Dysfunction of PINK1 causes familial Parkinson's disease. Recent work suggested that accumulation of PINK1 on damaged mitochondria is a critical step for mitophagy. It was not clear, however, how PINK1 is stabilized. PINK1 forms a complex with SARM1 and TRAF6, which is important for stabilization of PINK1 and induction of mitophagy.
Abstract
Mutations in PTEN-induced putative kinase 1 (PINK1) or parkin cause autosomal recessive forms of Parkinson's disease. Recent work suggests that loss of mitochondrial membrane potential stabilizes PINK1 and that accumulated PINK1 recruits parkin from the cytoplasm to mitochondria for elimination of depolarized mitochondria, which is known as mitophagy. In this study, we find that PINK1 forms a complex with sterile α and TIR motif containing 1 (SARM1) and tumor necrosis factor receptor–associated factor 6 (TRAF6), which is important for import of PINK1 in the outer membrane and stabilization of PINK1 on depolarized mitochondria. SARM1, which is known to be an adaptor protein for Toll-like receptor, binds to PINK1 and promotes TRAF6-mediated lysine 63 chain ubiquitination of PINK1 at lysine 433. Down-regulation of SARM1 and TRAF6 abrogates accumulation of PINK1, followed by recruitment of parkin to damaged mitochondria. Some pathogenic mutations of PINK1 reduce the complex formation and ubiquitination. These results indicate that association of PINK1 with SARM1 and TRAF6 is an important step for mitophagy.
INTRODUCTION
Parkinson's disease (PD) is a common neurodegenerative movement disorder. Although most cases are sporadic, several genes have been linked to familial PD, including SNCA (Polymeropoulos et al., 1997), DJ-1 (Bonifati et al., 2003), OMI/HTRA2 (Strauss et al., 2005), leucine-rich repeat kinase 2 (LRRK2; Zimprich et al., 2004), parkin (Kitada et al., 1998), and PTEN-induced putative kinase 1 (PINK1; Valente et al., 2004). Approximately 50% of recessive early-onset familial PD cases are caused by homozygous mutations in parkin (Kitada et al., 1998; Hattori and Mizuno, 2004). Homozygous mutations in PINK1 are the second-most-common cause of early-onset recessive PD (Valente et al., 2004; Bonifati et al., 2005). Studies on their functions have provided important insights into molecular mechanisms of the pathogenesis of PD in general. This is possible because symptoms observed in familial PD patients with genetic abnormalities are very similar to those of sporadic idiopathic PD patients (Hardy et al., 2006).
The PINK1 gene encodes a 581–amino acid putative mitochondrial serine/threonine kinase that is expressed in many tissues and cell types, including dopaminergic neurons (Nakajima et al., 2003; Valente et al., 2004; Gandhi et al., 2006). Overexpression of PINK1 protects neuronal cells against various stresses (Haque et al., 2008), whereas down-regulation of PINK1 sensitizes the cells to them (Deng et al., 2005). PINK1 has kinase activity, which is required for its cytoprotective function. Phosphorylation of Akt is enhanced by overexpression of PINK1, and Akt activation is crucial for protection of neuronal SH-SY5Y cells from cytotoxic agents (Murata et al., 2011a, b). Some pathogenic mutations of PINK1 result in reduction in phosphorylation levels of Akt.
On the other hand, accumulating evidence indicates that PINK1 is involved in mitochondrial maintenance in collaboration with parkin, a PD-linked E3 ubiquitin ligase. Genetic analysis of Drosophila showed a significant link between PINK1 and parkin (Clark et al., 2006; Park et al., 2006), and human patients with mutations in either of these proteins display very similar clinical symptoms (Abeliovich and Flint Beal, 2006). Narendra et al. (2008) first demonstrated that parkin translocates from the cytoplasm to dysfunctional mitochondria having low membrane potential. The translocated parkin ubiquitinates mitochondrial outer membrane proteins such as voltage-dependent anion channel 1 (VDAC1) and mitofusin 1 and 2 (MFN1/2; Gegg et al., 2010; Geisler et al., 2010; Poole et al., 2010; Tanaka et al., 2010; Ziviani et al., 2010; Glauser et al., 2011; Rakovic et al., 2011). The damaged mitochondria carrying ubiquitinated proteins on the surface are then degraded via the proteasome and autophagy (Narendra et al., 2008; Chan et al., 2011; Yoshii et al., 2011). As an initial step of this process, PINK1 is stabilized and accumulates on the damaged mitochondria and subsequently recruits parkin to damaged mitochondria (Matsuda et al., 2010; Narendra et al., 2010; Vives-Bauza et al., 2010). The exact mechanism by which PINK1 is stabilized on damaged mitochondria is unknown. Therefore we sought insight into regulation of the functional level of PINK1 by identifying and analyzing new proteins interacting with PINK1. We found new interacting proteins of PINK1—sterile α and Toll/interleukin 1 receptor (TIR) motif containing 1 (SARM1) and tumor necrosis factor receptor–associated factor 6 (TRAF6)—and showed that PINK1 is ubiquitinated and stabilized on damaged mitochondria through interaction with SARM1 and TRAF6. The complex formation is required for recruitment of parkin to damaged mitochondria. The results provide evidence supporting a link between the deregulation of complex formation and PD pathogenesis.
RESULTS
PINK1 interacts with SARM1
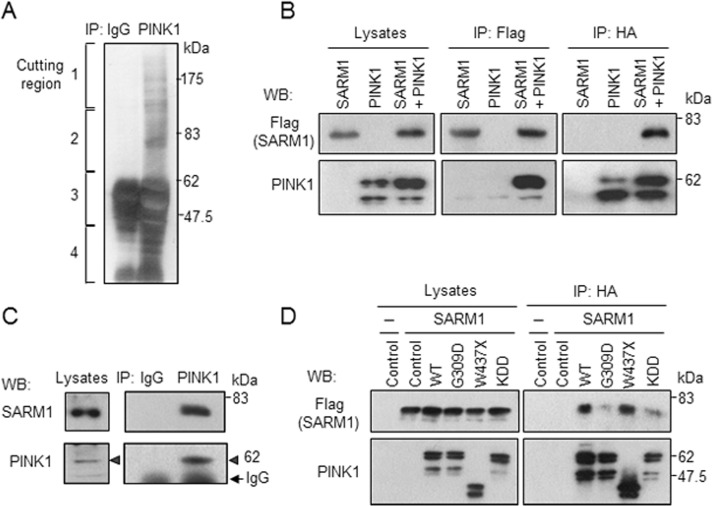
To identify proteins that interact with endogenous PINK1, we performed immunoprecipitation experiments using an anti-PINK1 antibody. The antibody was raised in our laboratory (Murata et al., 2011a) because commercially available antibodies against PINK1 were not suitable for immunoprecipitation. Silver staining of the affinity-purified and electrophoresed PINK1 immune complexes revealed the presence of several proteins. Mass spectrometry analysis identified a number of candidate proteins, among them SARM1, a TIR adaptor protein (12 total spectra, 8 unique peptides for 16% coverage of SARM1 with 100% probability). The protein was recovered from region 2 of the silver-staining gel (Figure 1A).1 We focused on the function of SARM1 because SARM1 was reported to be preferentially expressed in neurons (Kim et al., 2007) and to regulate neuronal function in Caenorhabditis elegans (Chuang and Bargmann, 2005; Chang et al., 2011) and mammals (Chen et al., 2011).
FIGURE 1:
Interaction of SARM1 with PINK1. (A) Identification of SARM1 as a protein interacting with PINK1. Cell lysates from HEK293 cells were subjected to immunoprecipitation with anti-PINK1 antibody or control rabbit IgG, followed by gel-electrophoretic separation and silver staining. The gels were cut into four pieces for mass spectrometric analysis. SARM1 was identified from region 2. PINK1 was contained in region 3. (B) Binding of PINK1 to SARM1. HEK293 cells expressing PINK1-HA and/or SARM1-Flag were immunoprecipitated with designated antibodies and analyzed by Western blotting. (C) Endogenous PINK1 interacts with SARM1 in SH-SY5Y cells. Cell lysates prepared from SH-SY5Y cells were subjected to immunoprecipitation with anti-PINK1 antibody or control rabbit IgG, followed by Western blot analysis. Arrowheads indicate immunoprecipitated PINK1. (D) Effects of PINK1 pathogenic mutations and kinase-dead mutation on PINK1-SARM1 interaction. Cell lysates from HEK293 cells cotransfected with SARM1-Flag and HA-tagged PINK1 variants were immunoprecipitated with anti-HA agarose and analyzed by Western blotting. WT, wild-type PINK1; G309D and W437X, respective pathogenic PINK1 mutants; KDD, catalytically inactive triple mutant (K219A/D362A/D384A) of PINK1.
To confirm the binding of PINK1 to SARM1, we transfected vectors expressing SARM1-Flag and PINK1-hemagglutinin (HA) to HEK293 cells. Immunoprecipitation of the lysates with anti-Flag antibody revealed that PINK1-HA coimmunoprecipitated with SARM1-Flag (Figure 1B, middle). In a reciprocal coimmunoprecipitation experiment, anti-HA antibody was able to coimmunoprecipitate SARM1-Flag with PINK1-HA (Figure 1B, right). To examine binding of endogenous proteins, we immunoprecipitated cell extracts prepared from intact SH-SY5Y cells with anti-PINK1 antibody. As shown in Figure 1C, endogenous SARM1 coprecipitated with endogenous PINK1, whereas a control experiment using preimmune immunoglobulin G (IgG) showed no bands, indicating specific interaction between endogenous PINK1 and SARM1. Next we assessed binding of SARM1 to PD-linked PINK1 mutants, G309D (Kessler et al., 2005) and W437X (Criscuolo et al., 2006), and with a synthetic kinase-dead PINK1 triple mutant, KDD (Beilina et al., 2005). We found that the W437X mutant retained the ability to interact with SARM1, whereas G309D and KDD mutants showed reduced binding capability (Figure 1D).
N-terminal region of SARM1 is required for interaction with PINK1 and localization to mitochondria
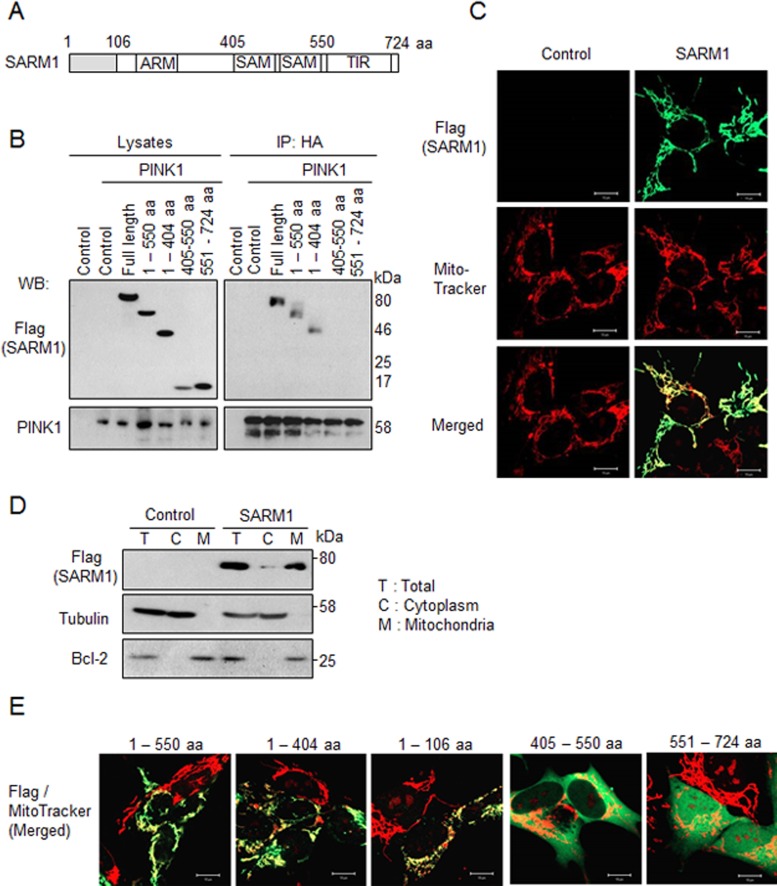
SARM1 contains HEAT/Armadillo (ARM) repeats, two sterile α motif (SAM) domains, and a TIR domain, all of which are involved in protein–protein interactions (Figure 2A; Kim et al., 2007). To identify the PINK1-binding region of SARM1, we made C-terminal Flag-tagged serial deletion mutants and examined their ability to bind to PINK1 by coimmunoprecipitation assays. The N-terminal region (1–404 amino acids) that contains the ARM domain but not the SAM (405–550 amino acids) and TIR (551–724 amino acids) domains was shown to be necessary for interaction with PINK1 (Figure 2B).
FIGURE 2:
The N-terminal region of SARM1 is required for the interaction with PINK1 and localization to mitochondria. (A) Schematic diagram of SARM1. The SARM1 gene encodes 724 amino acids. ARM, HEAT/Armadillo repeats; SAM, sterile α motif domain; TIR, Toll/interleukin 1 receptor domain. (B) PINK1 binds to the N-terminal region of SARM1. Cell lysates from SH-SY5Y cells cotransfected with PINK1-HA and SARM1 deletion mutants tagged with Flag were immunoprecipitated with anti-HA agarose and analyzed by Western blotting. (C) Intracellular localization of SARM1. Forty-eight hours after transfection with SARM1-Flag, SH-SY5Y cells were immunostained with anti-Flag antibody (green). The cells were costained with MitoTracker Orange for identifying mitochondria (red). (D) Forty-eight hours after transfection with SARM1-Flag, SH-SY5Y cells were fractionated. Tubulin and Bcl-2 were used as markers of the cytoplasm and mitochondria, respectively. (E) For localization to mitochondria, 1–106 amino acids of SARM1 are required. SH-SY5Y cells were transfected with designated constructs tagged with Flag. The experiment was performed under conditions similar to those in C.
To examine intracellular localization of SARM1, we transfected full-length SARM1-Flag to SH-SY5Y cells and costained the cells with anti-Flag antibody and MitoTracker Orange CMTMRos for visualization of mitochondria. As shown in Figure 2C, SARM1 colocalized with mitochondria. This result was corroborated by a subcellular fractionation experiment (Figure 2D). Tubulin and Bcl-2 were used as markers of the cytoplasm and mitochondria, respectively. The band of SARM1 was mainly detected in mitochondrial fraction. The mitochondrial association of SARM1 was also confirmed in another cell line, HEK293 cells (Supplemental Figure S1A). To identify the specific region of SARM1 responsible for the association with mitochondria, we used several C-terminal Flag-tagged SARM1 deletion mutants. The N-terminal region (1–106 amino acids) was able to localize to mitochondria (Figure 2E). This region was predicted as a mitochondrial targeting sequence from the Center for Biological Sequence TargetP 1.1 Server (www.cbs.dtu.dk/services/TargetP/), and the region of 1–12 amino acids of SARM1 was conserved from Xenopus to human (Supplemental Figure S1B). These data suggest that PINK1 interacts with SARM1 on mitochondria through the N-terminal region of SARM1.
SARM1 binds to and recruits TRAF6 into PINK1 complexes
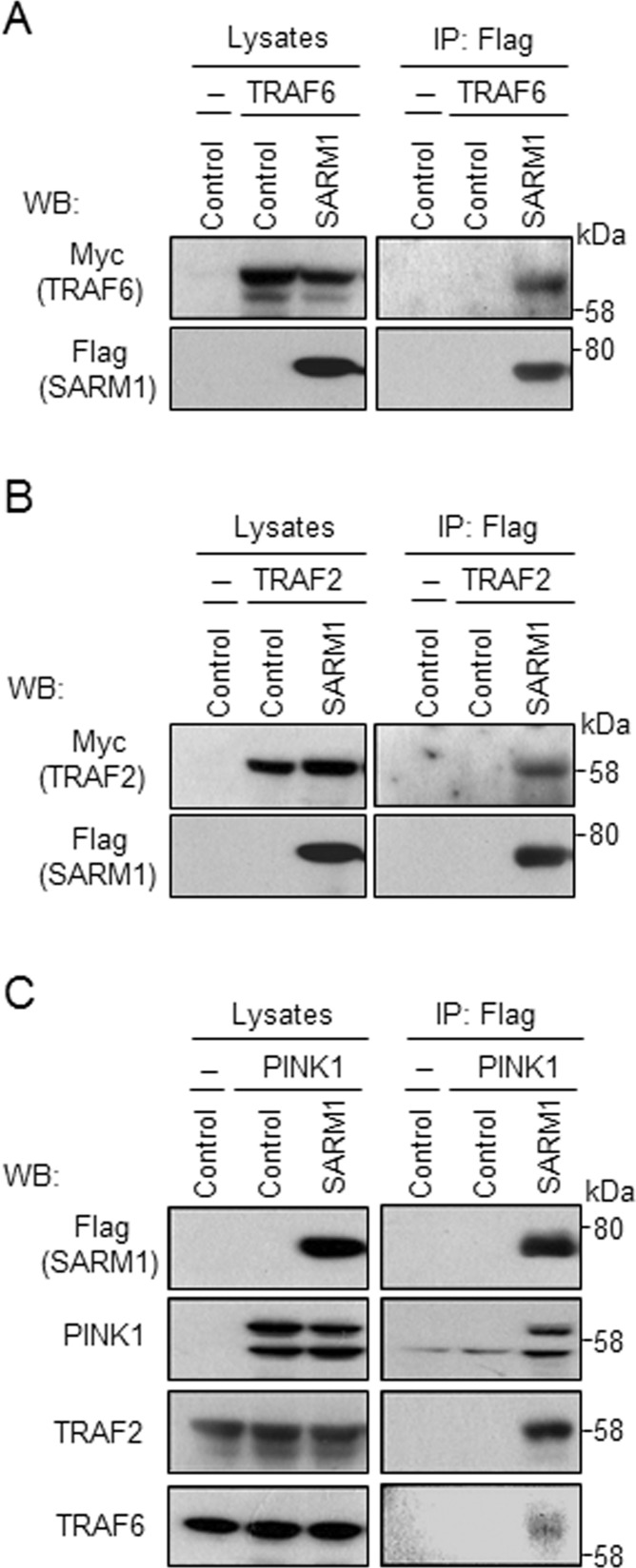
To gain insight into the potential biological significance of the PINK1–SARM1 interaction, we sought suggestive functional domains and found a putative TRAF6-binding motif, P-x-E-x-x-aromatic/acidic amino acid (Mansell et al., 2004), in the sequence of SARM1 at the 50– to 55–amino acid position (PREVSP). TRAF6 plays a critical role in innate and adaptive immunity, bone metabolism, and development of the nervous system (Wu and Arron, 2003). To determine the association of SARM1 and TRAF6, we performed coimmunoprecipitation analysis using HEK293 cells transfected with vectors expressing SARM1-Flag and/or TRAF6-Myc. Immunoprecipitation of the lysates with anti-Flag antibody resulted in recovery of TRAF6-Myc (Figure 3A). TRAF2, another TRAF family protein, has a similar binding motif with TRAF6 (P-x-Q-x-x-D; Ye et al., 1999). We therefore checked whether SARM1 binds to TRAF2 in HEK293 cells under similar conditions. TRAF2-Myc coimmunoprecipitated with SARM1-Flag (Figure 3B). In addition, endogenous TRAF2 and TRAF6 were recovered from immunoprecipitates for SARM1 (Figure 3C), indicating specific interaction between PINK1, SARM1, and TRAF2/6.
FIGURE 3:
SARM1 binds to and recruits TRAF2/6 into PINK1 complexes. (A, B) Cell lysates from HEK293 cells cotransfected with vectors expressing TRAF6/2-Myc and SARM1-Flag, respectively, were immunoprecipitated with anti-Flag antibody and analyzed by Western blotting. (C) Cell lysates from HEK293 cells cotransfected with vectors expressing PINK1-HA and/or SARM1-Flag were immunoprecipitated with anti-Flag antibody and analyzed by Western blotting. Endogenous TRAF2 and TRAF6 were recovered in the immune complex of PINK1–SARM1.
SARM1 promotes K63-linked ubiquitination of PINK1 by TRAF6
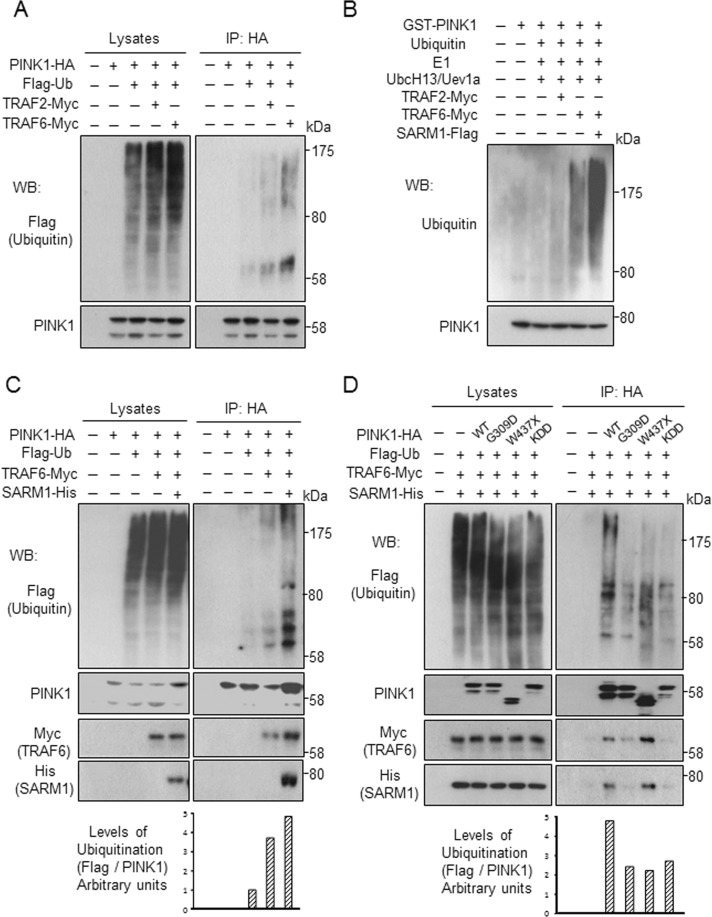
TRAF2 and TRAF6 are E3 ubiquitin ligases. The results shown in Figure 3 prompted us to test whether TRAF2 and TRAF6 ubiquitinated PINK1. To address this question, we transfected tagged PINK1 and ubiquitin with either TRAF2 or TRAF6 to SH-SY5Y cells. As shown in Figure 4A, TRAF6 stimulated ubiquitination of PINK1. TRAF2 also appeared to stimulate ubiquitination of PINK1, but to a far lesser extent.
FIGURE 4:
SARM1 promotes TRAF6-mediated ubiquitination of PINK1. (A) Ubiquitination of PINK1 by TRAF6 in cells. Cell lysates from SH-SY5Y cells transfected with PINK1-HA and Flag-ubiquitin (Ub) along with TRAF2-Myc or TRAF6-Myc were immunoprecipitated with anti-HA agarose and analyzed by Western blotting. (B) An in vitro ubiquitination assay was performed by incubation of purified GST-PINK1 with Flag-tagged SARM1, Myc-tagged TRAF2, or Myc-tagged TRAF6 in the presence of E1, E2 (UbcH13/Uev1a), and ubiquitin. Ubiquitination of PINK1 was detected by pull down with glutathione-agarose beads, followed by Western blot analysis for ubiquitin. (C) SARM1 promotes TRAF6-mediated PINK1 ubiquitination in SH-SY5Y cells. SH-SY5Y cells were transfected with designated constructs. Levels of PINK1 ubiquitination were determined by immunoprecipitation with anti-HA agarose, followed by Western blot analysis. (D) Ubiquitination levels are reduced in PINK1 mutants. The experiment was performed under conditions similar to those in C.
We then determined the linkage of TRAF6-mediated PINK1 ubiquitination using ubiquitin mutants Ub-K48R and Ub-K63R, which contain a single lysine-to-arginine mutation at positions 48 and 63, respectively. The Ub-K48R and Ub-K63R mutants are expected to specifically disrupt the assembly of respective K48- and K63-linked polyubiquitin chains. TRAF6 is known to catalyze K63-linked ubiquitination. As expected, transfection of K63R mutant ubiquitin, but not that of K48R mutant, resulted in abrogation of PINK1 ubiquitination in HEK293 cells (Supplemental Figure S2A). Furthermore, the TRAF6 C70A mutant lacking E3 ligase activity (Lamothe et al., 2007) failed in ubiquitination of PINK1 and showed reduced binding capability with PINK1 and SARM1 (Supplemental Figure S2, B and C).
To obtain further evidence supporting the K63-linked ubiquitination of PINK1, we performed in vitro ubiquitination assays using GST-partial PINK1 (containing 112–581 amino acids of PINK1). We found that TRAF6 was able to facilitate in vitro ubiquitination of PINK1 in cooperation with the UbcH13/Uev1a E2 enzyme (Figure 4B). SARM1 significantly promoted TRAF6-mediated ubiquitination of PINK1 in vitro. Ubiquitination of PINK1 was enhanced by co-overexpression of TRAF6 and SARM1 in vivo as well (Figure 4C). Next we assessed ubiquitination of PD-linked PINK1 mutants by TRAF6 and SARM1. Levels of ubiquitination of the PINK1 mutants were distinctly lower than that of the wild type (Figure 4D).
PINK1 is stabilized and ubiquitinated by TRAF6 on mitochondrial depolarization
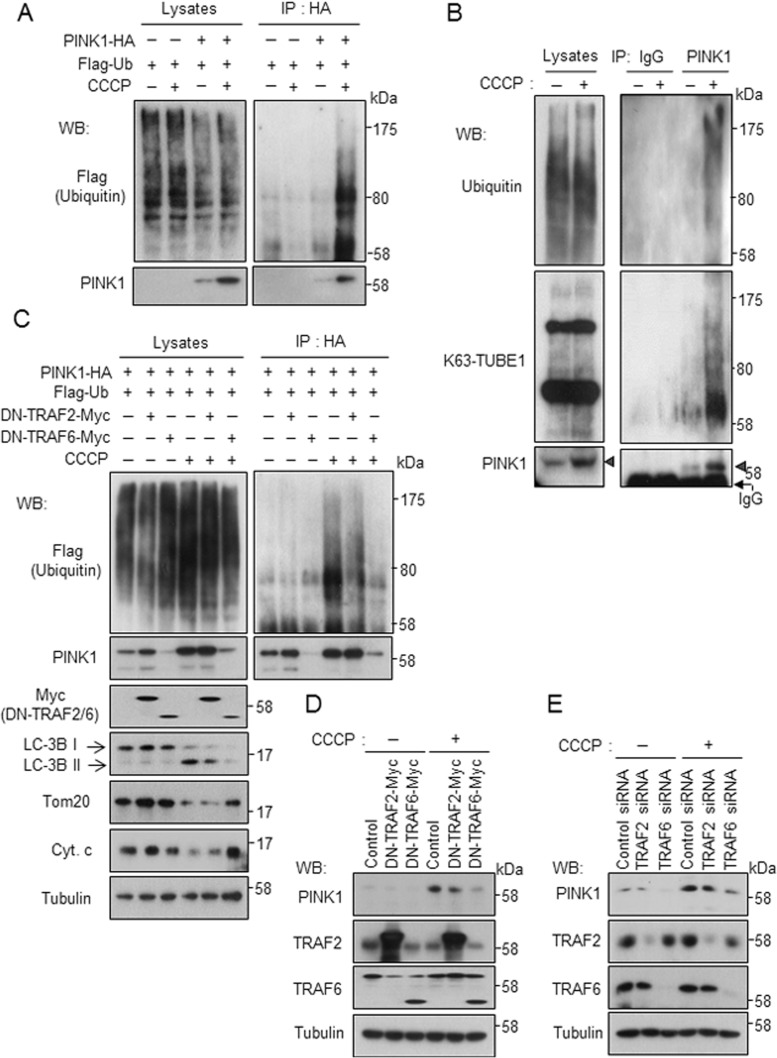
Next we investigated the biological relevance of the ternary complex formation and eventual ubiquitination of PINK1. We noted that PINK1 was stabilized under conditions in which the ternary complex was formed (Figure 4C). PINK1 plays a protective role for mitochondrial functions against various physical and chemical insults. PINK1 accumulates on mitochondria in a mitochondrial membrane potential–dependent manner and triggers mitophagy of damaged mitochondria (Matsuda et al., 2010; Narendra et al., 2010). Our finding that TRAF6 and SARM1 enhance the stability of PINK1 raises the possibility that TRAF6 and SARM1 may function together with PINK1 for protection of mitochondria. To test this possibility, we examined the stabilization and/or ubiquitination of PINK1 with various mitochondrial stress inducers. When SH-SY5Y cells were exposed to carbonyl cyanide m-chlorophenyl hydrazine (CCCP), a mitochondrial uncoupler that functions as a H+ uniporter, transfected PINK1 was stabilized and ubiquitinated at 1 and 10 μM but not 50 μM CCCP (Figure 5A and Supplemental Figure S3A). The amount and ubiquitination levels changed in parallel. Valinomycin, another type of mitochondrial uncoupler that functions as a K+ uniporter, had a similar effect (Supplemental Figure S3B). In contrast to mitochondrial depolarization with CCCP and valinomycin, other mitochondrial stress inducers, such as rotenone, which impairs mitochondrial complex I, ethidium bromide (EtBr), which preferentially impairs replication and transcription of the mitochondrial genome, and H2O2, which is reactive oxygen species, did not induce stabilization of PINK1 (Supplemental Figure S3, C–E). To examine the stabilization and ubiquitination of endogenous PINK1, we treated SH-SY5Y cells with CCCP and immunoprecipitated PINK1 with anti-PINK1 antibody. Endogenous PINK1 was stabilized and ubiquitinated with mitochondrial depolarization (Figure 5B). The ubiquitination type of endogenous PINK1 was the K63-linked linkage since the band was detected by K63-TUBE1, which recognizes K63-linked ubiquitination. Overexpression of dominant-negative TRAF6 lacking the N-terminal RING finger domain (289–522 amino acids; Cao et al., 1996) resulted in abrogation of stabilization of PINK1 and induction of mitophagy (Figure 5C). Production of LC3B-II, a marker of autophagy, and degradation of mitochondrial proteins such as Tom 20 and cytochrome c were suppressed by dominant-negative TRAF6. Stabilization of endogenous PINK1 was also inhibited by overexpression of dominant-negative TRAF6 or knockdown of endogenous TRAF6 (Figure 5, D and E). Overexpression of dominant-negative TRAF2 lacking the N-terminal RING finger domain (87–501 amino acids; Hsu et al., 1996) or knockdown of endogenous TRAF2 also appeared to inhibit stabilization of PINK1, but to a far lesser extent. These results indicate that TRAF6-mediated ubiquitination is involved in stabilization of PINK1 and induction of mitophagy.
FIGURE 5:
PINK1 is ubiquitinated by TRAF6 upon impairment of mitochondria. (A) PINK1 is ubiquitinated under the condition of low membrane potential of mitochondria. SH-SY5Y cells were transfected with designated constructs for 48 h, followed by treatment with 10 μM CCCP for 6 h. Ubiquitination of PINK1 was determined by immunoprecipitation with anti-HA agarose, followed by Western blot analysis. (B) SH-SY5Y cells were treated with 10 μM CCCP for 6 h. Ubiquitination of endogenous PINK1 was determined by immunoprecipitation with anti-PINK1 antibody, followed by Western blot analysis. K63-linked ubiquitination of PINK1 was detected using biotinylated K63-TUBE1 and streptavidin-HRP. Arrowheads indicate immunoprecipitated PINK1. (C, D) Overexpression of dominant-negative TRAF6 destabilizes exogenous and endogenous PINK1 as shown in C and D, respectively. The experiment was performed under conditions similar to those in A. (E) Knockdown of TRAF6 inhibits stabilization of endogenous PINK1. SH-SY5Y cells were transfected with designated siRNAs for 72 h, followed by treatment with 10 μM CCCP for 6 h.
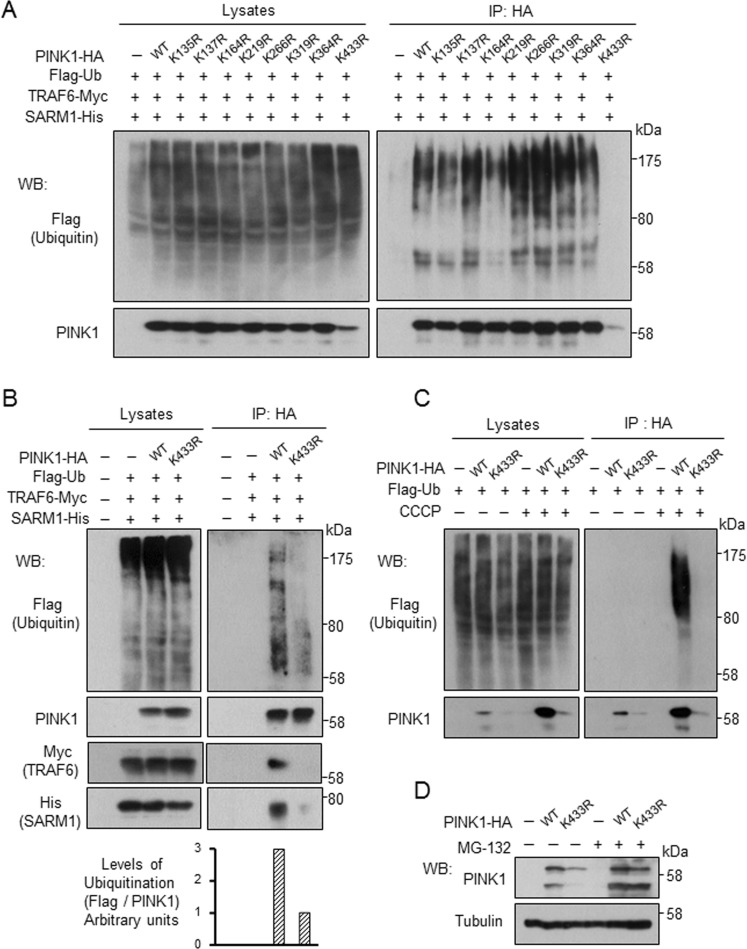
PINK1 is ubiquitinated at Lys-433 by TRAF6
PINK1 contains 21 lysine residues in its 581 amino acids, and eight lysine residues are evolutionarily conserved. To identify the ubiquitination site of PINK1 by TRAF6, we replaced each of the eight conserved lysine residues with arginine residues. We transfected C-terminal HA-tagged PINK1 wild type (WT) and mutants with different lysine-to-arginine mutations into SH-SY5Y cells along with Flag-ubiquitin, TRAF6-Myc, and SARM1-His. TRAF6 failed in ubiquitination of the K433R mutant but not of others (Figure 6A). The lower ubiquitination level of K433R mutant was also observed under conditions in which PINK1 protein level was adjusted at the same levels (Figure 6B). In addition, K433R mutant showed reduced binding capability with SARM1 and TRAF6 (Figure 6B and Supplemental Figure S4A). Ubiquitination and stabilization of the K433R mutant was not promoted by CCCP treatment (Figure 6C). Even under normal conditions, the stability of K433R mutant was lower than that of WT. Instability of K433R mutant was rescued by treatment with MG-132, an inhibitor of proteasome (Figure 6D). Furthermore, application of cycloheximide resulted in more rapid degradation of K433R mutant than WT (Supplemental Figure S4B).
FIGURE 6:
TRAF6 ubiquitinates PINK1 at Lys433. SH-SY5Y cells were transfected with designated constructs along with C-terminal HA-tagged PINK1 wild type or mutants with different lysine-to-arginine mutations for 48 h. (A) Ubiquitination of PINK1 was determined by immunoprecipitation with anti-HA agarose, followed by Western blot analysis. (B) The experiment was performed under conditions similar to those in A. For K433R, double amount of plasmid was used compared with that of PINK1-WT to attain the same protein levels of PINK1. (C, D) SH-SY5Y cells transfected with designated constructs were treated with 10 μM CCCP for 6 h (C) or 0.5 μM MG-132 for 12 h (D).
Complex formation of PINK1 with SARM1 and TRAF6 promotes its interaction with TOM20, an outer mitochondrial membrane protein
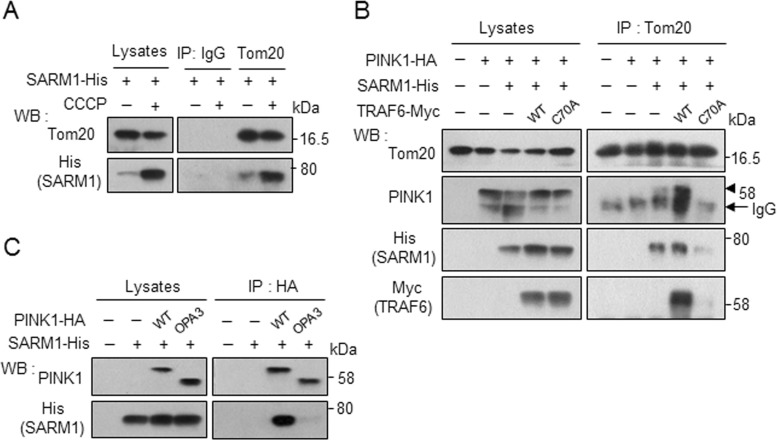
The results hitherto described indicate that ubiquitination at K433 of PINK1 through complex formation with SARM1 and TRAF6 somehow contributes to stabilization of PINK1. At the same time, it should be noted that the major fraction of PINK1 was apparently not ubiquitinated even under conditions in which PINK1 was stabilized and ubiquitinated by ternary complex formation (e.g., see Figure 4C). One possible interpretation is that ubiquitination is necessary at a specific step of stabilization of PINK1. Lazarou et al. (2012) showed that PINK1 forms a 700 kDa complex with the translocase of the outer membrane (TOM) selectively on depolarized mitochondria. We examined whether SARM1 interacts with TOM. Under mitochondrial depolarization conditions, binding of SARM1 with Tom20, a component of TOM, increased (Figure 7A). Of interest, the protein level of SARM1 increased like that of PINK1 on treatment with CCCP (Figure 5, A and B). The association of PINK1 with Tom20 was promoted by cooverexpression of SARM1 and TRAF6-WT but not ubiquitination-defective TRAF6-C70A mutant (Figure 7B). To determine the intracellular location at which SARM1 interacts with PINK1, we used PINK1Δ1-110 fused to the N-terminal mitochondrial anchor of OPA3 at the 1– to 30–amino acid sequence (OPA3-PINK1). OPA3-PINK1 is stably localized on the outer membrane (Narendra et al., 2010). As shown in Figure 7C, OPA3-PINK1 showed reduced binding capability with SARM1. These results suggest that PINK1 interacts with SARM1 in the intermembrane space under normal conditions and that ternary complex formation and ubiquitination contribute to import PINK1 in the outer membrane with Tom20 under depolarized conditions.
FIGURE 7:
SARM1 and TRAF6 promote the translocation of PINK1 to the outer membrane of mitochondria. (A) SH-SY5Y cells were transfected with SARM1-His for 48 h, followed by treatment with 10 μM CCCP for 2 h. The cell lysates were immunoprecipitated with anti-Tom20 antibody or control mouse IgG, followed by Western blot analysis. (B) SARM1 and TRAF6 promote the association of PINK1 with Tom20. SH-SY5Y cells were transfected with designated constructs for 48 h and analyzed under the conditions similar to those in A. Arrowhead indicates immunoprecipitated PINK1. (C) PINK1-WT but not OPA3-PINK1 interacts with SARM1. Cell lysates from SH-SY5Y cells transfected with SARM1-His along with PINK1-WT-HA or OPA3-PINK1-HA (PINK1 1-110 fused to the N-terminal mitochondrial anchor of OPA3 at amino acid positions 1–30) were immunoprecipitated with anti-HA agarose and analyzed by Western blotting.
SARM1 and TRAF6 are required for translocation of parkin to damaged mitochondria
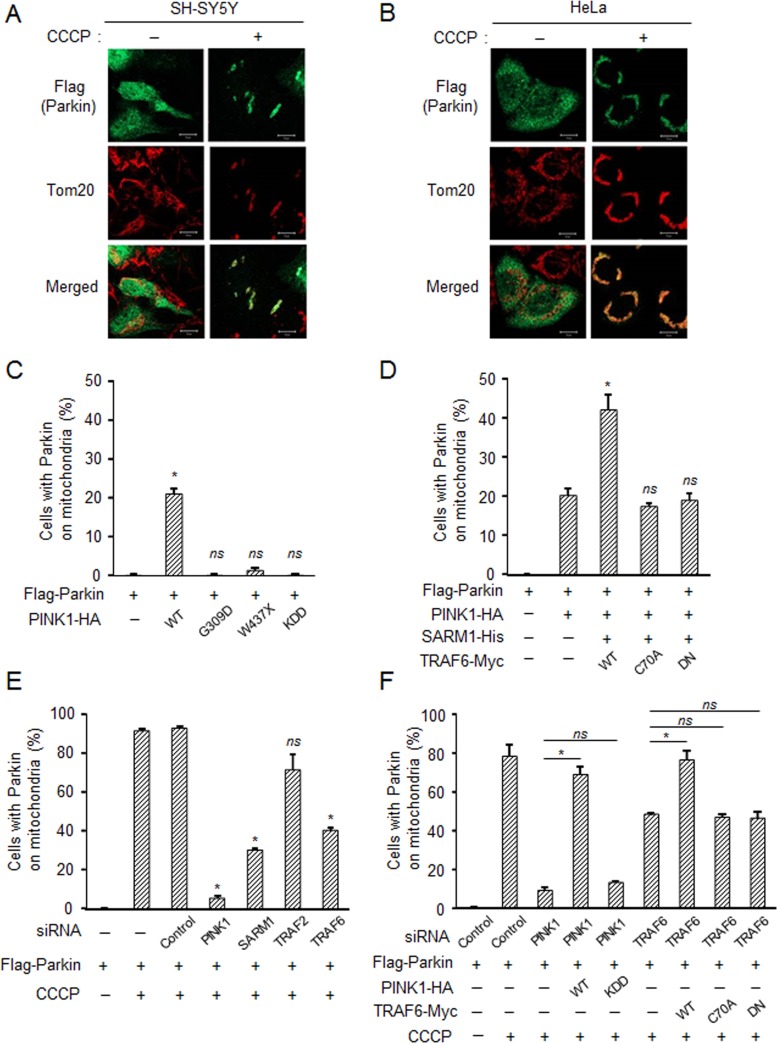
Parkin, another causative gene product for early-onset PD, is recruited to and promotes mitophagy of damaged mitochondria and thus protects cells (Geisler et al., 2010; Matsuda et al., 2010; Narendra et al., 2010). We investigated the possible role of SARM1 and TRAF6 in the mitochondrial recruitment of parkin. Flag-parkin transfected to SH-SY5Y and HeLa cells was diffusely localized throughout the cytoplasm under normal conditions, whereas upon treatment with CCCP, parkin was remarkably recruited to mitochondria (Figure 8, A and B). Because translocation of parkin in HeLa cells was clearer than that in SH-SY5Y cells, we tried to confirm the role of PINK1, SARM1, and TRAF6 using HeLa cells. Overexpression of PINK1-WT but not PINK1 mutants induced the translocation of parkin to mitochondria regardless of depolarization (Figure 8C), as reported previously (Narendra et al., 2010). Co-overexpression of SARM1 and TRAF6-WT but not TRAF6 mutants with PINK1 promoted this effect (Figure 8D). Small interfering RNAs (siRNAs) targeting PINK1, SARM1, TRAF2, or TRAF6 were transfected into HeLa cells along with Flag-parkin. In contrast to control siRNA, knockdown of PINK1 significantly abolished parkin recruitment to mitochondria after 6 h of CCCP treatment (Figure 8E). Knockdown of SARM1 and TRAF6, but not that of TRAF2, partially inhibited the translocation of parkin from the cytosol to mitochondria (Figure 8E). Overexpression of PINK1-WT or TRAF6-WT restored the translocation of parkin after knockdown of endogenous PINK1 or TRAF6 (Figure 8F). Transfection with PINK1-KDD or TRAF6 mutants, however, did not show this effect. These results indicate that complex formation of PINK1 with SARM1 and TRAF6 is important for the mitochondrial recruitment of parkin.
FIGURE 8:
SARM1 and TRAF6 are required for recruitment of parkin to damaged mitochondria. SH-SY5Y (A) and HeLa cells (B) were transfected with Flag-parkin for 48 h, followed by treatment with 10 μM CCCP for 6 h and then immunostained with the indicated antibodies. (C–F) HeLa cells were transfected with designated constructs for 48 h. The number of cells with parkin-positive mitochondria among the parkin-expressing cells. Three independent experiments were performed, and at least 100 cells were counted for each experiment. Error bars represent mean ± SD. *Significantly different from the control group (p < 0.01); ns, not significant. (C) Cells were transfected with PINK1 variants. (D) Effect of cotransfection with SARM1 and TRAF6. (E) Effect of knockdown of endogenous PINK1, SARM1, TRAF2, and TRAF6. (F) A rescue experiment for down-regulated endogenous proteins with overexpression of the respective proteins.
DISCUSSION
In this study, we demonstrated that PINK1 forms a complex with SARM1 and TRAF6, which is important for localization of PINK1 in the outer membrane and stabilization of PINK1 on depolarized mitochondria. TRAF6 ubiquitinates PINK1 at Lys-433 in a K63-linked manner via interaction with SARM1.
We identified SARM1 as an interacting protein of PINK1. SARM1 is one of the TIR adaptor proteins with an important role in the immune system and Toll-like receptor (TLR) signaling (O'Neill and Bowie, 2007). Signaling from TLRs involves five adaptor proteins: MyD88, TIRAP, TRIF, TRAM, and SARM1. We and other groups (Kim et al., 2007; Panneerselvam et al., 2012) showed that SARM1 is distinct from other adaptor proteins, in that SARM1 preferentially localizes in mitochondria. SARM1 is evolutionally most ancient, being the only member of the family to have a clear orthologue in C. elegans (Liberati et al., 2004). SARM1 is also conserved in Drosophila (Mink et al., 2001), zebrafish (Meijer et al., 2004), horseshoe crab (Belinda et al., 2008), and mouse (Kim et al., 2007). These homologues share a common domain constitution of ARMs, two SAM domains, and a TIR domain. The unique combination of three protein–protein interaction domains in SARM1 suggests that among the family of TLR adaptors, SARM1 probably functions differently from the other adaptor molecules (Dalod, 2007). In fact, SARM1 is preferentially expressed in neurons (Kim et al., 2007), in contrast to ubiquitous expression of the other proteins. In C. elegans, TIR-1, a homologue of SARM1, was reported to regulate neuronal asymmetry (Chuang and Bargmann, 2005; Chang et al., 2011). In mice, SARM1 was reported to mediate stress-induced neuronal toxicity (Kim et al., 2007) and axonal degeneration (Osterloh et al., 2012). It would be interesting to determine whether PINK1 is involved in these neuronal regulations with SARM1.
The ubiquitination of PINK1 by TRAF6 is a K63-linked type. Of the several types of ubiquitin chain topologies, K63-linked ubiquitination is the only type known to fulfill diverse proteasome-independent roles, including DNA repair (Hofmann and Pickart, 1999), endocytosis (Mukhopadhyay and Riezman, 2007), and NFκB signaling (Wang et al., 2001). Lee et al. (2012) reported that PINK1 stimulates interleukin-1β–mediated NFκB signaling via TRAF6. In this study, we found that K63-linked ubiquitination contributed to stabilization of PINK1 and activation of mitophagy. On the other hand, it is believed that PINK1 is also modified with K48-linked ubiquitination for proteasome degradation because PINK1 is degraded under steady-state conditions and is accumulated by treatment with a proteasome inhibitor (Figure 6D; Muqit et al., 2006). It would be intriguing to determine the E3 ligase and ubiquitination site(s) of PINK1 for K48-linked ubiquitination.
Lazarou et al. (2012) showed that PINK1 forms a 700-kDa complex with TOM selectively on depolarized mitochondria. We found that SARM1 and TRAF6 promoted the association of PINK1 with Tom20 and that the protein level of SARM1 increased upon mitochondrial depolarized conditions the same as PINK1. These results suggest that PINK1 and SARM1 are inserted in the outer membrane under depolarized conditions and then TRAF6 is recruited in the complex and ubiquitinates PINK1. We showed that the PINK1-ubiquitination defective mutant K433R is degraded earlier than PINK1-WT (Figure 6D and Supplemental Figure S4B). The inner membrane rhomboid protease PARL cleaves PINK1, depending on mitochondrial membrane potential for proteolytic destabilization (Jin et al., 2010; Deas et al., 2011; Meissner et al., 2011; Greene et al., 2012). These results suggest that ubiquitination at the cytosolic face of the outer membrane prevents PINK1 from translocation into the intermembrane space, thereby preventing access to PARL. Given that the ubiquitination levels of PD-linked PINK1 mutants are reduced compared with that of PINK1-WT (Figure 4D), impairment of PINK1 ubiquitination may be involved in the pathogenesis of PD. The kinase-dead mutant of PINK1 also shows reduced levels of ubiquitination. These results suggest that the kinase activity of PINK1 is required for the ubiquitination of PINK1. We and other groups showed that PINK1 has autophosphorylation activity (Nakajima et al., 2003; Silvestri et al., 2005; Kondapalli et al., 2012; Okatsu et al., 2012). It is possible that PINK1 autophosphorylates upon depolarization of mitochondria and that SARM1 and TRAF6 bind to phosphorylated PINK1, resulting in ubiquitination and stabilization of PINK1.
Parkin-mediated ubiquitination is also important for the process of mitophagy. VDAC1 and MFN1/2 are substrates of ubiquitination by parkin (Gegg et al., 2010; Geisler et al., 2010; Poole et al., 2010; Tanaka et al., 2010; Ziviani et al., 2010; Glauser et al., 2011; Rakovic et al., 2011). This ubiquitination leads to the sequestration and elimination of damaged mitochondria. In this study, we found that TRAF6 but not parkin ubiquitinates PINK1 through interaction with SARM1. Thus ubiquitination is linked to multiple steps of mitophagy and mitochondrial quality control. Our finding of a mitochondrial SARM1-TRAF6-PINK1 pathway for mitophagy provides new insights into the pathogenic mechanisms of PD and suggests novel targets for therapeutic intervention.
MATERIALS AND METHODS
Cells, chemicals, and antibodies
SH-SY5Y (ECACC; Salisbury, Wiltshire, United Kingdom), HeLa, and HEK293 cells were cultured in D/F medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum. CCCP, valinomycin, rotenone, EtBr, H2O2, MG-132, and cycloheximide were purchased from Sigma-Aldrich (St. Louis, MO). Biotinylated K63-TUBE1 was purchased from LifeSensors (Malvern, PA; UM304). Streptavidin–horseradish peroxidase (HRP) was purchased from R&D Systems (Minneapolis, MN).
The antibodies used were as follows: rabbit polyclonal antibody against PINK1 (BC100-494; Novus Biologicals, Littleton, CO); rabbit polyclonal antibody against PINK1 raised in our previous study (Murata et al., 2011a); antibodies against TRAF6 (rabbit monoclonal, 8028), LC-3B (rabbit polyclonal, 2775), Flag (rabbit polyclonal, 2368), Myc (mouse monoclonal, 2276), and HRP-labeled anti-mouse and anti-rabbit secondary antibodies (Cell Signaling Technologies, Danvers, MA); antibodies against TRAF2 (rabbit polyclonal, 592), Flag (rabbit polyclonal, PM020 for immunocytochemistry), and histidine (rabbit polyclonal, PM032) (MBL, Woburn, MA); rabbit polyclonal antibody against SARM1 (NB100-94383; Novus Biologicals); antibodies against ubiquitin (mouse monoclonal, sc-8017), Tom20 (mouse monoclonal, sc-17764), Tom20 (rabbit polyclonal, sc-11415), and Bcl-2 (mouse monoclonal, sc-7382; Santa Cruz Biotechnology, Santa Cruz, CA); antibodies against ubiquitin (mouse monoclonal, 550944) and cytochrome c (mouse monoclonal, 556433; BD PharMingen, San Jose, CA); antibodies against tubulin (mouse monoclonal, T5168) and HA-agarose (mouse monoclonal, A2095 for immunoprecipitation; Sigma-Aldrich); and HRP-labeled TrueBlot anti-mouse IgG (18-8817-33; eBioscience, San Diego, CA).
Plasmid constructs
Conventional molecular biological techniques were used to generate the following expression constructs: C-terminal HA- or hexahistidine (6His)-tagged human wild-type PINK1, pathogenic PINK1 G309D and W437X mutants, and PINK1 catalytically inactive KDD triple mutant (K219A/D362A/D384A; Beilina et al., 2005); C-terminal Flag- or 6His-tagged SARM1 and SARM1-deletion mutants; C-terminal Myc-tagged TRAF2, N-terminal–deleted TRAF2 mutant (DN-TRAF2), TRAF6, N-terminal–deleted TRAF6 mutant (DN-TRAF6), and TRAF6 catalytically inactive mutant (C70A); N-terminal 6His- or Flag-tagged wild-type ubiquitin (Ub), Ub-K48R, and Ub-K63R; and N-terminal Flag-tagged parkin. All expression constructs were sequenced to ensure that the fusion was in the correct reading frame and there were no additional mutations.
Cell transfection
For plasmid transfection, cells were transfected with the indicated plasmids using FuGENE-HD (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions.
For RNA interference, siRNA targeting PINK1 (2742364, NM_032409; Qiagen, Valencia, CA), TRAF6 (2698952, NM_004620; Qiagen), SARM1 (M-008076-01-0005, NM_015077; Thermo Scientific Dharmacon, Lafayette, CO) or TRAF2 (M-005198-00-0005, NM_021138; Thermo Scientific Dharmacon) was transfected into cells using Lipofectamine RNAiMAX (Invitrogen). The siRNAs for PINK1 and TRAF6 were targeted to 3′-untranslated region to enable a rescue experiment. A control siRNA with no known mammalian homology (siGENONE nontargeting siRNA pool #1, Thermo Scientific Dharmacon) was used as a negative control.
Western blot analysis and immunoprecipitation
Western blot analysis was performed under conventional conditions after lysing cells with M-PER Mammalian Protein Extraction Reagent (Thermo Scientific, Waltham, MA) with PhosphoSTOP (Roche Applied Science). For immunoprecipitation, cells were lysed in ice-cold lysis buffer (40 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.5, 120 mM NaCl, 1 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, 50 mM NaF, 1 mM orthovanadate, and 0.3% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate [CHAPS]). One-tenth of cell lysates was used for direct Western blot analysis, and the remaining fractions were used for immunoprecipitation. They were incubated with 4 μg of respective antibodies for 2 h and precipitated after another incubation with 30 μl of 50% slurry of Dynabeads protein G (Invitrogen) or monoclonal anti–HA-agarose (Sigma-Aldrich). Immunoprecipitates were washed three times with the lysis buffer before elution. The band intensity was quantified by ImageJ (National Institutes of Health, Bethesda, MD).
Mass spectrometry
HEK293 (1 × 107 cells) were lysed in ice-cold lysis buffer (40 mM HEPES, pH 7.5, 120 mM NaCl, 1 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, 50 mM NaF, 1 mM orthovanadate, and 0.3% CHAPS). PINK1–protein complexes were harvested by immunoprecipitation with 4 μg of anti-PINK1 antibody and 30 μl of 50% slurry of Dynabeads protein G. The complexes containing PINK1 protein were pooled and resolved on SDS–PAGE, followed by silver staining. Protein bands were excised from the SDS–PAGE gel, digested by trypsin, and analyzed by 1100LC/MSD TRAP XCT Ultra (Agilent Technologies, Santa Clara, CA).
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde and permeabilized with 100% ethanol for 1 h at −20°C or with 0.01% digitonin for 10 min at room temperature. After incubation with respective first antibodies, the samples were incubated with Alexa Fluor 488 and 594 goat anti-mouse and/or rabbit IgG antibody (Invitrogen). Mitochondria were stained with MitoTracker Orange CMTMRos (Invitrogen). The specimens were observed using a confocal laser-scanning microscope (model LSM 510; Carl Zeiss, Jena, Germany).
In vitro assays for protein binding and ubiquitination
For in vitro assay, glutathione S-transferase (GST)–PINK1 (112–581 amino acids) protein was purified from the bacterial lysates of BL21-competent cells transformed with pGEX6P1-PINK1 using glutathione Sepharose beads (GE Healthcare, Piscataway, NJ) according to the manufacturer's standard procedures. SARM1-Flag, TRAF2-Myc, and TRAF6-Myc proteins were purified from the cell lysates of HEK293 cells transfected with plasmids using anti-Flag antibody or anti-Myc antibody with Dynabeads protein G. An in vitro ubiquitination assay was performed by incubation of purified GST-PINK1 and SARM1-Flag and TRAF2-Myc or TRAF6-Myc at 37°C for 2 h in 60 μl of reaction buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 2 mM dithiothreitol, and 2 mM ATP) containing 50 μM ubiquitin (Boston Biochem, Cambridge, MA), 50 nM E1 (Boston Biochem), and 500 nM E2 ubiquitin-conjugating enzyme (UbcH13/Uev1a; Boston Biochem). Ubiquitination of GST-PINK1 was detected by affinity purification with glutathione Sepharose beads, followed by Western blot analysis using anti-ubiquitin antibody.
Statistical analysis
Before statistical analysis, each experiment was repeated there times. The results are expressed as means ± SD For comparison, analysis of variance (ANOVA) was used. If the ANOVA showed a significant difference, the Bonferroni procedure was used as a post hoc test. p values of <0.01 were considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported in part by grants to H.M. from the Japan Science and Technology Agency (AS232Z00635F), the Ministry of Education, Culture, Sports, Science, and Technology of Japan (25860216), and the Okayama Medical Foundation.
Abbreviations used:
- ARM
HEAT/Armadillo
- CCCP
carbonyl cyanide m-chlorophenyl hydrazine
- HA
hemagglutinin
- MFN1/2
mitofusin 1 and 2
- PD
Parkinson's disease
- PINK1
PTEN-induced putative kinase 1
- SAM
sterile α motif
- SARM1
sterile α and TIR motif containing 1
- TIR
Toll/interleukin 1 receptor
- TRAF6
tumor necrosis factor receptor–associated factor 6
- VDAC1
voltage-dependent anion channel 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-01-0016) on July 24, 2013.
1The proteins identified by liquid chromatography/mass spectrometry analysis in fraction 2 are vimentin, SARM1, heat-shock 70-kDa protein 1A/1B, and RBI-inducible coiled-coil protein 1.
REFERENCES
- Abeliovich A, Flint Beal M. Parkinsonism genes: culprits and clues. J Neurochem. 2006;99:1062–1072. doi: 10.1111/j.1471-4159.2006.04102.x. [DOI] [PubMed] [Google Scholar]
- Beilina A, Van Der Brug M, Ahmad R, Kesavapany S, Miller DW, Petsko GA, Cookson MR. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc Natl Acad Sci USA. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinda LW, Wei WX, Hanh BT, Lei LX, Bow H, Ling DJ. SARM: a novel Toll-like receptor adaptor, is functionally conserved from arthropod to human. Mol Immunol. 2008;45:1732–1742. doi: 10.1016/j.molimm.2007.09.030. [DOI] [PubMed] [Google Scholar]
- Bonifati V, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- Bonifati V, et al. Early-onset parkinsonism associated with PINK1 mutations: frequency, genotypes, and phenotypes. Neurology. 2005;65:87–95. doi: 10.1212/01.wnl.0000167546.39375.82. [DOI] [PubMed] [Google Scholar]
- Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Hsieh YW, Lesch BJ, Bargmann CI, Chuang CF. Microtubule-based localization of a synaptic calcium-signaling complex is required for left-right neuronal asymmetry in C. elegans. Development. 2011;138:3509–3518. doi: 10.1242/dev.069740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Lin CW, Chang CY, Jiang ST, Hsueh YP. Sarm1, a negative regulator of innate immunity, interacts with syndecan-2 and regulates neuronal morphology. J Cell Biol. 2011;193:769–784. doi: 10.1083/jcb.201008050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Bargmann CI. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 2005;19:270–281. doi: 10.1101/gad.1276505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Criscuolo C, et al. PINK1 homozygous W437X mutation in a patient with apparent dominant transmission of parkinsonism. Mov Disord. 2006;21:1265–1267. doi: 10.1002/mds.20933. [DOI] [PubMed] [Google Scholar]
- Dalod M. Studies of SARM1 uncover similarities between immune and neuronal responses to danger. Sci STKE. 2007;2007:pe73. doi: 10.1126/stke.4172007pe73. [DOI] [PubMed] [Google Scholar]
- Deas E, et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Jankovic J, Guo Y, Xie W, Le W. Small interfering RNA targeting the PINK1 induces apoptosis in dopaminergic cells SH-SY5Y. Biochem Biophys Res Commun. 2005;337:1133–1138. doi: 10.1016/j.bbrc.2005.09.178. [DOI] [PubMed] [Google Scholar]
- Gandhi S, et al. PINK1 protein in normal human brain and Parkinson's disease. Brain. 2006;129:1720–1731. doi: 10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Glauser L, Sonnay S, Stafa K, Moore DJ. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J Neurochem. 2011;118:636–645. doi: 10.1111/j.1471-4159.2011.07318.x. [DOI] [PubMed] [Google Scholar]
- Greene AW, Grenier K, Aguileta MA, Muise S, Farazifard R, Haque ME, McBride HM, Park DS, Fon EA. Mitochondrial processing peptidase regulates PINK1 processing, import and parkin recruitment. EMBO Rep. 2012;13:378–385. doi: 10.1038/embor.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque ME, Thomas KJ, D'Souza C, Callaghan S, Kitada T, Slack RS, Fraser P, Cookson MR, Tandon A, Park DS. Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc Natl Acad Sci USA. 2008;105:1716–1721. doi: 10.1073/pnas.0705363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Cai H, Cookson MR, Gwinn-Hardy K, Singleton A. Genetics of Parkinson's disease and parkinsonism. Ann Neurol. 2006;60:389–398. doi: 10.1002/ana.21022. [DOI] [PubMed] [Google Scholar]
- Hattori N, Mizuno Y. Pathogenetic mechanisms of parkin in Parkinson's disease. Lancet. 2004;364:722–724. doi: 10.1016/S0140-6736(04)16901-8. [DOI] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler KR, Hamscho N, Morales B, Menzel C, Barrero F, Vives F, Gispert S, Auburger G. Dopaminergic function in a family with the PARK6 form of autosomal recessive Parkinson's syndrome. J Neural Transm. 2005;112:1345–1353. doi: 10.1007/s00702-005-0281-9. [DOI] [PubMed] [Google Scholar]
- Kim Y, Zhou P, Qian L, Chuang JZ, Lee J, Li C, Iadecola C, Nathan C, Ding A. MyD88–5 links mitochondria, microtubules, and JNK3 in neurons and regulates neuronal survival. J Exp Med. 2007;204:2063–2074. doi: 10.1084/jem.20070868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Kondapalli C, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates parkin E3 ligase activity by phosphorylating serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J Biol Chem. 2007;282:4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase parkin. Dev Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Jang SH, Kim H, Yoon JH, Chung KC. PINK1 stimulates interleukin-1beta-mediated inflammatory signaling via the positive regulation of TRAF6 and TAK1. Cell Mol Life Sci. 2012;69:3301–3315. doi: 10.1007/s00018-012-1004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, Ausubel FM. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci USA. 2004;101:6593–6598. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansell A, Brint E, Gould JA, O'Neill LA, Hertzog PJ. Mal interacts with tumor necrosis factor receptor-associated factor (TRAF)-6 to mediate NF-kappaB activation by toll-like receptor (TLR)-2 and TLR4. J Biol Chem. 2004;279:37227–37230. doi: 10.1074/jbc.C400289200. [DOI] [PubMed] [Google Scholar]
- Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits parkin to damaged mitochondria and activates latent parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AH, Gabby Krens SF, Medina Rodriguez IA, He S, Bitter W, Ewa Snaar-Jagalska B, Spaink HP. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol Immunol. 2004;40:773–783. doi: 10.1016/j.molimm.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Meissner C, Lorenz H, Weihofen A, Selkoe DJ, Lemberg MK. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem. 2011;117:856–867. doi: 10.1111/j.1471-4159.2011.07253.x. [DOI] [PubMed] [Google Scholar]
- Mink M, Fogelgren B, Olszewski K, Maroy P, Csiszar K. A novel human gene (SARM) at chromosome 17q11 encodes a protein with a SAM motif and structural similarity to Armadillo/beta-catenin that is conserved in mouse, Drosophila, and Caenorhabditis elegans. Genomics. 2001;74:234–244. doi: 10.1006/geno.2001.6548. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Muqit MM, et al. Altered cleavage and localization of PINK1 to aggresomes in the presence of proteasomal stress. J Neurochem. 2006;98:156–169. doi: 10.1111/j.1471-4159.2006.03845.x. [DOI] [PubMed] [Google Scholar]
- Murata H, Sakaguchi M, Jin Y, Sakaguchi Y, Futami J, Yamada H, Kataoka K, Huh NH. A new cytosolic pathway from a Parkinson disease-associated kinase, BRPK/PINK1: activation of AKT via mTORC2. J Biol Chem. 2011a;286:7182–7189. doi: 10.1074/jbc.M110.179390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata H, Sakaguchi M, Kataoka K, Huh NH. Multiple functions of PINK1 at different intracellular locations: beyond neurodegenerative diseases. Cell Cycle. 2011b;10:1518–1519. doi: 10.4161/cc.10.10.15445. [DOI] [PubMed] [Google Scholar]
- Nakajima A, Kataoka K, Hong M, Sakaguchi M, Huh NH. BRPK, a novel protein kinase showing increased expression in mouse cancer cell lines with higher metastatic potential. Cancer Lett. 2003;201:195–201. doi: 10.1016/s0304-3835(03)00443-9. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- Okatsu K, et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for parkin recruitment to damaged mitochondria. Nat Commun. 2012;3:1016. doi: 10.1038/ncomms2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterloh JM, et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam P, Singh LP, Ho B, Chen J, Ding JL. Targeting of pro-apoptotic TLR adaptor SARM to mitochondria: definition of the critical region and residues in the signal sequence. Biochem J. 2012;442:263–271. doi: 10.1042/BJ20111653. [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovic A, Grunewald A, Kottwitz J, Bruggemann N, Pramstaller PP, Lohmann K, Klein C. Mutations in PINK1 and parkin impair ubiquitination of mitofusins in human fibroblasts. PLoS One. 2011;6:e16746. doi: 10.1371/journal.pone.0016746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri L, Caputo V, Bellacchio E, Atorino L, Dallapiccola B, Valente EM, Casari G. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet. 2005;14:3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- Strauss KM, et al. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson's disease. Hum Mol Genet. 2005;14:2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, et al. PINK1-dependent recruitment of parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Wu H, Arron JR. TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. Bioessays. 2003;25:1096–1105. doi: 10.1002/bies.10352. [DOI] [PubMed] [Google Scholar]
- Ye H, Park YC, Kreishman M, Kieff E, Wu H. The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol Cell. 1999;4:321–330. doi: 10.1016/s1097-2765(00)80334-2. [DOI] [PubMed] [Google Scholar]
- Yoshii SR, Kishi C, Ishihara N, Mizushima N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem. 2011;286:19630–19640. doi: 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.