Abstract
Listeria monocytogenes is a Gram-positive foodborne pathogen and the causative agent of listerosis a disease that manifests predominately as meningitis in the non-pregnant individual or infection of the fetus and spontaneous abortion in pregnant women. Common-source outbreaks of foodborne listeriosis are associated with significant morbidity and mortality. However, relatively little is known concerning the mechanisms that govern infection via the oral route. In order to aid functional genetic analysis of the gastrointestinal phase of infection we designed a novel signature-tagged mutagenesis (STM) system based upon the invasive L. monocytogenes 4b serotype H7858 strain. To overcome the limitations of gastrointestinal infection by L. monocytogenes in the mouse model we created a H7858 strain that is genetically optimised for oral infection in mice. Furthermore our STM system was based upon a mariner transposon to favour numerous and random transposition events throughout the L. monocytogenes genome. Use of the STM bank to investigate oral infection by L. monocytogenes identified 21 insertion mutants that demonstrated significantly reduced potential for infection in our model. The sites of transposon insertion included lmOh7858_0671 (encoding an internalin homologous to Lmo0610), lmOh7858_0898 (encoding a putative surface-expressed LPXTG protein homologous to Lmo0842), lmOh7858_2579 (encoding the HupDGC hemin transport system) and lmOh7858_0399 (encoding a putative fructose specific phosphotransferase system). We propose that this represents an optimised STM system for functional genetic analysis of foodborne/oral infection by L. monocytogenes.
Introduction
Listeria monocytogenes is a significant food-borne pathogen that is commonly used as a model Gram-positive pathogen for infection and immunity studies. L. monocytogenes causes the disease listeriosis which is acquired by ingesting contaminated food. The disease primarily affects pregnant women, the newborn and the immunocompromised. While L. monocytogenes infections are not frequent they have a high mortality rate (20-30%) therefore making them one of the most deadly food-borne infections [1] However, very little information is available concerning the means by which gastrointestinal colonisation and persistence occur prior to invasive disease [2]. Furthermore, it is clear that L. monocytogenes strains differ in their ability to cause disease with serotype 4b strains responsible for the majority of disease epidemics [2].
Therefore to investigate the early stages of intragastric L. monocytogenes infection we utilised the powerful molecular tool of signature-tagged mutagenesis (STM). STM is an effective technique for functional genetic analysis of microbial factors involved in the infection and colonization of a host [3]. The approach is based upon random transposon mutagenesis followed by in vivo selection to compare input and output mutant pools for mutants with impaired survival. Unlike sequence-based analytical approaches such as TraDIS (transposon directed insertion-site sequencing) it allows parallel physiological analysis of isolated mutant strains [4]. In STM each mutant is tagged with a unique DNA sequence to permit co-amplification of all tags from the DNA of a mixed population of mutants by a single PCR reaction [3,5]. It was initially developed to identify virulence genes in Salmonella enteric serovar typhimurium but has subsequently been used in screens in many other bacterial species [3,6,7].
The mariner family of transposable elements are widespread in nature and are members of the IS630 family of Insertion sequences [8,9]. Mos1 is the most frequently used marnier transposon in eukaryotes while Himar1 has been extensively used for mutagenesis in bacteria [8]. Himar1 was originally derived from the horn-fly Haematobia irritans and is member of the Tc1/mariner superfamily of transposable elements [9,10]. The Himar1-based transposon system has many advantages compared to previous transposon systems used in L. monocytogenes. Firstly they do not require species-specific host factors for efficient transposition and they only require the dinucelotide TA for insertion into the chromosome which is relatively common in the low-GC L. monocytogenes [8,9,10]. Furthermore, while previous transposon systems such as Tn917 have a tendency to target hot-spots this is not the case with recently developed mariner transposon pJZ037 [11,12,13,14]. Finally transformation with mariner elements usually leads to 10-fold more mutants when compared to the Tn917-based vectors in L. monocytogenes [12].
Our STM bank was created in the L. monocytogenes 4b strain H7858. The L. monocytogenes strain H7858 is a serotype 4b frankfurter isolate from the multi-state outbreak of 1998-1999 in the USA [15]. L. monocytogenes serotype 4b strains are responsible for 33 to 50 percent of sporadic human cases worldwide and for all major foodborne outbreaks in Europe and North America since the 1980’s [16,17,18]. It is well established that mice offer a poor model for the analysis of oral infection by L. monocytogenes. Commonly used inbred strains of mice (e.g. BALB/c or C57Bl/6) require administration of exceptionally high oral doses of the pathogen in order to achieve a significant invasive infection [19]. To overcome the limitations of the mouse model we created a H7858 strain that is genetically optimised for oral infection in mice. The construction of this murinised H7858 (H7858m) strain was based on the previous Lmo-InlAm strain created by Wollert and colleagues [20]. Our data shows that this H7858m has an increased ability to infect by the oral route and will enhance the sensitivity of the STM screen, most likely through enhanced dissemination from the GI tract to mesenteric lymph nodes [21]. We have therefore created a novel STM system for use in L. monocytogenes which utilises a mariner-based transposon system and a murinised host strain for enhanced infection of mice via the oral route.
Materials and Methods
Ethics Statement
All animal procedures were approved by the University Animal Experimental Ethics Committee (AEEC) in University College Cork (approval ID 2008/32) and were carried out in a specialized facility. Work was carried out under license from the Irish Department of Health.
Bacterial strains, growth media and reagents
Bacterial strains, plasmids and primers used in this study are listed in Table 1 and Table S1 . All Escherichia coli strains were routinely grown in LB media shaking at 180 rpm at 37°C. All strains of L. monocytogenes were grown in brain heart infusion broth (BHI, Oxoid) or vegetable peptone broth (Oxoid) shaking at 180 rpm at 37°C. Defined media (DM) was made following the protocol of Premarante [22]. For growth curves in high salt environment 7.5% NaCl was added to BHI. Where appropriate antibiotics were added at the following concentrations: for E. coli 200 µg ml-1 carbenicillin, 15 µg ml-1 chloramphenicol and for L. monocytogenes erythromycin (ERY) 8 µg ml-1 and 7.5 µg ml-1 chloramphenicol.
Table 1. Strains and plasmids used in this study.
| Strains and plasmids | Description | Reference or source |
|---|---|---|
| Listeria monocytogenes | ||
| H7858 | Wild-type strain | ATCC |
| H7858m | H7858ΔinlA with inlA locus recreated containing S192N and Y369S in this chromosome | This study |
| Escherichia coli | ||
| XL1-Blue | hsdR17, supE44, recA1, endA1, gyrA46, thi, relA1, lac/F′[proAB + , lacI q, lacZ M15::Tn10(tet r)] | Stratagene |
| EC10B | E. coli DH10B derivative, with repA integrated into the glgB gene. Kanr | [24] |
| Plasmids | ||
| NZ9000+pNZ8048binlA m | Internalin A containing S192N and Y369S in pNZ8048b. | [23] |
| pORI280 | RepA- gene replacement vector, constitutive lacZ, 5.3 kb, Emr | [70] |
| pVE6007 | Temperature-sensitive helper plasmid, supplies RepA in trans | [71] |
| pORI280-inlAm | Internalin A containing S192N and Y369S mutation | [23] |
| pJZ037 | Himar1-based transposon delivery system with pSpac(hly) promoter | [14] |
Creation of murinized H7858m and non-polar mutants
A 2 Kb fragment was PCR amplified (primers IM466 and IM490) from the appropriate mutated pNZ8048binlA plasmid, with primer design incorporating the first 16 nt upstream of the inlA GTG start codon [23]. The amplimers were digested with NcoI/PstI, ligated into complementary digested pORI280 and transformed into E. coli strain EC10B (Table 1). The plasmids pORI280 and pVE6007 were co-transformed into H7858ΔinlA and mutagenesis preformed as described previously [24]. The reconstruction of the inlA locus was identified by colony PCR (primers IM317 and IM318) with the integrity of the gene confirmed by DNA sequencing.
Caco-2 invasion assays
Human (Caco-2) colonic epithelial cell lines (originally obtained from the American Type Culture Collection, Rockville, MD) were routinely cultured at 37°C in 5% CO2. Media was composed of DMEM glutamax, 10% FBS, Pen/Strep and 1% non-essential amino acids with all cell culture media purchased from Gibco. An overnight culture of L. monocytogenes was diluted down to OD600 0.1 and grown to OD600 0.8-1.0 and diluted down to cfu ml-1 1 x 107. Caco-2 cells were seeded at 1 × 105 cells, until confluency in 24 well plates (Falcon) and L. monocytogenes was infected at MOI of 10:1. On the day prior to use, antibiotics were removed from the media. On the day of use, cells were washed twice with DMEM to remove FBS. Both cell types were subjected to bacterial invasion for 1 h at 37°C in 5% CO2, washed once with Dulbecco’s PBS (Sigma) and then overlaid with DMEM containing 10 µg ml-1 gentamicin for 1 h. Monolayers were washed a further three times with PBS to remove residual antibiotic and then lysed with 1 ml of ice cold sterile water. Bacterial cells were enumerated by serial dilution in PBS and plated on BHI agar.
Production of the STM tags
A pool of single stranded 99 bp DNA molecules containing a unique 40 bp region flanked by two invariant repeats were generated by oligonucleotide synthesis (MWG-Eurofins). The oligonucleotide tag was similar to RT1 designed by Hensel et al., except that XhoI was introduced at the either end of the sequence and the variable portion was flanked by Nar1 restriction sites [3]. Double stranded DNA tags were generated by PCR amplification using RT1 as the template and J3 and J4 as primers. The PCR was carried out in a final volume of 100 µl containing 200 pg of RT1, a 100 pmol of primers and was amplified using Go-Taq® Green master mix (Promega) under the same conditions described by Hensel et al. [3], PCR products were PCR purified (Qiagen) and digested with XhoI (Roche). The plasmid pJZ037 was also digested with XhoI and PCR purified after digestion. The PCR product was ligated into pJZ037 using T4-DNA ligase (Roche) and was introduced into E. coli XL1-Blue (Stratagene) by electroporation according to the manufactures instructions. Clones carrying tagged pJZ037 were screened by colony PCR by using primers pJZ037FP and pJZ037RP. A series of 60 randomly chosen tagged plasmids were checked by sequencing (MWG-Eurofins) using pJZ037FP and confirmed the hypervariability of the 40 bp central portion (data not shown).
Generation of STM mutant banks
Electrocompetent L. monocytogenes organisms were prepared as previously described with the exception that vegetable peptone broth (Oxoid) was used instead of BHI to increase electroporation efficiency [25]. Approximately 1.5 µg of pJZ037 containing the STM tag was used to electroporate each 50-µl aliquot of electrocompetent cells. Bacteria were recovered in 1 ml of vegetable peptone broth-0.5 M sucrose left for 1 hour at 30°C and plated onto BHI plates containing 8 µg ml-1 ERY. Plates were incubated for 48 h at 30°C (the permissive temperature) and then replica plated onto BHI ERY plates and incubated overnight at 42°C (the nonpermissive temperature) to cure the plasmid.
Infection of mice
The pools were prepared in two steps. First 48 mutants were grown individually in 120 µl of BHI-ERY at 37°C with agitation in 96-well plates. Then, a 100 µl fraction from each mutant was collected and mixed into 100 ml of BHI-ERY and grown at 37°C at 180 rpm overnight. For oral inoculation, overnight cultures were centrifuged (7000xg for 5 minutes), washed twice with PBS and resuspended at 5x1010 cfu ml-1 in PBS containing 100 mg ml-1 CaCO3. Balb/C mice were intragastrically gavaged with 100 µl inoculum. Mice were euthanized after 1 day with the mesenteric lymph nodes, spleen and livers aseptically removed. The organs were homogenized and half was used to inoculate an overnight culture containing BHI-ERY and left grow at 37°C at 180 rpm. This was then used for chromosomal DNA preparation. Chromosomal DNA was prepared using the Gene Elute Bacterial Genomic DNA kit (Sigma-Aldrich). Once attenuated mutants had been identified a second screen was carried out to verify these results but a smaller pool size was used of only 24 mutants per pool.
Identification of attenuated mutants
Chromosomal DNA from each culture generated was extracted prior to infection of the mice for the input pool. The attenuated mutants were identified by carrying out 2 rounds of PCR. The first round used primers pJZ037 FP and pJZ037 RP which amplified at 250 bp region on the plasmid which contained the unique 40 bp region. This PCR product was then used as the template for the second round of PCR which amplified a 200 bp region. The primers used were pJZ037 FP and a unique primer specific to each STM. The primers were designed based on the sequence data from the 60 STM analysed (MWG-Eurofins), they were designed to have the same annealing temperature and the same sized PCR product.
Identification of the transposon insertion site in the Listeria genome
Chromosomal DNA of 1.5 ml overnight culture was extracted using the Gene Elute Bacterial Genomic DNA kit (Sigma-Aldrich). To identify the sites of transposon insertion, we initially performed arbitrary PCR to amplify the DNA sequences flanking the transposon based on the method by Cao and colleagues [12]. DNA was amplified from either end of the transposon with a series of two rounds of PCR with Taq polymerase in the first round and KOD High Fidelity polymerase (Novagen) in the second round. In each round, a transposon-specific primer and an arbitrary primer were used. In the first round, DNA fragments from the right end of the transposon were amplified with primer pairs Marq207/JZ-001. For the second round, 1 µl of the first round of PCR was used in a 25-µl reaction. DNA fragments from the right end of the transposon were amplified with primer pairs Marq208/JZ-002 or Marq208/JZ-003, respectively. The PCR products were PCR purified (Qiagen) and transformed into TOPO plasmid pCR2.1 following the manufactures instructions (Invitrogen). The plasmid was purified and was sequenced using M13 reverse primer (MWG Eurofins). The sequence data was analyzed by both BLASTn and BLASTx at the National Centre for Biotechnology (NCBI). To verify the results from the BLAST analysis the mutants were amplified using a primer from the gene of interest and JZ-184 or JZ-185 primer corresponding to a region on the mariner insertion site.
Bile growth experiments
For bile broth assays, overnights were grown in BHI shaking at 180 rpm at 37°C. Cells were then washed twice in PBS and inoculated into BHI containing 1% bovine bile (pH 5.5) at an approximate level of 2 x 105 cfu ml-1. Cell growth was determined using viable cell counts by diluting cultures in PBS solution and enumeration on BHI agar. Where bile was used as the growth medium, all growth curves were carried out using manual plate counts after 8 hours of growth.
Survival in synthetic gastric fluid
To determine the ability to survive the gastric environment, overnights were grown in BHI shaking at 180 rpm at 37°C. Cells were then washed twice in PBS and resuspended in the same volume of synthetic gastric fluid (pH 2.5) [8.3 g l−1 proteose peptone, 3.5 g l−1d-glucose, 2.05 g l−1 NaCl, 0.6 g l−1 KH2PO4, 0.11 g l−1 CaCl2, 0.37 g l−1 KCl, 0.05 g l−1 bile, 0.1 g l−1 lysozyme and 13.3 mg l−1 pepsin; adjusted to pH 2.5 with 1 N HCl [26]. Cell survival was determined using viable cell counts by diluting cultures in PBS solution and enumeration on BHI agar. Samples were taken after 2 hours of exposure.
Statistics
Statistical analysis of data was performed using unpaired student t-tests to compare datasets with individual controls as appropriate.
Results and Discussion
Creation of a murinized H7858 strain with increased ability to infect mice by the oral route
Prior to creating the STM bank we sought to improve the ability of our strain to infect mice by the oral route. We chose the 4b strain H7858 for the STM background as 4b serotypes are the most common strains associated with outbreaks and sporadic cases of listeriosis [27]. The murinized H7858 (H7858m) strain was created using the same alterations as previously described by Wollert and colleagues except that we utilised preferred Listeria codons for the mutated 192Asn and 369Ser as described by Monk et al. [20,23]. To ensure the InlA alterations had the same effect as previously reported in the EGDe background we tested its ability to infect mice by the oral route by competitive index (CI) assays. Enumeration of livers and spleens 3-days post-infection confirmed that the H7858m had an increased ability to infect by the oral route compared to the wild-type strain (Figure 1A ). The H7858m exhibited a 1-log increase in the number of bacteria recovered from the liver and 2-log increase in the CFU recovered from the spleen (Figure 1A ). However the H7858m strain did not demonstrate enhanced invasion into Caco-2 cell line but had a decreased ability to invade when compared to the wild-type background (Figure 1B ). This is similar to findings in the recreated L. monocytogenes EGDe InlAm* strain by Monk and colleagues [23]. The reason for this decrease is not known but it does not seem to affect the ability of the strain to infect mice by the oral route.
Figure 1. Analysis of murinized H7858 L. monocytogenes.
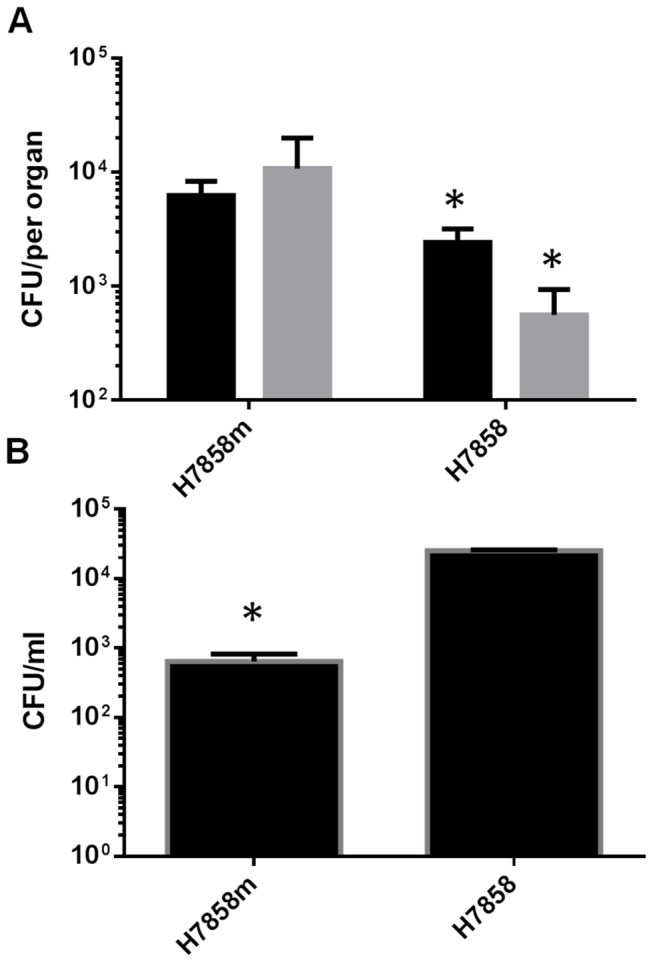
(A) The murinized H7858 strain has a greater ability to infect the mouse by the oral route compared to the wild-type strain. BALB/c mice were orally infected with 1 x 1010 CFU with either the murinized and wild-type H7858 strain. Bacterial CFU in the liver (black bars) and spleen (grey bars) were enumerated at 3 days post-infection. N=5 mice per group and the values are the mean and standard deviation. (B) Invasion assay of Caco2 cell line by wild-type and murinized H7858. Under our conditions tested the murinized strain had a decreased ability to invade the Caco2 cell line. This was carried out in triplicate and the values are the mean and standard deviation. * indicates P<0.05 relative to control strain.
Construction of STM mutant bank in H7858m and In vivo screening
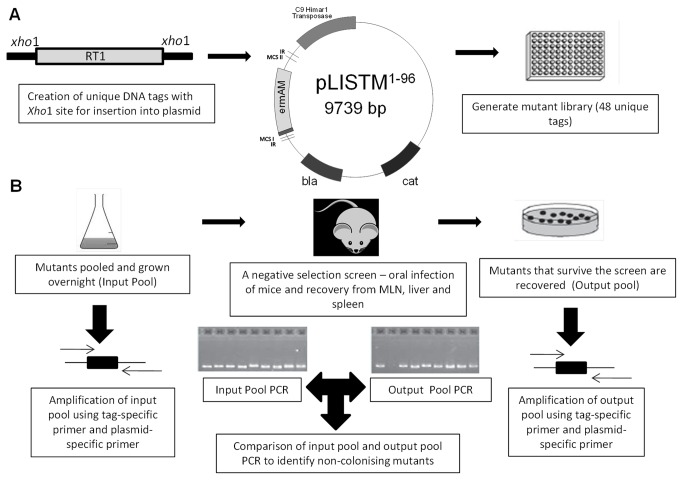
We used the Himar-1 based transposon delivery system, pJZ037 to construct the STM system in L. monocytogenes. We used a mariner based transposon as it requires no factors for transposition. Rather it requires the dinucelotide TA for insertion and this minimises the potential for multiple insertions within the same region [12,14]. Double-stranded DNA tags were cloned into the Xho1 site of pJZ037, this site was chosen as this is the region that inserts into the host genome. The recombinant clones in E. coli were screened by colony PCR using primers flanking the Xho1 insertion site. In total 96 tags were created to ensure as much variability in the sequences as possible. They were introduced into L. monocytogenes by electroporation, thus generating 96 banks of L . monoctyogenes mutants (Figure 2 ). A preliminary screen was performed to determine which size bank was required to ensure all STMs were equally represented. A STM bank size of 72, 48 and 24 were pooled and infected into mice as described below and from this it was determined that a bank size of 48 was sufficient to ensure all mutants were fairly represented.
Figure 2. Overview of the STM system.
(A) A unique STM tag was created with Xho1 restriction enzyme sites and integrated into the mariner plasmid pJZ037. In total there were 48 unique tags created in an E. coli background and then transformed into the L. monocytogenes H7858m strain. (B) The mutants were pooled and screened in BALB/c mice where the liver, spleen and mesenteric lymph nodes were removed at 1 day post-infection. The IP and OP pools were analysed by PCR to identify non-colonising mutants.
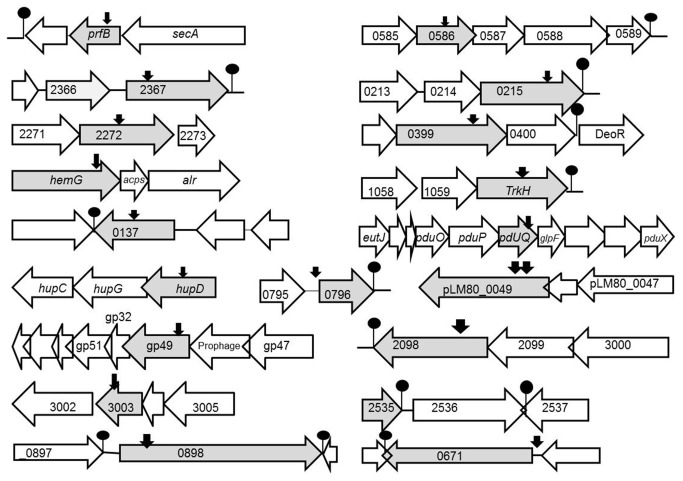
In this study we used STM to identify transposon insertions that decrease early in vivo survival of L. monocytogenes 4b serotype H7858 following oral infection. a total of 960 mariner mutants were screened by oral inoculation of 6-8 week old Balb/C mice with recovery of output pools at 1-day post-infection. 225 mutants were initially identified as being absent in the output pool of the liver, spleen or mesenteric lymph nodes. These mutants were re-organised into a new pools and 24 mutants were re-infected into the mice per pool. From the second screen 25 mutants were identified as absent in the output pool of the liver, spleen or mesenteric lymph node. The mutation leading to the attenuation was identified by arbitrary PCR, sequencing and comparison with L . moncytogenes H7858 genome (TIGR or NCBI). To verify that the insertion site was correctly identified a PCR was carried out using primer corresponding to the transposon insertion and primer for gene of interest in all cases (data not shown). Seven mutants were assessed by Southern blot analysis and all demonstrated single transposon insertions in line with previous studies of this system [14,28]. The sequence of the 25 attenuated clones corresponded to 21 distinct loci (Table 2 and Figure 3 ). Three of the insertion sites corresponded to a mutation in the internalin A (inlA) gene. This is to be expected as inlA is important for oral infection and listerial uptake into intestinal epithelial cells [29]. Furthermore this demonstrates the robustness of the STM screen. In line with previous STM mutant studies in L. monocytogenes [6] and other pathogens [3,4] we provide a table of the insertion sites for mutants identified in our study (Table 2 ) and a brief discussion of the potential role of individual genes in oral infection follows. Physiological analysis of individual mutants was used to provide clues as to stress-related defects which may impact upon gut colonisation. Future work in our laboratory will analyse the impact of precise mutations in these candidate 20 loci upon oral pathogenesis of L. monocytogenes.
Table 2. Overview of mutants identified from STM mouse screen.
| Gene name | Size (kb) | Transposon insertion site | Initial Screen Liver | Initial Screen Spleen | Initial Screen Mesenteric Lymph Nodes | Annotation | Function |
|---|---|---|---|---|---|---|---|
| lmOh7858_0215 | 1.209 | 855 bp/621bp | Present | Present | Not Detected | ABC transport protein | permease activity |
| lmOh7858_2367 | 1.371 | 572 bp | Present | Present | Not Detected | CBS domain protein | unknown function |
| lmOh7858_pLM80_0049 | 0.990 | 115 bp/150 bp | Present | Present | Not Detected | pil0073 | unknown function |
| lmOh7858_2579 | 0.972 | 520 bp | Not Detected | Not Detected | Present | iron ABC transporter | iron ion transport |
| lmOh7858_0944 (hemG) | 1.3 | 1197 bp | Present | Present | Not Detected | protoporphyrinogen oxidase | porphyrin biosynthetic process |
| lmOh7858_3003 | 0.69 | 356 bp | Present | Not Detected | Present | transcriptional regulator | regulatory functions |
| lmOh7858_2658 (prfB) | 0.984 | 390 bp | Present | Present | Not Detected | peptide chain release factor 2 | translational termination |
| lmOh7858_0796 | 0.405 | -100 bp | Present | Present | Not Detected | conserved hypothetical protein | unknown function |
| lmOh7858_2449 (gp49) | 0.915 | 57 bp | Present | Not Detected | Present | protein gp49 homolog | energy metabolism: electron transport |
| lmOh7858_0586 | 1.497 | 941 bp | Present | Present | Not Detected | conserved hypothetical protein | unknown function |
| lmOh7858_0398 | 1.395 | 1011 bp | Present | Present | Not Detected | phosphotransferase system | Fructose specific IIB subunit family |
| lmOh7858_0671 | 1770 | -0.05bp | Not Detected | Not Detected | Present | LPXTG-motif cell wall anchor domain protein | |
| lmOh7858_0498 (inlA) | 1.3 | 2394 bp | Present | Present | Not Detected | internalin A | Pathogenesis |
| lmOh7858_2272 | 1.236 | 690bp | Not Detected | Present | Present | ABC transport, permease | transport and binding proteins |
| lmOh7858_0137 | 0.421 | 696bp | Not Detected | Not Detected | Present | transcriptional regulator | Regulatory functions:DNA interactions |
| lmOh7858_1239 | 1.119 | 842bp | Present | Not Detected | Present | propanol dehydrogenase | Energy metabolism; propanediol utilization |
| lmOh7858_1060 | 1.326 | 720bp | Not Detected | Present | Present | cation transport protein | cation transmembrane transporter activity |
| lmOh7858_2098 | 1.097 | 250bp | Present | Present | Not Detected | DNA-damage-inducible protein P | |
| lmOh7858_0898 | 6.084 | 300bp | Present | Not Detected | Present | cell wall surface anchor f protein peptidoglycan-based cell wall biogenesis | |
| lmOh7858_2535 | 0.474 | -0.5bp | Present | Not Detected | Not Detected | Conserved hypothetical protein | Degradation of proteins and peptides |
Figure 3. Insertion sites of transposon mutants identified in the GI STM screen.
The diagram was drawn approximately to scale using Listeria monocytogenes H7858 genome sequence data (TIGR). Open reading frames (shaded in grey) are genes with transposon insertion. Black arrowheads represent the approximate location of transposon insertion. White open reading frames are flanking genes. Lollipops indicate predicted terminator locations. The number correspond to the lmOh7858 annotated numbers in the H7858 genome.
Genes encoding internalins
In the H7858 4b strain there are a total of 26 genes encoding putative internalins. From the in vivo STM screen in mice two internalins genes were identified as having a role in oral infection, inlA and lmOh7858_0671. InlA is the best characterized member of the internalin family and mediates recognition and invasion of epithelial cells through specific interaction with host E-cadherin (Ecad) [30]. Therefore the identification of inlA from our screen corresponds with earlier findings of the importance of inlA for oral infection and verifies that the conditions we used for our screen were appropriate for identification of virulence loci in L. monocytogenes.
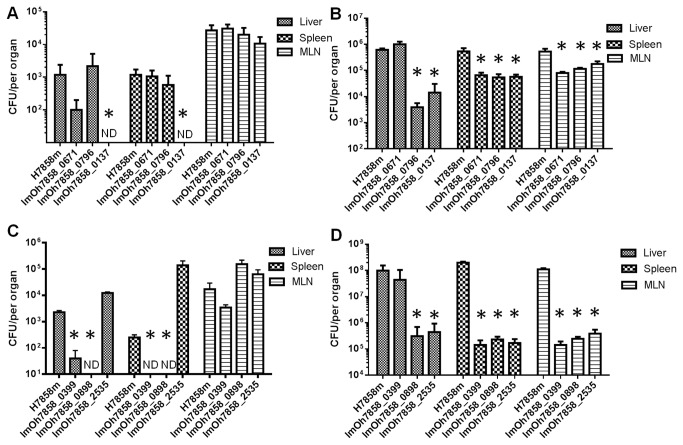
The second internalin identified by the screen was lmOh7858_0671 (Figure 3 ). This gene is 82% homologous to the EGDe gene lmo0610. This is a LPXTG internalin that contains several other regions such as a signal peptide, 8 leucine rich repeats (LRR), 2 PKD domains and a sorting signal (Figure S1 ). Previous microarray analysis identified that the EGDe homologue lmo0610 is regulated by the alternative sigma factor σB and contains a σB promoter sequence upstream from the gene [31]. Clustal W analysis identified that the same σB promoter sequence was present upstream from the lmOh7858_0671 start site in H7858 indicating that it is most likely regulated by this sigma factor. Significantly, σB is known to be activated by conditions encountered in the gastrointestinal tract [32]. To verify the results from the initial STM screen this mutant was orally infected into mice and compared to the wild-type H7858m. This mutant showed a 1-log decreased infection of the liver at one day post-infection (Figure 4A ) and had 1-log decrease in survival in spleen and MLN on day 3 (Figure 4B ).
Figure 4. In vivo analyses of individual Tn mutants after oral infection.
The kinetics of infection was analyzed on day 1 (A) (C) and day 3 (B) (D) post infection. Bacterial infection was monitored in the liver, spleen and mesenteric lymph nodes. Values are the mean and standard deviation of 5 mice and CFU per organ. ND, not detected. * indicates P<0.05 relative to wild-type control.
lmOh7858_0898
Another transposon insertion mutant identified in the screen was in lmOh7858_0898 (Figure 3 ). This gene encodes a cell wall surface anchor family protein that contains a LPXTG motif, which is the signature sequence that is recognized by the sortase enzyme for localization to the cell wall (Figure S1 ). As well as the LPXTG motif this gene also contains 8 Bacterial-like Ig, which is mostly likely a PKD domain, but it does not contain a LRR region (Figure S1 ). In addition upstream from the start site is a putative PrfA box (TTAAAAATTACTAA) indicating this gene could be regulated by PrfA (Figure S1 ). Interestingly, the homologue of this gene in EGDe (lmo0842) has previously been shown to be upregulated in the host compared to stationary growth in BHI [33]. Furthermore the homologue of this gene was downregulated when grown in soil after 15, 30 minutes and 18 hours (10-fold decreased expression) of exposure to soil [34]. Piveteau and colleagues postulate that virulence associated genes are downregulated due to stimuli in the soil which result in decreased expression of virulence associated genes [34]. When this mutant was subsequently used to orally infect Balb/C mice it had a reduced ability to proliferate in the liver and spleen on day 1 and day 3 post-infection compared to the wild-type strain (Figure 4 C,D ).
Peptide chain release factor (prfB)
One of the transposon insertion sites identified in the screen was prfB a gene encoding a putative peptide chain release factor (RF2) (Figure 3 ). RF2 recognizes the translational stop sites UAA and UGA and is itself regulated through RNA frameshifting events [35]. Recent data suggests that RF2 is important for survival and colonization of the gut by the E. coli K12 strain [36,37]. An RF2 mutation in E. coli leads to growth inhibition, presumably due to aberrant translational termination events and this may also prevent the strain from being able to colonize the gut [36]. While we did not identify a growth defect in BHI (data not shown) the prfB mutant was unable to grow to the same degree as the wild-type in the presence of BHI and high salt (7.5% NaCl) (Figure 5A ). This phenotype may account for the inability of our mutant to survive GI infection, as increased osmolarity of the upper small intestine (equivalent to 0.3 M NaCl) would provide an in vivo challenge for this mutant [38].
Figure 5. In vitro analysis of virulence-attenuated Tn mutants.
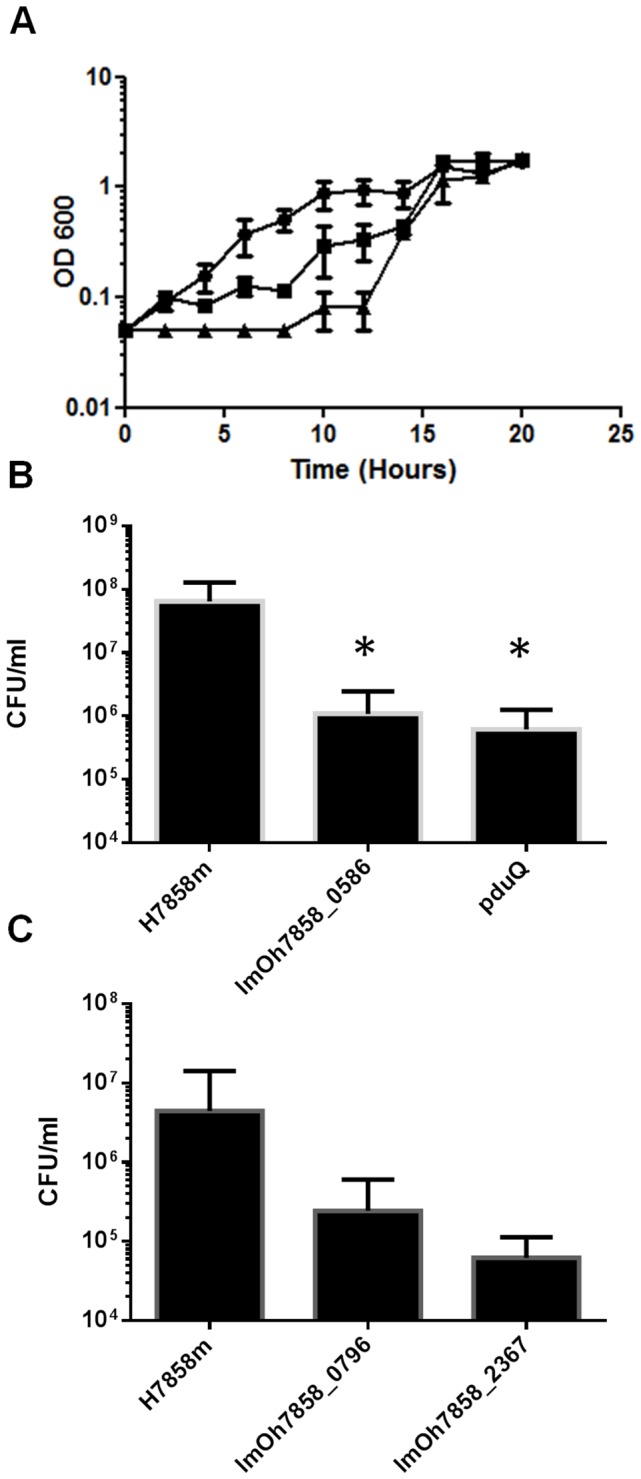
All mutants were subjected to in vitro stress analysis as outlined in Materials and Methods. Shown are mutants which differ from the wild-type in an aspect of their stress resistance profile. For clarity mutants with similar profiles to the wild-type are not shown. (A) Growth kinetics of mariner mutants compared to wild-type H7858m grown in BHI + 7.5% NaCl. The mariner mutants in prfB and lmoH7858_0137 have a decreased ability to grow in a high salt environment compared to the wild-type strain. (B) Survival of wild-type and mariner mutants exposed to synthetic gastric fluid (pH 2.5) for 2 hours. The mutants in lmOh7858_0586 and pduQ had 2-log less survival compared to wild-type strain. (C) Survival of wild-type and mariner mutants in BHI containing 1% bovine bile at pH 5.5. The insertion mutants in lmOh7858_0796 and lmOh7858_2367 exhibited decreased survival compared to the wild-type strain after 6 hours of exposure. All experiments were carried out in triplicate three independent times. The values are the mean and standard deviation. * indicates P<0.05 relative to control.
lmOh7858_0586
Another interesting locus identified in the STM screen was lmOh7858_0586. This gene is part of a putative operon ranging from lmOh7858_0585 to lmOh7858_0589 (Figure 3). The LmOh7858_0586 gene has 89% homology to the EGDe gene lmo0528, which encodes a hypothetical secreted protein. We show that a transposon insertion in lmOh7858_0586 results in decreased survival in synthetic gastric fluid (SGF) (Figure 5B ). This mutant exhibited a 2-log decrease in survival after 2 hours of exposure to SGF compared to the wild-type H7858m strain [22].
lmOh7858_2367
Another gene identified from the STM screen was lmOh7858_2367, which encodes a cystathionine-β-synthase (CBS) domain (Figure 3 ). Bioinformatic analysis demonstrates that this particular CBS domain is most likely associated with CorC_HlyC transport. This small domain is found in Na+/H+ antiporters, in proteins involved in magnesium and cobalt efflux, and in association with some proteins of unknown function. When this mutant was exposed to 1% bovine bile at pH 5.5 it resulted in a 1-log decrease in survival compared to the wild-type after 6 hours of exposure (Figure 5C ). There was no effect on the mutant when it was exposed to bile at pH7 (data not shown). This indicates that a transposon insertion at this site affects survival in bile under conditions similar to those encountered in the duodenum where bile mixes with chyme from the stomach [28,39,40].
ABC transporters
From the STM screen several of the isolates were identified as being ABC transporters. A transposon insertion into the gene lmOh7858_2272 which encodes a putative ABC transporter was identified as affecting gastrointestinal pathogenesis (Figure 3 ). This gene is part of an operon with lmOh7858_2271 which is an ABC transporter with an ATP binding protein. From InterPro Scan analysis this ABC transporter is a member of the ABC-2 family of transporters. There are 9 members of this family, which mostly consists of domains of unknown function. LmOh7858_2272 has 93.4% homology to its L. monocytogenes EGDe homologue lmo2140.
Transposon insertions into lmOh7858_0215 were independently identified twice in our STM screen. This gene is part of a three-gene operon ranging from lmOh7858_0213 to lmOh7858_0215 (Figure 3). LmOh7858_0215 is an ABC transporter with an FtxS-like permease, which is one of a family of predicated permeases and hypothetical transmembrane proteins that have been shown to transport lipids targeted to the outer membrane (OM) across inner membrane (IM) in Gram negative bacteria.
A transposon insertion in lmOh7858_2579 was identified as reducing oral infection in our mouse model. This gene is part of a three-gene operon (lmOh7858_2579-lmOh7858_2577) that encodes an iron (hemin) ABC transport system (Figure 3 ). The gene lmOh7858_2579 has 89.5% homology to the EGDe gene lmo2431 recently demonstrated to be part of the hupDGC operon [41,42]. It was established that a mutation in this operon prevents uptake of iron from hemoglobin and hemin and results in significant attenuation in systemic infection in mice [41]. Most iron regulation is under the control of ferric uptake regulator (FUR) and a Fur box is present upstream from hupD [43]. An identical FUR box is found upstream from lmOh7858_2579 (Figure S2 ). It was also previously found that the EGDe homologue is upregulated in the host compared to stationary growth in BHI indicating that this gene plays a role in vivo [33].
lmOh7858_0399
lmOh7858_0399 is annotated as a fructose specific IIB subunit (Figure 3) and a component of a putative phosphoenolypyruvate-dependent phosphotransferase (PTS) system [44]. Fructose metabolism has been linked to virulence in other pathogens [45,46,47]. This operon is usually regulated by FruR, which belongs to the DeoR family of transcriptional regulators. Directly upstream from lmOh7858_0400 is a DeoR transcriptional regulator (Figure 3 ). More work would have to be carried out to determine how the PTSFru system may be involved in survival during GI phase of infection. To verify the results from the STM screen this transposon mutant was orally infected into Balb/C mice and shown to have significantly decreased survival on day 1 and day 3 (Figure 4 C,D ). During the early phase of infection there were no detectable mutant bacteria detected in the spleen and a 2-log difference in the amount of bacteria present in the liver compared to the H7858m wild-type. Furthermore this transposon mutant had a decreased ability to proliferate in the spleen and MLN during the late stage of GI infection.
Protoporphyrinogen oxidase (hemG)
The hemG gene (Figure 3 ) is a protoporphyrinogen oxidase that is involved in the penultimate step in heme biosynthesis [48,49]. L. monocytogenes has all the necessary genes for biosynthesis of heme from glutamate via the C5 pathway [50]. In E. coli and Bacillus subtilis a mutation in hemG renders the bacteria heme defective [51,52].
lmOh7858_1060 (trkH)
On the TIGR website lmOh7858_1060 (Figure 3 ) is annotated as a cation transport protein but CDART and InterPro Scan results demonstrate that it has homology to TrkH, a key component in potassium transport in many bacteria [53]. In prokaryotes, K+ is essential for the activation of enzymes, for turgor pressure homeostasis, maintaining intracellular pH and for salt tolerance [54,55]. The transposon insertion in lmOh7858_1060 did not affect growth at elevated NaCl concentrations (data not shown). A recent publication identified a trkH homologue in the facultative intracellular pathogen Francisella tularensis that is involved in systemic dissemination in mice [56].
lmOh7858_0137
The gene lmOh7858_0137 encodes a protein annotated as a member of the Crp/Fnr family of transcriptional regulators (Figure 3 ). Members of the Crp/Fnr superfamily are involved in a vast range of physiological functions such as metabolism, anaerobic and aerobic respiration, resistance to oxidative stress and virulence [57]. A mutant in the lmOh7858_0137 homologue in L. monocytogenes strain F2365 (LMOf2365_0130) was previously exposed to several stresses (oxidative stress, regulation of carbohydrate utilization, low temperature, heat resistance) in order to determine its function but it was not affected under any of the conditions tested [57,58]. We carried out similar experiments and found that a transposon insertion in lmOh7858_0137 led to a growth defect in a high salt environment (Figure 5A ). In vivo analyses in mice indicated that this mutant was not detectable in liver and spleen on day 1 post-infection (Figure 4A ) and on day 3 it had a 3-log difference in survival in liver and 1-log difference in spleen and MLN compared to wild-type (Figure 4B ).
pduQ
The gene lmOh7858_1239 encodes pduQ and a transposon insertion into this gene was identified in our STM screen as impacting upon virulence (Figure 3). PduQ is involved in degradation of 1,2-propanediol (1,2-PD). It is a propanol dehydrogenase that converts propionaldehyde to propanol [59]. The genes for degradation of 1,2-PD are conserved in three significant food-borne pathogens (L. monocytogenes, Clostridium perfringens and Salmonella typhimurium ) but are absent in almost all other species [60]. Korbel and colleagues have postulated that 1,2-PD is a crucial genomic determinant of pathogenicity associated with food poisoning, by promoting anaerobic growth both in the host and in processed food [60]. In Salmonella 1,2-PD was shown to play a role in pathogenesis and a deletion of the pdu genes specifically impairs growth in the host [61].
Our data demonstrate that a transposon insertion in pduQ results in a 2-log decrease in survival in SGF compared to the wild-type strain indicating the 1,2-PD may be important for survival within the stomach (Figure 5b). Recent work in Salmonella has demonstrated that a pduA mutant has low colonization of the chicken cecum which is weakly acidic (pH 6.5) [62]. In addition their work demonstrated increased expression of pdu genes in the chicken intestine after infection with Salmonella indicating the importance of these genes in Salmonella virulence [62].
lmOh7858_2098
lmOh7858 _2098 (Figure 3) is annotated as a DNA-damage-inducible protein P and is homologous to the dinB gene originally identified in E. coli. However dinB mutation in other bacteria such as E. coli and Mycobacterium failed to exhibit a clear phenotype with respect to survival following exposure to DNA-damaging stressors [63,64]. Similarly when we exposed the transposon mutant to these stresses in vitro it did not demonstrate any alteration in survival compared to wild-type strain (data not shown). Further work is needed to fully determine the effect of mutation upon survival in vivo.
Miscellaneous genes
From our STM screen the location of two transposon insertions corresponded to lmOh7858_pLM80_0049 (Figure 3 ). This gene is present on the plasmid pLM80 found in L. monocytogenes H7858. This plasmid is approximately 80 kb in size and contains several different transposable elements that are not present on the chromosome suggesting that the plasmid is a recent acquisition [65]. The plasmid has a high level of sequence and gene organization homology to the L. innocua CLIP 11262 plasmid pLI100 and the B. anthracis plasmid pXO2 [66]. The gene in question has a homologue on the pLI100 plasmid from L. innocua (pil0073). Both genes are classified as conserved hypothetical genes with no known function. This gene is also part of a 3-gene operon and these genes are also annotated as conserved hypothetical genes (Figure 3 ). The mutant was exposed to several environmental stresses (low pH, bile and high salt) and did not demonstrate any discernible phenotype (data not shown). Therefore it is difficult to determine how this gene may play a role in the GI phase of infection.
The gene lmOh7858_2449 was identified in the STM screen (Figure 3 ). This gene has homology to gp49 from the Listeria bacteriophage A118. The function of the Gp49 protein is predicted to involve endonuclease VII activity, which is the first step in the mismatch repair pathway in T4 bacteriophage [67]. This gene has 62.5% homology to the DNaD gene in the L. monocytogenes strain F6854 and the gene is required for replication initiation. When this mutant was exposed to environmental stress (low pH, bile at low pH, high salt) it did not demonstrate any decrease in survival or growth (data not shown).
Transposon insertion into lmOh7858_0796 was identified by the STM screen as affecting virulence. This gene is a hypothetical gene with homologues in other L. monocytogenes strains as well as L . welshimeri and L. innocua. Our mutant had decreased survival in BHI containing 1% bovine bile (pH 5.5) (Figure 5C ). Compared to the wild-type the lmOh7858_0796 transposon mutant had a 2-log reduced level of survival after 6 hours of exposure to bile. In vivo analyses of this mutant demonstrated that it had decreased survival in liver, spleen and MLN 3-days post-infection compared to H7858m (Figure 4B ). The greatest decrease was seen in the liver with a 3-log decrease in infection.
lmOh7858_3003 (Figure 3 ) is classified as belonging to the Sir2 family of transcriptional regulators. Silent information regulator-like proteins (Sir/sirutins) were first identified in Saccharomyces cerevisiae and shown to function as transcriptional repressors of telomeres, the silent mating-type loci and ribosomal DNA [68].
From the STM screen two independently isolated mutants of interest corresponded to transposon insertions into lmOh7858_2535. This gene is not on an operon and is classified as having homology to B. subtilis YuiD protein (Figure 3 ). From bioinformatic analysis it was determined that this gene is related to the acid phosphatase/vanadium-dependent haloperoxidase whose function is currently uncharacterized but it is thought may play a role in phospholipid metabolism [69]. This gene shares 99.4% homology to the EGDe gene lmo2485. From a previous microarray analysis this gene was shown to upregulated more than 2-fold in the host compared to stationary and exponential growth in BHI [33]. Furthermore the gene was classified as being involved in the stress response [33]. When we infected mice with this mutant via the oral route it demonstrated a decreased ability to survive and proliferate in the liver, spleen and MLN during the late stage of GI infection (Figure 4D ).
Conclusions
We have engineered an improved STM system for the analysis of genetic loci required for intragastric infection by L. monocytogenes in the mouse model. The basis of the approach is a mariner transposon system and the method employed a murinized strain of serotype 4b L. monocytogenes that is optimized for oral infection in mice. Very recent sequence-based approaches for functional genetic analysis of mutant banks (such as TraDIS) offer great potential for large-scale mutant screening [7]. However these approaches also currently have limitations such as the requirement for complete unbiased transposon coverage, the need for an animal model capable of extremely efficient gastrointestinal colonization/infection, high costs associated with sequencing input and output banks and the inability to work with individual mutants isolated using the system [7]. In contrast STM offers the ability to tailor the size of the input pool to overcome any limitations associated with the animal model and to analyse individual mutants in vitro subsequent to the screen [4,7]. Here we demonstrate that our novel system has identified transposon insertion mutants that are compromised for infection via the oral route. In an approach used previously in V. cholerae we also performed analysis of our mutants for resistance to physico-chemical stressors encountered in vivo [4]. Some of the mutants identified using our screen were also analyzed for individual infection dynamics in subsequent infection studies. The approach identified an insertion into known virulence-related loci (inlA, hupDGC) as well as transposon insertions into genes which encode another internalin, a transcriptional regulator and genes putatively involved in metabolic processes (including (putatively) fructose metabolism and propanol metabolism). Analysis of the roles of these loci in pathogenesis will form the basis of further study.
Supporting Information
Characterisation of internalins from STM screen. (a) Genomic organization of inlA and insertion site in transposon mutants identified in STM screen in mouse model of infection. The diagram was drawn approximately to scale using Listeria monocytogenes H7858 genome sequence data (TIGR). Open reading frames (shaded in gray) are genes with transposon insertion. Black arrowheads represent the approximate location of transposon insertion. White open reading frames are flanking genes. Lollipops indicate predicted terminator locations. (b) Schematic domain organisation of internalin lmOh7858_0671 based on EGDe homologue lmo0610 and InterPro Scan. Black box represent the signal peptide, pink box the 8 LRR, green region 2 PKD domains, yellow arrow sorting signal and yellow box the LPXTG motif. Upstream from start site is the σB promoter region at 61 bp and 82 bp from start site. (c) Schematic domain organization of lmOh7858_0898 based on Interpro Scan results. Black box represents a domain of hypothetical protein PA1324 superfamily, green box 8 PKD and yellow box represents LPXTG domain. Approximatley 199 bp upstream from start site there is a putative PrfA box.
(PPTX)
Clustal W analysis of FUR box found upstream of lmOh7858_2579. This region was compared to FUR box found in hupD homologue in EGDe and found to be completely identical to FUR box found in hupD region.
(PPTX)
Primers used in this study.
(DOCX)
Acknowledgments
We thank Marc McCarthy for technical assistance and Dr. Ian Monk for providing initial advice.
Funding Statement
This work was funded by a grant from Science Foundation Ireland under the Research Frontiers Programme (08/RFP/GEN1320). The authors also acknowledge the funding received from Science Foundation Ireland under the Centers for Science Engineering and Technology programme which funds the Alimentary Pharmabiotic Centre. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G et al. (2001) Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev 14: 584-640. doi:10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gahan CG, Hill C (2005) Gastrointestinal phase of Listeria monocytogenes infection. J Appl Microbiol 98: 1345-1353. doi:10.1111/j.1365-2672.2005.02559.x. [DOI] [PubMed] [Google Scholar]
- 3. Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E et al. (1995) Simultaneous identification of bacterial virulence genes by negative selection. Science 269: 400-403. doi:10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 4. Merrell DS, Hava DL, Camilli A (2002) Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae . Mol Microbiol 43: 1471-1491. doi:10.1046/j.1365-2958.2002.02857.x. [DOI] [PubMed] [Google Scholar]
- 5. Mazurkiewicz P, Tang CM, Boone C, Holden DW (2006) Signature-tagged mutagenesis: bar coding mutants for genome-wide screens. Nat Rev Genet 7: 929-939. doi:10.1038/ni1375. [DOI] [PubMed] [Google Scholar]
- 6. Autret N, Dubail I, Trieu-Cuot P, Berche P, Charbit A (2001) Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis. Infect Immun 69: 2054-2065. doi:10.1128/IAI.69.4.2054-2065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cummins J, Gahan CG (2012) Signature tagged mutagenesis in the functional genetic analysis of gastrointestinal pathogens. Gut Microbes 3: 93-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Picardeau M (2009) Transposition of fly mariner elements into bacteria as a genetic tool for mutagenesis. Genetica 138: 551-558. [DOI] [PubMed] [Google Scholar]
- 9. Plasterk RH, Izsvak Z, Ivics Z (1999) Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet 15: 326-332. doi:10.1016/S0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 10. Lampe DJ, Churchill ME, Robertson HM (1996) A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J 15: 5470-5479. [PMC free article] [PubMed] [Google Scholar]
- 11. Camilli A, Portnoy A, Youngman P (1990) Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J Bacteriol 172: 3738-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cao M, Bitar AP, Marquis H (2007) A mariner-based transposition system for Listeria monocytogenes . Appl Environ Microbiol 73: 2758-2761. doi:10.1128/AEM.02844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garsin DA, Urbach J, Huguet-Tapia JC, Peters JE, Ausubel FM (2004) Construction of an Enterococcus faecalis Tn917-mediated-gene-disruption library offers insight into Tn917 insertion patterns. J Bacteriol 186: 7280-7289. doi:10.1128/JB.186.21.7280-7289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zemansky J, Kline BC, Woodward JJ, Leber JH, Marquis H et al. (2009) Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J Bacteriol 191: 3950-3964. doi:10.1128/JB.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anonymous. (1998) Mulitstate outbreak of Listeriosis-United States. MMWR Weekly 47: 1085-1086. [PubMed] [Google Scholar]
- 16. Anonymous. (2001) Programme, Proposed Draft Guidelines for the Control of Listeria monocytogenes in Foods. WHO Health Organization Joint FAO/WHO Food Standards. Technical Report No. Agenda Item 6 Technical Report No. Agenda Item 6 [Google Scholar]
- 17. Dharmarha V (2008) A Focus on Listeria Monocytogenes . National Agricultural Library, Food Safety Research Information Office. [Google Scholar]
- 18. Ward TJ, Gorski L, Borucki MK, Mandrell RE, Hutchins J et al. (2004) Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes . J Bacteriol 186: 4994-5002. doi:10.1128/JB.186.15.4994-5002.2004. PubMed: 15262937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sleator RD, Watson D, Hill C, Gahan CG (2009) The interaction between Listeria monocytogenes and the host gastrointestinal tract. Microbiology 155: 2463-2475. doi:10.1099/mic.0.030205-0. [DOI] [PubMed] [Google Scholar]
- 20. Wollert T, Pasche B, Rochon M, Deppenmeier S, van den Heuvel J et al. (2007) Extending the host range of Listeria monocytogenes by rational protein design. Cell 129: 891-902. doi:10.1016/j.cell.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 21. Bou Ghanem EN, Jones GS, Myers-Morales T, Patil PD, Hidayatullah AN et al. (2012) InlA promotes dissemination of Listeria monocytogenes to the mesenteric lymph nodes during food borne infection of mice. PLOS Pathog 8: e1003015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Premaratne RJ, Lin WJ, Johnson EA (1991) Development of an improved chemically defined minimal medium for Listeria monocytogenes . Appl Environ Microbiol 57: 3046-3048. PubMed: 1746963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Monk IR, Casey PG, Hill C, Gahan CG (2010) Directed evolution and targeted mutagenesis to murinize Listeria monocytogenes internalin A for enhanced infectivity in the murine oral infection model. BMC Microbiol 10: 318. doi:10.1186/1471-2180-10-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Monk IR, Gahan CG, Hill C (2008) Tools for functional postgenomic analysis of Listeria monocytogenes . Appl Environ Microbiol 74: 3921-3934. doi:10.1128/AEM.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Park SF, Stewart GS (1990) High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94: 129-132. doi:10.1016/0378-1119(90)90479-B. [DOI] [PubMed] [Google Scholar]
- 26. Beumer RR, de Vries J, Rombouts FM (1992) Campylobacter jejuni non-culturable coccoid cells. Int J Food Microbiol 15: 153-163. doi:10.1016/0168-1605(92)90144-R. PubMed: 1622752. [DOI] [PubMed] [Google Scholar]
- 27. Anonymous. (2001) World Health Orgnaziation Joint FAO/WHO Food Standards.
- 28. Dowd GC, Joyce SA, Hill C, Gahan CG (2011) Investigation of the mechanisms by which Listeria monocytogenes grows in porcine gallbladder bile. Infect Immun 79: 369-379. doi:10.1128/IAI.00330-10. PubMed: 20937762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gaillard JL, Berche P, Frehel C, Gouin E, Cossart P (1991) Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65: 1127-1141. doi:10.1016/0092-8674(91)90009-N. PubMed: 1905979. [DOI] [PubMed] [Google Scholar]
- 30. Bierne H, Sabet C, Personnic N, Cossart P (2007) Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes . Microbes Infect 9: 1156-1166. doi:10.1016/j.micinf.2007.05.003. PubMed: 17764999. [DOI] [PubMed] [Google Scholar]
- 31. McGann P, Raengpradub S, Ivanek R, Wiedmann M, Boor KJ (2008) Differential regulation of Listeria monocytogenes internalin and internalin-like genes by sigmaB and PrfA as revealed by subgenomic microarray analyses. Foodborne Pathog Dis 5: 417-435. doi:10.1089/fpd.2008.0085. PubMed: 18713061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Begley M, Sleator RD, Gahan CG, Hill C (2005) Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes . Infect Immun 73: 894-904. doi:10.1128/IAI.73.2.894-904.2005. PubMed: 15664931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Camejo A, Buchrieser C, Couve E, Carvalho F, Reis O et al. (2009) In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLOS Pathog 5: e1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Piveteau P, Depret G, Pivato B, Garmyn D, Hartmann A (2011) Changes in gene expression during adaptation of Listeria monocytogenes to the soil environment. PLOS ONE 6: e24881. doi:10.1371/journal.pone.0024881. PubMed: 21966375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Betney R, de Silva E, Krishnan J, Stansfield I (2010) Autoregulatory systems controlling translation factor expression: thermostat-like control of translational accuracy. Rna 16: 655-663. doi:10.1261/rna.1796210. PubMed: 20185543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dreyfus M, Heurgue-Hamard V (2011) Termination troubles in Escherichia coli K12. Mol Microbiol 79: 288-291. doi:10.1111/j.1365-2958.2010.07468.x. PubMed: 21219451. [DOI] [PubMed] [Google Scholar]
- 37. Bachmann B (1996) Derivations and genotypes of some mutant derivatives of Escherichia coli K12. In Neidhardt FC, Escherichiacoli and Salmonella: Cellular and Molecular Biology. District of Columbia: Washington; pp. 2460–2488. [Google Scholar]
- 38. Sleator RD, Clifford T, Hill C (2007) Gut osmolarity: a key environmental cue initiating the gastrointestinal phase of Listeria monocytogenes infection? Med Hypotheses 69: 1090-1092. [DOI] [PubMed] [Google Scholar]
- 39. Campbell I (2009) The mouth, stomach and intestines. Anaesthesia. Intensive Care Med 10: 336-338. doi:10.1016/j.mpaic.2009.04.018. [Google Scholar]
- 40. Watson BW, Meldrum SJ, Riddle HC, Brown RL, Sladen GE (1972) pH profile of gut as measured by radiotelemetry capsule. BMJ 2: 104-106. doi:10.1136/bmj.2.5805.104. PubMed: 5018285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin B, Newton SM, Shao Y, Jiang X, Charbit A et al. (2006) Iron acquisition systems for ferric hydroxamates, haemin and haemoglobin in Listeria monocytogenes . Mol Microbiol 59: 1185-1198. doi:10.1111/j.1365-2958.2005.05015.x. [DOI] [PubMed] [Google Scholar]
- 42. Xiao Q, Jiang X, Moore KJ, Shao Y, Pi H et al. (2011) Sortase independent and dependent systems for acquisition of haem and haemoglobin in Listeria monocytogenes . Mol Microbiol 80: 1581-1597. doi:10.1111/j.1365-2958.2011.07667.x. PubMed: 21545655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ledala N, Sengupta M, Muthaiyan A, Wilkinson BJ, Jayaswal RK (2010) Transcriptomic response of Listeria monocytogenes to iron limitation and Fur mutation. Appl Environ Microbiol 76: 406-416. doi:10.1128/AEM.01389-09. PubMed: 19933349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saier MH Jr., Reizer J (1994) The bacterial phosphotransferase system: new frontiers 30 years later. Mol Microbiol 13: 755-764. doi:10.1111/j.1365-2958.1994.tb00468.x. PubMed: 7815935. [DOI] [PubMed] [Google Scholar]
- 45. Gaurivaud P, Danet JL, Laigret F, Garnier M, Bove JM (2000) Fructose utilization and phytopathogenicity of Spiroplasma citri . Mol Plant Microbe Interact 13: 1145-1155. doi:10.1094/MPMI.2000.13.10.1145. PubMed: 11043476. [DOI] [PubMed] [Google Scholar]
- 46. Loo CY, Mitrakul K, Voss IB, Hughes CV, Ganeshkumar N (2003) Involvement of an inducible fructose phosphotransferase operon in Streptococcus gordonii biofilm formation. J Bacteriol 185: 6241-6254. doi:10.1128/JB.185.21.6241-6254.2003. PubMed: 14563858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stoll R, Goebel W (2010) The major PEP-phosphotransferase systems (PTSs) for glucose, mannose and cellobiose of Listeria monocytogenes, and their significance for extra- and intracellular growth. Microbiology 156: 1069-1083. doi:10.1099/mic.0.034934-0. PubMed: 20056707. [DOI] [PubMed] [Google Scholar]
- 48. Corradi HR, Corrigall AV, Boix E, Mohan CG, Sturrock ED et al. (2006) Crystal structure of protoporphyrinogen oxidase from Myxococcus xanthus and its complex with the inhibitor acifluorfen. J Biol Chem 281: 38625-38633. doi:10.1074/jbc.M606640200. PubMed: 17046834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koch M, Breithaupt C, Kiefersauer R, Freigang J, Huber R et al. (2004) Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis. EMBO J 23: 1720-1728. doi:10.1038/sj.emboj.7600189. PubMed: 15057273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Panek H, O’Brian MR (2002) A whole genome view of prokaryotic haem biosynthesis. Microbiology 148: 2273-2282. PubMed: 12177321. [DOI] [PubMed] [Google Scholar]
- 51. Homuth G, Rompf A, Schumann W, Jahn D (1999) Transcriptional control of Bacillus subtilis hemN and hemZ . J Bacteriol 181: 5922-5929. PubMed: 10498703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sasarman A, Letowski J, Czaika G, Ramirez V, Nead MA et al. (1993) Nucleotide sequence of the hemG gene involved in the protoporphyrinogen oxidase activity of Escherichia coli K12, Can. J Microbiol 39: 1155-1161. [DOI] [PubMed] [Google Scholar]
- 53. Nakamura T, Yamamuro N, Stumpe S, Unemoto T, Bakker EP (1998) Cloning of the trkAH gene cluster and characterization of the Trk K(+)-uptake system of Vibrio alginolyticus . Microbiology 144(8): 2281-2289. doi:10.1099/00221287-144-8-2281. PubMed: 9720051. [DOI] [PubMed] [Google Scholar]
- 54. Csonka L Na E, W (1996) Osmoregulation. In Escherichia coli and Salmonella: Cellular and Molecular Biology. others FCNa, editor. Washington D.C.: American Society for Microbiology; . 1210-1223 p [Google Scholar]
- 55. Stumpe SB, [!(surname)!] (1996) K+ circulation across the prokaryotic cell membrane: K+ uptake systems. WN Konings HRKaJSL Amsterdam: Elesevier; pp. 474-499. [Google Scholar]
- 56. Alkhuder K, Meibom KL, Dubail I, Dupuis M, Charbit A (2010) Identification of trkH, encoding a potassium uptake protein required for Francisella tularensis systemic dissemination in mice. PLOS ONE 5: e8966. doi:10.1371/journal.pone.0008966. PubMed: 20126460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Uhlich GA, Wonderling LD, Luchansky JB (2006) Analyses of the putative Crp/Fnr family of transcriptional regulators of a serotype 4b strain of Listeria monocytogenes . Food Microbiol 23: 300-306. doi:10.1016/j.fm.2005.03.007. PubMed: 16943018. [DOI] [PubMed] [Google Scholar]
- 58. Bayles DO, Uhlich GA (2006) Inactivation of the Crp/Fnr family of regulatory genes in Listeria monocytogenes strain F2365 does not alter its heat resistance at 60 degrees C. J Food Protect 69: 2758-2760. [DOI] [PubMed] [Google Scholar]
- 59. Bobik TA, Ailion M, Roth JR (1992) A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol 174: 2253-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Korbel JO, Doerks T, Jensen LJ, Perez-Iratxeta C, Kaczanowski S et al. (2005) Systematic association of genes to phenotypes by genome and literature mining. PLOS Biol 3: e134. doi:10.1371/journal.pbio.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Conner CP, Heithoff DM, Julio SM, Sinsheimer RL, Mahan MJ (1998) Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci U S A 95: 4641-4645. doi:10.1073/pnas.95.8.4641. PubMed: 9539791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Harvey PC, Watson M, Hulme S, Jones MA, Lovell M et al. Salmonella enterica serovar Typhimurium colonizing the lumen of the chicken intestine are growing slowly and up-regulate a unique set of virulence and metabolism genes. Infect Immun. [DOI] [PMC free article] [PubMed]
- 63. Fuchs RP, Fujii S, Wagner J (2004) Properties and functions of Escherichia coli: Pol IV and Pol V. Adv Protein Chem 69: 229-264. doi:10.1016/S0065-3233(04)69008-5. [DOI] [PubMed] [Google Scholar]
- 64. Kana BD, Abrahams GL, Sung N, Warner DF, Gordhan BG et al. Role of the DinB homologs Rv1537. and Rv3056 in Mycobacterium tuberculosis . J Bacteriol 192: 2220-2227. doi:10.1128/JB.01135-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT et al. (2004) Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res 32: 2386-2395. doi:10.1093/nar/gkh562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Read TD, Salzberg SL, Pop M, Shumway M, Umayam L et al. (2002) Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296: 2028-2033. doi:10.1126/science.1071837. [DOI] [PubMed] [Google Scholar]
- 67. Seeberg E, Eide L, Bjoras M (1995) The base excision repair pathway. Trends Biochem Sci 20: 391-397. doi:10.1016/S0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 68. Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ et al. (1997) Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev 11: 255-269. doi:10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 69. Neuwald AF (1997) An unexpected structural relationship between integral membrane phosphatases and soluble haloperoxidases. Protein Sci 6: 1764-1767. doi:10.1002/pro.5560060817. PubMed: 9260289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J et al. (1996) A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet 253: 217-224. doi:10.1007/s004380050315. PubMed: 9003306. [DOI] [PubMed] [Google Scholar]
- 71. Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A (1992) New thermosensitive plasmid for gram-positive bacteria. J Bacteriol 174: 5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterisation of internalins from STM screen. (a) Genomic organization of inlA and insertion site in transposon mutants identified in STM screen in mouse model of infection. The diagram was drawn approximately to scale using Listeria monocytogenes H7858 genome sequence data (TIGR). Open reading frames (shaded in gray) are genes with transposon insertion. Black arrowheads represent the approximate location of transposon insertion. White open reading frames are flanking genes. Lollipops indicate predicted terminator locations. (b) Schematic domain organisation of internalin lmOh7858_0671 based on EGDe homologue lmo0610 and InterPro Scan. Black box represent the signal peptide, pink box the 8 LRR, green region 2 PKD domains, yellow arrow sorting signal and yellow box the LPXTG motif. Upstream from start site is the σB promoter region at 61 bp and 82 bp from start site. (c) Schematic domain organization of lmOh7858_0898 based on Interpro Scan results. Black box represents a domain of hypothetical protein PA1324 superfamily, green box 8 PKD and yellow box represents LPXTG domain. Approximatley 199 bp upstream from start site there is a putative PrfA box.
(PPTX)
Clustal W analysis of FUR box found upstream of lmOh7858_2579. This region was compared to FUR box found in hupD homologue in EGDe and found to be completely identical to FUR box found in hupD region.
(PPTX)
Primers used in this study.
(DOCX)