Abstract
We propose and demonstrate a dark-field imaging technique capable of automated identification of individual bacteria. An 87-channel multispectral system capable of angular and spectral resolution was used to measure the scattering spectrum of various bacteria in culture smears. Spectra were compared between various species and between various preparations of the same species. A 15-channel system was then used to prove the viability of bacterial identification with a relatively simple microscope system. A simple classifier was able to identify four of six bacterial species with greater than 90% accuracy in bacteria-by-bacteria testing.
OCIS codes: (170.0180) Microscopy, (170.4580) Optical diagnostics for medicine, (290.5820) Scattering measurements
1. Introduction
Accurate identification of bacteria is vital for many applications such as infectious disease diagnosis, food contamination monitoring, and microbiological research. Common methods of bacterial identification include culture, PCR, and microscopy. Culture and PCR are methods that require a laboratory setting and significant resources to perform. In addition, bacteria culturing remains a time-consuming process, taking days – or in the case of slow-growing bacteria weeks – to achieve sufficient quantities of bacteria for reliable identification. While microscopy is a well-established method in microbiology, it relies on skilled microscopists to identify key morphological traits and stains that generally lack genus-level specificity. This skill requirement has restricted the adoption of microscopy in resource-limited environments such as field clinics for malaria and tuberculosis diagnosis [1]. Outside of maintained laboratories there is often a lack of necessary equipment, consumables, and experience to effectively detect and identify bacterial contaminants or pathogens using culture, PCR, or microscopy [2]. Accurate, onsite bacterial identification becomes particularly important for the diagnosis of diseases that require medical intervention such as drug administration, as patient follow-up is often difficult in resource-limited settings.
Scattering measurements have been used to identify bacteria and other cells for decades [3–6]. First proposed in the 1960’s, flow cytometer systems capable of fine angular resolution scattering measurements have been shown to produce effective identification for certain bacterial species. In addition, the theory behind extracting particle information from angle-resolved scattering measurements is well-explored [7–9]. Angle-resolved flow cytometry systems are inherently complex and expensive, requiring complex optics to achieve sufficient angular resolution. More recently, systems with fewer channels have been explored in conjunction with more sophisticated machine learning algorithms [10].
Dark-field (DF) microscopy has seen relatively little use in clinical microscopy applications [11–13]. DF has been used to image bacterial flagella [14] and is an established procedure for the diagnosis of syphilis infections [13]. DF imaging is an inherently scattering-based method that can be used to determine optical characteristics that are generally unobservable in bright-field. While DF microscopy is relatively simple to implement, the numerical aperture necessary to resolve individual bacteria makes the fine angular resolution necessary for scattering-based identification cumbersome.
Recent methods have combined DF microscopy with hyperspectral cameras for nanoparticle imaging in biological applications [15, 16] and hyperspectral dark-field microscopes are commercially available. The present work explores the capabilities of a dark-field microscopy system capable of both angular and spectral resolution, and its potential for identifying bacterial species without exogenous contrast agents. This method will be referred to as multispectral angular-resolved dark-field imaging (MARDI). We will present the scattering spectra (resolved in wavelength and angle) for various bacteria and identify which intra-species variations can affect the scattering spectra. We also present a simple computer algorithm for bacteria identification based on scattering spectrum.
2. Method
2.1 Microscope system
To create a MARDI microscope, a monochromator was inserted between the lamp and the illuminator optics of a transmission-mode upright microscope. The monochomator had a FWHM of 10 nm and was varied from 420 nm to 700 nm in 10 nm increments. The Abbe condenser (dry, NA = 0.9) was fitted with a carousel containing three DF aperture rings to produce different angles of illumination (shown in Fig. 1(a) ). Light was collected with a dry 40x long working distance objective (NA = 0.45). Images were captured with an Andor Luca-R EMCCD camera. The electron-multiplier gain of the EMCCD was kept at a constant value for all experiments, which was within the linear operation range of the EMCCD. This optical configuration is shown in Fig. 1(b). 87 grey-scale images (29 wavelengths times 3 DF rings) were normalized by imaging 2 µm silica spheres (n = 1.45) suspended in cytoseal (Richard Allan, Kalamazoo, MI, n = 1.48) and normalizing to expected values calculated using Mie theory. 2 µm silica spheres were chosen to simulate the size and shape of bacteria and were easily resolved by the microscope.
Fig. 1.
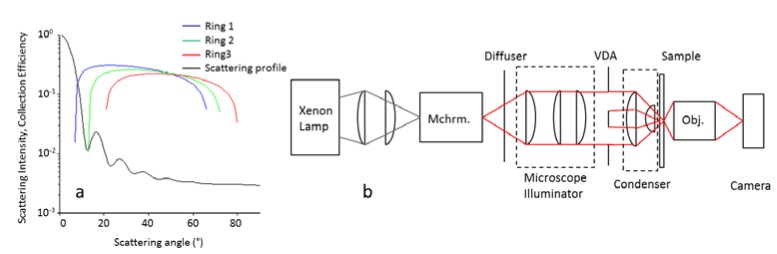
(a) Theoretical collection efficiencies of the three rings of the VDA. The black outline is the unpolarized scattering intensity of a 2 µm silica sphere in cytoseal at 550 nm. (b) Schematic for the 87-channel MARDI microscope. Mchrm. = monochromator, VDA = variable dark-field aperture, Obj. = objective.
A 15-channel MARDI system was also used. This system was identical to the previously described system with the exception of the monochomator being omitted and the EMCCD camera being replaced by a Leica EC3 RGB camera. 15 channels were comprised of three colors (RGB) at five illumination angles. Normalization for this system was performed by white balancing on a white target.
2.2 Bacterial samples
Haemophilus influenzae type b (Hib) 10211 was cultured on chocolate agar and in tryptic soy broth (TSB; Fluka, St. Louis, MO) in the presence of 10% CO2 for 24 h. Pseudomonas aeruginosa 15442 and Staphylococcus aureus 6558 were cultured on tryptic soy agar (TSA; Fluka) and TSB for 12-24 hours. Mycobacterium smegmatis mc2155 was cultured in Middlebrook 7H9 broth (Fluka) supplemented with AODC (Beckon Dickenson, Sparks MD) and Middlebrook 7H10 agar (BD, Franklin Lakes, NJ) with AODC growth supplement and grown for 24-48 hours. Mycobacterium bovis BCG 19274 was grown in 7H9 broth and 7H11 agar supplemented with AODC and Sodium Pyruvate substituted for glycerol for 3-4 weeks. All incubations were performed at 37°C. All cultured bacteria strains were purchased from ATCC, (Manassas, VA). Escherichia coli, additional S. aureus, and Streptococcus pneumoniae culture smears were purchased as prepared slides (Ward’s Natural Science, Rochester, NY). H. influenzae, S. pneumoniae, P. aeruginosa, and S. aureus are species commonly found in sputum and leading causes of bacterial pneumonia [17–20]. M. smegmatis and M. bovis BCG are closely related to Mycobacterium tuberculosis (M.tb).
To test the effect of culture growth stage, samples were prepared from a single culture of M. smegmatis at culture ages of 6, 12, 24, and 48 hours. Using plate counts and optical absorption spectroscopy the 6 and 12-hour-old samples were found to be in log phase, the 24-hour-old sample in stationary phase, and the 48-hour-old sample in death phase. An additional step of centrifugation and rinsing was used to concentrate these samples; denoted by C6, C12, C24, and C48, respectively. The stationary phase culture smears (C24) were used to compare sample preparation methods to the standard preparation of M. smegmatis (with samples denoted by N). As a third sample preparation for M. smegmatis, cultures were prepared using the same culture medium and sample preparation process as M. bovis BCG (this involved a modified culture medium and the use of dispersants to break up bacterial agglomerations before smearing). These samples are denoted by SP.
To test the effect of cell wall and membrane on scattering the outer membrane of Gram-negative P. aeruginosa was stripped using a five-minute exposure to 70% ethanol solution after smearing. Scattering spectra were measured on unstained samples, which were then Gram stained to confirm successful membrane stripping.
2.2 Image processing and classifier algorithm
Figure 2 shows a diagram of MARDI post processing. Significant objects in the original image stacks were identified using background subtraction and a dynamic threshold. Identified objects were then pre-filtered for area A and compactness c = P 2/A, where P is the perimeter. Pre-filtering was designed to screen objects too small or large to be bacteria. Because the same size filter was used for all samples, clusters of smaller bacteria regularly passed the pre-filter and were included in subsequent testing. Once thumbnails of significant objects were extracted from the original images, backgrounds were subtracted and the intensity was summed over all pixels belonging to the object. This resulted in a single 87-dimensional vector (29 wavelengths, three angles) to characterize each identified object, which was then normalized for overall brightness and background subtracted. This vector is referred to as the object’s scattering spectrum .
Fig. 2.
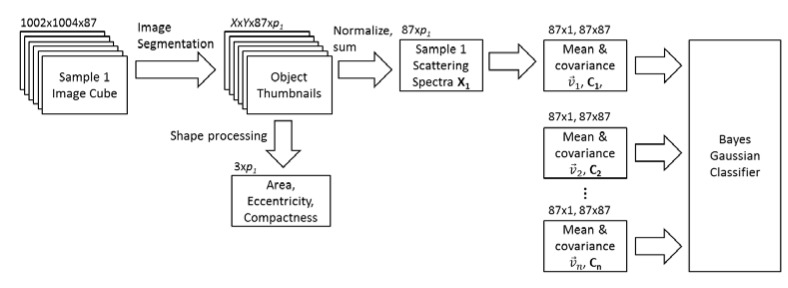
Diagram of MARDI post processing and classification algorithm.
In addition to the scattering spectrum, area, eccentricity, and compactness were recorded for each object. While algorithms exist for shape identification in microscope images, including M.tb identification [21, 22], these were omitted from our process to isolate the discriminatory capabilities of the scattering spectrum.
To test the usefulness of the bacterial scattering spectra for species identification a Bayes-Gaussian classifier [23] was used. Briefly, these classifiers use the covariance matrix C and mean vector from n training classes to train a classifier that can then be used to classify a test set. For each test vector to be classified the classifier produces n likelihood scores, one for each of the training classes, using the follow criterion [23]:
| (1) |
where dj is the likelihood score for the jth class and P(wj) is the probability of the jth class occurring. The classification decision is made by designating the test vector to the class with the highest likelihood score. All classifier results reported in this work were produced using leave one out cross validation. In all tests an uninformative prior was chosen, so P(wj) was set as equal for all classes. Sensitivity and positive predictive value (PPV) were used as performance metrics for the classifier. Sensitivity is defined as the likelihood of a bacterium of known class being accurately identified by the classifier. PPV is defined as the likelihood that a bacterium classified as a specific class was in fact a bacterium of that class.
Principle component analysis (PCA) – implemented using singular value decomposition – was used to visualize cluster separation. In all PCA tests an equal number of test vectors were used for each test class.
3. Results
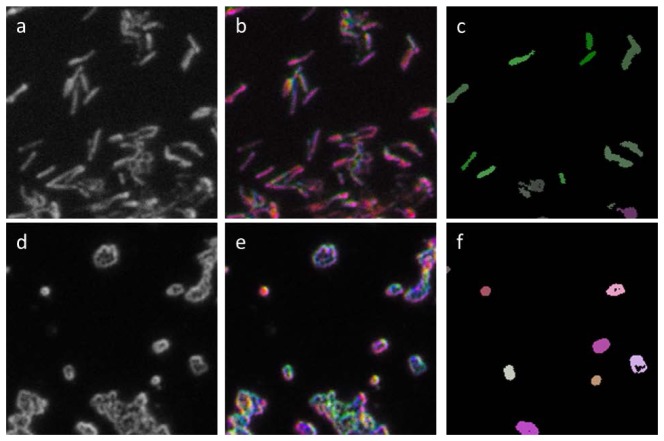
Figure 3 shows example MARDI images. MARDI produces dark-field images as seen in Fig. 3(a) and 3(d), with bacteria appearing light and on a dark background. Each pixel has a corresponding 87-dimensional scattering spectrum. Figures 3(b) and 3(e) show the common method of dimensional reduction based on PCA scores [24]. This pixel-by-pixel PCA method is computationally expensive and principal components were generally dominated by the chromatic aberrations of the MARDI system, thus this method was of limited use for identification.
Fig. 3.
Sample MARDI images of M. smegmatis (a-c) and S. aureus (d-f). (a) and (d) show a portion of a single channel without post processing. Each frame is 80x80 µm. (b) and (e) show the same frames colored by assigning RGB values to pixel-by-pixel PCA score for three modes. (c) and (f) show the same field as (a) and (d), respectively, with object identification and summing prior to PCA. RGB values are assigned to three PCA mode scores. In all cases mode scores were chosen to maximize the visible difference between species.
Figure 3(c) and 3(f) are a graphic representation of the method used for classification described in Fig. 2. Instead of pixel-by-pixel PCA, significant objects were identified and their pixels averaged before extracting the scattering spectrum. This method was found to eliminate the effects of chromatic aberrations, thus producing consistent scattering spectra over full fields of view. The colors in Fig. 3(c) and 3(f) were then assigned based on the PCA scores of each object’s scattering spectrum.
3.1 Intra-species
M. smegmatis and S. aureus were used to study the variation of scattering spectra within populations of a single species. Figure 4 shows PCA score values for several sets of samples. In all cases each test group is composed of 100 bacteria chosen randomly from multiple slides. Figure 4(a) shows the first principal component scores for M. smegmatis samples of different culture ages. Note the poor separation of clusters. In Fig. 4(b) the first principal components are plotted for S. aureus from two lab culture slides and three purchased slides. Figure 4(c) shows the comparative effect of differing sample preparations.
Fig. 4.
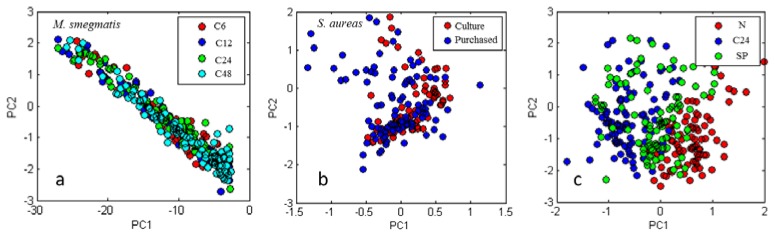
Principal components score plots for various intra-species tests. (a) M. smegmatis prepared with method C6, C12, C24, and C48, (b) S. aureas from purchased slides and slides prepared from lab culture, (c) M. smegmatis prepared with methods N, C24, and SP.
A Bayes-Gaussian classifier was used to quantify the potential classification accuracy for distinguishing culture age and sample preparation methods. Results are shown in Tables 1 and 2 , respectively. In these and all following tables correct classifications are marked in gray on the diagonal and common misclassifications are marked in different shades of red according to their likelihood. Sensitivity and PPV are shown for each sample set. All classifier results shown in this paper were obtained using between N = 250 and N = 4576 individual bacteria from two to four different slides.
Table 1. Classifier results for culture age test with M. smegmatis .
| Classification (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Truth | C6 | C12 | C24 | C48 | Sens (%) | ||||
| C6 | 78.5 | 3.2 | 8.5 | 9.7 | 78.5 | ||||
| C12 | 4.3 | 57.0 | 18.8 | 19.9 | 57.0 | ||||
| C24 | 6.5 | 14.2 | 65.0 | 14.2 | 65.0 | ||||
| C48 | 3.6 | 13.5 | 12.5 | 70.4 | 70.4 | ||||
| PPV (%) | 84.5 | 64.8 | 62.0 | 61.6 | |||||
Table 2. Classifier results for culture preparation tests of M. smegmatis .
| Classification (%) | |||||||
|---|---|---|---|---|---|---|---|
| Truth | N | C24 | SP | M. bov. | Sens (%) | ||
| N | 88.3 | 5.9 | 1.8 | 4.0 | 88.3 | ||
| C24 | 10.5 | 70.4 | 10.5 | 8.6 | 70.4 | ||
| SP | 3.9 | 15.4 | 66.8 | 13.9 | 66.8 | ||
| M. bovis | 6.1 | 7.2 | 18.7 | 68.0 | 68.0 | ||
| PPV(%) | 81.1 | 71.2 | 68.3 | 72.0 | |||
The way the sample was cultured and prepared can be seen to affect the scattering spectrum. Of particular interest is that M. smegmatis prepared using the M. bovis culture method (M. smegmatis SP) is much more often misclassified as M. bovis than it is misclassified as M. smegmatis cultured in the recommended way (M. smegmatis N). Note that the difference in culture method between M. smegmatis N and SP entailed only substituting glycerol for sodium pyruvate in the culture medium and the addition of surfactants. In contrast, the age of the culture when the sample was prepared was found to be relatively unimportant, although the 6-hour old preparation (C6) was the easiest to distinguish.
The effect of cell membrane on the scattering spectrum was tested by stripping the outer membrane of Gram-negative P. aeruginosa. Outer membranes were successfully stripped as shown in Fig. 5(b) and 5(c). The score plot for stripped and non-stripped P. aeruginosa is shown in Fig. 5(a). Note the poor cluster separation, implying that the presence or lack of outer membrane has limited effect on bacterial scattering. Despite the poor cluster separation in Fig. 5, the Bayes-Gaussian classifier distinguished the two classes with 90% accuracy.
Fig. 5.
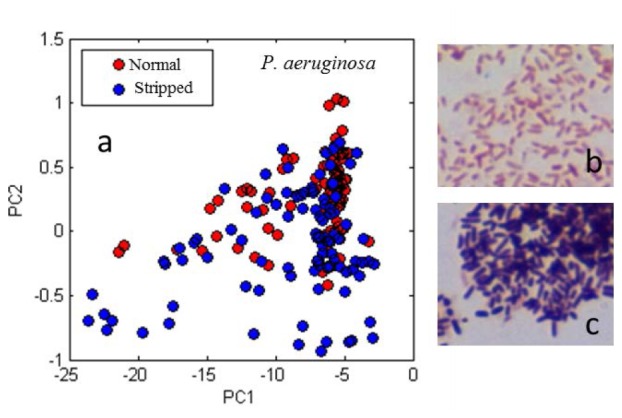
(a) Principal components score plot for two samples each of normally prepared and membrane stripped P. aeruginosa. (b, c) Micrographs of Gram stained P. aeruginosa without (b) and with (c) membrane stripped.
The age of the smear on the slide was found to not affect the scattering spectrum of the bacteria; no changes in spectrum were observed even after several months at room temperature.
3.2 Interspecies
The scattering spectra of various bacterial species are shown in Fig. 6 . As expected for particles of bacteria’s size and dielectric properties, the scattering spectra lack sharp features. The spectra do have distinguishing features, however, when averaged over thousands of bacteria.
Fig. 6.
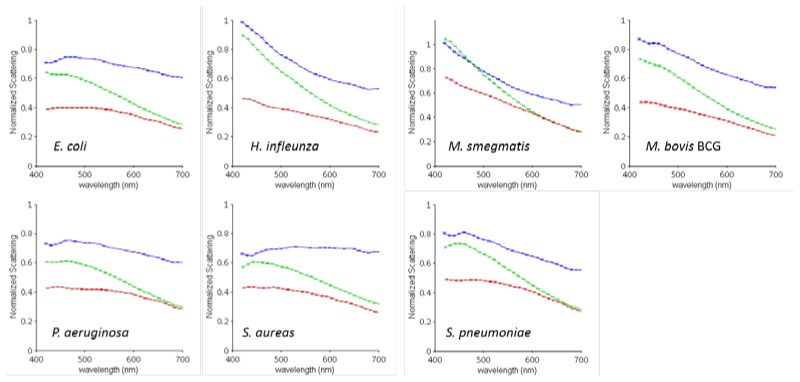
Scattering spectra of the eight tested bacteria species. The three dark-field aperture setting are denoted by colors corresponding to Fig. 1(b) (ring 1 = blue, ring 2 = green, ring 3 = red). Error bars show the standard error of the mean for sample sizes ranging from N = 346 for S. pneumoniae to N = 2172 for P. aeruginosa.
Figure 6 shows that specific scattering characteristics exist for bacterial species. These scattering characteristics enable individual bacteria to be identified. Some principal components are shown in Fig. 7 . Figure 7(a) shows H. influenzae and S. aureus. H. influenzae and S. aureus represent a pair of species that are easily distinguished in classifier tests. Figure 7(b) shows two preparations of M. smegmatis (N and SP) with M. bovis BCG. Note that as expected from results in Table 2 M. smegmatis SP clusters more closely with M. bovis than with M. smegmatis N. Table 3 contains the Bayes-Gaussian classifier results for different bacterial species.
Fig. 7.
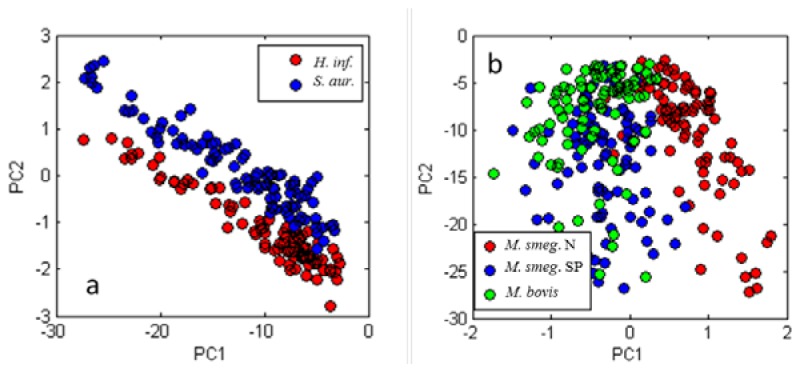
Principal components score plots for various inter-species tests. (a) H. influenzae and S. aureas. (b) two preparations of M. smegmatis versus M. bovis BCG.
Table 3. Classifier results for interspecies tests with 87 channel system.
| Classification (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| E. coli | H. inf. | M. sm. | M. bo. | P. aer. | S. aur. | S. pne. | Sens(%) | ||
| Truth | E. coli | 85.1 | 0.8 | 1.9 | 1.3 | 3.9 | 3.0 | 4.0 | 85.1 |
| H. inf. | 0.2 | 95.9 | 1.6 | 1.9 | 0.1 | 0.4 | 0.0 | 95.9 | |
| M. sme. | 3.2 | 4.1 | 81.1 | 7.2 | 1.6 | 0.9 | 2.0 | 81.1 | |
| M. bov. | 0.3 | 3.0 | 4.0 | 88.7 | 0.6 | 0.3 | 3.2 | 88.7 | |
| P. aer. | 1.7 | 0.4 | 1.6 | 0.5 | 92.8 | 1.3 | 1.8 | 92.8 | |
| S. aur. | 2.0 | 0.9 | 0.6 | 0.5 | 0.2 | 94.3 | 1.5 | 94.3 | |
| S. pne. | 4.3 | 1.5 | 3.2 | 4.3 | 3.8 | 4.3 | 78.6 | 78.6 | |
| PPV (%) | 88.0 | 90.1 | 86.3 | 84.9 | 90.2 | 90.2 | 86.2 | ||
Certain bacteria species, including S. aureas and H. infleunzae, were distinguished on a bacterium-by-bacterium basis with greater than 90% accuracy, even from a pool of seven possible species. Other species were much less discernible, such as S. pneumoniae, which was difficult to tell apart from the pool in general. The most common misclassification was between M. smegmatis and M. bovis, which were the most closely related species in the test.
3.3 15-Channel system
The results in Tables 1 through 3 were found utilizing an 87-dimensional observation of individual bacteria. Such a system is likely too slow and complex for use in realistic diagnostic situations. To show the potential for a simpler diagnostic device a 15-channel system utilizing an RGB camera for spectral resolution was tested.
In general, the 15-channel system gave more consistent and accurate results than the 87-channel system. This is attributed primarily to a simpler acquisition process leading to a reduction in systematic noise. For the 15-channel system PCA results show cluster separation superior to the 87-channel system for most species (Fig. 8(a) ). Table 4 also shows higher sensitivities than Table 3 and misclassifications concentrated in only two interspecies mistakes (E. coli – P. aeruginosa and M. smegmatis – S. aureas). Interestingly, S. pneumonia, which had the lowest sensitivity with the 87-channel system, had the highest sensitivity with the 15-channel system.
Fig. 8.
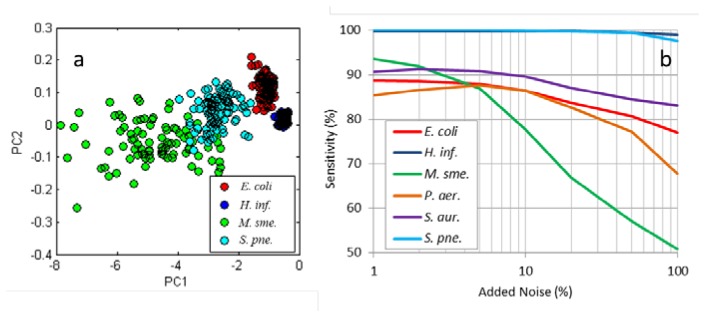
(a) PCA scores for four species scanned using the 15 channel system, showing good cluster separation. (b) Sensitivity for classification of different species as a function of added Gaussian noise.
Table 4. Classifier results for interspecies tests with 15 channel system.
| Classification (%) | ||||||||
|---|---|---|---|---|---|---|---|---|
| E. Coli | H. inf. | M. sme. | P. aer. | S. aur. | S. pne. | Sens (%) | ||
| Truth | E. coli | 88.7 | 0.0 | 0.2 | 10.9 | 0.0 | 0.2 | 88.7 |
| H. inf. | 0.0 | 99.7 | 0.3 | 0.0 | 0.0 | 0.0 | 99.7 | |
| M. sme. | 0.0 | 0.0 | 93.5 | 0.0 | 6.5 | 0.0 | 93.5 | |
| P. aer. | 14.6 | 0.0 | 0.0 | 85.4 | 0.0 | 0.0 | 85.4 | |
| S. aur. | 0.0 | 0.0 | 9.3 | 0.0 | 90.7 | 0.0 | 90.7 | |
| S. pne. | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 100.0 | 100.0 | |
| PPV (%) | 85.8 | 100.0 | 90.5 | 88.7 | 93.3 | 99.8 | ||
To test the robustness of the spectral features used in classification, sensitivity was monitored as Gaussian noise was added to the spectra used for training and testing the Bayes-Gaussian classifier. This had the additional effect of showing whether systematic noise was being used by the classifier to distinguish species. Gaussian noise was added multiplicatively as the most likely sources of systematic noise in the system are from focus errors, white balance, and exposure; all of which introduce multiplicative noise to the scattering spectrum. Results are shown in Fig. 8(b). With the exception of M. smegmatis, all species maintain their sensitivity until at least 10% added noise. Sensitivity roll off above 10% noise is due to increased misclassifications of M. smegmatis to S. aureas and E. coli to P. aeruginosa.
4. Discussion
Different bacterial species were found to have unique scattering spectra. The origin of such unique spectral features can be inferred by referring to previous works. Differences in scattering profile can be due to a bacterium’s size, shape, bulk refractive index, any present pigment, and internal variations of refractive index. The coarse angular resolution of the MARDI system strongly implies that gathered light is mainly from the main lobe of the bacterium’s scattering profile. This is a good indication that unique spectral features are likely due to the size, shape, and bulk refractive index of the bacteria [7, 8, 10]. This is confirmed in part by the result that stripping the cell membrane of P. aeruginosa had only minor effects on scattering spectrum (Fig. 4). The cell wall structure has often been cited as the most likely source of internal index of refraction variation [3, 9]. There was no clear correlation between bacterial shape or cell wall structure and scattering spectra. It should be noted, however, that referenced works measured bacteria in aqueous suspension.
The data in Tables 3 and 4 and Fig. 8(b) show the results of computer classification algorithms that utilize only the scattering spectra of the test bacteria. Were a MARDI system to be used for bacteria identification applications, shape, size, and clustering pattern observations could be easily incorporated into the computer classification algorithm. Using shape and size observations could help add robustness to the algorithm for identifying bacteria of similar scattering spectra, such as M. smegmatis and S. aureas, which are significantly different in shape (see Fig. 3).
5. Conclusion and future work
The results of the previous sections must be considered in context of potential applications for MARDI identification. While we have shown that scattering spectra can be used to identify bacterial species, scattering spectra were also shown to vary based on the environmental conditions surrounding the bacteria (in this case the culture medium) and how samples were prepared. Therefore any diagnostic test would require a rigid process of sample preparation. In one foreseeable application MARDI identification could be used in conjunction with cell culture to classify bacteria. This would ensure sufficient control over the bacteria’s growth medium and sample preparation.
While MARDI combined with cell culture could potentially cut culture time and complexity, the greater potential of MARDI is perhaps best realized in using direct environmental or clinical samples. The development of a simplified MARDI system could enable the use of samples such as ground water, sputum, blood, or feces for fast on-site identification of pathogenic species, which is not currently possible for PCR, flow cytometry, or culture. This raises additional challenges for sample preparation to ensure that the resulting smear is relatively free of environmental debris and necessitates classification algorithms that account for environmental contaminants on the same size order as bacteria. Such methods are currently under development for sputum samples for use in diagnosis of TB and pneumonia.
Acknowledgments
The authors thank Bill and Melinda Gates for their active support of this work and their sponsorship through the Global Good Fund. Support with M. bovis BCG culture was provided by Christine Cosma at the University of Washington Department of Microbiology. Josh Proctor, Grace Huynh, and Sarah Walters supplied valuable support to this project at Intellectual Ventures Laboratory.
References and links
- 1.International Union against Tuberculosis and Disease, “Priorities for tuberculosis bacteriology services in low income countries,” (2007).
- 2.The New Diagnostics Working Group of the Stop TB Partnership , “Pathways to better diagnostics for tuberculosis” (2009).
- 3.Wyatt P. J., “Differential light scattering: a physical method for identifying living bacterial cells,” Appl. Opt. 7(10), 1879–1896 (1968). 10.1364/AO.7.001879 [DOI] [PubMed] [Google Scholar]
- 4.Wyatt P. J., Phillips D. T., “Structure of single bacteria from light scattering,” J. Theor. Biol. 37(3), 493–501 (1972). 10.1016/0022-5193(72)90087-2 [DOI] [PubMed] [Google Scholar]
- 5.Bartholdi M., Salzman G. C., Hiebert R. D., Kerker M., “Differential light scattering photometer for rapid analysis of single particles in flow,” Appl. Opt. 19(10), 1573–1581 (1980). 10.1364/AO.19.001573 [DOI] [PubMed] [Google Scholar]
- 6.Loken M. R., Sweet R. G., Herzenberg L. A., “Cell discrimination by multiangle light scattering,” J. Histochem. Cytochem. 24(1), 284–291 (1976). 10.1177/24.1.1254923 [DOI] [PubMed] [Google Scholar]
- 7.Kerker M., Chew H., McNulty P. J., Kratohvil J. P., Cooke D. D., Sculley M., Lee M. P., “Light scattering and fluorescence by small particles having internal structure,” J. Histochem. Cytochem. 27(1), 250–263 (1979). 10.1177/27.1.438501 [DOI] [PubMed] [Google Scholar]
- 8.Mullaney P. F., Dean P. N., “Cell sizing: a small-angle light-scattering method for sizing particles of low relative refractive index,” Appl. Opt. 8(11), 2361–2362 (1969). 10.1364/AO.8.002361 [DOI] [PubMed] [Google Scholar]
- 9.Morris V. J., Jennings B. R., “Light scattering by bacteria. I. Angular dependence of the scattered intensity,” Proc. Roy. Soc. A: Math. Phys. Eng. Sci. 338(1613), 197–208 (1974). 10.1098/rspa.1974.0081 [DOI] [Google Scholar]
- 10.Rajwa B., Venkatapathi M., Ragheb K., Banada P. P., Hirleman E. D., Lary T., Robinson J. P., “Automated classification of bacterial particles in flow by multiangle scatter measurement and support vector machine classifier,” Cytometry A 73(4), 369–379 (2008). 10.1002/cyto.a.20515 [DOI] [PubMed] [Google Scholar]
- 11.Jamjoom G. A., “Dark-field microscopy for detection of malaria in unstained blood films,” J. Clin. Microbiol. 17(5), 717–721 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson B. K., Behrend M. R., Horning M. P., Hegg M. C., “Detection of malarial byproduct hemozoin utilizing its unique scattering properties,” Opt. Express 19(13), 12190–12196 (2011). 10.1364/OE.19.012190 [DOI] [PubMed] [Google Scholar]
- 13.S. A. Larsen, B. M. Steiner, A. H. Rudolph, S. A. Larsen, and B. M. Steiner, “Laboratory diagnosis and interpretation of tests for syphilis,” Clin. Microbio. Rev. 8, (1995). [DOI] [PMC free article] [PubMed]
- 14.Macnab R. M., “Examination of bacterial flagellation by dark-field examination of bacterial flagellation by dark-field microscopy,” J. Clin. Microbio. 4, (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seekell K., Crow M. J., Marinakos S., Ostrander J., Chilkoti A., Wax A., “Hyperspectral molecular imaging of multiple receptors using immunolabeled plasmonic nanoparticles,” J. Biomed. Opt. 16(11), 116003 (2011). 10.1117/1.3646529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aaron J., Travis K., Harrison N., Sokolov K., “Dynamic imaging of molecular assemblies in live cells based on nanoparticle plasmon resonance coupling,” Nano Lett. 9(10), 3612–3618 (2009). 10.1021/nl9018275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erb-Downward J. R., Thompson D. L., Han M. K., Freeman C. M., McCloskey L., Schmidt L. A., Young V. B., Toews G. B., Curtis J. L., Sundaram B., Martinez F. J., Huffnagle G. B., “Analysis of the lung microbiome in the “healthy” smoker and in COPD,” PLoS ONE 6(2), e16384 (2011). 10.1371/journal.pone.0016384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pye A., Hill S. L., Bharadwa P., Stockley R. A., “Effect of storage and postage on recovery and quantitation of bacteria in sputum samples,” J. Clin. Pathol. 61(3), 352–354 (2007). 10.1136/jcp.2007.051490 [DOI] [PubMed] [Google Scholar]
- 19.Monroe P. W., Muchmore H. G., Felton F. G., Pirtle J. K., “Quantitation of microorganisms in sputum,” Appl. Microbiol. 18(2), 214–220 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.UNICEF, WHO, “Pneumonia: the forgotten killer of children,” (2006).
- 21.Forero M. G., Sroubek F., Cristóbal G., “Identification of tuberculosis bacteria based on shape and color,” Real-Time Imag. 10(4), 251–262 (2004). 10.1016/j.rti.2004.05.007 [DOI] [Google Scholar]
- 22.Sadaphal P., Rao J., Comstock G. W., Beg M. F., “Image processing techniques for identifying Mycobacterium tuberculosis in Ziehl-Neelsen stains,” Int. J. Tuberc. Lung Dis. 12(5), 579–582 (2008). [PMC free article] [PubMed] [Google Scholar]
- 23.R. C. Gonzalez, R. E. Woods, and S. L. Eddins, Digital Image Processing using MATLAB: 2nd Ed. (Gatesmark Publishing 2009), Chap, 13. [Google Scholar]
- 24.H. F. Grahan and P. Geladi, Techniques and Applications of Hyperspectral Image Analysis (John Wiley and Sons, 2007). [Google Scholar]