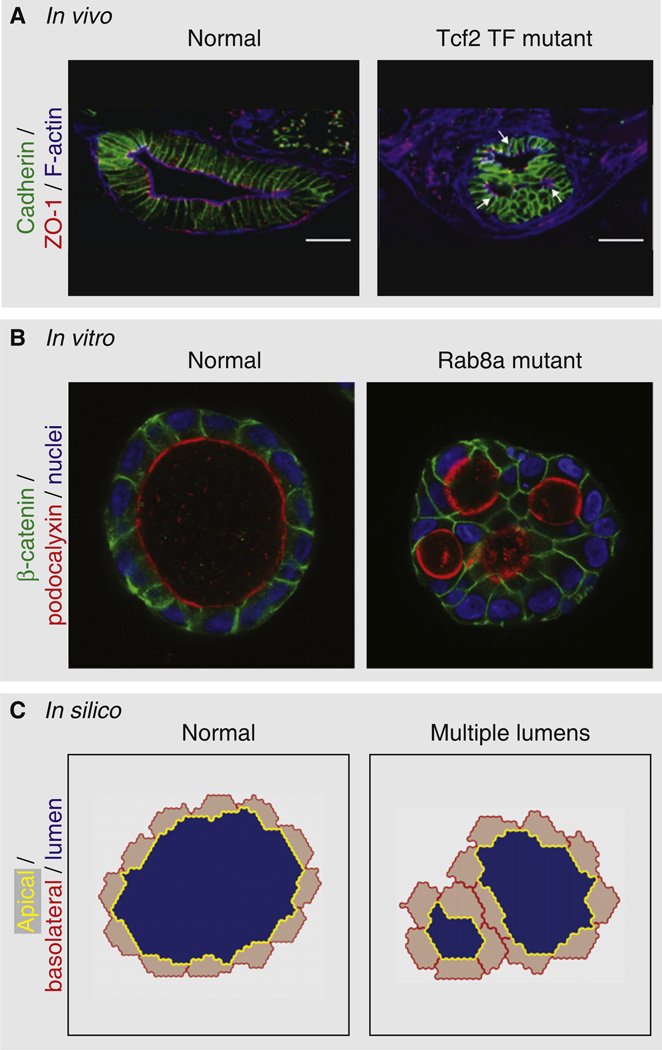
Figure 2. In vivo, in vitro and in silico models of lumen formation.
Unraveling molecular mechanisms of lumen formation requires modeling the 3D organization of lumen-containing structures, precluding the use of traditional monolayer culture of cells on glass, plastic or Transwell filters. Thus, a combination of analyses from (A) in vivo, genetically engineered model organisms, (B) in vitro, 3D cysts of cultured cells, and (c) in silico models have begun to elucidate common design principles underpinning this process. (A) In vivo analysis of model organisms, such as during zebrafish gut development, presents as a powerful method, via both forward and reverse genetic analysis, to identify physiologically relevant regulators of tubular epithelium formation. Though genetically tractable, in vivo analysis is limited to a small number of molecular alterations due to the time-scale required to generate mutant organisms. (B) In vitro, 3D cyst analysis, which involves various techniques of growing cells in ECM-enriched conditions to allow self-organization into lumen-containing structures, complements in vivo analysis by allowing for rapid reverse genetic approaches. 3D cultures enable dissection of large molecular networks regulating lumenogenesis, using combinations of knockdown and protein overexpression technologies, such as the effect of compromised apical exocytosis (Rab8a knockdown) on lumenogene-sis. (C) In silico analysis has recently emerged as a further complementary approach to understanding lumenogenesis [134], facilitating derivation of common, and fundamental, design principles required to form polarized structures with a lumen. As such models develop, future aims should include creation of in vitro and in silico models of complex luminal networks, such as the hierarchical and modular lung branching program [135]. Presented are images from (A) in vivo analyses of a Tcf2 transcription factor (TF) mutant (the image is a cross-section through the developing intestine in zebrafish, and is reproduced from [92]), (B) in vitro analyses of an apical transport mutant (knockdown of Rab8a GTPase in MDCK cysts [35]), and (C) in silico analyses showing multilumen cysts under conditions in which cysts polarize later during cystogenesis [136,137], each of which disrupts single lumen formation, causing multiple lumens
Similarly, in MDCK 3D cultures, a laminin–β1-integrin– Rac1 module controls apical-basal polarization [10,20]. β1-integrin deficiency perturbs the normal orientation of the apical surface to a central region between cells, through inappropriate activation of a RhoA–ROCKI–myosin II pathway [21], suggesting that ECM-derived signals can influence lumenogenesis via regulating cytoskeletal tension [22]. Lumen positioning (apical versus lateral) in MDCK is also controlled by regulation of the cytoskeleton by the Par protein Par1b [23–25]. In Drosophila laminin mutants, development of most organs, including the gut, airway and nervous system, is defective [26]. Thus, the ECM, β1 integrins, and polarity proteins are key regulators of apical surface and lumen orientation.
Sensing neighboring cells occurs via a multitude of adhesion receptors, including cadherins and nectins [27]. Defining individual roles of these molecules during lumenogenesis has been complicated by the partial redundancy of multiple family members. For instance, N-cadherin- and VE-cadherin-null mice display varying developmental defects, including aortic and vascular luminal perturbations [28,29]. While global E-cadherin knockout mice are not viable, tissue-specific knockouts, such as in thyroid follicles, reveal that lumens are present, but often smaller [30]. Similarly, epithelial polarization occurs in DE-cadherin-null Drosophila [31]. Thus, although fundamental roles for adhesion molecules in generating tissue polarization have been postulated from decades of studies in 2D culture, if and how these molecules actually regulate lumenogenesis in vivo still remains largely unclear.
Establishment of Apical-Basal Polarity
Once newly polarizing cells recognize the ECM and their neighbors, luminal space can be generated, either by hollowing, cavitation, or focalized contact and repulsion (Figure 1). Each mechanism requires the spatial and temporal coordination of cellular processes such as directional vesicle trafficking (hollowing), luminal cell death (cavitation) and localized formation of cell–cell contact and repulsion (focalized contact). Recent data suggest that these processes may not be mutually exclusive; if one process is perturbed, compensatory induction of the other may ensue [32].
Creating an Apical Surface de Novo
To generate a luminal domain de novo, neighboring cells must coordinate delivery of apical membrane components to a common site (Figure 1B). One possible mechanism may be to utilize a molecular landmark on the cell surface, common between neighboring cells. Such a landmark may form at the midbody during mitosis, as occurs in budding yeast [33]. During vegetative growth of Saccharomyces cerevisiae, the ‘bud scar’, retained from a previous cytokinetic event, provides a landmark for anchorage of the cytoskeleton and for localized membrane growth leading to the new bud formation. Recent studies [34,35] indicate that in mammalian cells, vesicles containing apical proteins are delivered to a discrete, common landmark between neighboring cells to initiate the lumen, a region termed the apical membrane initiation site (AMIS [35]).
A cohesive picture of the regulatory networks that control lumen initiation has begun to emerge (Figure 3). Prior to lumen initiation (in MDCK cysts), the polarity protein Crumbs3a and the apical glycoprotein podocalyxin/gp135 accumulate in Rab11a-positive vesicles [34–36]. Similar subapical vesicles, containing the apical glycoprotein Muc1, are observed in vivo at the onset of murine pancreas lumenogenesis [37]. Rab11a initiates a GTPase cascade, recruiting the Rab guanine nucleotide exchange factor (GEF) Rabin8 to sub-apical vesicles, in turn activating Rab8a/b at this locale [35]. This Rab cascade drives vesicle surface delivery, possibly by activating motor proteins such as myosin-5B [38–40]. Transport and docking of these vesicles with the AMIS is promoted by the hetero-octameric exocyst complex [35]. Fusion of apical vesicles with the plasma membrane to create an apical surface de novo is likely to occur via SNARE proteins, with syntaxin-3 acting as one likely key regulatory SNARE [41].
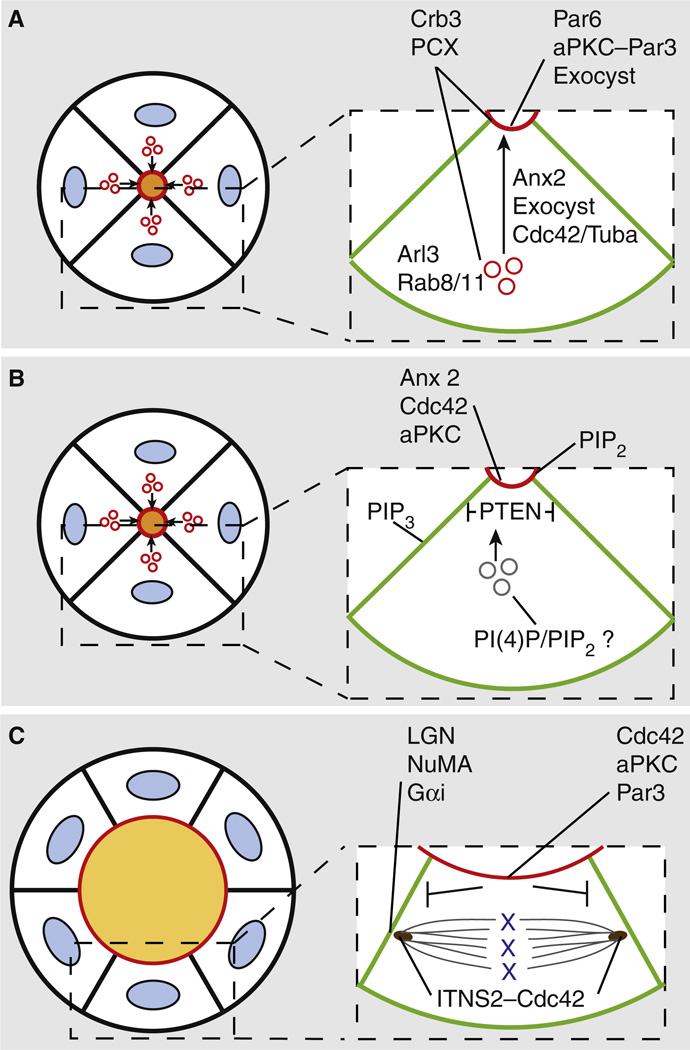
Figure 3. Molecular control of lumen generation and maintenance.
(A) Exocytosis of apical membrane-initiating proteins (such as Crumbs3a (Crb3), podocalyxin (PCX) and Muc1) to the cell surface induces formation of the nascent lumen. These proteins are transported via Rab8/11-positive vesicles, in conjunction with the PI(4,5)P2-binding protein annexin2 (Anx2), both of which are required for Cdc42 activation on these vesicles via the GEF protein Tuba. Delivery and docking of these vesicles with the cell surface at the apical membrane initiation site (AMIS) requires the concerted function of Arl3, the exocyst and Par3–aPKC complexes. The Cdc42– Par6 complex is required for efficient delivery of apical proteins, such as Crumbs. (B) As nascent lumens are formed, phosphoinositides also become asymmetrically distributed. PI(4,5)P2 (PIP2) becomes enriched at the lumen, while PI(3,4,5)P3 (PIP3) is localized to the basolateral membrane. Apically localized PTEN excludes PIP3 from this domain by dephosphorylating PIP3, converting it to PIP2. Anx2 associates with Cdc42, which in turn directs aPKC localization, with all three components acting to generate and maintain the PIP2-enriched apical membrane. The phospholipid content of exocytic vesicles destined for the lumen is not clear, although PI(4)P and PIP2 are likely candidates. (C) Once a single lumen has been established, this polarized architecture is maintained during acini/tissue growth by orienting cell division events, in which Cdc42 plays a key role. Apically localized Cdc42, in conjunction with the Par3, recruits aPKC, which in turn phosphorylates LGN, excluding the spindle-orientating LGN–NuMA complex from the apical surface. This ensures mitosis occurs only in the plane of the monolayer. Cdc42, in concert with the GEF protein Intersectin-2 (ITSN2), also localizes to centrosomes during mitosis, and participates in spindle orientation. Discrete pools of active Cdc42, such as at apical versus centrosomal regions, are apparently controlled via the GEFs Tuba (A) and ITSN2, respectively
The AMIS is demarcated by a polarity complex comprising Par3 and atypical protein kinase C (aPKC) and the exocyst subunit Sec8 (though other polarity proteins and exocyst members also overlap with both the AMIS and non-AMIS regions of the cell–cell contact) [35]. Notably, an AMIS-like structure has been observed in mouse aorta lumenogenesis [34,42], zebrafish neuroepithelial lumen formation [43], Drosophila pupal photoreceptor and tracheal tube intercellular lumenogenesis [7,44], and during the formation of the intracellular lumen of Drosophila terminal tracheal cells [45], suggesting it as a common de novo lumen-initiating structure both in 3D culture and in vivo. Targeting of apical vesicles, the exocyst and Par3–aPKC complexes to the AMIS is mutually interdependent and, moreover, requires the upstream Rab8–Rab11 cascade [35], and in Drosophila, the Arf-like3 (Arl3) GTPase (which localizes to Rab11 endosomes) [46,47]. Studies in Drosophila trachea also suggest that cadherin-mediated adhesions may be prerequisite for AMIS formation [7,44]. All of these components are likely required to form a single lumen. This suggests that the apical exocytosis and polarity machinery operate in a positive feedback loop to establish and expand an apical domain during lumen initiation.
In yeast, Cdc42-directed networks play a critical role in targeting of vesicles to the new bud site [48]. Unlike unicellular yeast polarization, however, metazoa usually require the contribution of multiple cells to form a lumen. Thus, the orientation of cell division must be coupled to apical surface generation. In mammalian cells, Cdc42 plays a critical role in both processes. Cdc42 is activated at the apical pole of cells by Rab8–Rab11, and the Cdc42–Par3–aPKC polarity complex, in conjunction with the phosphatidylinositol (4,5) bisphosphate (PI(4,5)P2)-binding protein annexin 2 (Anx2), controls exocytosis to the AMIS [35,49].
The AMIS matures into a ‘pre-apical patch’ (PAP), an early apical domain between cells where the luminal space has not yet expanded [34,35]. Here, the Par3–aPKC polarity complex, and the plasma-membrane-localized exocyst subunits (Sec8–Sec10) relocalize to tight junction regions. How is this lumen maturation step controlled? In MDCK cysts, Par3 and aPKC kinase activity are required for apical trafficking to the AMIS to expand to a PAP [34,35]. In Drosophila, aPKC-mediated Par3 phosphorylation disrupts Par3–Pals1 association [50], presumably to allow formation of the apical Crumbs–Pals1–PatJ complex, which is then involved in dissociation of Par3 from Par6 and aPKC [51,52]. Crumbs3a delivery to the nascent lumen may similarly exclude Par3 and other junction proteins from this region, helping to establish and expand the nascent apical domain. To this end, during Drosophila photoreceptor development, Cdc42, in conjunction with Par6, recruits Crumbs to the apical membrane, facilitating restriction of Par3 to the boundary between the apical membrane and the nascent adherens junction [52]. Thus, different combinations of polarity complexes form during the development of apical polarity, regulating maturation and expansion of the apical domain itself.
A critical design principle for making a single lumen is that, once the AMIS lumen is formed, subsequent divisions should reinforce, not disrupt, this polarized architecture (Figures 2C and 3C). Accordingly, Cdc42, in conjunction with Par3, also controls the correct orientation of the mitotic spindle [43,53–55] via apical recruitment of aPKC [56]. aPKC phosphorylates and excludes the spindle-orientating LGN– NuMA complex from the apical surface, consequently preventing division in the apical-basal axis and maintaining growth only in the plane of the monolayer [56,57]. Recent reports identify Tuba and Intersectin-2 as the sole Cdc42 GEFs that control localized Cdc42 activation during lumenogenesis [54,55]. Notably, while depletion of either GEF disrupts spindle orientation, only Tuba regulates apical exocytosis [35]. Whether Tuba directly regulates spindle orientation or acts indirectly through modulating exocytosis remains to be elucidated. Nonetheless, the Cdc42–Par6– aPKC–Par3 module co-operates with a Rab11-directed network to integrate apical exocytosis, apical polarity complex maturation, and cell division orientation — all processes fundamental for de novo lumenogenesis.
Generating Apical-Basal Identity
The breaking of cortical symmetry in cells during AMIS formation results in the differential apical-basal polarization not only of proteins, but also of lipids, particularly phosphoinositides (Figure 3B). Moreover, certain phospholipids themselves specify membrane identity; PI(4,5)P2 is enriched at, and specifies, apical/luminal membrane identity, whereas phosphatidylinositol (3,4,5)-trisphosphate (PIP3) localizes solely to, and specifies, basolateral membrane identity [58]. Addition of exogenous PIP3 to the apical surface of cells induces basal-membrane-like protrusions from the apical surface [59], while conversely PI(4,5)P2 addition to the basal surface relocalizes apical proteins to this domain [32]. Exclusion of PIP3 from the apical surface during lumen initiation is controlled by the lipid phosphatase PTEN, a pool of which localizes to the apical membrane and metabolizes PIP3 into PI(4,5)P2, a process crucial for lumenogenesis [32] (Figure 3B). What regulates PTEN localization during de novo lumen formation is not yet clear, but direct binding to Par3 may contribute at early stages [60,61] (although Par3 localizes to tight junctions, whereas PTEN is apical once lumens form). Notably, direct binding of Par3 to phosphoinositides contributes to its plasma membrane localization (though exactly how this occurs is controversial) [62,63]. As Par3 is both a target for phosphoinositides and a regulator of their metabolism (through PTEN), the Par complex might be a master regulator of both protein and phosphoinositide asymmetry. Accordingly, aPKC controls PI(4,5)P2/PIP3 asymmetry during development of MDCK monolayers [64].
Whether PI(4,5)P2 at the lumen is solely generated by PTEN is unknown. For instance, PI(4,5)P2 can also be generated from either PI(4)P or PI(5)P by a combination of type-I, -II, and -III phosphatidylinositol kinases (PIKs) [65]. Notably, PI4K-IIIβ generates PI(4)P at, and recruits Rab11a to, exocytic vesicles [66,67], with PI(4)P required for subsequent recruitment of the Rabin8 homologue, Sec2p, in yeast [68]. Rab8a/11a vesicles, which control delivery of apical proteins to initiate the lumen (see above), are enriched for both PI(4)P and PI(4,5)P2 [69]. Notably, the PI(4,5)P2-binding protein Anx2 is present on, and regulates exocytic traffic of, Rab8a–Rab11a vesicles to the lumen [35,49]. Furthermore, Anx2 binds Cdc42 and regulates its localization. This suggests that PI(4,5)P2 may have a key role in regulating the apical exocytic machinery [70]. However, whether PI(4,5)P2 is actually delivered to the AMIS via Rab11 vesicles remains to be demonstrated. Furthermore, the identity of the key targets of PI(4,5)P2/PIP3 in effecting protein asymmetry are unknown, although the exocyst complex, some subunits of which directly bind phosphoinositides, is one likely candidate [71–73].
Hollowing and Lumen Initiation
A design principle underlying hollowing lumenogenesis is that once apical membrane is delivered to the cell surface, extracellular space must be generated between neighboring adherent cells. How is space initiated? Along with mucin 1 (Muc1) and Crumbs, the CD34 family of anti-adhesins, including podocalyxin, are some of the earliest known proteins to localize at nascent lumens, both in 3D cysts and in vivo [34,42]. Notably, these proteins are extensively glycosylated and/or sialylated, resulting in highly negatively charged extracellular domains [74,75] that can act as anti-adhesive molecules [76,77]. In developing mouse aorta, electrostatic repulsion of podocalyxin from apposing endothelial plasma membranes provides the key initiating step necessary for subsequent endothelial lumen expansion [78]. Whether the Crumbs3 extracellular domain plays a similar role is not yet clear, but loss of podocalyxin or Crumbs3 impairs generation of this intercellular space [42,78–80]. The intracellular domains of these proteins also play key roles in lumen initiation, with the cytoplasmic tail of Crumbs recruiting ezrin–radixin–moesin (ERM) family members, and the polarity proteins Par6 and aPKC [81]. Podocalyxin also controls subapical recruitment of an F-actin–ERM–RhoA–myosin-II network, which may generate force for lumen expansion and maintenance [75]. Accordingly, ezrin knockout mice have defects in the formation of mouse intestinal lumens [82]. This apical actin network, in cooperation with the Diaphanous family of formins, may also be required for subsequent secretion during lumen maturation [38]. Thus, Crumbs and CD34 family molecules may participate in extracellular and intracellular remodeling events required for de novo lumen formation; however, whether a core requirement for such molecules exists in all lumen-forming tissues is yet to be elucidated.
Tubular Polarity by Cavitation
An alternative design principle for lumenogenesis is that there must be a mechanism to form luminal space when large clusters of cells, many of which may not be in contact, are present. This must also begin with sensing ECM and neighboring cells. Cells in the cluster periphery receive ECM-derived polarization and survival cues; those in the interior die by anoikis (loss of ECM contact) — a process termed cavitation [83] (Figure 1C).
Cavitation is the predominant lumenogenesis mode during mammary branching and salivary gland development, where highly proliferative ductal outgrowths form multilayered terminal end buds [5,84]. Bim and Bmf, pro-apoptotic members of the Bcl-2 family, regulate luminal apoptosis in vitro [85,86]. Bim-null mammary glands show transient lumen filling, but the lumens eventually clear through caspase-independent mechanisms, indicating the presence of alternative cavitation pathways [87]. The role of Bmf in mammary morphogenesis in vivo is unclear [87]. Cells undergoing anoikis during mammary morphogenesis also strongly upregulate autophagy (self-eating) pathways [5]. Surprisingly, autophagy suppresses, rather than promotes, apoptosis [88], suggesting that our understanding of luminal clearance is incomplete. Notably, in MDCK cysts, which normally undergo lumenogenesis via hollowing, these cells switch to cavitation as an alternative lumenogenesis mechanism when rapid polarization is disrupted [32]. In contrast, in 3D prostate cultures lumenogenesis is driven by hydrostatic pressure rather than cell death [89]. While the exact molecular details are only recently coming to light, it is important to note that, although multiple lumenogenesis mechanisms occur, built-in redundancy between these alternative mechanisms ensures a lumen eventually occurs.
Focalized Contact and Repulsion
A variant on the hollowing method is to combine focalized cell–cell contacts with active membrane repulsion, such as is employed in the developing Drosophila cardiac tube [90,91] (Figure 1A). Here, non-contacting myoendothelial cell rows line up along the midline, forming cadherin-mediated adhesions only at the ventral-most, then dorsal-most regions. That the luminal membranes do not form contacts is ensured by a gradient of secreted Slit protein in the intercellular space, acting on the Robo receptor to induce active membrane repulsion. The combined actions of focalized adhesive and repulsive cues, which must be facilitated by differential polarized membrane trafficking of cadherins and Slit/Robo to these sites, allows generation of the intercellular luminal space de novo. It remains to be demonstrated whether the molecules that break symmetry of the plasma membrane to allow such differential polarized exocytosis to adhesive/repulsive sites are the same as those in epithelial/endothelial cells (i.e. Par complexes).
Lumen Expansion
Once lumens are formed they must expand to their mature, functional size. Hydrostatic pressure, regulated by apical delivery and activation of pumps and channels, is thought to account for part of luminal expansion in most tissues [3]. In addition, roles for an apical ‘matrix’ are becoming clear. Expansion of the luminal network may also involve division of cells in the wall of the epithelium.
Role of Pumps and Channel Proteins
During the development of several organs in a number of species, multiple smaller ‘micro lumens’ normally coalesce to form a single lumen [2,37,82,92,93]. Key roles for the Na-K-ATPase and the claudin family of tight-junction proteins have emerged in this lumenogenesis step. Loss of the Tcf2 transcription factor in the gut strongly inhibits Na-K-ATPase and claudin-15 expression, resulting in multiple lumens. Parallel MDCK cyst analysis revealed that chloride-channel-mediated ion transport and Na-K-ATPase-mediated fluid transport are essential for lumen expansion, with claudin-15 forming a paracellular pore regulating these channel activities [92]. Similarly, in zebrafish ventricle lumen expansion claudin-5a regulates paracellular permeability across the neuroepithelial barrier and is crucial for ventricle lumen expansion [94]. Claudin-4 and −6 play similar roles in mouse blastocyst lumen expansion, while the Drosophila claudin Kune-Kune controls tracheal tube size [95,96]. Na-K-ATPase (Atp1α1) expression is also required for brain ventricular lumen expansion, but whether its pump activity is required is controversial [97,98]. Nonetheless, lumen expansion seems to occur in multiple organs via a conserved interplay between claudin-regulated paracellular permeability, and Na-K-ATPase-modulated luminal hydrostatic pressure.
Control of chloride transport through the cystic fibrosis transmembrane conductance regulator (CFTR) also appears to be essential for regulating fluid transport into epithelial lumens [99]. Chloride currents are controlled by protein kinase A (PKA)-dependent CFTR phosphorylation events in response to signals promoting local increases in cyclic AMP levels [100]. Pharmacological hyper-activation of CFTR-dependent fluid transport results in overexpansion of the gut lumen in developing zebrafish [101], and also in MDCK cysts [102]. A recent genetic screen identified Cse1l as an inhibitor of CFTR function during zebrafish gut development [101]. Notably, Cse1l loss-of-function leads to lumen overexpansion in both zebrafish gut and MDCK cysts through CFTR hyperactivation. Whether cyclic AMP/CFTR-dependent signaling affects lumen formation solely through chloride secretion, or through as yet unappreciated mechanisms, remains unclear.
Luminal Matrix in Lumen Expansion
Luminal matrices are increasingly being appreciated as key regulators of lumen expansion. For example, the Drosophila trachea contains a remarkable set of interconnected tubes formed by both intercellular and intracellular lumens [103]. Maturation of this lumen is a multistep process involving secretion of a luminal matrix, followed by rapid clearing and the initiation of gas exchange [104]. The matrix transiently secreted into the lumen is composed of fibrillar chitin and is required for lumen expansion [105]. Interestingly, the two putative chitin deacetylases, Vermiform and Serpentine, which are secreted into the lumen and assumed to regulate clearance of chitin to allow mature lumen function, appear to regulate tube length but not diameter [106,107], suggesting that our understanding of the role of the chitin matrix in lumen morphogenesis is far from complete. Instead, tracheal-tube expansion appears to be a cell-autonomous process whereby endoplasmic reticulum (ER)–Golgi transport pathways contribute to apical membrane expansion, cell flattening and luminal cuticle formation [108], although the mechanisms behind this process remain largely unclear. In contrast, lumen expansion in the Drosophila photoreceptor requires apical secretion of the proteoglycan Eyes Shut (Eys) [109], perhaps acting to induce membrane repulsion, similar to podocalyxin or Muc1. Notably, mutations in Eys in humans are associated with autosomal recessive retinitis pigmentosa [110,111], suggesting conservation in the mechanisms of retinal lumen morphogenesis between insects and humans. We suggest that, while a chitin-based extracellular matrix appears not to play a role in vertebrate lumen development, an analogous luminal matrix consisting of the ‘glycocalyx’ [112] provided by luminal proteins such as podocalyxin, Muc1 or Eys may instead be involved. Our understanding of these events, however, is still in its infancy.
ER–Golgi Transport and Lumen Expansion
As mentioned above, bidirectional ER-Golgi transport has somewhat surprisingly emerged as a key regulator of apical transport and lumen morphogenesis, rather than being a generalized membrane transport step. The coat protein complex II (COPII), comprising Sar1 (regulatory GTPase), Sec23–Sec24 (cargo-binding subunits), and Sec13–Sec31 (coat components), regulates anterograde ER–Golgi transport, while the COPI complex regulates retrograde transport (Golgi-to-ER) [113]. Most subunits exist as multiple isoforms, allowing different combinatorial complexes [114], such as for general versus apical secretion. Perturbation of Sar1 or Sec24 (COPII complex) in Drosophila attenuates apical secretion, luminal matrix deposition, and luminal expansion without affecting apical-basal polarization [104,108]. Sec24B mutant mice fail to complete neural tube closure (and thus lumen formation), due to defective transport of the planar cell polarity regulator Vangl2 [115]. Loss of function of γCOP (COPI complex) also disrupts luminal secretion during Drosophila tracheal tube [116] and salivary gland maturation [117]. Notably, Sec23A mutant mice are defective in proteoglycan and collagen secretion [118]. This suggests that isoform-specific COP-complex-regulated secretion of matrix proteins (structural and proteoglycan) may be a fundamental step in lumen morphogenesis in diverse tissues and organisms. Whether these pathways only control matrix secretion or also control apical transport of expansion-regulating pumps and channels remains to be demonstrated [119].
Polarity Proteins in Lumen Expansion
Members of the conserved polarity complexes regulate apical-basal polarization and lumen size, but how these regulate the latter is poorly understood. Generally, apical and basolateral polarity complexes are thought to act in mutual opposition, negatively regulating the overexpansion of one domain into the other [120]. The basolateral complex comprising Scribble, Discs large (Dlg) and Lethal giant larvae (Lgl) negatively controls lumen expansion in the Drosophila trachea [121]. However, of these, only Lgl loss abrogates luminal matrix deposition, while Scribble and Dlg control lumen size in a matrix-independent pathway (the precise mechanism of which remains to be elucidated) [121]. The loss of another basolateral polarity protein Yurt leads to tracheal tube enlargement. In contrast, Crumbs overexpression results in apical membrane overexpansion [81]. As Yurt negatively regulates apical Crumbs, lumen enlargement in Yurt mutants may be due to an imbalance of the mutual inhibition between basolateral and apical polarity complexes [121,122]. The apical polarity proteins Crumbs, Par6γb, and aPKC also regulate zebrafish brain ventricle size [123–125], with embryos lacking these proteins having brain ventricle expansion defects.
As aforementioned, fusion of multiple rudimentary lumens into a single lumen occurs normally during development of several vertebrate organs [2,82,92]. During zebrafish gut lumen formation, multiple clusters of actin-enriched foci form and eventually fuse, in an aPKC-dependent manner, at a single focal point prior to lumen expansion [123]. A similar phenomenon occurs during mouse pancreatic tube formation, where multiple microlumens fuse to form a central lumen. This fusion is dependent on Cdc42 acting upstream of aPKC and loss of either Cdc42 or aPKC leads to multilumen phenotypes [37]. The apical ERM protein ezrin similarly controls progression from multiple to single lumens in the mouse gut [82]. Thus, Cdc42,as part of the apical polarity complexes, and probably together with ezrin-mediated effects on the apical cytoskeleton, regulates multiple steps in lumen formation and maturation. How changes in these polarity proteins and apical cytoskeleton influence cellular mechanisms that regulate lumen expansion remain poorly understood, but may involve modulation of membrane transport pathways [9,35]. Indeed, in Drosophila, Diaphanous formins modulate apical actin networks to facilitate myosin-5B-directed apical secretion during tracheal morphogenesis [38]. Thus, polarity proteins likely control lumen formation by acting as an interface node between ECM-derived signaling networks, cytoskeletal organization, and membrane transport.
Integrating Morphogens, Polarity, and Membrane Transport
A fundamental design principle for developmental morphogen systems specifying epithelial or endothelial cell fate is that these signals must induce polarization networks to form a lumen. How do traditional ‘fate-generating’ signals induce such cellular behaviors?
Recent studies reveal that transcription factors regulating epithelial differentiation modulate transcription of apical polarity and transport factors (Table 1). In particular, the Rab11 GTPase family (Rab11a/b and Rab25), required for lumen morphogenesis in diverse systems [35,36,40, 126,127], appears to be a key transcriptional target. For instance, Snail,a transcription factor that induces the epithelial–mesenchymal transition, directly represses Crumbs3, Rab25 and PTEN transcription [128–130], all of which are required for lumen formation [35,49]. In Drosophila, loss of the transcription factor Ribbon decreases Rab11a expression and consequently apical Crumbs levels, resulting in impaired expansion of tracheal and salivary gland lumens [131]. Rab11a similarly regulates apical Crumbs3a and polarity protein delivery in MDCK cysts [35,80]. Notably, the homeobox transcription factor Cdx2, which specifies intestinal fate, regulates lumenogenesis via the key transcriptional targets Rab11a and the kinesin-II subunit, Kif3b [132] (which is linked to Rab11 via its effector Rab11–FIP5 [133]). Similarly, during Drosophila airway branching morphogenesis, interplay between Wingless and Decapentaplegic morphogen gradients controls the expression of Rab11a and Rip11, affecting the type of epithelial lumen that forms [127]. These studies reveal that apical transport and polarity proteins are key targets of morphogen systems regulating epithelial differentiation.
Table 1.
Selected transcription factors regulating development of tubular epithelial networks.
| Transcription factor | Species | Tissue affected | Function/target genes | Lumen defect | Ref. |
|---|---|---|---|---|---|
| Caudal-type homeobox protein 2 (Cdx2) |
Mouse | Intestine | Endocytic/exocytic proteins Pumps and channels Apical trafficking proteins (Rab11a, Kif3b) |
Multiple lumens Disrupted apical polarity and surface morphology |
[132] |
| Transcription factor CP2-like1 (Tcfcp2l1), related to Grainyhead |
Mouse | Salivary gland, kidney |
Channel, pumps and ion exchanger proteins |
Dilated salivary gland lumen Defective renal tubule maturation |
[140] |
| Grainyhead (Grhl1) | Drosophila | Trachea | Septate junction proteins (Fasciclin, Coracle, Sinuous) |
Elongated/tortuous tracheal branches |
[141,142] |
| Hairy (h) | Drosophila | Salivary gland | Represses Huckebein and Klarsicht | Branched and enlarged salivary gland lumen |
[143] |
| Huckebein (hkb) | Drosophila | Salivary gland | Apical polarity (Crumbs) Microtubule transport (Klarsicht) |
Dome-shaped glands, small lumens | [143] |
| Krüuppel-like factor 4 (Klf4) | Mouse | Sertoli cells | Exocytosis (sec 8l1) Lysosomal trafficking (Hps5) Apoptosis (Htatip2) |
Delayed lumen formation (defect is rescued later in development) |
[144] |
| No Tail-a (ntla)/Brachyury (T) | Zebrafish | Kupffer’s vesicle | Mesenchymal to epithelial transition of dorsal forerunner cells to form Kupffer’s vesicle |
Uninflated Kupffer’s vesicle lumen | [145] |
| Ribbon (rib) | Drosophila | Salivary gland | Apical polarity (Crumbs) Apical cytoskeleton (Moesin) |
Short salivary gland tubes Decreased Rab11a expression Excessive microvilli |
[131] |
| Spalt major/Spalt (salm) | Drosophila | Trachea | Inhibits tracheal dorsal trunk intercalation Membrane recycling (Rab11a, Rip11) |
Excessive tracheal branch intercalation Disrupted Rab11a vesicle organization |
[127] |
| HNF1 homeobox Ba (hnf1ba/tcf2) | Zebrafish | Gut | Ion pumps (Na/K-ATPase) Paracellular pores (Claudin 15) |
Unexpanded, multiple lumens | [92] |
| Tramtrack (ttk) | Drosophila | Trachea | Septate junctions Cuticle formation |
Increased tube size Impaired branching |
[146] |
Conclusions and Future Directions
Despite the seeming multitudes of morphogenetic processes governing de novo lumen formation, several key design principles have emerged in the last few years, which we have described here. More recently, key common molecular regulators of these processes have been elucidated: β1 integrin, which transduces ECM-derived signals [10,18,20,21]; Rab11a, which directs apical transport [35,36]; Cdc42, which functions in apical polarity, membrane transport, and cell division [35,49,53–55]; and the Par3– aPKC complex, which integrates all of these signals together into polarity- and lumen-generating modules [18,35,49]. Several questions remain. How are these key polarization molecules regulated, both transcriptionally and via upstream regulators? How is such regulation differentially controlled to give rise to different luminal structures during diverse morphogenetic events? Parallel analysis of such questions using in vivo, in vitro and in silico models (Figure 2) should allow us to uncover further regulators of this complex morphogenetic event, a key requirement for improving human health (Box 1).
Box 1. Lumen formation and human health.
Lumen formation was a crucial step in metazoan evolution, enabling essential functions such as nutrient uptake, gas exchange, and circulation. The dysfunction of luminal networks is often fatal. Hyperdilated tubules associated with reduced renal function occur in polycystic kidney diseases, which is caused by mutation in numerous genes, and can also be induced by long-term renal dialysis [138]. Such dilation may therefore be a final common pathway resulting from perturbation of the finely balanced control of lumen diameter. Stenosis, or reduction of lumen size, is associated with vascular diseases such as hypertension [4]. Defective brain ventricle closure or expansion leads to anencephaly, schizencephaly and hydrocephalus [139]. Early stages of many epithelial cancers display luminal filling, such as in ductal carcinomas in situ [83]. Understanding the molecular mechanisms controlling formation and maintenance of lumens is therefore key to effectively treating such common human diseases.
Acknowledgements
We thank Jesse Engelberg and Michel Bagnat for images and Ross Metzger for discussion. We apologize to those authors whose original work was not directly cited due to space constraints. Our work is supported by a DOD Lung Cancer Concept Award (A.D.), a Susan G. Komen Foundation Fellowship (D.M.B.), and NIH R01DK083330, R01DK074398, R01AI25144 and P01AI53194 (K.E.M.).
References
- 1.Lubarsky B, Krasnow MA. Tube morphogenesis. Making and shaping biological tubes. Cell. 2003;112:19–28. doi: 10.1016/s0092-8674(02)01283-7. [DOI] [PubMed] [Google Scholar]
- 2.Hogan BL, Kolodziej PA. Organogenesis: molecular mechanisms of tubulogenesis. Nat. Rev. Genet. 2002;3:513–523. doi: 10.1038/nrg840. [DOI] [PubMed] [Google Scholar]
- 3.Bryant DM, Mostov KE. From cells to organs: building polarized tissue. Nat. Rev. Mol. Cell Biol. 2008;9:887–901. doi: 10.1038/nrm2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iruela-Arispe ML, Davis GE. Cellular and molecular mechanisms of vascular lumen formation. Dev. Cell. 2009;16:222–231. doi: 10.1016/j.devcel.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mailleux AA, Overholtzer M, Brugge JS. Lumen formation during mammary epithelial morphogenesis: insights from in vitro and in vivo models. Cell Cycle. 2008;7:57–62. doi: 10.4161/cc.7.1.5150. [DOI] [PubMed] [Google Scholar]
- 6.Andrew DJ, Ewald AJ. Morphogenesis of epithelial tubes: Insights into tube formation, elongation, and elaboration. Dev. Biol. 2010;341:34–55. doi: 10.1016/j.ydbio.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baer MM, Chanut-Delalande H, Affolter M. Cellular and molecular mechanisms underlying the formation of biological tubes. Curr. Top. Dev. Biol. 2009;89:137–162. doi: 10.1016/S0070-2153(09)89006-6. [DOI] [PubMed] [Google Scholar]
- 8.Kucera T, Strilic B, Regener K, Schubert M, Laudet V, Lammert E. Ancestral vascular lumen formation via basal cell surfaces. PLoS ONE. 2009;4:e4132. doi: 10.1371/journal.pone.0004132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mellman I, Nelson WJ. Coordinated protein sorting, targeting and distribution in polarized cells. Nat. Rev. Mol. Cell Biol. 2008;9:833–845. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Brien LE, Jou TS, Pollack AL, Zhang Q, Hansen SH, Yurchenco P, Mostov KE. Rac1 orientates epithelial apical polarity through effects on basolateral laminin assembly. Nat. Cell Biol. 2001;3:831–838. doi: 10.1038/ncb0901-831. [DOI] [PubMed] [Google Scholar]
- 11.Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. The organizing principle: microenvironmental influences in the normal and malignant breast. Differentiation. 2002;70:537–546. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Brien LE, Zegers MM, Mostov KE. Opinion: Building epithelial architecture: insights from three-dimensional culture models. Nat. Rev. Mol. Cell Biol. 2002;3:531–537. doi: 10.1038/nrm859. [DOI] [PubMed] [Google Scholar]
- 13.Levi BP, Ghabrial AS, Krasnow MA. Drosophila talin and integrin genes are required for maintenance of tracheal terminal branches and luminal organization. Development. 2006;133:2383–2393. doi: 10.1242/dev.02404. [DOI] [PubMed] [Google Scholar]
- 14.Fassler R, Meyer M. Consequences of lack of beta 1 integrin gene expression in mice. Genes Dev. 1995;9:1896–1908. doi: 10.1101/gad.9.15.1896. [DOI] [PubMed] [Google Scholar]
- 15.Wu W, Kitamura S, Truong DM, Rieg T, Vallon V, Sakurai H, Bush KT, Vera DR, Ross RS, Nigam SK. Beta1-integrin is required for kidney collecting duct morphogenesis and maintenance of renal function. Am. J. Physiol. Renal Physiol. 2009;297:F210–F217. doi: 10.1152/ajprenal.90260.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson TR, Hu H, Braren R, Kim YH, Wang RA. Cell-autonomous requirement for beta1 integrin in endothelial cell adhesion, migration and survival during angiogenesis in mice. Development. 2008;135:2193–2202. doi: 10.1242/dev.016378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei L, Liu D, Huang Y, Jovin I, Shai SY, Kyriakides T, Ross RS, Giordano FJ. Endothelial expression of beta1 integrin is required for embryonic vascular patterning and postnatal vascular remodeling. Mol. Cell Biol. 2008;28:794–802. doi: 10.1128/MCB.00443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zovein AC, Luque A, Turlo KA, Hofmann JJ, Yee KM, Becker MS, Fassler R, Mellman I, Lane TF, Iruela-Arispe ML. Beta1 integrin establishes endothelial cell polarity and arteriolar lumen formation via a Par3-dependent mechanism. Dev. Cell. 2010;18:39–51. doi: 10.1016/j.devcel.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sacharidou A, Koh W, Stratman AN, Mayo AM, Fisher KE, Davis GE. Endothelial lumen signaling complexes control 3D matrix-specific tubulogenesis through interdependent Cdc42- and MT1-MMP-mediated events. Blood. 2010;115:5259–5269. doi: 10.1182/blood-2009-11-252692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu W, Datta A, Leroy P, O’Brien LE, Mak G, Jou TS, Matlin KS, Mostov KE, Zegers MM. Beta1-integrin orients epithelial polarity via Rac1 and laminin. Mol. Biol. Cell. 2005;16:433–445. doi: 10.1091/mbc.E04-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu W, Shewan AM, Brakeman P, Eastburn DJ, Datta A, Bryant DM, Fan QW, Weiss WA, Zegers MM, Mostov KE. Involvement of RhoA, ROCK I and myosin II in inverted orientation of epithelial polarity. EMBO Rep. 2008;9:923–929. doi: 10.1038/embor.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat. Rev. Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen D, Brennwald PJ, Rodriguez-Boulan E, Musch A. Mammalian PAR-1 determines epithelial lumen polarity by organizing the microtubule cytoskeleton. J. Cell Biol. 2004;164:717–727. doi: 10.1083/jcb.200308104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen D, Rodriguez-Boulan E, Musch A. Par-1 promotes a hepatic mode of apical protein trafficking in MDCK cells. Proc. Natl. Acad. Sci. USA. 2004;101:13792–13797. doi: 10.1073/pnas.0403684101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen D, Tian Y, Musch A. Par1b promotes hepatic-type lumen polarity in Madin Darby canine kidney cells via myosin II- and E-cadherin-dependent signaling. Mol. Biol. Cell. 2007;18:2203–2215. doi: 10.1091/mbc.E07-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urbano JM, Torgler CN, Molnar C, Tepass U, Lopez-Varea A, Brown NH, de Celis JF, Martin-Bermudo MD. Drosophila laminins act as key regulators of basement membrane assembly and morphogenesis. Development. 2009;136:4165–4176. doi: 10.1242/dev.044263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris TJ, Tepass U. Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 2010;11:502–514. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- 28.Carmeliet P, Lampugnani MG, Moons L, Breviario F, Compernolle V, Bono F, Balconi G, Spagnuolo R, Oosthuyse B, Dewerchin M, et al. Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell. 1999;98:147–157. doi: 10.1016/s0092-8674(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 29.Luo Y, Radice GL. N-cadherin acts upstream of VE-cadherin in controlling vascular morphogenesis. J. Cell Biol. 2005;169:29–34. doi: 10.1083/jcb.200411127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cali G, Zannini M, Rubini P, Tacchetti C, D’Andrea B, Affuso A, Wintermantel T, Boussadia O, Terracciano D, Silberschmidt D, et al. Conditional inactivation of the E-cadherin gene in thyroid follicular cells affects gland development but does not impair junction formation. Endocrinology. 2007;148:2737–2746. doi: 10.1210/en.2006-1344. [DOI] [PubMed] [Google Scholar]
- 31.Harris TJ, Peifer M. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J. Cell Biol. 2004;167:135–147. doi: 10.1083/jcb.200406024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Belmonte F, Yu W, Rodriguez-Fraticelli AE, Ewald A, Werb Z, Alonso MA, Mostov K. Cell-polarity dynamics controls the mechanism of lumen formation in epithelial morphogenesis. Curr. Biol. 2008;18:507–513. doi: 10.1016/j.cub.2008.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson WJ. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422:766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrari A, Veligodskiy A, Berge U, Lucas MS, Kroschewski R. ROCK-mediated contractility, tight junctions and channels contribute to the conversion of a preapical patch into apical surface during isochoric lumen initiation. J. Cell Sci. 2008;121:3649–3663. doi: 10.1242/jcs.018648. [DOI] [PubMed] [Google Scholar]
- 35.Bryant DM, Datta A, Rodriguez-Fraticelli AE, Peranen J, Martin-Belmonte F, Mostov KE. A molecular network for de novo generation of the apical surface and lumen. Nat. Cell Biol. 2010;12:1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schluter MA, Pfarr CS, Pieczynski J, Whiteman EL, Hurd TW, Fan S, Liu CJ, Margolis B. Trafficking of Crumbs3 during cytokinesis is crucial for lumen formation. Mol. Biol. Cell. 2009;20:4652–4663. doi: 10.1091/mbc.E09-02-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kesavan G, Sand FW, Greiner TU, Johansson JK, Kobberup S, Wu X, Brakebusch C, Semb H. Cdc42-mediated tubulogenesis controls cell specification. Cell. 2009;139:660–662. doi: 10.1016/j.cell.2009.08.049. [DOI] [PubMed] [Google Scholar]
- 38.Massarwa R, Schejter ED, Shilo BZ. Apical secretion in epithelial tubes of the Drosophila embryo is directed by the Formin-family protein Diaphanous. Dev. Cell. 2009;16:877–888. doi: 10.1016/j.devcel.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 39.Wakabayashi Y, Dutt P, Lippincott-Schwartz J, Arias IM. Rab11a and myosin Vb are required for bile canalicular formation in WIF-B9 cells. Proc. Natl. Acad. Sci. USA. 2005;102:15087–15092. doi: 10.1073/pnas.0503702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li BX, Satoh AK, Ready DF. Myosin V, Rab11, and dRip11 direct apical secretion and cellular morphogenesis in developing Drosophila photoreceptors. J. Cell Biol. 2007;177:659–669. doi: 10.1083/jcb.200610157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma N, Low SH, Misra S, Pallavi B, Weimbs T. Apical targeting of syntaxin 3 is essential for epithelial cell polarity. J. Cell Biol. 2006;173:937–948. doi: 10.1083/jcb.200603132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strilic B, Kucera T, Eglinger J, Hughes MR, McNagny KM, Tsukita S, Dejana E, Ferrara N, Lammert E. The molecular basis of vascular lumen formation in the developing mouse aorta. Dev. Cell. 2009;17:505–515. doi: 10.1016/j.devcel.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 43.Tawk M, Araya C, Lyons DA, Reugels AM, Girdler GC, Bayley PR, Hyde DR, Tada M, Clarke JD. A mirror-symmetric cell division that orchestrates neuroepithelial morphogenesis. Nature. 2007;446:797–800. doi: 10.1038/nature05722. [DOI] [PubMed] [Google Scholar]
- 44.Uv A, Cantera R, Samakovlis C. Drosophila tracheal morphogenesis: intricate cellular solutions to basic plumbing problems. Trends Cell Biol. 2003;13:301–309. doi: 10.1016/s0962-8924(03)00083-7. [DOI] [PubMed] [Google Scholar]
- 45.Gervais L, Casanova J. In vivo coupling of cell elongation and lumen formation in a single cell. Curr. Biol. 2010;20:359–366. doi: 10.1016/j.cub.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 46.Jiang L, Rogers SL, Crews ST. The Drosophila Dead end Arf-like3 GTPase controls vesicle trafficking during tracheal fusion cell morphogenesis. Dev. Biol. 2007;311:487–499. doi: 10.1016/j.ydbio.2007.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kakihara K, Shinmyozu K, Kato K, Wada H, Hayashi S. Conversion of plasma membrane topology during epithelial tube connection requires Arf-like 3 small GTPase in Drosophila. Mech. Dev. 2008;125:325–336. doi: 10.1016/j.mod.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 48.Harris KP, Tepass U. Cdc42 and vesicle trafficking in polarized cells. Traffic. 2010;11:1272–1279. doi: 10.1111/j.1600-0854.2010.01102.x. [DOI] [PubMed] [Google Scholar]
- 49.Martin-Belmonte F, Gassama A, Datta A, Yu W, Rescher U, Gerke V, Mostov K. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell. 2007;128:383–397. doi: 10.1016/j.cell.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krahn MP, Buckers J, Kastrup L, Wodarz A. Formation of a Bazooka-Stardust complex is essential for plasma membrane polarity in epithelia. J. Cell Biol. 2010;190:751–760. doi: 10.1083/jcb.201006029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morais-de-Sa E, Mirouse V, St Johnston D. aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell. 2010;141:509–523. doi: 10.1016/j.cell.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walther RF, Pichaud F. Crumbs/DaPKC-dependent apical exclusion of Bazooka promotes photoreceptor polarity remodeling. Curr. Biol. 2010;20:1065–1074. doi: 10.1016/j.cub.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 53.Jaffe AB, Kaji N, Durgan J, Hall A. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J. Cell Biol. 2008;183:625–633. doi: 10.1083/jcb.200807121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qin Y, Meisen WH, Hao Y, Macara IG. Tuba, a Cdc42 GEF, is required for polarized spindle orientation during epithelial cyst formation. J. Cell Biol. 2010;189:661–669. doi: 10.1083/jcb.201002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez-Fraticelli AE, Vergarajauregui S, Eastburn DJ, Datta A, Alonso MA, Mostov K, Martin-Belmonte F. The Cdc42 GEF Intersectin 2 controls mitotic spindle orientation to form the lumen during epithelial morphogenesis. J. Cell Biol. 2010;189:725–738. doi: 10.1083/jcb.201002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hao Y, Du Q, Chen X, Zheng Z, Balsbaugh JL, Maitra S, Shabanowitz J, Hunt DF, Macara IG. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical pins. Curr. Biol. 2010;20:1809–1818. doi: 10.1016/j.cub.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng Z, Zhu H, Wan Q, Liu J, Xiao Z, Siderovski DP, Du Q. LGN regulates mitotic spindle orientation during epithelial morphogenesis. J. Cell Biol. 2010;189:275–288. doi: 10.1083/jcb.200910021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin-Belmonte F, Mostov K. Phosphoinositides control epithelial development. Cell Cycle. 2007;6:1957–1961. doi: 10.4161/cc.6.16.4583. [DOI] [PubMed] [Google Scholar]
- 59.Gassama-Diagne A, Yu W, ter Beest M, Martin-Belmonte F, Kierbel A, Engel J, Mostov K. Phosphatidylinositol-3,4,5-trisphosphate regulates the formation of the basolateral plasma membrane in epithelial cells. Nat. Cell Biol. 2006;8:963–970. doi: 10.1038/ncb1461. [DOI] [PubMed] [Google Scholar]
- 60.von Stein W, Ramrath A, Grimm A, Muller-Borg M, Wodarz A. Direct association of Bazooka/PAR-3 with the lipid phosphatase PTEN reveals a link between the PAR/aPKC complex and phosphoinositide signaling. Development. 2005;132:1675–1686. doi: 10.1242/dev.01720. [DOI] [PubMed] [Google Scholar]
- 61.Feng W, Wu H, Chan LN, Zhang M. Par-3-mediated junctional localization of the lipid phosphatase PTEN is required for cell polarity establishment. J. Biol. Chem. 2008;283:23440–23449. doi: 10.1074/jbc.M802482200. [DOI] [PubMed] [Google Scholar]
- 62.Krahn MP, Klopfenstein DR, Fischer N, Wodarz A. Membrane targeting of Bazooka/PAR-3 is mediated by direct binding to phosphoinositide lipids. Curr. Biol. 2010;20:636–642. doi: 10.1016/j.cub.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 63.Wu H, Feng W, Chen J, Chan LN, Huang S, Zhang M. PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol. Cell. 2007;28:886–898. doi: 10.1016/j.molcel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 64.Takahama S, Hirose T, Ohno S. aPKC restricts the basolateral determinant PtdIns(3,4,5)P3 to the basal region. Biochem. Biophys. Res. Commun. 2008;368:249–255. doi: 10.1016/j.bbrc.2008.01.083. [DOI] [PubMed] [Google Scholar]
- 65.Giudici ML, Hinchliffe KA, Irvine RF. Phosphatidylinositol phosphate kinases. J. Endocrinol. Invest. 2004;27:137–142. [PubMed] [Google Scholar]
- 66.de Graaf P, Zwart WT, van Dijken RA, Deneka M, Schulz TK, Geijsen N, Coffer PJ, Gadella BM, Verkleij AJ, van der Sluijs P, et al. Phosphatidylinositol 4-kinasebeta is critical for functional association of rab11 with the Golgi complex. Mol. Biol. Cell. 2004;15:2038–2047. doi: 10.1091/mbc.E03-12-0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Polevoy G, Wei HC, Wong R, Szentpetery Z, Kim YJ, Goldbach P, Steinbach SK, Balla T, Brill JA. Dual roles for the Drosophila PI 4-kinase four wheel drive in localizing Rab11 during cytokinesis. J. Cell Biol. 2009;187:847–858. doi: 10.1083/jcb.200908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mizuno-Yamasaki E, Medkova M, Coleman J, Novick P. Phosphatidylinositol 4-phosphate controls both membrane recruitment and a regulatory switch of the Rab GEF Sec2p. Dev. Cell. 2010;18:828–840. doi: 10.1016/j.devcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jovic M, Kieken F, Naslavsky N, Sorgen PL, Caplan S. Eps15 homology domain 1-associated tubules contain phosphatidylinosi-tol-4-phosphate and phosphatidylinositol-(4,5)-bisphosphate and are required for efficient recycling. Mol. Biol. Cell. 2009;20:2731–2743. doi: 10.1091/mbc.E08-11-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guerriero CJ, Weixel KM, Bruns JR, Weisz OA. Phosphatidylinositol 5-kinase stimulates apical biosynthetic delivery via an Arp2/3-dependent mechanism. J. Biol. Chem. 2006;281:15376–15384. doi: 10.1074/jbc.M601239200. [DOI] [PubMed] [Google Scholar]
- 71.Liu J, Zuo X, Yue P, Guo W. Phosphatidylinositol 4,5-bisphosphate mediates the targeting of the exocyst to the plasma membrane for exocytosis in mammalian cells. Mol. Biol. Cell. 2007;18:4483–4492. doi: 10.1091/mbc.E07-05-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang X, Orlando K, He B, Xi F, Zhang J, Zajac A, Guo W. Membrane association and functional regulation of Sec3 by phos-pholipids and Cdc42. J. Cell Biol. 2008;180:145–158. doi: 10.1083/jcb.200704128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fabian L, Wei HC, Rollins J, Noguchi T, Blankenship JT, Bellamkonda K, Polevoy G, Gervais L, Guichet A, Fuller MT, et al. Phosphatidylinositol 4,5-bisphosphate directs spermatid cell polarity and exocyst localization in Drosophila. Mol. Biol. Cell. 2010;21:1546–1555. doi: 10.1091/mbc.E09-07-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kilic A, Klose S, Dobberstein B, Knust E, Kapp K. The Drosophila Crumbs signal peptide is unusually long and is a substrate for signal peptide peptidase. Eur. J. Cell Biol. 2010;89:449–461. doi: 10.1016/j.ejcb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 75.Nielsen JS, McNagny KM. Novel functions of the CD34 family. J. Cell Sci. 2008;121:3683–3692. doi: 10.1242/jcs.037507. [DOI] [PubMed] [Google Scholar]
- 76.Takeda T, Go WY, Orlando RA, Farquhar MG. Expression of podocalyxin inhibits cell-cell adhesion and modifies junctional properties in Madin-Darby canine kidney cells. Mol. Biol. Cell. 2000;11:3219–3232. doi: 10.1091/mbc.11.9.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wesseling J, van der Valk SW, Hilkens J. A mechanism for inhibition of E-cadherin-mediated cell-cell adhesion by the membrane-associated mucin episialin/MUC1. Mol. Biol. Cell. 1996;7:565–577. doi: 10.1091/mbc.7.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Strilic B, Eglinger J, Krieg M, Zeeb M, Axnick J, Babal P, Muller DJ, Lammert E. Elecrostatic cell surface repulsion initiates lumen formation in developing blood vessels. Curr. Biol. 2010;20:2003–2009. doi: 10.1016/j.cub.2010.09.061. [DOI] [PubMed] [Google Scholar]
- 79.Meder D, Shevchenko A, Simons K, Fullekrug J. Gp135/podocalyxin and NHERF-2 participate in the formation of a preapical domain during polarization of MDCK cells. J. Cell Biol. 2005;168:303–313. doi: 10.1083/jcb.200407072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schluter MA, Margolis B. Apical lumen formation in renal epithelia. J. Am. Soc. Nephrol. 2009;20:1444–1452. doi: 10.1681/ASN.2008090949. [DOI] [PubMed] [Google Scholar]
- 81.Bulgakova NA, Knust E. The Crumbs complex: from epithelial-cell polarity to retinal degeneration. J. Cell Sci. 2009;122:2587–2596. doi: 10.1242/jcs.023648. [DOI] [PubMed] [Google Scholar]
- 82.Saotome I, Curto M, McClatchey AI. Ezrin is essential for epithelial organization and villus morphogenesis in the developing intestine. Dev. Cell. 2004;6:855–864. doi: 10.1016/j.devcel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 83.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat. Rev. Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 84.Jaskoll T, Melnick M. Submandibular gland morphogenesis: stage-specific expression of TGF-alpha/EGF, IGF, TGF-beta, TNF, and IL-6 signal transduction in normal embryonic mice and the phenotypic effects of TGF-beta2, TGF-beta3, and EGF-r null mutations. Anat. Rec. 1999;256:252–268. doi: 10.1002/(SICI)1097-0185(19991101)256:3<252::AID-AR5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 85.Reginato MJ, Mills KR, Becker EB, Lynch DK, Bonni A, Muthuswamy SK, Brugge JS. Bim regulation of lumen formation in cultured mammary epithelial acini is targeted by oncogenes. Mol. Cell Biol. 2005;25:4591–4601. doi: 10.1128/MCB.25.11.4591-4601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schmelzle T, Mailleux AA, Overholtzer M, Carroll JS, Solimini NL, Lightcap ES, Veiby OP, Brugge JS. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proc. Natl. Acad. Sci. USA. 2007;104:3787–3792. doi: 10.1073/pnas.0700115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mailleux AA, Overholtzer M, Schmelzle T, Bouillet P, Strasser A, Brugge JS. BIM regulates apoptosis during mammary ductal morphogenesis, and its absence reveals alternative cell death mechanisms. Dev. Cell. 2007;12:221–234. doi: 10.1016/j.devcel.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol. Biol. Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pearson JF, Hughes S, Chambers K, Lang SH. Polarized fluid movement and not cell death, creates luminal spaces in adult prostate epithelium. Cell Death Differ. 2009;16:475–482. doi: 10.1038/cdd.2008.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Santiago-Martinez E, Soplop NH, Patel R, Kramer SG. Repulsion by Slit and Roundabout prevents Shotgun/E-cadherin-mediated cell adhesion during Drosophila heart tube lumen formation. J. Cell Biol. 2008;182:241–248. doi: 10.1083/jcb.200804120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Medioni C, Astier M, Zmojdzian M, Jagla K, Semeriva M. Genetic control of cell morphogenesis during Drosophila melanogaster cardiac tube formation. J. Cell Biol. 2008;182:249–261. doi: 10.1083/jcb.200801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bagnat M, Cheung ID, Mostov KE, Stainier DY. Genetic control of single lumen formation in the zebrafish gut. Nat. Cell Biol. 2007;9:954–960. doi: 10.1038/ncb1621. [DOI] [PubMed] [Google Scholar]
- 93.Gutzman JH, Sive H. Epithelial relaxation mediated by the myosin phosphatase regulator Mypt1 is required for brain ventricle lumen expansion and hindbrain morphogenesis. Development. 2010;137:795–804. doi: 10.1242/dev.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang J, Piontek J, Wolburg H, Piehl C, Liss M, Otten C, Christ A, Willnow TE, Blasig IE, Abdelilah-Seyfried S. Establishment of a neuroepithelial barrier by Claudin5a is essential for zebrafish brain ventricular lumen expansion. Proc. Natl. Acad. Sci. USA. 2010;107:1425–1430. doi: 10.1073/pnas.0911996107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moriwaki K, Tsukita S, Furuse M. Tight junctions containing claudin 4 and 6 are essential for blastocyst formation in preimplantation mouse embryos. Dev. Biol. 2007;312:509–522. doi: 10.1016/j.ydbio.2007.09.049. [DOI] [PubMed] [Google Scholar]
- 96.Nelson KS, Furuse M, Beitel GJ. The Drosophila Claudin Kune-kune is required for septate junction organization and tracheal tube size control. Genetics. 2010;185:831–839. doi: 10.1534/genetics.110.114959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krupinski T, Beitel GJ. Unexpected roles of the Na-K-ATPase and other ion transporters in cell junctions and tubulogenesis. Physiology. 2009;24:192–201. doi: 10.1152/physiol.00008.2009. [DOI] [PubMed] [Google Scholar]
- 98.Lowery LA, Sive H. Initial formation of zebrafish brain ventricles occurs independently of circulation and requires the nagie oko and snakehead/atp1a1a.1 gene products. Development. 2005;132:2057–2067. doi: 10.1242/dev.01791. [DOI] [PubMed] [Google Scholar]
- 99.Riordan JR. CFTR function and prospects for therapy. Annu. Rev. Biochem. 2008;77:701–726. doi: 10.1146/annurev.biochem.75.103004.142532. [DOI] [PubMed] [Google Scholar]
- 100.Li C, Naren AP. CFTR chloride channel in the apical compartments: spatiotemporal coupling to its interacting partners. Integr. Biol. 2010;2:161–177. doi: 10.1039/b924455g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bagnat M, Navis A, Herbstreith S, Brand-Arzamendi K, Curado S, Gabriel S, Mostov K, Huisken J, Stainier DY. Cse1l is a negative regulator of CFTR-dependent fluid secretion. Curr. Biol. 2010;20:1840–1845. doi: 10.1016/j.cub.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang B, Sonawane ND, Zhao D, Somlo S, Verkman AS. Small-molecule CFTR inhibitors slow cyst growth in polycystic kidney disease. J. Am. Soc. Nephrol. 2008;19:1300–1310. doi: 10.1681/ASN.2007070828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ghabrial A, Luschnig S, Metzstein MM, Krasnow MA. Branching morphogenesis of the Drosophila tracheal system. Annu. Rev. Cell Dev. Biol. 2003;19:623–647. doi: 10.1146/annurev.cellbio.19.031403.160043. [DOI] [PubMed] [Google Scholar]
- 104.Tsarouhas V, Senti KA, Jayaram SA, Tiklova K, Hemphala J, Adler J, Samakovlis C. Sequential pulses of apical epithelial secretion and endocytosis drive airway maturation in Drosophila. Dev. Cell. 2007;13:214–225. doi: 10.1016/j.devcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 105.Swanson LE, Beitel GJ. Tubulogenesis: an inside job. Curr. Biol. 2006;16:R51–R53. doi: 10.1016/j.cub.2006.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Luschnig S, Batz T, Armbruster K, Krasnow MA. serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol. 2006;16:186–194. doi: 10.1016/j.cub.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 107.Wang S, Jayaram SA, Hemphala J, Senti KA, Tsarouhas V, Jin H, Samakovlis C. Septate-junction-dependent luminal deposition of chitin deacetylases restricts tube elongation in the Drosophila trachea. Curr. Biol. 2006;16:180–185. doi: 10.1016/j.cub.2005.11.074. [DOI] [PubMed] [Google Scholar]
- 108.Forster D, Armbruster K, Luschnig S. Sec24-dependent secretion drives cell-autonomous expansion of tracheal tubes in Drosophila. Curr. Biol. 2010;20:62–68. doi: 10.1016/j.cub.2009.11.062. [DOI] [PubMed] [Google Scholar]
- 109.Husain N, Pellikka M, Hong H, Klimentova T, Choe KM, Clandinin TR, Tepass U. The agrin/perlecan-related protein eyes shut is essential for epithelial lumen formation in the Drosophila retina. Dev. Cell. 2006;11:483–493. doi: 10.1016/j.devcel.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 110.Abd El-Aziz MM, O’Driscoll CA, Kaye RS, Barragan I, El-Ashry MF, Borrego S, Antinolo G, Pang CP, Webster AR, Bhattacharya SS. Identification of novel mutations in the ortholog of Drosophila eyes shut gene (EYS) causing autosomal recessive retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 2010;51:4266–4272. doi: 10.1167/iovs.09-5109. [DOI] [PubMed] [Google Scholar]
- 111.Collin RW, Littink KW, Klevering BJ, van den Born LI, Koenekoop RK, Zonneveld MN, Blokland EA, Strom TM, Hoyng CB, den Hollander AI, et al. Identification of a 2 Mb human ortholog of Drosophila eyes shut/spacemaker that is mutated in patients with retinitis pigmentosa. Am. J. Hum. Genet. 2008;83:594–603. doi: 10.1016/j.ajhg.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Van Teeffelen JW, Brands J, Stroes ES, Vink H. Endothelial glycocalyx: sweet shield of blood vessels. Trends Cardiovasc. Med. 2007;17:101–105. doi: 10.1016/j.tcm.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 113.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 114.Wendeler MW, Paccaud JP, Hauri HP. Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep. 2007;8:258–264. doi: 10.1038/sj.embor.7400893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Merte J, Jensen D, Wright K, Sarsfield S, Wang Y, Schekman R, Ginty DD. Sec24b selectively sorts Vangl2 to regulate planar cell polarity during neural tube closure. Nat. Cell Biol. 2010;12(sup):41–46. doi: 10.1038/ncb2002. 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grieder NC, Caussinus E, Parker DS, Cadigan K, Affolter M, Luschnig S. gammaCOP is required for apical protein secretion and epithelial morphogenesis in Drosophila melanogaster. PLoS ONE. 2008;3:e3241. doi: 10.1371/journal.pone.0003241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jayaram SA, Senti KA, Tiklova K, Tsarouhas V, Hemphala J, Samakovlis C. COPI vesicle transport is a common requirement for tube expansion in Drosophila. PLoS ONE. 2008;3:e1964. doi: 10.1371/journal.pone.0001964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Townley AK, Feng Y, Schmidt K, Carter DA, Porter R, Verkade P, Stephens DJ. Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal craniofacial development. J. Cell Sci. 2008;121:3025–3034. doi: 10.1242/jcs.031070. [DOI] [PubMed] [Google Scholar]
- 119.Bryant D, Mostov Development: inflationary pressures. Nature. 2007;449:549–550. doi: 10.1038/449549a. [DOI] [PubMed] [Google Scholar]
- 120.McCaffrey LM, Macara IG. Widely conserved signaling pathways in the establishment of cell polarity. Cold Spring Harb. Perspect. Biol. 2009;1:a001370. doi: 10.1101/cshperspect.a001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Laprise P, Paul SM, Boulanger J, Robbins RM, Beitel GJ, Tepass U. Epithelial polarity proteins regulate Drosophila tracheal tube size in parallel to the luminal matrix pathway. Curr. Biol. 2010;20:55–61. doi: 10.1016/j.cub.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Laprise P, Lau KM, Harris KP, Silva-Gagliardi NF, Paul SM, Beronja S, Beitel GJ, McGlade CJ, Tepass U. Yurt, Coracle, Neurexin IV and the Na(+), K(+)-ATPase form a novel group of epithelial polarity proteins. Nature. 2009;459:1141–1145. doi: 10.1038/nature08067. [DOI] [PubMed] [Google Scholar]
- 123.Horne-Badovinac S, Lin D, Waldron S, Schwarz M, Mbamalu G, Pawson T, Jan Y, Stainier DY, Abdelilah-Seyfried S. Positional cloning of heart and soul reveals multiple roles for PKC lambda in zebrafish organogenesis. Curr. Biol. 2001;11:1492–1502. doi: 10.1016/s0960-9822(01)00458-4. [DOI] [PubMed] [Google Scholar]
- 124.Munson C, Huisken J, Bit-Avragim N, Kuo T, Dong PD, Ober EA, Verkade H, Abdelilah-Seyfried S, Stainier DY. Regulation of neurocoel morphogenesis by Pard6 gamma b. Dev. Biol. 2008;324:41–54. doi: 10.1016/j.ydbio.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Omori Y, Malicki J. oko meduzy and related crumbs genes are determinants of apical cell features in the vertebrate embryo. Curr. Biol. 2006;16:945–957. doi: 10.1016/j.cub.2006.03.058. [DOI] [PubMed] [Google Scholar]
- 126.Desclozeaux M, Venturato J, Wylie FG, Kay JG, Joseph SR, Le HT, Stow JL. Active Rab11 and functional recycling endo-some are required for E-cadherin trafficking and lumen formation during epithelial morphogenesis. Am. J. Physiol. Cell Physiol. 2008;295:C545–C556. doi: 10.1152/ajpcell.00097.2008. [DOI] [PubMed] [Google Scholar]
- 127.Shaye DD, Casanova J, Llimargas M. Modulation of intracellular trafficking regulates cell intercalation in the Drosophila trachea. Nat. Cell Biol. 2008;10:964–970. doi: 10.1038/ncb1756. [DOI] [PubMed] [Google Scholar]
- 128.De Craene B, Gilbert B, Stove C, Bruyneel E, van Roy F, Berx G. The transcription factor snail induces tumor cell invasion through modulation of the epithelial cell differentiation program. Cancer Res. 2005;65:6237–6244. doi: 10.1158/0008-5472.CAN-04-3545. [DOI] [PubMed] [Google Scholar]
- 129.Whiteman EL, Liu CJ, Fearon ER, Margolis B. The transcription factor snail represses Crumbs3 expression and disrupts apicobasal polarity complexes. Oncogene. 2008;27:3875–3879. doi: 10.1038/onc.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Escriva M, Peiro S, Herranz N, Villagrasa P, Dave N, Montserrat-Sentis B, Murray SA, Franci C, Gridley T, Virtanen I, et al. Repression of PTEN phosphatase by Snail1 transcriptional factor during gamma radiation-induced apoptosis. Mol. Cell Biol. 2008;28:1528–1540. doi: 10.1128/MCB.02061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kerman BE, Cheshire AM, Myat MM, Andrew DJ. Ribbon modulates apical membrane during tube elongation through Crumbs and Moesin. Dev. Biol. 2008;320:278–288. doi: 10.1016/j.ydbio.2008.05.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gao N, Kaestner KH. Cdx2 regulates endo-lysosomal function and epithelial cell polarity. Genes Dev. 2010;24:1295–1305. doi: 10.1101/gad.1921510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schonteich E, Wilson GM, Burden J, Hopkins CR, Anderson K, Goldenring JR, Prekeris R. The Rip11/Rab11-FIP5 and kinesin II complex regulates endocytic protein recycling. J. Cell Sci. 2008;121:3824–3833. doi: 10.1242/jcs.032441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Grant M, Mostov K, Tlsty T, Hunt CA. Simulating properties of in vitro epithelial cell morphogenesis. PLoS Comp. Biol. 2006;2:e129. doi: 10.1371/journal.pcbi.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Engelberg JA, Kim M, Mostov KE, Hunt CA. An agent-based model of epithelial cystogenesis implemented with a cellular Potts model. J. Crit. Care. 2009;24:e32–e33. [Google Scholar]
- 137.Kim SH, Park S, Mostov K, Debnath J, Hunt CA. Computational investigation of epithelial cell dynamic phenotype in vitro. Theor. Biol. Med. Model. 2009;6:8. doi: 10.1186/1742-4682-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Harris PC, Torres VE. Polycystic kidney disease. Annu. Rev. Med. 2009;60:321–337. doi: 10.1146/annurev.med.60.101707.125712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lowery LA, Sive H. Totally tubular: the mystery behind function and origin of the brain ventricular system. Bioessays. 2009;31:446–458. doi: 10.1002/bies.200800207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yamaguchi Y, Yonemura S, Takada S. Grainyhead-related transcription factor is required for duct maturation in the salivary gland and the kidney of the mouse. Development. 2006;133:4737–4748. doi: 10.1242/dev.02658. [DOI] [PubMed] [Google Scholar]
- 141.Hemphala J, Uv A, Cantera R, Bray S, Samakovlis C. Grainy head controls apical membrane growth and tube elongation in response to Branchless/FGF signalling. Development. 2003;130:249–258. doi: 10.1242/dev.00218. [DOI] [PubMed] [Google Scholar]
- 142.Narasimha M, Uv A, Krejci A, Brown NH, Bray SJ. Grainy head promotes expression of septate junction proteins and influences epithelial morphogenesis. J. Cell Sci. 2008;121:747–752. doi: 10.1242/jcs.019422. [DOI] [PubMed] [Google Scholar]
- 143.Myat MM, Andrew DJ. Epithelial tube morphology is determined by the polarized growth and delivery of apical membrane. Cell. 2002;111:879–891. doi: 10.1016/s0092-8674(02)01140-6. [DOI] [PubMed] [Google Scholar]
- 144.Godmann M, Katz JP, Guillou F, Simoni M, Kaestner KH, Behr R. Kruppel-like factor 4 is involved in functional differentiation of testicular Sertoli cells. Dev. Biol. 2008;315:552–566. doi: 10.1016/j.ydbio.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Amack JD, Wang X, Yost HJ. Two T-box genes play independent and cooperative roles to regulate morphogenesis of ciliated Kupffer’s vesicle in zebrafish. Dev. Biol. 2007;310:196–210. doi: 10.1016/j.ydbio.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 146.Araujo SJ, Cela C, Llimargas M. Tramtrack regulates different morphogenetic events during Drosophila tracheal development. Development. 2007;134:3665–3676. doi: 10.1242/dev.007328. [DOI] [PubMed] [Google Scholar]