Abstract
Small noncoding RNAs (sRNAs) are ubiquitous posttranscriptional regulators of gene expression. Using the model plant-pathogenic bacterium Xanthomonas campestris pv. vesicatoria (Xcv), we investigated the highly expressed and conserved sRNA sX13 in detail. Deletion of sX13 impinged on Xcv virulence and the expression of genes encoding components and substrates of the Hrp type III secretion (T3S) system. qRT-PCR analyses revealed that sX13 promotes mRNA accumulation of HrpX, a key regulator of the T3S system, whereas the mRNA level of the master regulator HrpG was unaffected. Complementation studies suggest that sX13 acts upstream of HrpG. Microarray analyses identified 63 sX13-regulated genes, which are involved in signal transduction, motility, transcriptional and posttranscriptional regulation and virulence. Structure analyses of in vitro transcribed sX13 revealed a structure with three stable stems and three apical C-rich loops. A computational search for putative regulatory motifs revealed that sX13-repressed mRNAs predominantly harbor G-rich motifs in proximity of translation start sites. Mutation of sX13 loops differentially affected Xcv virulence and the mRNA abundance of putative targets. Using a GFP-based reporter system, we demonstrated that sX13-mediated repression of protein synthesis requires both the C-rich motifs in sX13 and G-rich motifs in potential target mRNAs. Although the RNA-binding protein Hfq was dispensable for sX13 activity, the hfq mRNA and Hfq::GFP abundance were negatively regulated by sX13. In addition, we found that G-rich motifs in sX13-repressed mRNAs can serve as translational enhancers and are located at the ribosome-binding site in 5% of all protein-coding Xcv genes. Our study revealed that sX13 represents a novel class of virulence regulators and provides insights into sRNA-mediated modulation of adaptive processes in the plant pathogen Xanthomonas.
Author Summary
Since the discovery of the first regulatory RNA in 1981, hundreds of small RNAs (sRNAs) have been identified in bacteria. Although sRNA-mediated control of virulence was demonstrated for numerous animal- and human-pathogenic bacteria, sRNAs and their functions in plant-pathogenic bacteria have been enigmatic. We discovered that the sRNA sX13 is a novel virulence regulator of Xanthomonas campestris pv. vesicatoria (Xcv), which causes bacterial spot disease on pepper and tomato. sX13 contributes to the Xcv-plant interaction by promoting the synthesis of an essential pathogenicity factor of Xcv, i. e., the type III secretion system. Thus, in addition to transcriptional regulation, sRNA-mediated posttranscriptional regulation contributes to virulence of plant-pathogenic xanthomonads. To repress target mRNAs carrying G-rich motifs, sX13 employs C-rich loops. Hence, sX13 exhibits striking structural similarity to sRNAs in distantly related human pathogens, e. g., Staphylococcus aureus and Helicobacter pylori, suggesting that structure-driven target regulation via C-rich motifs represents a conserved feature of sRNA-mediated posttranscriptional regulation. Furthermore, sX13 is the first sRNA shown to control the mRNA level of hfq, which encodes a conserved RNA-binding protein required for sRNA activity and virulence in many enteric bacteria.
Introduction
The survival and prosperity of bacteria depends on their ability to adapt to a variety of environmental cues such as nutrient availability, osmolarity and temperature. Besides the adaptation to the environment by transcriptional regulation of gene expression bacteria express regulatory RNAs that modulate expression on the posttranscriptional level [1], [2]. Small regulatory RNAs (sRNAs; ∼50–300 nt) have been intensively studied in the enterobacteria Escherichia coli and Salmonella spp. and, in most cases, regulate translation and/or stability of target mRNAs through short and imperfect base-pairing (10 to 25 nucleotides) [1], [3], [4], [5]. The majority of characterized sRNAs inhibits translation of target mRNAs by pairing near or at the ribosome-binding site (RBS) [1], [6]. In addition, sRNAs can promote target mRNA translation, e. g., the sRNAs ArcZ, DsrA and RprA activate translation of sigma factor RpoS [7], [8], [9]. Regulation of multiple rather than single genes has emerged as a major feature of sRNAs affecting processes like iron homeostasis, carbon metabolism, stress responses and quorum sensing (QS) [1], [2], [6]. In numerous cases, sRNAs are under transcriptional control of two-component systems (TCS), which themselves are often controlled by sRNAs [10]. The activity and stability of most enterobacterial sRNAs requires the hexameric RNA-binding protein Hfq, which facilitates the formation of sRNA-mRNA duplexes and their subsequent degradation by the RNA degradosome [1], [11]. Hfq is present in approximately 50% of all bacterial species and acts in concert with sRNAs to regulate stress responses and virulence of a number of animal- and human-pathogenic bacteria [5], [12].
To date, little is known about sRNAs in plant-pathogenic bacteria. Only recently, high throughput RNA-sequencing approaches uncovered potential sRNAs in the plant-pathogenic α-proteobacterium Agrobacterium tumefaciens [13], the γ-proteobacteria Pseudomonas syringae pv. tomato [14] and Xanthomonas campestris pv. vesicatoria (Xcv) [15], [16]. Additional studies identified four and eight sRNAs in X. campestris pv. campestris (Xcc) and X. oryzae pv. oryzae (Xoo), respectively [17], [18]. So far, only few sRNAs of plant-pathogenic bacteria were characterized with respect to potential targets. Examples include the A. tumefaciens antisense RNA RepE and the sRNA AbcR1, which regulate Ti-plasmid replication and the expression of ABC transporters, respectively [19], [20]. RNAs involved in virulence of plant-pathogenic bacteria were so far only reported for Erwinia spp. and Xcv. In Erwinia, the protein-binding RNA RsmB modulates the activity of the translational repressor protein RsmA, which impacts on QS, the production of extracellular enzymes and virulence [21], [22], [23]. In Xcv, sX12 was reported to be required for full virulence [16].
Xanthomonads are ubiquitous plant-pathogenic bacteria that infect approximately 120 monocotyledonous and 270 dicotyledonous plant species, many of which are economically important crops [24], [25]. These pathogens, usually only found in association with plants and plant parts, differ from most other Gram-negative bacteria in their high G+C content (∼65%), and high numbers of TonB-dependent transporters and signaling proteins [26]. Pathogenicity of most Xanthomonas spp. and other Gram-negative plant- and animal-pathogenic bacteria relies on a type III secretion (T3S) system which translocates bacterial effector proteins into the eukaryotic host cell [27], [28]. In addition, other protein secretion systems, degradative enzymes and QS regulation contribute to virulence of Xanthomonas spp. [29], [30].
One of the models to study plant-pathogen interactions is Xcv, the causal agent of bacterial spot disease on pepper and tomato [31], [32]. The T3S system of Xcv is encoded by the hrp− [hypersensitive response (HR) and pathogenicity] gene cluster and translocates effector proteins into the plant cell where they interfere with host cellular processes to the benefit of the pathogen [29], [33], [34]. The mutation of hrp-genes abolishes bacterial growth in the plant tissue and the induction of the HR in resistant plants. The HR is a local and rapid programmed plant cell death at the infection site and coincides with the arrest of bacterial multiplication [33], [35], [36]. The expression of the T3S system is transcriptionally induced in the plant and in the synthetic medium XVM2, and is controlled by the key regulators HrpG and HrpX [37], [38], [39], [40]. The OmpR-type regulator HrpG induces transcription of hrpX which encodes an AraC-type activator [39], [41]. HrpG and HrpX control the expression of hrp, type III effector and other virulence genes [16], [29], [40], [42], [43]. Recently, dRNA-seq identified 24 sRNAs in Xcv strain 85-10, expression of eight of which is controlled by HrpG and HrpX, including the aforementioned sX12 sRNA [15], [16].
In this study, we aimed at a detailed characterization of sX13 from Xcv strain 85-10, which is an abundant and HrpG-/HrpX-independently expressed sRNA [16]. Using mutant and complementation studies, we discovered that sX13 promotes the expression of the T3S system and contributes to virulence of Xcv. Microarray and quantitative reverse transcription PCR (qRT-PCR) analyses identified a large sX13 regulon and G-rich motifs in presumed sX13-target mRNAs. Selected putative targets were analyzed by site-directed mutagenesis of sX13 and mRNA::gfp fusions. Furthermore, we provide evidence that sX13 acts Hfq-independently. Our study provides the first comprehensive characterization of a trans-encoded sRNA that contributes to virulence of a plant-pathogenic bacterium.
Results
sX13 contributes to bacterial virulence
The sRNA sX13 (115 nt; [16]) is encoded in a 437-bp intergenic region of the Xcv 85-10 chromosome, i. e., 175 bp downstream of the stop codon of the DNA polymerase I-encoding gene polA and 148 bp upstream of the translation start site (TLS) of XCV4199, which encodes a hypothetical protein. Sequence comparisons revealed that the sX13 gene is exclusively found in members of the Xanthomonadaceae family, i. e., in the genomes of plant-pathogenic Xanthomonas and Xylella species, the human pathogen Stenotrophomonas maltophilia and non-pathogenic bacteria of the genus Pseudoxanthomonas. Interestingly, sX13 homologs are highly conserved [16] and always located downstream of polA. By contrast, sX13-flanking sequences are highly variable.
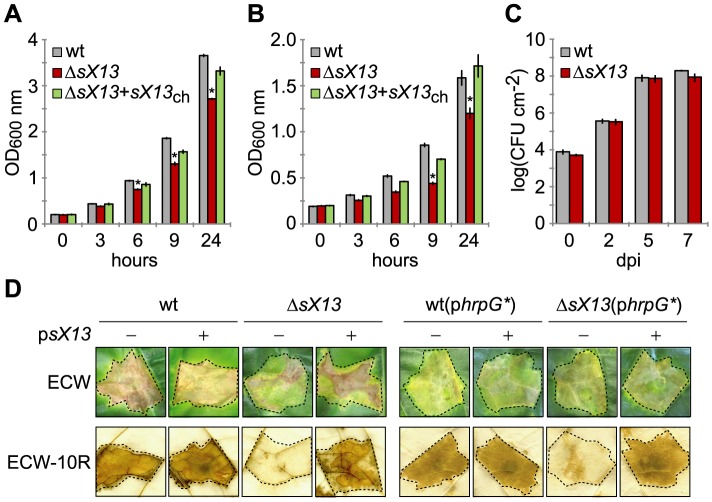
To characterize sX13 in Xcv strain 85-10, we introduced an unmarked sX13 deletion into the chromosome (see ‘Materials and Methods’). Analysis of bacterial growth of the sX13 mutant strain (ΔsX13) revealed a significantly reduced stationary-phase density compared to the Xcv wild-type strain 85-10 in complex medium (NYG; Figure 1A) and in minimal medium A (MMA; Figure 1B). The mutant phenotype of XcvΔsX13 was complemented by chromosomal re-integration of sX13 into the sX13 locus, termed ΔsX13+sX13 ch (Figure 1A, B; see ‘Materials and Methods’).
Figure 1. sX13 contributes to bacterial growth in culture and virulence.
Growth of Xcv wild type 85-10 (wt), the sX13 deletion mutant (ΔsX13) and ΔsX13 containing chromosomally re-integrated sX13 (ΔsX13+sX13 ch) in (A) complex medium NYG and (B) minimal medium MMA, respectively. Error bars represent standard deviations. Asterisks indicate statistically significant differences compared to wt (t-test; P<0.05). (C) Growth of Xcv 85-10 (wt) and ΔsX13 in leaves of susceptible ECW pepper plants. Data points represent the mean of three different samples from three different plants of one experiment. Standard deviations are indicated by error bars. (D) Plant infection assay. Xcv strains 85-10 (wt) and ΔsX13 carrying the empty vector (pB) or the sX13 expression construct (psX13) and strains additionally expressing HrpG* (phrpG*) were inoculated at a density of 4×108 (left panel) and 108 cfu/ml (right panel), respectively, into leaves of susceptible ECW and resistant ECW-10R pepper plants. Disease symptoms in ECW were photographed 9 days post inoculation (dpi). The HR was visualized by ethanol bleaching of the leaves 3 dpi (left panel) and 18 hours post inoculation (right panel), respectively. Dashed lines indicate the inoculated areas. All experiments were performed at least three times with similar results.
To address a potential role of sX13 in virulence, we performed plant infection assays. As shown in Figure 1C, the Xcv strains 85-10 and ΔsX13 grew similarly in leaves of susceptible pepper plants (ECW). Strikingly, infection with the sX13 mutant resulted in strongly delayed disease symptoms in susceptible and a delayed HR in resistant pepper plants (ECW-10R) (Figure 1D). Ectopic expression of sX13 under control of the lac promoter (psX13), which is constitutive in Xcv [38], and re-integration of sX13 into the ΔsX13 locus fully complemented the mutant phenotype of XcvΔsX13 (Figure 1D; data not shown). Strain Xcv 85-10 carrying psX13 induced an accelerated HR in comparison to the wild type (data not shown).
Deletion of sX13 derogates hrp-gene expression
As the HR induction in ECW-10R plants depends on the recognition of the bacterial type III effector protein AvrBs1 by the plant disease resistance gene Bs1 [44], [45], the in planta phenotype of XcvΔsX13 suggested a reduced activity of the T3S system. To address this question, we investigated protein accumulation of selected components of the T3S system. Given that T3S apparatus proteins are not detectable in NYG-grown bacteria, we incubated the bacteria for 3.5 hours in the hrp-gene inducing XVM2 medium [38], [40]. Western blot analysis revealed reduced amounts of the translocon protein HrpF, the T3S-ATPase HrcN and the T3S-apparatus component HrcJ in XcvΔsX13 compared to the wild type, ΔsX13(psX13) (Figure 2A) and strain ΔsX13+sX13 ch (selectively tested for HrcJ; Figure 2B). Thus, sX13 positively affects the synthesis of T3S components.
Figure 2. Deletion of sX13 derogates virulence gene expression.
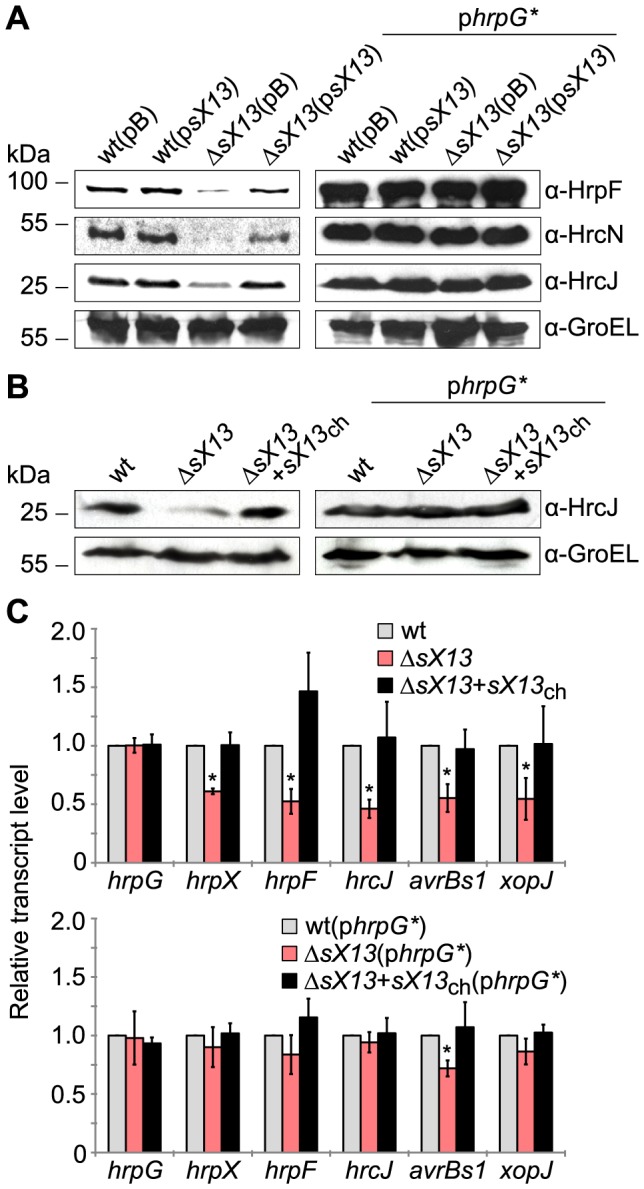
(A) Xcv strains 85-10 (wt) and the sX13 deletion mutant (ΔsX13) carrying the empty vector (pB) or the sX13 expression construct (psX13) and strains additionally expressing HrpG* (phrpG*) were incubated for 3.5 hours in hrp-gene inducing medium XVM2. Total protein extracts were analyzed by immunoblotting using antibodies directed against HrpF, HrcN, HrcJ and GroEL. The experiment was repeated twice with similar results. (B) Xcv 85-10 (wt), ΔsX13 and ΔsX13+sX13 ch and strains additionally expressing HrpG* were incubated for 3.5 hours in hrp-gene inducing medium XVM2. Total protein extracts were analyzed by immunoblotting using antibodies directed against HrcJ and GroEL. The experiment was repeated twice with similar results. (C) Indicated genes were analyzed by qRT-PCR using RNA from cultures described in (B). The amount of each RNA in Xcv 85-10 was set to 1. Data points and error bars represent mean values and standard deviations obtained with three independent biological samples. Asterisks indicate statistically significant differences compared to wt (t-test; P<0.03).
As HrpG controls the expression of the hrp-regulon [39], we analyzed whether the reduced virulence of strain ΔsX13 is due to a reduced activity of HrpG. Therefore, we ectopically expressed a constitutively active version of HrpG (HrpG*; phrpG*; [41]) in XcvΔsX13 and performed plant-infection assays. The disease symptoms induced by XcvΔsX13 and the wild type were comparable in presence of phrpG*, whereas with low inoculum of Xcv 85-10ΔsX13 the HR was slightly delayed (Figure 1D). This suggests that HrpG* suppresses the 85-10ΔsX13 phenotype. HrpF, HrcN and HrcJ protein accumulation in strain ΔsX13(phrpG*) was identical to the wild type suggesting full complementation (Figure 2A, B).
To investigate whether the reduced protein amounts of T3S-system components in XcvΔsX13 are due to altered mRNA levels, we performed qRT-PCR analyses. mRNA accumulation of hrpF, hrcJ and the type III effector genes avrBs1 and xopJ was two-fold lower in XcvΔsX13 than in the wild type and the complemented strain ΔsX13+sX13 ch (Figure 2C). In addition, the mRNA amount of hrpX, but not of hrpG, was reduced in the sX13 mutant (Figure 2C). In presence of phrpG*, comparable mRNA amounts of hrpG, hrpX, hrpF, hrcJ and xopJ were detected in Xcv 85-10, ΔsX13 and ΔsX13+sX13 ch, whereas the avrBs1 mRNA accumulation was significantly reduced in strain 85-10ΔsX13 (Figure 2C). Taken together, our data suggest that the reduced virulence of the 85-10ΔsX13 mutant is caused by a lower expression of components and substrates of the T3S system (Figure 1D; Figure 2A–C).
The deletion and chromosomal re-insertion of sX13 in XcvΔsX13 and ΔsX13+sX13 ch, respectively, were verified by Northern blot using an sX13-specific probe (Figure S1). The sX13 abundance was not affected by expression of HrpG*, which confirms our previous findings [16] and suggests that expression of sX13 is independent of HrpG and HrpX (Figure S1).
sX13 accumulates under stress conditions
The expression of known bacterial sRNAs depends on a variety of environmental stimuli, which often reflect the physiological functions of sRNAs [2], [46], e. g., the E. coli sRNA Spot42 is repressed in the absence of glucose and regulates carbon metabolism [47], [48]. Northern blots revealed similar sX13 amounts in bacteria incubated in NYG medium at 30°C (standard condition), in presence of H2O2, at 4°C and in NYG medium lacking nitrogen (Figure 3A). By contrast, sX13 accumulation was increased in presence of high salt (NaCl), 37°C and in MMA (Figure 3A). Hence, sX13 is differentially expressed in different growth conditions and might contribute to environmental adaptation of Xcv.
Figure 3. sX13 accumulation is altered under stress conditions in Xcv 85-10.
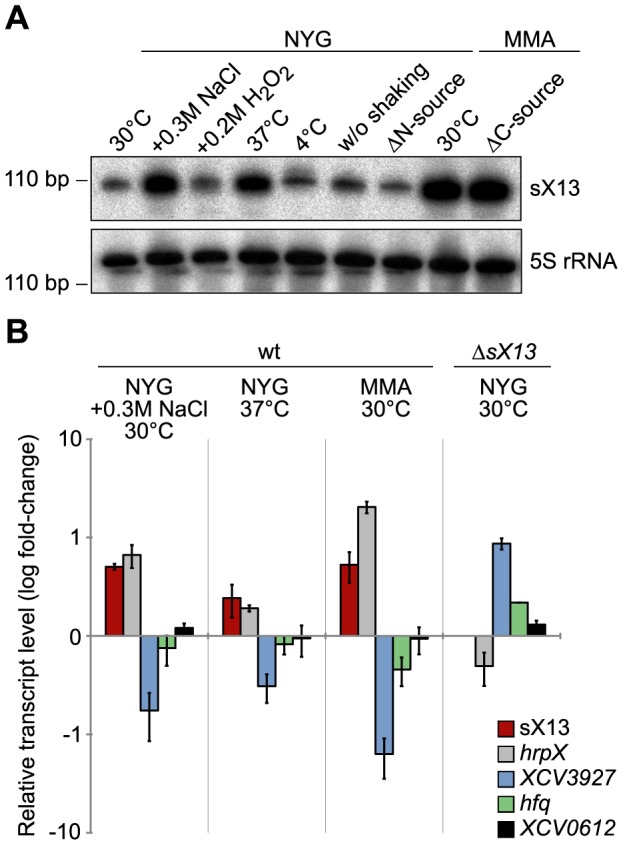
(A) Northern blot analysis of sX13. Exponential phase cultures of NYG-grown Xcv 85-10 were transferred to NYG medium or MMA containing the indicated additives or lacking a nitrogen or carbon source (ΔN; ΔC). Cultures were shaken for three hours at 30°C unless otherwise indicated. 5S rRNA was probed as loading control. (B) sX13 and selected sX13-regulated genes (see Table 1) were analyzed by qRT-PCR using RNA from Xcv 85-10 (wt) cultures shown in (A) and NYG-grown ΔsX13. Bars represent fold-changes (log10) of mRNA amounts compared to Xcv 85-10 grown in NYG at 30°C. Experiments were performed twice with similar results.
Microarray analyses suggest a large sX13 regulon
To gain an insight into the sX13 regulon we performed microarray analyses. For this, cDNA derived from Xcv strains 85-10 and ΔsX13 grown in NYG and MMA, respectively, was used as a probe. The hybridization data were evaluated using EMMA 2.8.2 [49] (see ‘Materials and Methods’). In XcvΔsX13 grown in NYG, 23 mRNAs were upregulated and 21 mRNAs were downregulated by a factor of at least 1.5 compared to the wild type (Table S2). In the MMA-grown sX13 mutant, 23 upregulated mRNAs were detected, four of which were also upregulated in NYG-grown bacteria, whereas no downregulated genes were identified (Table S2). With respect to both growth conditions, 42 and 21 genes were upregulated and downregulated, respectively, in the sX13 mutant. qRT-PCR analyses of 11 selected upregulated and four downregulated genes confirmed the microarray data (Table 1; Figure 4).
Table 1. Selected sX13-regulated genes validated by qRT-PCR analysis.
| Locusa | Annotated gene productb | 4G-motifc | Microarray – Fold-change (ΔsX13/wt)d | qRT-PCR – Fold-change (ΔsX13/wt)e | ||
| NYG | MMA | NYG | MMA | |||
| Upregulated genes (Δ sX13 /wt) | ||||||
| XCV0678 | AlgR; two-component system regulatory protein | a,a,a | 1.8 | — | 2.5±0.23 | n.t. |
| XCV1768 | Hfq; host factor-I protein | b | 1.6 | — | 2.4±0.08 | 1.6±0.31 |
| XCV2186 | methyl-accepting chemotaxis protein | a | 7.7 | — | 2.1±0.34 | 10.2±4.63 |
| XCV2814 | PilE; type IV pilus pilin | — | 2.8 | — | 3.3±0.36 | n.t. |
| XCV2819 | type IV pilus assembly protein PilW | a | 3.7 | 4.0 | 3.4±0.37 | 5.5±3.0 |
| XCV2821 | type IV pilus assembly protein FimT | a | 4.3 | 7.4 | 4.2±0.32 | 3.4±1.27 |
| XCV3067 | PilU; type IV pilus assembly protein ATPase | a | 1.8 | — | 1.7±0.29 | n.t. |
| XCV3096 | ComEA-related DNA uptake protein | — | — | 4.2 | n.t. | 1.9±0.12 |
| XCV3233 | PilG; type IV pilus response regulator | a,b | — | 2.0 | 2.3±0.26 | 4.1±1.71 |
| XCV3500 | PilN; type IV pilus assembly protein | — | 2.7 | — | 2.7±0.16 | n.t. |
| XCV3927 | putative secreted protein | a | — | 1.7 | 5.6±0.45 | 8.3±4.54 |
| Downregulated genes (Δ sX13 /wt) | ||||||
| XCV1315 | HrpX; AraC-type transcriptional regulator | — | 0.6 | — | 0.6±0.01 | 0.7±0.13 |
| XCV1957 | CheY; chemotaxis response regulator | — | 0.4 | — | 0.1±0.04 | n.t. |
| XCV2022 | FliC; flagellin and related hook-associated proteins | — | 0.2 | — | 0.06±0.03 | 1.0±0.39 |
| XCV3572 | TonB-dependent outer membrane receptor | a | 0.2 | — | 0.2±0.04 | 0.9±0.24 |
| Additional genes tested by qRT-PCR | ||||||
| XCV0173 | putative secreted protein | a,b,b,b | — | — | 1.9±0.19 | 0.8±0.26 |
| XCV0612 | ATPase of the AAA+ class | a | — | — | 1.0±0.06 | 0.8±0.26 |
| XCV1533 | AsnB2; asparagine synthase | b | — | — | 1.0±0.04 | 1.0±0.17 |
| XCV3232 | PilH; type IV pilus response regulator | a | — | — | 2.2±0.07 | 1.9±0.67 |
| XCV3573 | putative transcriptional regulator, AraC family | a | — | — | 0.2±0.11 | n.t. |
| XCV0324 | type III effector protein XopS | — | — | — | 0.6±0.05 | n.t. |
, bold letters indicate genes with known TSS [16].
, refers to Thieme et al. (2005) [32].
, presence of a 4G-motif within the 5′-UTR or 100 bp upstream of translation start codon if TSS is unknown (a) and within 100 bp downstream of start codon (b) (see Figure S4).
, genes not detected as expressed are marked with —.
, values represent mean fold-change and standard deviation (see Figure 4);
n.t. - not tested.
Figure 4. qRT-PCR analysis of sX13-regulated genes.

Selected sX13-regulated genes (see Table 1) were analyzed by qRT-PCR using RNA from NYG- and MMA-grown Xcv strains 85-10 (wt) and ΔsX13. The amount of each mRNA in the wt was set to 1. Bars represent fold-changes of mRNA amounts in strain ΔsX13 compared to 85-10 on a logarithmic scale (log10). Data points and error bars represent mean values and standard deviations obtained with at least three independent biological samples. Asterisks denote statistically significant differences (t-test; P<0.05). Dashed lines indicate a 1.5-fold change. Transcripts not detected in the microarray analyses are marked with ‘a’.
sX13 negatively affects hfq and type IV pilus-biosynthesis mRNAs
Based on the annotated genome sequence of Xcv 85-10 [32], genes upregulated in XcvΔsX13 can be grouped (Table S2): 18 genes encode proteins with unknown function, e. g., the putative LysM-domain protein XCV3927. 14 genes encode proteins involved in type IV pilus (Tfp) biogenesis, e. g., the putative Tfp assembly protein XCV2821, the pilus component PilE and the TCS response regulator PilG. Tfp enable twitching motility, i. e., adhesion to and movement on solid surfaces [50], [51]. Three genes encode proteins assigned to signal transduction, i. e., the TCS regulator AlgR, the GGDEF-domain protein XCV2041 and the chemotaxis regulator XCV2186. Moreover, hfq mRNA accumulation was two-fold increased in XcvΔsX13.
The microarray data suggested that most upregulated genes in XcvΔsX13 were only expressed in NYG- or MMA-grown bacteria (Table S2), which might be explained by the P-value and signal-strength thresholds applied for data evaluation. qRT-PCR analyses showed that the mRNA accumulation of hfq, XCV2186, pilG and XCV3927 was increased in both the NYG- and MMA-grown sX13 mutant compared to the wild type (Figure 4; Table 1). qRT-PCR analyses also revealed an upregulation of pilH in the NYG- and MMA-grown XcvΔsX13 compared to the wild type (Figure 4; Table 1). Because pilH is the second gene in the pilG operon and was not detected as expressed in the microarray data, the number of mRNAs affected by sX13 deletion might be higher than suggested by the microarray data.
sX13 positively affects hrpX and chemotaxis-regulating mRNAs
Five of 21 genes downregulated in XcvΔsX13 presumably encode proteins involved in flagellum-mediated chemotaxis, e. g., the sensor kinase CheA1, the corresponding response regulator CheY and the flagellum components FliD and FliC (Table S2). qRT-PCR analyses revealed 17-fold lower fliC mRNA abundance in XcvΔsX13 grown in NYG compared to the wild type, whereas the accumulation in MMA-grown cells was identical (Figure 4; Table 1). Similarly, XCV3572, which encodes a TonB-dependent receptor, was downregulated in NYG- but not in MMA-grown XcvΔsX13 (Figure 4; Table 1). Gene XCV3573, which is encoded adjacent to XCV3572 and encodes an AraC-type regulator, was also downregulated (Figure 4; Table 1). As mentioned above, sX13 positively affected the mRNA accumulation of hrpX in XVM2 medium (see Figure 2C), which was also true for bacteria grown in NYG and MMA (Figure 4; Table 1). Since HrpX controls the expression of many type III effector genes, we analyzed xopS [52] by qRT-PCR and detected similarly decreased levels in NYG-grown XcvΔsX13 as for hrpX (Figure 4; Table 1). Taken together, our data suggest that the sX13 regulon comprises genes involved in signal transduction, motility, transcriptional and posttranscriptional regulation and virulence.
Accumulation of potential target mRNAs correlates with sX13 abundance
To address whether differential expression of sX13 under different conditions (see Figure 3A) affects the mRNA abundance of sX13-regulated genes, we performed qRT-PCR. We detected elevated sX13 levels in Xcv strain 85-10 cultivated in high salt conditions, at 37°C and in MMA compared to standard conditions and an increased hrpX and decreased XCV3927 mRNA accumulation (Figure 3B). In addition, low amounts of the hfq mRNA were detected in presence of high sX13 levels, whereas the abundance of the sX13-independent XCV0612 mRNA (see Table 1) was not altered (Figure 3B).
sX13 activity does not require Hfq
The hfq mRNA accumulation in XcvΔsX13 (Figure 3B; Figure 4; Table 1) prompted us to test whether sX13 activity depends on the RNA-binding protein Hfq. For this, we introduced a frameshift mutation into the hfq gene of Xcv strains 85-10 and 85-10ΔsX13. Northern blot analyses revealed comparable sX13 accumulation in both strains and the complemented hfq mutant, which ectopically expressed Hfq (phfq) (Figure 5A). By contrast, the accumulation of the sRNA sX14 [16] was strongly reduced in the hfq mutant; this was restored by phfq (Figure 5A). Unexpectedly, the hfq mutant strain was not altered in the induction of in planta phenotypes, i. e., in virulence (Figure 5B).
Figure 5. sX13 activity does not require Hfq.
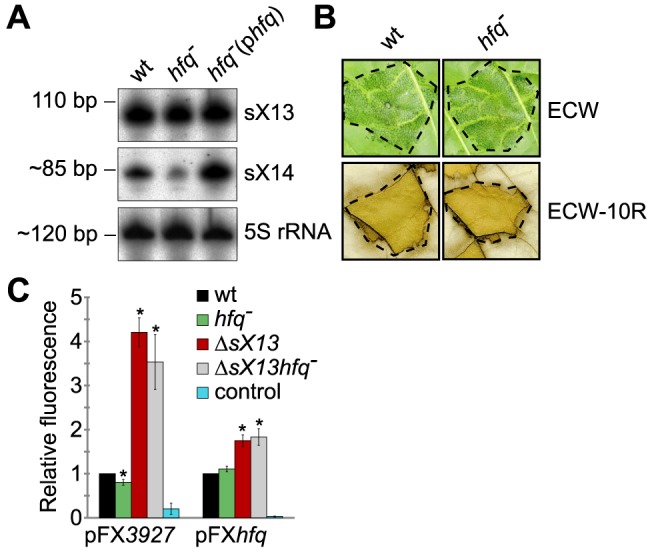
(A) Northern blot analysis. Total RNA from NYG-grown Xcv strains 85-10 (wt), the hfq frameshift mutant (hfq−) and the hfq mutant ectopically expressing Hfq (phfq) was analyzed using sX13- or sX14-specific probes. 5S rRNA was probed as loading control. The experiment was performed twice with two independent mutants and with similar results. (B) Plant infection assay. The Xcv wild-type 85-10 (wt) and hfq mutant strain (hfq−) were inoculated at 2×108 cfu/ml into leaves of susceptible ECW and resistant ECW-10R plants. Disease symptoms were photographed 6 dpi. The HR was visualized 2 dpi by ethanol bleaching of the leaves. Dashed lines indicate the inoculated areas. The experiment was repeated three times with similar results. (C) GFP fluorescence of NYG-grown Xcv 85-10 (wt), the hfq mutant (hfq−), the sX13 deletion mutant (ΔsX13) and the double mutant (ΔsX13hfq−) carrying pFX3927 or pFXhfq. Xcv autofluorescence was determined by Xcv 85-10 carrying pFX0 (control). Data points and error bars represent mean values and standard deviations obtained with at least four independent experiments. GFP fluorescence of the wt was set to 1. Asterisks denote statistically significant differences (t-test; P<0.01).
To investigate whether sX13 affects translation of putative target mRNAs, we established a GFP-based in vivo reporter system for Xcv similar to the one described for E. coli [53]. The promoterless broad-host range plasmid pFX-P permits generation of translational gfp fusions in a one-step restriction-ligation reaction (Golden Gate cloning [54]; see ‘Materials and Methods’). We cloned the promoter, 5′-UTRs, and 10 and 25 codons of XCV3927 and hfq, respectively, into pFX-P resulting in pFX3927 and pFXhfq. XCV3927 was selected because of a strongly increased mRNA accumulation in XcvΔsX13 compared to the wild type (see Table 1). In presence of pFX3927 or pFXhfq, fluorescence of XCV3927::GFP or Hfq::GFP fusion proteins was comparable in the Xcv wild type and hfq mutant (Figure 5C). The XCV3927::GFP and Hfq::GFP fluorescence was about 4-fold and 2-fold increased, respectively, in XcvΔsX13 compared to strain 85-10 (Figure 5C), suggesting that the synthesis of the fusion proteins is repressed by sX13. Interestingly, the XCV3927::GFP and Hfq::GFP fluorescence was similarly increased in XcvΔsX13 and the sX13hfq double mutant (Figure 5C). As abundance and activity of sX13 were not affected by the hfq mutation, we assume that sX13 acts Hfq-independently.
sX13 activity in planta depends on C-rich loop motifs
The predicted secondary structure of sX13 obtained by mfold [55] displays an unstructured 5′-region and three stable stem-loops, termed stem 1 to 3, and loop 1 to 3 (Figure 6A). Interestingly, loop 1 and loop 2 contain a ‘CCCC’ (4C) motif, whereas loop 3 harbors a ‘CCCCC’ (5C) motif (Figure 6A). To experimentally verify the predicted structure, we performed structure analyses of in vitro transcribed and radioactively-labeled sX13 treated with RNase V1 or RNase T1. While RNase T1 cleaves single-stranded RNA with a preference for G residues, RNase V1 randomly cleaves double-stranded RNA. We detected RNase T1-cleavage products for the 5′-region and RNase V1-cleavage products for stem 1 and 2, which is in good agreement with the predicted structure (Figure 6A; Figure S2). Moreover, RNase V1-cleavage products were less abundant for the 4C-motif of loop 1 and loop 2, suggesting single-stranded sequences (Figure 6A; Figure S2). The results did not allow conclusions about stem 3 and loop 3 structures.
Figure 6. sX13 loops impact on Xcv virulence.
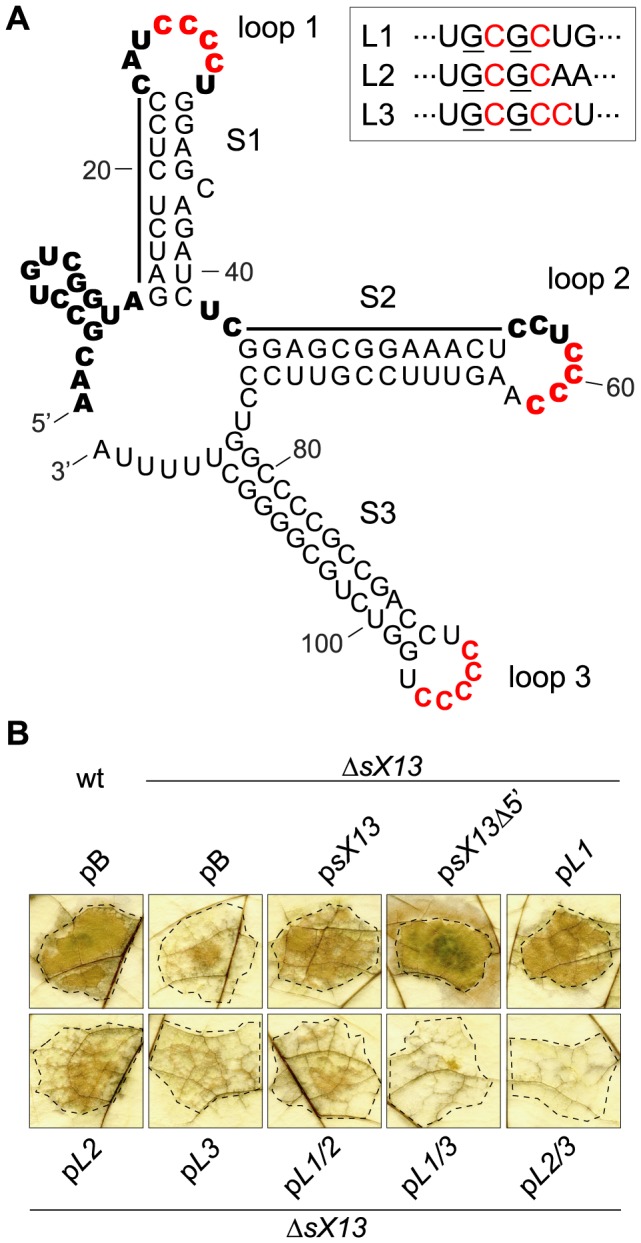
(A) Secondary structure of sX13 based on prediction and probing (see Figure S2). sX13 consists of an unstructured 5′-, three double-stranded regions (S1; S2; S3) and three loops (loop 1–3). 4C-/5C-motifs are highlighted in red. Bold letters indicate unpaired bases and bars mark double-stranded regions deduced from structure probing. Mutations in loops are boxed, exchanged nucleotides are underlined. (B) Derivatives mutated in loops 2 and 3 fail to complement the plant phenotype of ΔsX13. Leaves of resistant ECW-10R plants were inoculated at 108 cfu/ml with Xcv 85-10 (wt) and ΔsX13 carrying pBRS (pB), psX13 or one of the following derivatives: sX13 lacking 14 terminal nucleotides (psX13Δ5′), sX13 mutated in single loops (pL1, pL2, pL3) or in two loops (pL1/2, pL1/3, pL2/3). The HR was visualized by ethanol bleaching of the leaves 2 dpi. Dashed lines indicate the inoculated areas. The experiment was performed four times with similar results.
To assess the contribution of the 4C-/5C-motifs to sX13 function, we mutated psX13 in loop 1 and 2, respectively, to ‘GCGC’, and the 5C-motif in loop 3 to ‘GCGCC’ resulting in pL1, pL2 and pL3 (Figure 6A). In addition, loop mutations were combined (pL1/2, pL1/3, pL2/3) and analyzed for their ability to complement the in planta phenotype of strain ΔsX13. As shown above, XcvΔsX13 induced a delayed HR, which was complemented by psX13 (Figure 1D). Similar phenotypes were observed with sX13 mutants carrying pL1 or psX13Δ5′, which encodes a 5′-truncated sX13 derivative lacking the terminal 14 nucleotides (Figure 6B). The HR induced by the sX13 mutant containing pL2 or pL1/2 was intermediate, whereas pL3, pL1/3 and pL2/3 failed to complement XcvΔsX13 (Figure 6B). Northern blot analyses revealed expression of all sX13-loop mutant derivatives (Figure S3). The different RNA species derived from ectopically expressed sX13 and derivatives compared to chromosomally encoded sX13 might be due to alternative transcription termination of plasmid-derived sX13 and derivatives.
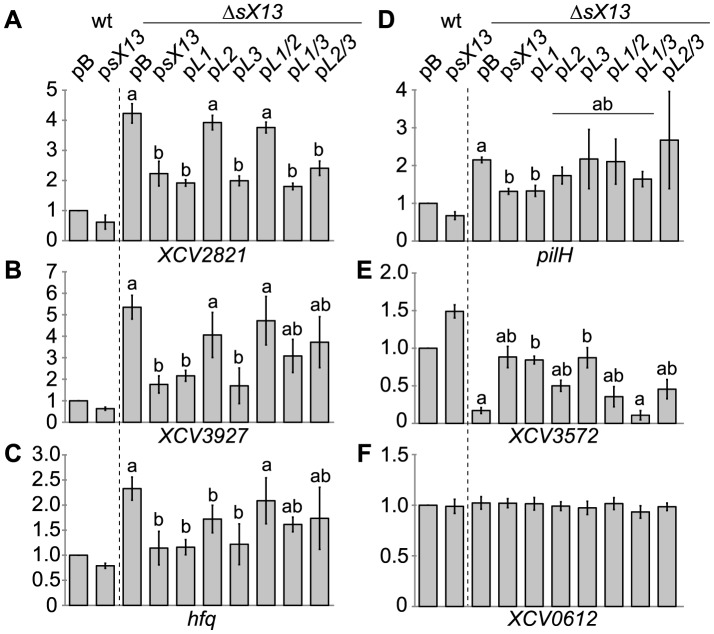
sX13 loops differentially contribute to mRNA accumulation
As mutation of sX13 loops impinged on Xcv virulence (Figure 6B), we addressed by qRT-PCR whether loop mutations affect the mRNA abundance of XCV2821, XCV3927, hfq and pilH, which were upregulated in XcvΔsX13 (see Figure 4; Table 1). In addition, we analyzed a downregulated gene, XCV3572, and XCV0612, which was not affected by sX13 deletion. As shown in Figure 7A–E, sX13 negatively affected the mRNA abundance of XCV2821, XCV3927, hfq and pilH, whereas sX13 promoted mRNA accumulation of XCV3572. Mutation of sX13 loops differentially affected the mRNA abundance of the tested genes: pL2 and pL1/2 failed to complement XcvΔsX13 with respect to the mRNA abundance of XCV2821, XCV3927 and hfq (Figure 7A–C). Intermediate mRNA amounts of XCV3927 and hfq were detected in XcvΔsX13 carrying pL1/3 or pL2/3 compared to pB and psX13 (Figure 7B, C). Taken together, the mRNA abundance of XCV2821, XCV3927 and hfq appears to be mainly controlled by sX13-loop 2. In contrast, pilH mRNA accumulation appears to depend on multiple sX13 loops as only psX13 and pL1 complemented XcvΔsX13 (Figure 7D). The reduced mRNA amount of XCV3572 in XcvΔsX13 was complemented by pL1 and pL3 but not by pL1/3 (Figure 7E), which suggests redundant roles of sX13-loops. In presence of pL2, pL1/2 or pL2/3 in XcvΔsX13, the XCV3572 mRNA levels were intermediate compared to XcvΔsX13 carrying pB or psX13 (Figure 7E). As expected, the mRNA abundance of XCV0612 was identical in the different strains (Figure 7F).
Figure 7. sX13 loops differentially contribute to abundance of putative mRNA targets.
Relative transcript levels of (A) XCV2821, (B) XCV3927, (C) hfq, (D) pilH, (E) XCV3572 and (F) XCV0612 were analyzed by qRT-PCR in total RNA of NYG-grown Xcv strains 85-10 (wt) and ΔsX13 carrying pBRS (pB), psX13 or mutated sX13-derivatives (see Figure 6). The mRNA abundance in the wt was set to 1. Data points and error bars represent mean values and standard deviations obtained with at least three independent biological samples. Statistically significant differences are indicated (t-test; P<0.015).
Identification of putative sX13-binding sites
To identify potential regulatory motifs in sX13-regulated mRNAs, a discriminative motif search was performed using DREME [56]. For this, sequences surrounding the TLSs of the 42 up- and 21 downregulated genes identified by microarray analyses (Table S2) were compared. More precisely, sequences spanning from known transcription start sites (TSSs) to 100 bp downstream of TLSs or, in case of unknown TSSs, 100 bp up- and 100 bp downstream of the TLS were inspected.
We found that up- and downregulated genes differ in the presence of ‘GGGG’ (4G) motifs. In the NYG-grown sX13 mutant, 15 out of 23 (65%) upregulated genes contain up to three 4G-motifs which are predominantly located upstream of the TLS (Figure S4A; Table S2). 70% of the genes upregulated in MMA (16 out of 23), but only 14% of the genes downregulated in NYG medium (3 out of 21) contain 4G-motifs (Figure S4A; Table S2). Thus, 4G-motifs appear to be enriched in sX13-repressed mRNAs. However, the position of the motifs and flanking nucleotides are not conserved among sX13-regulated genes. Note that the term ‘4G-motif’ also refers to motifs containing more than four G-residues in a row. The complementarity of C-rich sX13-loop sequences and G-rich mRNA motifs suggests sX13-mRNA interactions via antisense-base pairing (Figure 6A; Table 1; Table S2).
Compared to the occurrence of 4G-motifs in approximately 70% of sX13-repressed genes, only 30.71% of all chromosomally encoded Xcv genes (1,378 out of 4,487) carry 4G-motifs in proximity of their TLS (Figure S4A). Interestingly, 4G-motifs in 241 of the chromosomally encoded genes (5.37%) are located between nucleotide position 8 and 15 upstream of the TLS (Figure S4B). This position corresponds to the presumed location of the RBS and suggests a role of 4G-motifs in translation control.
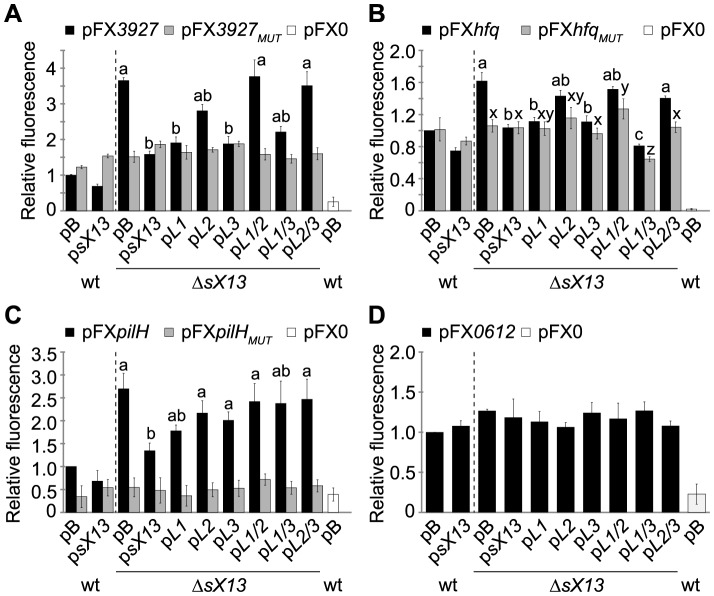
sX13 dependency of target::GFP synthesis requires both 4C- and 4G-motifs
To study the effect of sX13 on translation of selected putative targets, i. e., XCV3927 and hfq, we used the above-mentioned GFP-reporter plasmids pFX3927 and pFXhfq. In addition, we generated pilH::gfp (pFXpilH) and XCV0612::gfp (pFX0612) fusions (see ‘Materials and Methods’). All mRNA::gfp fusions contain a G-rich motif in the proximity of their TLS which is complementary to C-rich sX13-loop regions (see ‘Materials and Methods’). The fluorescence of the sX13 deletion mutant carrying pFX3927, pFXhfq and pFXpilH was about 3.5-, 1.6- and 2.5-fold higher, respectively, compared to the Xcv wild type (Figure 8A–C). In presence of psX13, pL1, pL3 or pL1/3 in XcvΔsX13, the XCV3927::GFP and Hfq::GFP fluorescence levels were comparable to the Xcv wild type (Figure 8A, B). By contrast, the XCV3927::GFP and Hfq::GFP fluorescence of strain ΔsX13 carrying pL2, pL1/2 or pL2/3 was similarly increased as compared to XcvΔsX13 carrying pB (Figure 8A, B). This suggests that the 4C-motif in sX13-loop 2 is required to repress XCV3927::GFP and Hfq::GFP synthesis. The increased PilH::GFP fluorescence of XcvΔsX13 was complemented by psX13 and pL1, in contrast to other sX13-loop mutant derivatives (Figure 8C). Fluorescence values of all analyzed Xcv strains carrying pFX0612 were comparable confirming sX13-independency (Figure 8D).
Figure 8. sX13-dependency of mRNA target::GFP synthesis requires a G-rich motif.
GFP fluorescence of NYG-grown Xcv strains 85-10 (wt) and ΔsX13 carrying pB, psX13 or mutated sX13-derivatives (see Figure 6) and carrying GFP-reporter plasmids (A) pFX3927, (B) pFXhfq, (C) pFXpilH or (D) pFX0612. pFXMUT derivatives contain a mutated 4G-motif. Xcv autofluorescence was determined using pFX0. GFP fluorescence of the wt was set to 1. Data points and error bars represent mean values and standard deviations obtained from at least four independent experiments. Statistically significant differences are indicated (t-test; P<0.015).
To address whether the G-rich motif in presumed target mRNAs is required for sX13 dependency of mRNA::gfp translation, we introduced compensatory mutations. i. e., mutated the motif to ‘GCGC’. Xcv strains carrying the resulting plasmids, pFX3927MUT, pFXhfqMUT or pFXpilHMUT, exhibited a similar fluorescence in absence and presence of sX13 and mutated sX13 derivatives (Figure 8A–C). This suggests that the G-rich motif is required for sX13-dependency of target::GFP synthesis. However, mutation of the C-rich motifs in sX13 and the G-rich motifs in mRNA::gfp fusions did not restore sX13 dependency (Figure 8A–C). Unexpectedly, the fluorescence detected for Xcv strains containing pFX3927MUT or pFXhfqMUT was comparable to the fluorescence of Xcv 85-10 carrying the non-mutated plasmids pFX3927 and pFXhfq, respectively (Figure 8A, B). The mutation of the 5G-motif in pilH abolished the fluorescence of strains containing pFXpilHMUT indicating an essential role of the motif in pilH translation (Figure 8C).
Because sX13 was more abundant in MMA- than NYG-grown bacteria (Figure 3), we also analyzed the fluorescence of MMA-grown Xcv strains containing pFX-derivatives. XCV3927::GFP and PilH::GFP synthesis in MMA was sX13-dependently repressed to a greater extent than in NYG (Figure S5; see Figure 8).
Because sX13 negatively affected both the mRNA accumulation of chromosomally encoded XCV3927, hfq and pilH genes and accumulation of the corresponding GFP-fusion proteins, we exemplarily analyzed whether this is due to an altered mRNA abundance. However, qRT-PCR analyses revealed that the XCV3927::gfp mRNA accumulation was sX13-independent suggesting that sX13 posttranscriptionally affects the synthesis of XCV3927::GFP (Figure S6).
To discriminate between transcriptional and posttranscriptional effects of sX13 on target::GFP synthesis we generated reporter fusions controlled by plac (see ‘Materials and Methods’). Note that the activity of the lac promoter is not affected by deletion of sX13 (data not shown). As shown in Figure S7, the fluorescence of XcvΔsX13 carrying pFXpl-3927 (XCV3927) and pFXpl-pilH (pilH) was 2.5- and 4-fold higher, respectively, compared to the Xcv wild type and the complemented sX13 mutant strain. Interestingly, mutation of the 4G-motif in the XCV3927 5′-UTR did not only abolish sX13-dependency but also led to a significantly reduced fluorescence compared to the Xcv wild type which carried the non-mutated reporter plasmid (Figure S7). This suggests that the 4G-motif in the XCV3927 5′-UTR promotes translation, i. e., acts as translational enhancer element. In presence of pFXpl-pilH, the fluorescence of the fusion protein was only detectable in the sX13 mutant but not in the wild type or complemented strain, confirming that PilH::GFP synthesis is repressed by sX13 (Figure S7). Overall, the data confirm that sX13 represses the synthesis of XCV3927 and PilH on the posttranscriptional level.
Discussion
sX13 controls Xcv virulence
This study provides a first insight into the posttranscriptional modulation of clade-specific adaptive processes in a plant-pathogenic γ-proteobacterium. We identified sX13 as a major regulator of Xcv virulence in that it promotes expression of genes in the hrp-regulon, i. e., components and substrates of the T3S system (Figure 2A–C). This finding is remarkable because the hrp-regulon is only expressed under certain conditions, whereas sX13 is constitutively expressed (Figure S1) [16]. The sX13 gene is exclusively found and highly conserved in members of the Xanthomonadaceae family of Gram-negative bacteria [16]. Intriguingly, several species with an sX13 homolog lack a T3S system, e. g., the plant pathogen X. fastidiosa and the opportunistic human pathogen S. maltophilia. This suggests a role of sX13 apart from regulation of the hrp-regulon in these organisms.
The expression of the hrp-regulon depends on HrpG and HrpX [39], [40]. HrpG is presumably posttranslationally activated in the plant and in XVM2 medium and induces the expression of hrpX [38], [39], [40], [41]. As the XVM2-grown sX13 mutant displayed decreased mRNA amounts of hrpX but not of hrpG (Figure 2C), we suppose that sX13 acts upstream of HrpG. This idea is supported by the finding that constitutively active HrpG (HrpG* [41]) suppressed the sX13 mutation with respect to virulence and the expression of hrpX and downstream genes (Figure 1D; Figure 2A–C). In addition, sX13 affected the basal expression level and, hence, the activity of HrpX under non-inducing conditions, which might impact on the efficiency of hrp-gene induction during infection. Based on the fact that HrpG::GFP and HrpX::GFP synthesis was sX13-independent (Figure S8) we assume that sX13 indirectly controls the expression of the hrp-regulon.
Physiological roles of sX13
Deletion of sX13 affected the mRNA abundance of more than 60 genes involved in signaling, motility, transcriptional and posttranscriptional regulation (Table S2). sX13 negatively regulated mRNAs involved in Tfp biogenesis but promoted the accumulation of mRNAs involved in flagellum-mediated chemotaxis in a growth-phase dependent manner (Table 1; Table S2). This, together with the fact that sX13 is differentially expressed under certain stress conditions (Figure 3), implies a central role of sX13 in the transduction of environmental signals into comprehensive cellular responses affecting virulence gene expression, motility and QS regulation. The latter is corroborated by the reduced stationary-phase cell density of the sX13 mutant compared to the Xcv wild type (Figure 1A, B) and the sX13-dependency of the XCV2041 mRNA (Table S2), which encodes a GGDEF-/EAL-domain protein. Such domains play a role in the control of cyclic-di-GMP levels and QS regulation [57]. Interestingly, XCV2041 shares 94% identity with the Xcc protein XC2226 which is a repressor of Tfp-mediated motility [58].
Another remarkable finding of our study was the sX13-dependent accumulation of the hfq mRNA. To the best of our knowledge, sX13 is the first sRNA which affects expression of this conserved RNA-binding protein (Table 1). The Xcv hfq mutant was unaltered in virulence on its host plant (Figure 5B), which is in good agreement with recent findings for Xoo [18]. By contrast, Hfq contributes to virulence in a number of other bacteria, including the plant pathogen A. tumefaciens, and is also involved in symbiotic plant interactions of Sinorhizobium meliloti [5], [12], [59], [60]. In Vibrio cholerae, four redundantly acting and Hfq-dependent sRNAs (Qrr) destabilize hapR mRNA, which encodes the master regulator of QS, the T3S system and other virulence genes [61], [62]. In the Gram-positive human pathogen Staphylococcus aureus, the Hfq-independent RNAIII is induced by the agr QS system and mediates the switch between the expression of surface proteins and excreted toxins through translational repression of Rot (repressor of toxins) [63], [64], [65].
sX13 activity depends on C-rich loop regions
Xcv sRNAs are strongly structured and lack extended single-stranded regions [15], [16], [32]. In contrast, enterobacterial sRNAs commonly exhibit a modular structure consisting of a single-stranded mRNA-targeting domain, often located at the 5′-end, an A/U-rich Hfq-binding site and a Rho-independent terminator [1]. The sX13 structure suggests that direct sRNA-mRNA interactions are energetically confined to the unstructured 5′-region and its three C-rich loops (Figure 6A). However, the 5′-region of sX13 was dispensable for full virulence of Xcv and sX13 activity appears to be exerted via loops 2 and 3 (Figure 6B). Although loops 1 and 2 just differ in the 3′-adjacent nucleotide (U/A) (Figure 6A), only loop 2 was required to repress the synthesis of XCV3927::GFP and Hfq::GFP, which might depend on the position of stem-loops in the sRNA and, thus, accessibility. By contrast, repression of PilH::GFP appears to depend on multiple sX13 regions (Figure 8C).
An important question is whether sX13 controls target gene expression on the level of mRNA stability or translation. On one hand, sX13-loop mutant derivatives affected the mRNA levels of presumed targets (Figure 7). On the other hand, protein levels, but not the mRNA level of an XCV3927::gfp fusion, harboring only the 5′-UTR and 10 codons of XCV3927, were sX13-dependent (Figure 8A; Figure S6). This suggests that the impact of sX13 on XCV3927 mRNA abundance and translation are separate events and hints at the presence of additional regulatory sites in the XCV3927 mRNA. It should be noted that the assessment of RNA stability by rifampicin treatment is hampered by the fact that our Xcv strains are rifampicin resistant.
The sX13 loops are reminiscent of regulatory RNAs in S. aureus, many of which contain ‘UCCC’-motifs in loops [66]. For example, RNAIII contains C-rich stem-loops, which interact with the RBS of target mRNAs [63], [65], [67]. RNAIII represses Rot synthesis through formation of kissing complexes between two stem-loops of each RNAIII and rot mRNA [64], [65]. Such multiple loop interactions are also employed by the E. coli sRNA OxyS to target fhlA [68]. In Helicobacter pylori, the sRNA HPnc5490 represses the synthesis of the chemotaxis regulator TlpB [69]. Interestingly, the central part of the HPnc5490-loop sequence is identical to the ‘UCCCCCU’-motif of loop 3 in sX13 [69].
G-rich enhancer motifs confer sX13-dependency of target mRNAs
Similarly to RNAIII targets in S. aureus and the tlpB mRNA in H. pylori [65], [69], mRNAs repressed by sX13 are enriched for G-rich motifs in proximity of the TLS (Figure S4; Table S2). The complementarity between these motifs and the 4C-/5C-motif in the sX13 loops suggests sX13-mRNA interactions through antisense base pairing. Our data emphasize that sX13 acts posttranscriptionally on target genes that contain G-rich motifs, as shown for XCV3927 and pilH (Figure S7). This idea is supported by the fact that mutation of the G-rich motifs, located in the mRNA of XCV3927, pilH and hfq, abolished sX13-dependency of protein synthesis (Figure 8; Figure S5; Figure S7). However, the presence of a G-rich motif does not necessarily confer regulation by sX13 (see XCV0612; Figure 7F; Figure 8D). Given that eight of 28 repressed and 4G-motif-containing mRNAs contain at least two 4G-motifs close to the TLSs (Figure S4), we assume that sX13 loops can interact with multiple 4G-motifs in certain mRNAs. As positively regulated mRNAs lack G-rich motifs, sX13 presumably acts indirectly on the corresponding genes (Figure S4; Table S2).
Direct sRNA-mRNA interactions are commonly validated by compensatory mutant studies [1]. However, in case of the E. coli sRNA RyhB, mutations were suggested to interfere with Hfq-binding and rendered compensatory mutants non-functional [70]. Here, mutation and deletion of sX13 increased the synthesis of XCV3927::GFP and Hfq::GFP fusions, whereas mutation of corresponding 4G-motifs resulted in similar fluorescence values as non-mutated mRNA::gfp fusions in Xcv wild type. In addition, the reduced fluorescence of mutated target::GFP fusions was unaffected by compensatory sX13-mutant derivatives (Figure 8). This suggests that G-rich motifs in sX13-repressed mRNAs play a role besides mediation of sRNA interactions. While Xanthomonas spp., like other G+C-rich bacteria, lack a consensus RBS [16], [71], 5% of the chromosomal Xcv coding sequences (241 of 4,487) contain a G-rich motif 8–15 nucleotides upstream of their TLS (Figure S4). As anticipated, mutation of the 5G-motif at the RBS position of pilH abolished translation (Figure 8; Figure S5; Figure S7). By contrast, the 4G-motifs in XCV3927 and hfq mRNAs, located 21 nucleotides upstream and nine nucleotides downstream of the AUG, respectively, confer sX13-dependency but were not essential for translation (Figure 8). Thus, G-rich motifs confer sX13-dependency and mRNA translation in a position-dependent manner. As mutation of the 4G-motif in XCV3927 reduced protein synthesis, the motif appears to function as translational enhancer (Figure S7). We suggest that sequestration of a G-rich motif by sX13 as well as mutation of the motif precludes the binding of an unknown factor, which promotes mRNA translation. Such a factor could be RNA, protein or the ribosome.
The presumed sX13 mode of action is reminiscent of the Salmonella sRNA GcvB, which inhibits translation of mRNAs by targeting C/A-rich enhancer elements [72], [73]. By increasing the ribosome-binding affinity, C/A-rich motifs enhance mRNA translation, irrespective of their localization upstream or downstream of the TLS [72], [74].
sRNAs encoded at the polA locus in other bacteria
Homologs of Xcv sRNAs are predominantly found in members of the Xanthomonadaceae family but not in other bacteria [15], [16]. The sX13 gene is located adjacent to the DNA polymerase I-encoding polA gene, which resembles a locus encoding the Spot42 sRNA in E. coli and members of the αr7 sRNA family in α-proteobacteria [75], [76], [77], [78]. In contrast to sX13, Spot42 requires Hfq and regulates targets involved in carbon metabolism [48], [79], e. g., the discoordinated expression of genes within the gal galactose utilization operon [47], which is absent in Xcv. Although sX13 lacks sequence similarity to Spot42 and αr7 sRNAs, the latter also contain three stem-loops and apical C-rich motifs [80] suggesting that sRNAs in distantly related bacteria evolved divergently but retained structural conservation. Thus, it will be interesting to see whether the polA locus in other bacteria also encodes sRNAs, and whether sX13 and structurally related sRNAs act in a similar manner.
Materials and Methods
Bacterial strains and growth conditions
For bacterial strains, plasmids and oligonucleotides used in this study see Table S1. E. coli strains were grown at 37°C in lysogeny broth (LB), Xcv strains at 30°C in nutrient-yeast-glycerol (NYG) [81], XVM2 [40] or minimal medium A (MMA) [82], which was supplemented with casamino acids (0.3%) and sucrose (10 mM). Plasmids were introduced into E. coli by chemical transformation and into Xcv by tri-parental conjugation, using pRK2013 as helper plasmid [83]. Antibiotics were added to the final concentrations: ampicillin, 100 µg/ml; gentamycin, 15 µg/ml; kanamycin, 25 µg/ml; rifampicin, 100 µg/ml; spectinomycin, 100 µg/ml.
Generation of constructs
To generate the sRNA-expression vector pBRS, a 28-bp fragment between the lac promoter and the EcoRI cloning site in pBBR1mod1 [84] was replaced by a truncated fragment, amplified by PCR from pBBR1mod1 using primers pBRS-EcoRI-fw and pBRS-NcoI-rev. For generation of constructs expressing sX13 (psX13) and sX13Δ5′ (psX13Δ5′; lacking 14 nt at the 5′-end), respective fragments were PCR-amplified from Xcv 85-10 using primers sX13-fw/-rev or sX13d5-fw/-rev. PCR products were digested with EcoRI/HindIII and cloned into pBRS. The sX13-mutant plasmids pL1, pL2, pL3 and pL2/3 were generated by PCR amplification of plasmid psX13 using primers L1-fw/-rev, L2-fw/-rev, L3-fw/-rev and L3-fw/L2/3-rev, respectively; plasmid pL1/3 was generated with primers L3-fw/-rev and pL1 as template; pL1/2 was generated with primers L2-fw/L1/2-rev and pL2 as template. For ectopic expression of hfq under control of its own promoter, a PCR product obtained from Xcv 85-10 using primers pMphfq-fw/-rev was cloned into the promoterless vector pBRM-P [84].
The GFP-based promoter-less reporter plasmid pFX-P permits BsaI-mediated cloning of PCR amplicons (Golden Gate cloning) in a one-step restriction-ligation reaction [54] and was generated as follows: pDSK602 [85] was digested with PstI/BamHI to remove the lac promoter and multiple-cloning site. To allow blue-white selection, a dummy module containing 5′- and 3′-BsaI recognition sites, plac and lacZ was PCR-amplified from pBRM-P [84] using primers pFX-lz-fw/-rev. A fragment containing both the gfp coding sequence without translation start codon and the rrnB terminator was PCR-amplified from pXG-1 [53] using primers pFXgfp-fw/-rev. After blunt-end ligation of dummy- and gfp-module, the fragment was digested with Mph1103I/BglII and ligated into the PstI/BamHI sites of the pDSK602 backbone, resulting in pFX-P. For generation of the GFP-control plasmid pFX0, a promoterless fragment (138 bp) of the sRNA gene sX6 [16] was PCR-amplified from Xcv 85-10 using primers pFX0-fw/-rev and cloned into pFX-P.
To generate mRNA::gfp expression constructs, fragments containing the promoter, 5′-UTR and 10 to 25 codons of the respective genes were PCR-amplified from Xcv 85-10 using corresponding pFX-fw/-rev primers (Table S1) and cloned into pFX-P. Plasmids pFX3927, pFXhfq and pFX0612 were generated by cloning of nucleotide sequences −98 to +30, −160 to +75 and −116 to +33 relative to the translation start codon of XCV3927, hfq and XCV0612, respectively. pFXpilH was constructed by cloning a fragment spanning nucleotides −99 upstream of the pilG translation start codon to nucleotide +60 within the coding sequence of pilH.
The 4G-motif in XCV3927::gfp and hfq::gfp is located 21–24 bp upstream and 9–12 bp downstream of the ATG, respectively. pilH::gfp and XCV0612::gfp contain a 5G-motif at nucleotide positions 10–14 and 8–12 upstream of the ATG, respectively. Plasmids pFXMUT were constructed as follows: to mutate the ‘GGGG’ motif to ‘GCGC’, sequences upstream and downstream of the motif were PCR-amplified from Xcv 85-10 using primers pFX-fw/pFX-mut-L-rev and pFX-mut-R-fw/pFX-rev, respectively. Primers pFX-mut-L-rev and pFX-mut-R-fw contain the mutation flanked by a BsaI-recognition site. pFX and corresponding pFXMUT derivatives only differ in the sequence of the G-rich motif at nucleotide positions relative to the translation start codons: −24/−22 in pFX3927MUT, +10/+12 in pFXhfqMUT and −12/−10 in pFXpilHMUT.
Plasmids pFXpl, which express plac-driven mRNA::gfp fusions, were constructed by cloning respective fragments into pFX-P: plac was PCR-amplified from pBRM-P [84] using primers plac-fw/rev; sequences −59 to +54 and −147 to +54 relative to the ATG of hrpX and hrpG, respectively, were PCR-amplified from Xcv 85-10 using primers pFXpl-hrpX-fw/-rev and pFXpl-hrpG-fw/-rev; fragments of XCV3927 and pilH were PCR-amplified from respective pFX and pFXMUT plasmids using primers pFXpl3927-fw/pFXpl3927mut-fw/pFX3927-rev and pFXplpilH-fw/pFXpilH-rev.
Generation of Xcv mutant strains
To generate a chromosomal sX13 deletion mutant, flanking sequences of ∼650 bp up- and downstream of sX13 [16] were amplified by PCR from Xcv 85-10 using primers d13L-fw/-rev and d13R-fw/-rev. PCR products were digested with BamHI/HindIII and HindIII/XbaI, respectively, and ligated into the suicide vector pOK1 [86]. XcvΔsX13+sX13 ch, which carries an sX13 copy at the ΔsX13 locus, was created as follows: two PCR fragments amplified from Xcv 85-10 using primers int13L-fw/rev and int13R-fw/rev were digested with Psp1406I, ligated and cloned into the BamHI/XbaI site of pOK1. Conjugation of pOKΔsX13 into Xcv 85-10 and pOKint13 into XcvΔsX13 and selection of the correct double crossing-over events were performed as described [86]. XcvΔsX13+sX13 ch was identified by PCR amplification of the sX13 locus and Psp1406I restriction.
To introduce a frameshift mutation into chromosomal hfq, PCR products obtained from Xcv 85-10 using primers hfqL-fw/-rev and hfqR-fw/-rev were digested with BamHI/BsaI and BsaI/XbaI, respectively, and cloned into pOK1. Conjugation of pOK-fshfq into Xcv and selection of double crossing-over events were performed as described [86]. The resulting hfq mutant strain carries a 4 bp deletion in an MnlI site (nucleotides 33–36 in hfq) and was identified by PCR using primers seqhfq-fw/-rev followed by digestion with MnlI.
Plant material and plant inoculations
Pepper (Capsicum annuum) plants of the near-isogenic cultivars ECW and ECW-10R [87] were grown at 25°C with 60–70% relative humidity and 16 hours light. For infection assays, Xcv bacteria were resuspended in 10 mM MgCl2 and inoculated with a needleless syringe into the intercellular spaces of leaves using concentrations of 1–4×108 colony-forming units (CFU) per ml for scoring plant reactions and 104 CFU/ml for in planta growth curves. For better visualization of the HR, leaves were bleached in 70% ethanol. In planta growth curves were performed as described [33]. All experiments were repeated at least two times.
Protein detection and measurement of GFP fluorescence in Xcv
Xcv cells grown overnight in NYG medium were washed, incoculated at OD600 = 0.2 into XVM2 medium and incubated for 3.5 hours at 30°C. Total cell extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting using specific polyclonal antibodies directed against HrpF [88], HrcN [89], HrcJ [89] and GroEL (Stressgen). A horseradish peroxidase-labeled anti-rabbit antibody (Amersham Pharmacia Biotech) was used as secondary antibody. Antibody reactions were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech).
To determine GFP fluorescence, bacteria were adjusted to OD600 = 1.0 in 10 mM MgCl2. Fluorescence was measured at 485-nm excitation and 535-nm emission using a microplate reader (SpectraFluor Plus; Tecan Trading AG).
In vitro transcription and structure probing
sX13 [16] was PCR-amplified from Xcv 85-10 using primers sX13T7-fw, containing the T7-promoter, and sX13T7-rev and cloned into pUC57 (Thermo Fisher Scientific), resulting in pUC-13T7. Template DNA for in vitro transcription was amplified from pUC-13T7 using primers sX13-ITC-fw/-rev. sX13 transcription and DNase treatment were performed according to manufacturer's instructions (MEGAscript®Kit; Invitrogen). RNA labeling using [γ-32P]-ATP, treatment with RNase T1 (1 Pharmacia unit; Ambion) or RNase V1 (0.01 to 0.0002 Pharmacia units; Ambion) and generation of nucleotide ladders were performed as described [90]. Samples were analyzed on 12% polyacrylamide gels containing 7 M urea. Signals were visualized with a phosphoimager (FLA-3000 Series; Fuji).
RNA preparation, Northern blot and qRT-PCR analysis
Bacteria were grown overnight in NYG and inoculated at OD600 = 0.2 into NYG or XVM2 medium. XVM2 cultures were incubated for 3.5 hours at 30°C. NYG-grown cells were harvested at exponential growth phase (OD600 = 0.5–0.7) or used to inoculate the following media at OD600 = 0.5: NYG containing 0.3 M NaCl, 0.2 M H2O2 or NYG lacking a nitrogen source, MMA or MMA lacking a carbon source followed by incubation for 3 hours.
RNA isolation and Northern blot hybridization was performed as described [16], [91]. Oligonucleotide probes for detection of sX13 and 5S rRNA are described in [16].
For qRT-PCR analyses, cDNA was synthesized using RevertAid H Minus First Strand cDNA-Synthesis Kit according to manufacturer's instructions (Fermentas). qRT-PCR was performed using 2 ng cDNA and ABsolute BlueSYBR Green Fluorescein (Thermo Scientific) and analyzed on MyiQ2 and CFX Connect systems (Bio-Rad). The efficiency and specificity of PCR amplifications was determined by standard curves derived from a dilution series of template cDNA and melting curve analysis, respectively. Mean transcript levels were calculated based on values obtained from technical duplicates of at least three independent biological replicates and the levels of 16S rRNA (reference gene) as described (ABI user bulletin 2; Applied Biosystems).
Microarray analysis
For isolation of total RNA, NYG-grown cells were harvested at exponential growth phase (OD600 = 0.5–0.7) or used to inoculate MMA at OD600 = 0.5 followed by incubation for 3 hours. Fluorescently labeled cDNA was prepared as described [92]. Starting from 10–15 µg total RNA, aminoallyl-modified first strand cDNA was synthesized by reverse transcription using random hexamer primers, Bioscript reverse transcriptase (Bioline) and 0.5 mM dNTP, dTTP∶aminoallyl-dUTP (1∶4). After hydrolysis and clean up using Nucleotide removal kit (Qiagen), Cy3- and Cy5-N-Hydroxysuccinimidyl ester dyes (GE Healthcare) were coupled to the aminoallyl-labeled first strand cDNA. Uncoupled dye was removed using the Nucleotide removal kit (Qiagen). For RNA from NYG- and MMA-grown bacteria, four and three microarray hybridizations were performed, respectively, using labeled cDNA obtained from independent bacterial cultures.
The genome-wide microarray for Xcv strain 85-10 (Xcv5KOLI) carried 50–70 nt unique oligonucleotides representing CDSs, with each oligonucleotide spotted in three technical replicates per microarray [93]. Preprocessing of microarrays was performed as described [94]. Hybridization was performed in EasyHyb hybridization solution (Roche) supplemented with sonicated salmon sperm DNA at 50 µg/ml in a final volume of 130 µl for 90 min at 45°C using the HS400 Pro hybridization station (Tecan Trading AG). Labeled cDNA samples were denatured for 5 min at 65°C prior hybridization. After hybridization microarrays were washed as described [94].
Mean signal and mean local background intensities were obtained for each spot on the microarray images using ImaGene 8.0 software for spot detection, image segmentation and signal quantification (Biodiscovery Inc.). Spots were flagged as empty if R≤0.5 in both channels, where R = (signal mean−background mean)/background standard deviation. Remaining spots were analyzed further. The log2 value of the ratio of intensities was calculated for each spot using the formula Mi = log2(R i/G i). R i = I ch1(i)-Bg ch1(i) and G i = I ch2(i)-Bg ch2(i), where I ch1(i) or I ch2(i) is the intensity of a spot in channel 1 or channel 2, and Bg ch1(i) or Bg ch2(i) is the background intensity of a spot in channel 1 or channel 2. The mean intensity was calculated for each spot, A i = log2(R i G i)0.5 [95]. For data normalization (Median), significance test (Holm) and t-statistics analysis, the EMMA 2.8.2 open source platform was used [49]. Genes were accounted as differentially expressed if P adjusted ≤0.05, A≥8, and if the ratio showed at least a 1.5-fold difference between the two experimental conditions.
Biocomputational analyses
Homology searches were performed using BLASTn and the NCBI genome database (http://blast.ncbi.nlm.nih.gov; http://www.ncbi.nlm.nih.gov/genome; date: 11/22/2012).
The secondary structure of sX13 [16] was predicted using Mfold version 3.5 (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form) and default folding parameters [55]. To identify putative regulatory motifs in the 5′-regions of sX13-regulated mRNAs, a discriminative motif search was performed using DREME version 4.9.0 (http://meme.nbcr.net/meme/cgi-bin/dreme.cgi) [56]. Sequences of regulated genes comprising nucleotide positions −100 to +100 relative to translation start codons or in case of known TSSs [16] (see Table S2), sequences comprising the 5′-UTR to position +100 downstream of translation start codons, were extracted from the genome of Xcv strain 85-10 (NC_007508 and NC_007507) [32]. DREME motif search was performed with negatively regulated genes as input and positively regulated genes as comparative sequences and an E-value of ≤5.
Accession numbers
YP_363045.1; YP_363046.1; YP_362142.1; YP_362163.1; YP_362160.1; YP_361663.1; YP_363499.1; YP_365931.1; YP_363887.1; YP_365930.1; YP_365658.1; YP_364552.1; YP_363772.1; YP_363917.1; YP_364964.1; YP_364963.1; YP_364545.1; YP_365303.1; YP_365304.1; YP_362343.1; YP_362302.1; YP_362409.1; YP_364550.1; YP_364798.1; YP_364827.1; YP_365231.1; YP_363688.1; YP_363753.1; YP_361904.1; YP_363264.1; YP_362055.1; YP_363957.1
Supporting Information
sX13 abundance is not affected by expression of HrpG*. Xcv 85-10 (wt), ΔsX13 and ΔsX13+sX13 ch and strains additionally expressing HrpG* were incubated for 3.5 hours in hrp-gene inducing medium XVM2 (see Figure 2B). Total RNA was analyzed by Northern blot using an sX13-specific probe. 5S rRNA was probed as loading control. The experiment was performed twice with similar results.
(EPS)
Structure probing of sX13. In vitro transcribed sX13 was 5′-labeled and treated with RNase T1 (T1) or alkaline hydroxyl (OH−) buffer for generation of nucleotide ladders and RNase V1 (V1) for mapping of base-paired regions. Lane ‘C’ contains untreated sX13; triangle indicates decreasing concentrations of RNase V1; ‘#G’ indicates positions of G residues; the deduced secondary structure is indicated on the right hand side (see Figure 6A).
(EPS)
Expression of sX13 derivatives. Total RNA of NYG-grown Xcv strains 85-10 (wt) and ΔsX13 carrying pBRS (pB), psX13 or expressing mutated sX13-derivatives (see Figure 6) was analyzed by Northern blot using an sX13-specific probe. 5S rRNA was probed as loading control. The experiment was performed twice with similar results.
(EPS)
Distribution of 4G-motifs among sX13-regulated genes and chromosomally encoded Xcv genes. (A) Percentage of sX13-regulated genes identified by microarray analyses (see Table S2) and chromosomal CDSs in Xcv containing one or more 4G-motifs in region −100 to +100 relative to the TLS or in case of known TSSs, in the sequence comprising the 5′-UTR to position +100. The number of genes analyzed (n) is given below. (B) Distribution of 4G-motifs found in region −100 to +100 bp relative to the TLSs of 1,378 chromosomal CDSs [see (A)].
(EPS)
sX13-dependency of mRNA target::GFP synthesis in MMA-grown Xcv strains. GFP fluorescence of MMA-grown Xcv strains 85-10 (wt) and ΔsX13 carrying pB or psX13 and carrying GFP-reporter plasmids pFX3927, pFX3927MUT, pFXpilH or pFXpilHMUT. pFX3927MUT and pFXpilHMUT contain a mutated 4G- and 5G-motif, respectively. Xcv autofluorescence was determined using pFX0. GFP fluorescence of the wt was set to 1. Data points and error bars represent mean values and standard deviations obtained from three independent experiments. Statistically significant differences compared to the wt are indicated by an asterisk (t-test; P<0.03). For comparison, see Figure 8A and C.
(EPS)
mRNA amount of XCV3927::gfp is sX13-independent. The XCV3927::gfp mRNA amount in NYG-grown Xcv strains 85-10 (wt) and ΔsX13 carrying pB, psX13 or mutated sX13-derivatives and containing pFX3927 or pFX3927MUT was analyzed by qRT-PCR using gfp-specific oligonucleotides. The RNA level in the wt was set to 1. Data points and error bars represent mean values and standard deviations obtained with three independent biological samples. For comparison, see Figure 7B and Figure 8A.
(EPS)
sX13 posttranscriptionally affects XCV3927::GFP and PilH::GFP synthesis. GFP fluorescence of NYG-grown Xcv strains 85-10 (wt), ΔsX13 and ΔsX13 containing chromosomally re-integrated sX13 (ΔsX13+sX13 ch); strains express XCV3927::gfp (pFXpl-3927) or pilH::gfp (pFXpl-pilH) under control of plac. pFXMUT derivatives contain a mutated 4G-motif. Xcv autofluorescence was determined using pFX0 and is indicated by dashed line. GFP fluorescence of the wt carrying pFXpl-3927 or pFXpl-pilH was set to 1. Data points and error bars represent mean values and standard deviations obtained from three independent experiments. Asterisks indicate statistically significant differences (t-test; P<0.03).
(EPS)
Translation of HrpG::GFP and HrpX::GFP is sX13-independent. GFP fluorescence of NYG-grown Xcv strains 85-10 (wt) and ΔsX13 expressing hrpG::gfp (pFXpl-hrpG) or hrpX::gfp (pFXpl-hrpX) under control of plac. Xcv autofluorescence was determined using pFX0 and is indicated by dashed line. GFP fluorescence of the wt was set to 1. Data points and error bars represent mean values and standard deviations obtained from three independent experiments. Differences were not statistically significant (t-test; P<0.03).
(EPS)
Bacterial strains, plasmids and oligonucleotides used in this study.
(PDF)
sX13-regulated genes identified by microarray and qRT-PCR analysis.
(PDF)
Acknowledgments
We are grateful to C. Wagner, B. Rosinsky, C. Kretschmer, W. Bigott, and B. Herte for technical assistance. We thank J. Vogel (Institute for Molecular Infection Biology, Würzburg, Germany) for providing pXG-1 and H. Berndt and D. Büttner for helpful comments on the manuscript.
Funding Statement
This work was supported by grants from the Deutsche Forschungsgemeinschaft to UB and AB. (SPP 1258; “Sensory and Regulatory RNAs in Prokaryotes”) and the “Graduiertenkolleg” (GRK 1591) to UB, and by the Bundesministerium für Bildung und Forschung (grant 0313105) to AB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Storz G, Vogel J, Wassarman KM (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43: 880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Richards GR, Vanderpool CK (2011) Molecular call and response: the physiology of bacterial small RNAs. Biochim Biophys Acta 1809: 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Waters LS, Storz G (2009) Regulatory RNAs in bacteria. Cell 136: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J (2009) Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol 16: 840–846. [DOI] [PubMed] [Google Scholar]
- 5. Papenfort K, Vogel J (2010) Regulatory RNA in bacterial pathogens. Cell Host Microbe 8: 116–127. [DOI] [PubMed] [Google Scholar]
- 6. Gottesman S, Storz G (2011) Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harbor Perspectives in Biology 3: a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, et al. (2009) Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol 74: 139–158. [DOI] [PubMed] [Google Scholar]
- 8. Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S (1998) DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A 95: 12462–12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA (2010) Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci U S A 107: 9602–9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Göpel Y, Görke B (2012) Rewiring two-component signal transduction with small RNAs. Curr Opin Microbiol 15: 132–139. [DOI] [PubMed] [Google Scholar]
- 11. Vogel J, Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chao Y, Vogel J (2010) The role of Hfq in bacterial pathogens. Curr Opin Microbiol 13: 24–33. [DOI] [PubMed] [Google Scholar]
- 13. Wilms I, Overlöper A, Nowrousian M, Sharma CM, Narberhaus F (2012) Deep sequencing uncovers numerous small RNAs on all four replicons of the plant pathogen Agrobacterium tumefaciens . RNA Biol 9: 446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Filiatrault MJ, Stodghill PV, Bronstein PA, Moll S, Lindeberg M, et al. (2010) Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J Bacteriol 192: 2359–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Findeiß S, Schmidtke C, Stadler PF, Bonas U (2010) A novel family of plasmid-transferred anti-sense ncRNAs. RNA Biol 7: 120–124. [DOI] [PubMed] [Google Scholar]
- 16. Schmidtke C, Findeiß S, Sharma CM, Kuhfuss J, Hoffmann S, et al. (2012) Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res 40: 2020–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang RP, Tang DJ, Chen XL, He YQ, Feng JX, et al. (2010) Identification of four novel small non-coding RNAs from Xanthomonas campestris pathovar campestris . BMC Genomics 11 10.1186/1471-2164-1111-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang H, Zhao YT, Zhang JQ, Wang XJ, Fang RX, et al. (2011) Identification and functional characterization of small non-coding RNAs in Xanthomonas oryzae pathovar oryzae . BMC Genomics 12 10.1186/1471-2164-1112-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chai Y, Winans SC (2005) A small antisense RNA downregulates expression of an essential replicase protein of an Agrobacterium tumefaciens Ti plasmid. Mol Microbiol 56: 1574–1585. [DOI] [PubMed] [Google Scholar]
- 20. Wilms I, Voss B, Hess WR, Leichert LI, Narberhaus F (2011) Small RNA-mediated control of the Agrobacterium tumefaciens GABA binding protein. Mol Microbiol 80: 492–506. [DOI] [PubMed] [Google Scholar]
- 21. Cui Y, Chatterjee A, Liu Y, Dumenyo CK, Chatterjee AK (1995) Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-L-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J Bacteriol 177: 5108–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Y, Cui Y, Mukherjee A, Chatterjee AK (1998) Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol Microbiol 29: 219–234. [DOI] [PubMed] [Google Scholar]
- 23. Cui Y, Chatterjee A, Yang H, Chatterjee AK (2008) Regulatory network controlling extracellular proteins in Erwinia carotovora subsp. carotovora: FlhDC, the master regulator of flagellar genes, activates rsmB regulatory RNA production by affecting gacA and hexA (lrhA) expression. J Bacteriol 190: 4610–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leyns F, De Cleene M, Swings JG, De Ley J (1984) The host range of the genus Xanthomonas . The Botanical Review 50: 308–356. [Google Scholar]
- 25. Chan JW, Goodwin PH (1999) The molecular genetics of virulence of Xanthomonas campestris . Biotechnol Adv 17: 489–508. [DOI] [PubMed] [Google Scholar]
- 26. Mhedbi-Hajri N, Darrasse A, Pigne S, Durand K, Fouteau S, et al. (2011) Sensing and adhesion are adaptive functions in the plant pathogenic xanthomonads. BMC Evol Biol 11 10.1186/1471-2148-1111-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang X, Xiao Y, Zhou JM (2006) Regulation of the type III secretion system in phytopathogenic bacteria. Mol Plant Microbe Interact 19: 1159–1166. [DOI] [PubMed] [Google Scholar]
- 28. Ghosh P (2004) Process of protein transport by the type III secretion system. Microbiol Mol Biol Rev 68: 771–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Büttner D, Bonas U (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol Rev 34: 107–133. [DOI] [PubMed] [Google Scholar]
- 30. Fouhy Y, Lucey JF, Ryan RP, Dow JM (2006) Cell-cell signaling, cyclic di-GMP turnover and regulation of virulence in Xanthomonas campestris . Res Microbiol 157: 899–904. [DOI] [PubMed] [Google Scholar]
- 31. Jones JB, Stall RE, Bouzar H (1998) Diversity among xanthomonads pathogenic on pepper and tomato. Annu Rev Phytopathol 36: 41–58. [DOI] [PubMed] [Google Scholar]
- 32. Thieme F, Koebnik R, Bekel T, Berger C, Boch J, et al. (2005) Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J Bacteriol 187: 7254–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bonas U, Schulte R, Fenselau S, Minsavage GV, Staskawicz BJ, et al. (1991) Isolation of a gene-cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol Plant Microbe Interact 4: 81–88. [Google Scholar]
- 34. White FF, Potnis N, Jones JB, Koebnik R (2009) The type III effectors of Xanthomonas . Mol Plant Pathol 10: 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klement Z (1982) Hypersensitivity. In: Mount MS, Lacy GH, editors. Phytopathogenic Prokaryotes. New York: Academic Press. pp. 149–177.
- 36. White FF, Yang B, Johnson LB (2000) Prospects for understanding avirulence gene function. Curr Opin Plant Biol 3: 291–298. [DOI] [PubMed] [Google Scholar]
- 37. Schulte R, Bonas U (1992) Expression of the Xanthomonas campestris pv. vesicatoria hrp gene cluster, which determines pathogenicity and hypersensitivity on pepper and tomato, is plant inducible. J Bacteriol 174: 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wengelnik K, Marie C, Russel M, Bonas U (1996) Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive reaction. J Bacteriol 178: 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wengelnik K, Van den Ackerveken G, Bonas U (1996) HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol Plant Microbe Interact 9: 704–712. [DOI] [PubMed] [Google Scholar]
- 40. Wengelnik K, Bonas U (1996) HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J Bacteriol 178: 3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wengelnik K, Rossier O, Bonas U (1999) Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J Bacteriol 181: 6828–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Noël L, Thieme F, Nennstiel D, Bonas U (2001) cDNA-AFLP analysis unravels a genome-wide hrpG-regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria . Mol Microbiol 41: 1271–1281. [DOI] [PubMed] [Google Scholar]
- 43. Guo Y, Figueiredo F, Jones J, Wang N (2010) HrpG and HrpX play global roles in coordinating different virulence traits of Xanthomonas axonopodis pv. citri . Mol Plant Microbe Interact 24: 649–661. [DOI] [PubMed] [Google Scholar]
- 44. Ronald PC, Staskawicz BJ (1988) The avirulence gene avrBs1 from Xanthomonas campestris pv. vesicatoria encodes a 50-kD protein. Mol Plant Microbe Interact 1: 191–198. [PubMed] [Google Scholar]
- 45. Escolar L, Van Den Ackerveken G, Pieplow S, Rossier O, Bonas U (2001) Type III secretion and in planta recognition of the Xanthomonas avirulence proteins AvrBs1 and AvrBsT. Mol Plant Pathol 2: 287–296. [DOI] [PubMed] [Google Scholar]
- 46. Beisel CL, Storz G (2010) Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev 34: 866–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P (2002) Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev 16: 1696–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Beisel CL, Storz G (2011) The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli . Mol Cell 41: 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dondrup M, Albaum SP, Griebel T, Henckel K, Jünemann S, et al. (2009) EMMA 2–a MAGE-compliant system for the collaborative analysis and integration of microarray data. BMC Bioinformatics 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mhedbi-Hajri N, Jacques MA, Koebnik R (2011) Adhesion mechanisms of plant-pathogenic Xanthomonadaceae . Adv Exp Med Biol 715: 71–89. [DOI] [PubMed] [Google Scholar]
- 51. Jarrell KF, McBride MJ (2008) The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6: 466–476. [DOI] [PubMed] [Google Scholar]
- 52. Schulze S, Kay S, Büttner D, Egler M, Eschen-Lippold L, et al. (2012) Analysis of new type III effectors from Xanthomonas uncovers XopB and XopS as suppressors of plant immunity. New Phytol 195: 894–911. [DOI] [PubMed] [Google Scholar]
- 53. Urban JH, Vogel J (2007) Translational control and target recognition by Escherichia coli small RNAs in vivo . Nucleic Acids Res 35: 1018–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Engler C, Kandzia R, Marillonnet S (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS One 3: e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bailey TL (2011) DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hengge R (2009) Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7: 263–273. [DOI] [PubMed] [Google Scholar]
- 58. Ryan RP, Fouhy Y, Lucey JF, Jiang BL, He YQ, et al. (2007) Cyclic di-GMP signalling in the virulence and environmental adaptation of Xanthomonas campestris . Mol Microbiol 63: 429–442. [DOI] [PubMed] [Google Scholar]
- 59. Wilms I, Möller P, Stock AM, Gurski R, Lai EM, et al. (2012) Hfq influences multiple transport systems and virulence in the plant pathogen Agrobacterium tumefaciens . J Bacteriol 194: 5209–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Barra-Bily L, Pandey SP, Trautwetter A, Blanco C, Walker GC (2010) The Sinorhizobium meliloti RNA chaperone Hfq mediates symbiosis of S. meliloti and alfalfa. J Bacteriol 192: 1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, et al. (2004) The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae . Cell 118: 69–82. [DOI] [PubMed] [Google Scholar]
- 62. Bardill JP, Zhao X, Hammer BK (2011) The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base pairing interactions. Mol Microbiol 80: 1381–1394. [DOI] [PubMed] [Google Scholar]
- 63. Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, et al. (1993) Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12: 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP (2006) Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol 61: 1038–1048. [DOI] [PubMed] [Google Scholar]
- 65. Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, et al. (2007) Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 21: 1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Geissmann T, Chevalier C, Cros MJ, Boisset S, Fechter P, et al. (2009) A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res 37: 7239–7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Benito Y, Kolb FA, Romby P, Lina G, Etienne J, et al. (2000) Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA 6: 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Argaman L, Altuvia S (2000) fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J Mol Biol 300: 1101–1112. [DOI] [PubMed] [Google Scholar]
- 69. Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, et al. (2010) The primary transcriptome of the major human pathogen Helicobacter pylori . Nature 464: 250–255. [DOI] [PubMed] [Google Scholar]
- 70. Desnoyers G, Morissette A, Prevost K, Masse E (2009) Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA . EMBO J 28: 1551–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nakagawa S, Niimura Y, Miura K, Gojobori T (2010) Dynamic evolution of translation initiation mechanisms in prokaryotes. Proc Natl Acad Sci U S A 107: 6382–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sharma CM, Darfeuille F, Plantinga TH, Vogel J (2007) A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev 21: 2804–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, et al. (2011) Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol 81: 1144–1165. [DOI] [PubMed] [Google Scholar]
- 74. Martin-Farmer J, Janssen GR (1999) A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli . Mol Microbiol 31: 1025–1038. [DOI] [PubMed] [Google Scholar]
- 75. del Val C, Rivas E, Torres-Quesada O, Toro N, Jimenez-Zurdo JI (2007) Identification of differentially expressed small non-coding RNAs in the legume endosymbiont Sinorhizobium meliloti by comparative genomics. Mol Microbiol 66: 1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sahagan BG, Dahlberg JE (1979) A small, unstable RNA molecule of Escherichia coli: spot 42 RNA. I. Nucleotide sequence analysis. J Mol Biol 131: 573–592. [DOI] [PubMed] [Google Scholar]
- 77. Rice PW, Dahlberg JE (1982) A gene between polA and glnA retards growth of Escherichia coli when present in multiple copies: physiological effects of the gene for spot 42 RNA. J Bacteriol 152: 1196–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Joyce CM, Grindley ND (1982) Identification of two genes immediately downstream from the polA gene of Escherichia coli . J Bacteriol 152: 1211–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Polayes DA, Rice PW, Garner MM, Dahlberg JE (1988) Cyclic AMP-cyclic AMP receptor protein as a repressor of transcription of the spf gene of Escherichia coli . J Bacteriol 170: 3110–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. del Val C, Romero-Zaliz R, Torres-Quesada O, Peregrina A, Toro N, et al. (2011) A survey of sRNA families in a-proteobacteria. RNA Biol 9: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Daniels MJ, Barber CE, Turner PC, Sawczyc MK, Byrde RJW, et al. (1984) Cloning of genes involved in pathogenicity of Xanthomonas campestris pv. campestris using the broad host range cosmid pLAFR1. EMBO J 3: 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, et al.. (1996) Current Protocols in Molecular Biology. New York: John Wiley & Sons.
- 83. Figurski DH, Helinski DR (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans . Proc Natl Acad Sci U S A 76: 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Szczesny R, Jordan M, Schramm C, Schulz S, Cogez V, et al. (2010) Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria . New Phytol 187: 983–1002. [DOI] [PubMed] [Google Scholar]
- 85. Murillo J, Shen H, Gerhold D, Sharma A, Cooksey DA, et al. (1994) Characterization of pPT23B, the plasmid involved in syringolide production by Pseudomonas syringae pv. tomato PT23. Plasmid 31: 275–287. [DOI] [PubMed] [Google Scholar]
- 86. Huguet E, Hahn K, Wengelnik K, Bonas U (1998) hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host-specific hypersensitive reaction. Mol Microbiol 29: 1379–1390. [DOI] [PubMed] [Google Scholar]
- 87. Minsavage GV, Dahlbeck D, Whalen MC, Kearny B, Bonas U, et al. (1990) Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria - pepper interactions. Mol Plant Microbe Interact 3: 41–47. [Google Scholar]
- 88. Büttner D, Nennstiel D, Klüsener B, Bonas U (2002) Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria. J Bacteriol 184: 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rossier O, Van den Ackerveken G, Bonas U (2000) HrpB2 and HrpF from Xanthomonas are type III-secreted proteins and essential for pathogenicity and recognition by the host plant. Mol Microbiol 38: 828–838. [DOI] [PubMed] [Google Scholar]
- 90. Waldminghaus T, Gaubig LC, Klinkert B, Narberhaus F (2009) The Escherichia coli ibpA thermometer is comprised of stable and unstable structural elements. RNA Biol 6: 455–463. [DOI] [PubMed] [Google Scholar]
- 91. Hartmann RK, Bindereif A, Schön A, Westhof E (2005) Handbook of RNA biochemistry. Wiley-VCH, Weinheim, Germany 2: 636–637. [Google Scholar]
- 92. DeRisi JL, Iyer VR, Brown PO (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686. [DOI] [PubMed] [Google Scholar]
- 93. Mayer L, Vendruscolo CT, Silva WP, Vorhölter FJ, Becker A, et al. (2011) Insights into the genome of the xanthan-producing phytopathogen Xanthomonas arboricola pv. pruni 109 by comparative genomic hybridization. J Biotechnol 155: 40–49. [DOI] [PubMed] [Google Scholar]
- 94. Serrania J, Vorhölter FJ, Niehaus K, Pühler A, Becker A (2008) Identification of Xanthomonas campestris pv. campestris galactose utilization genes from transcriptome data. J Biotechnol 135: 309–317. [DOI] [PubMed] [Google Scholar]
- 95. Dudoit S, Yang YH, Callow MJ, Speed TP (2002) Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat Sin 12: 111–140. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sX13 abundance is not affected by expression of HrpG*. Xcv 85-10 (wt), ΔsX13 and ΔsX13+sX13 ch and strains additionally expressing HrpG* were incubated for 3.5 hours in hrp-gene inducing medium XVM2 (see Figure 2B). Total RNA was analyzed by Northern blot using an sX13-specific probe. 5S rRNA was probed as loading control. The experiment was performed twice with similar results.
(EPS)
Structure probing of sX13. In vitro transcribed sX13 was 5′-labeled and treated with RNase T1 (T1) or alkaline hydroxyl (OH−) buffer for generation of nucleotide ladders and RNase V1 (V1) for mapping of base-paired regions. Lane ‘C’ contains untreated sX13; triangle indicates decreasing concentrations of RNase V1; ‘#G’ indicates positions of G residues; the deduced secondary structure is indicated on the right hand side (see Figure 6A).
(EPS)
Expression of sX13 derivatives. Total RNA of NYG-grown Xcv strains 85-10 (wt) and ΔsX13 carrying pBRS (pB), psX13 or expressing mutated sX13-derivatives (see Figure 6) was analyzed by Northern blot using an sX13-specific probe. 5S rRNA was probed as loading control. The experiment was performed twice with similar results.
(EPS)
Distribution of 4G-motifs among sX13-regulated genes and chromosomally encoded Xcv genes. (A) Percentage of sX13-regulated genes identified by microarray analyses (see Table S2) and chromosomal CDSs in Xcv containing one or more 4G-motifs in region −100 to +100 relative to the TLS or in case of known TSSs, in the sequence comprising the 5′-UTR to position +100. The number of genes analyzed (n) is given below. (B) Distribution of 4G-motifs found in region −100 to +100 bp relative to the TLSs of 1,378 chromosomal CDSs [see (A)].
(EPS)
sX13-dependency of mRNA target::GFP synthesis in MMA-grown Xcv strains. GFP fluorescence of MMA-grown Xcv strains 85-10 (wt) and ΔsX13 carrying pB or psX13 and carrying GFP-reporter plasmids pFX3927, pFX3927MUT, pFXpilH or pFXpilHMUT. pFX3927MUT and pFXpilHMUT contain a mutated 4G- and 5G-motif, respectively. Xcv autofluorescence was determined using pFX0. GFP fluorescence of the wt was set to 1. Data points and error bars represent mean values and standard deviations obtained from three independent experiments. Statistically significant differences compared to the wt are indicated by an asterisk (t-test; P<0.03). For comparison, see Figure 8A and C.
(EPS)
mRNA amount of XCV3927::gfp is sX13-independent. The XCV3927::gfp mRNA amount in NYG-grown Xcv strains 85-10 (wt) and ΔsX13 carrying pB, psX13 or mutated sX13-derivatives and containing pFX3927 or pFX3927MUT was analyzed by qRT-PCR using gfp-specific oligonucleotides. The RNA level in the wt was set to 1. Data points and error bars represent mean values and standard deviations obtained with three independent biological samples. For comparison, see Figure 7B and Figure 8A.
(EPS)
sX13 posttranscriptionally affects XCV3927::GFP and PilH::GFP synthesis. GFP fluorescence of NYG-grown Xcv strains 85-10 (wt), ΔsX13 and ΔsX13 containing chromosomally re-integrated sX13 (ΔsX13+sX13 ch); strains express XCV3927::gfp (pFXpl-3927) or pilH::gfp (pFXpl-pilH) under control of plac. pFXMUT derivatives contain a mutated 4G-motif. Xcv autofluorescence was determined using pFX0 and is indicated by dashed line. GFP fluorescence of the wt carrying pFXpl-3927 or pFXpl-pilH was set to 1. Data points and error bars represent mean values and standard deviations obtained from three independent experiments. Asterisks indicate statistically significant differences (t-test; P<0.03).
(EPS)
Translation of HrpG::GFP and HrpX::GFP is sX13-independent. GFP fluorescence of NYG-grown Xcv strains 85-10 (wt) and ΔsX13 expressing hrpG::gfp (pFXpl-hrpG) or hrpX::gfp (pFXpl-hrpX) under control of plac. Xcv autofluorescence was determined using pFX0 and is indicated by dashed line. GFP fluorescence of the wt was set to 1. Data points and error bars represent mean values and standard deviations obtained from three independent experiments. Differences were not statistically significant (t-test; P<0.03).
(EPS)
Bacterial strains, plasmids and oligonucleotides used in this study.
(PDF)
sX13-regulated genes identified by microarray and qRT-PCR analysis.
(PDF)