GRK2 is localized to centrosomes and regulates EGF receptor–promoted separation of duplicated centrosomes. This pathway depends on Mst2 and Nek2A and involves GRK2-mediated phosphorylation and activation of Mst2. Thus GRK2 plays a central role in mitogen-promoted centrosome separation via its ability to phosphorylate Mst2.
Abstract
G protein–coupled receptor kinases (GRKs) play a central role in regulating receptor signaling, but recent studies suggest a broader role in modulating normal cellular functions. For example, GRK5 has been shown to localize to centrosomes and regulate microtubule nucleation and cell cycle progression. Here we demonstrate that GRK2 is also localized to centrosomes, although it has no role in centrosome duplication or microtubule nucleation. Of interest, knockdown of GRK2 inhibits epidermal growth factor receptor (EGFR)–mediated separation of duplicated centrosomes. This EGFR/GRK2-mediated process depends on the protein kinases mammalian STE20-like kinase 2 (Mst2) and Nek2A but does not involve polo-like kinase 1. In vitro analysis and dominant-negative approaches reveal that GRK2 directly phosphorylates and activates Mst2. Collectively these findings demonstrate that GRK2 is localized to centrosomes and plays a central role in mitogen-promoted centrosome separation most likely via its ability to phosphorylate Mst2.
INTRODUCTION
G protein–coupled receptor kinases (GRKs) are a family of seven protein kinases that phosphorylate agonist-occupied G protein–coupled receptors (GPCRs), thereby linking agonist binding with regulatory processes such as desensitization and internalization (Moore et al., 2007). Of interest, many studies revealed that GRKs have additional functions beyond regulating GPCRs (Gurevich et al., 2012). For example, GRKs phosphorylate and regulate the function of many additional proteins, including tubulin (Carman et al., 1998; Pitcher et al., 1998), NHERF (Hall et al., 1999), synucleins (Pronin et al., 2000), phosducin (Ruiz-Gomez et al., 2000), platelet-derived growth factor and epidermal growth factor receptors (Freedman et al., 2002), ezrin (Cant and Pitcher, 2005), p38 (Peregrin et al., 2006), NFκB1 p105 (Parameswaran et al., 2006), HDAC5 (Martini et al., 2008), IκBα (Patial et al., 2009), β-arrestin-1 (Barthet et al., 2009), p53 (Chen et al., 2010), Hip (Barker and Benovic, 2012), nucleophosmin (So et al., 2012), and many others. GRKs have also been linked with a number of diseases, including various neurological disorders (Suo et al., 2007; Bychkov et al., 2008, 2011; Garcia-Sevilla et al., 2010), cardiovascular disease (Ungerer et al., 1993; Gros et al., 1997; Huang et al., 2012), and cancer (Matsubayashi et al., 2008; Tiedemann et al., 2010; Kim et al., 2012; Woerner et al., 2012).
Whereas GRK-mediated phosphorylation by GPCRs occurs at the plasma membrane, GRKs are also localized in other areas of the cell, such as the nucleus (Yi et al., 2002; Johnson et al., 2004), mitochondria (Fusco et al., 2012), and centrosomes (Michal et al., 2012). In the nucleus, GRK5 mediates histone deacetylation to regulate the activity of the transcription factor MEF2 (Martini et al., 2008), whereas in the mitochondria, GRK2 controls ATP accumulation and tolerance to ischemia in skeletal muscle (Fusco et al., 2012). GRK5 localization in the centrosome regulates the G2/M-phase transition of the cell cycle by phosphorylating p53 (Michal et al., 2012).
A number of protein kinases localized in the centrosome are critical for the G2 and M phases of the cell cycle by properly orienting centrosomes to regulate the mitotic spindle (Bettencourt-Dias and Glover, 2007). Centrosome-localized kinases also regulate activities of the centrosomes themselves. For example, Nek2A controls a process at G2 known as centrosomal disjunction or separation. In this process, a protein linker, made up of rootletin and C-Nap1, connects the proximal end of the two mother centrioles of the duplicated centrosomes. During late G2 phase, polo-like kinase 1 (Plk1) phosphorylates the proapoptotic protein kinase mammalian STE20-like kinase 2 (Mst2), which then phosphorylates and activates Nek2A, resulting in inhibition of PP1γ interaction with the Mst2-Nek2A complex (Mardin et al., 2011). Activated Nek2A then phosphorylates C-Nap1, initiating a mechanism that untethers duplicated centrosomes (Mardin et al., 2010). After disjunction, centrosomes separate and move to opposite poles through the activity of the microtubule-dependent motor protein Eg5 and form the mitotic spindle (Smith et al., 2011). Errors in this process can lead to monopolar and multipolar mitotic spindles. For example, cells with incomplete spindle pole separation exhibit higher rates of kinetochore misattachment and chromosome missegregation than cells with properly timed and complete centrosome separation (Silkworth et al., 2012). These defects can cause genomic instability (Ganem et al., 2009) and the development of cancer (Basto et al., 2008).
In this article, we analyze the cellular localization and function of GRK2. GRK2 is localized to centrosomes and plays a critical role in mediating epidermal growth factor receptor (EGFR)–promoted separation of duplicated centrosomes. This separation also uses the kinases Nek2A and Mst2 but is independent of Plk1 function. We propose a model in which activated EGFR leads to GRK2 activation, which phosphorylates Mst2, resulting in activation of Nek2A, phosphorylation of C-Nap1, and centrosome separation.
RESULTS
Because GRK5 was previously shown to be localized in centrosomes and play a role in cell cycle regulation (Michal et al., 2012), we were interested in determining whether GRK2 is also localized in centrosomes. Initial analysis of GRK2 expression in HeLa, HEK293, and MDA-MB-231 cells demonstrated that all three lines express GRK2, with HEK293 cells having the highest level and HeLa cells the lowest (Figure 1A). To assess whether any endogenous GRK2 is localized in centrosomes, we first preextracted cells with detergent for 30 s and then fixed, permeabilized, incubated them with specific antibodies and analyzed them by immunofluorescence microscopy. These studies revealed that GRK2 colocalizes with the centrosomal marker γ-tubulin in ∼80–90% of cells (∼90% for HEK293 and MDA-MB-231; ∼80% for HeLa), suggesting that some GRK2 is localized in centrosomes (Figure 1B). To confirm these observations, we fractionated HEK293 cell lysates on a sucrose density gradient to isolate centrosomes and then probed them for GRK2 and α-tubulin by immunoblotting. GRK2 was found in the same fractions as α-tubulin, supporting its association with centrosomes (Figure 1C). To verify that the GRK2 antibody was specifically detecting endogenous GRK2, we stably knocked down GRK2 expression in HeLa cells by short hairpin RNA (shRNA) and then analyzed them by immunofluorescence. The ∼75% decrease in GRK2 expression detected by immunoblotting in GRK2 shRNA cells resulted in a nearly complete loss of GRK2 staining in centrosomes (Figure 1D). Further verification of GRK2 localization in centrosomes was achieved using a polyclonal GRK2-selective antibody (Figure 1E). Overall these data demonstrate that a portion of the endogenous GRK2 is localized in centrosomes.
FIGURE 1:
Localization of endogenous GRK2 to centrosomes. (A) Representative immunoblot demonstrating the expression of GRK2 in HeLa, HEK293, and MDA-MB-231 cells using a GRK2-specific monoclonal antibody (3A10). ERK2 was used as a loading control. (B) HEK293, HeLa, and MDA-MB-231 cells were fixed and stained with anti-GRK2/3 monoclonal and γ-tubulin polyclonal antibodies. Representative images of cells in interphase are shown from n > 80 cells. Arrows denote inset regions and colocalization. Scale bar, 5 μm. (C) Whole-cell (WC) HEK293 lysates and various fractions of the centrosomal preparation (30–70% sucrose) were electrophoresed by SDS–PAGE and immunoblotted for α-tubulin and GRK2 using the 3A10 monoclonal. (D) Top, control shRNA and GRK2 shRNA stably transfected HeLa cells were harvested and analyzed for GRK2 expression using an anti-GRK2/3 monoclonal antibody. Bottom, analysis of GRK2 localization in GRK2 shRNA cells. Cells were fixed and stained with anti-GRK2/3 monoclonal and γ-tubulin polyclonal antibodies. Note the loss of GRK2 staining in these cells. Representative images. Arrow denotes inset regions and colocalization. Scale bar, 5 μm. (E) Immunofluorescence performed using a polyclonal GRK2-specific antibody in control and GRK2 shRNA HeLa cells.
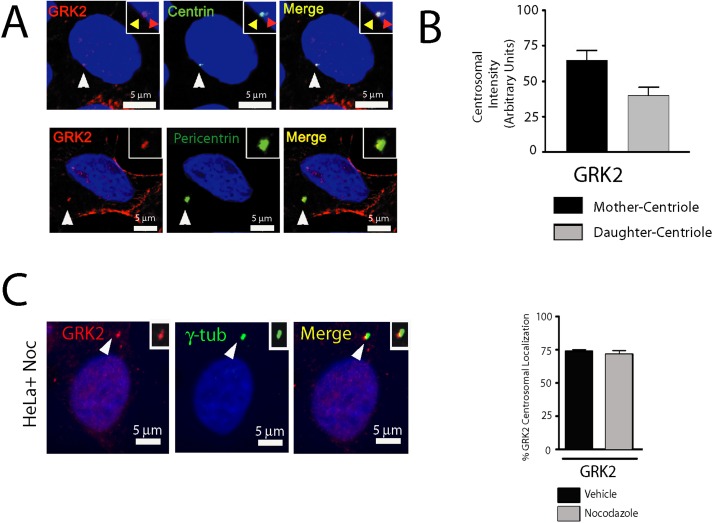
We next evaluated the characteristics of centrosomal GRK2. First, we studied more detailed centrosomal localization (centriolar vs. pericentriolar). GRK2 was localized to both centrioles, detected by green fluorescent protein (GFP)–centrin (Figure 2A, top), as well as to pericentriolar material, indicated by pericentrin staining (Figure 2A, bottom). GRK2 was also predominantly localized to mother centrioles compared with daughter centrioles, as determined by the differential extent of GFP-centrin staining (Figure 2B). To assess whether microtubules had a role in GRK2 localization to centrosomes, we treated cells with nocodazole to disrupt microtubules. Nocodazole treatment did not affect GRK2 centrosomal localization (Figure 2, C and D), suggesting that a microtubule-independent process is involved in this localization.
FIGURE 2:
GRK2 is a bona fide centrosomal component. (A) Top, GFP-centrin–expressing HeLa cells were fixed and stained using the polyclonal GRK2 antibody. Note that GRK2 is colocalized with centrin. Representative images of cells in interphase. Inset, yellow arrow denotes mother centriole; orange arrow denotes daughter centriole. Bottom, HeLa cells were fixed and stained with the monoclonal GRK2 (3A10) and polyclonal pericentrin antibodies. Representative images of cells in interphase. (B) Quantitation of GRK2 localization with the mother (centriole with greater GFP-centrin expression) vs. the daughter centriole (n > 30 cells from two independent experiments). (C) HeLa cells pretreated with nocodazole to depolymerize microtubules were fixed and stained for GRK2 and γ-tubulin (n > 85 cells from two independent experiments). Note that colocalization remains even after microtubule depolymerization. Quantitation of the number of cells displaying centrosomal localization after nocodazole treatment. For all images, arrows denote inset regions. Scale bar, 5 μm.
Next we determined whether GRK2 has any role in the regulation of centrosomal functions such as microtubule nucleation, centrosome duplication at S phase, or separation of duplicated centrosomes during the cell cycle. Stable knockdown of GRK2 expression in HeLa cells showed no discernible effect on microtubule nucleation, as demonstrated by no differences in aster growth between GRK2-knockdown and control cells (Supplemental Figure S1, A and B). These findings are in contrast to GRK5, for which knockdown revealed an important role for GRK5 in microtubule nucleation (Michal et al., 2012). Additional studies revealed no effect on centrosome duplication in either GRK2 shRNA or GRK5 shRNA cells compared with control cells (Supplemental Figure S1, C and D). Furthermore, no differences were observed in centrosome separation in asynchronized control, GRK2, or GRK5 shRNA cells (Supplemental Figure S1E). Centrosome separation was also determined after synchronization in G1/S by double-thymidine block (t = 0 h), followed by progression into G2/M (t = 6–8 h). As cells progress through the cell cycle, the percentage of cells showing separated centrosomes or cells in mitosis (separated centrosomes with condensed DNA) were unchanged in GRK2 shRNA cells compared with control cells (Supplemental Figure S1F). There does, however, appear to be a trend toward an increase in the number of cells in mitosis during the time course in the GRK2 shRNA HeLa cells compared with control cells. This may reflect increased cell cycle progression due to decreased GRK2 levels, as previously reported (Penela et al., 2010).
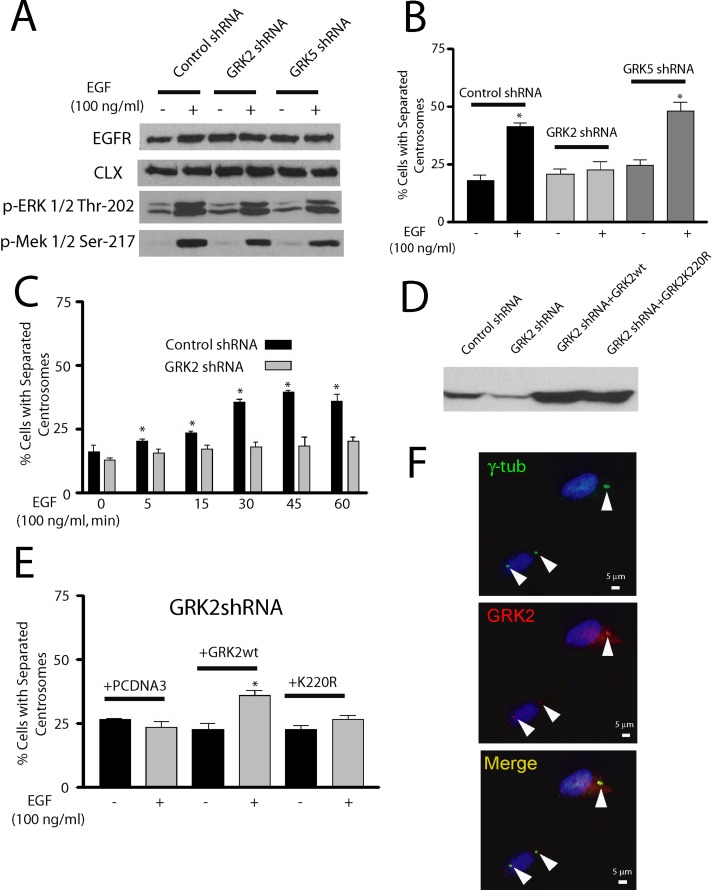
Among the many activities mediated by epidermal growth factor receptors (EGFRs) is the ability to promote rapid separation of duplicated centrosomes (Sherline and Mascardo, 1982a). Although this function may play a role in coordinately regulating mitogen-promoted centrosome separation and DNA synthesis (Sherline and Mascardo, 1984), the mechanism of this process is poorly understood. Because EGFR activation promotes the phosphorylation and activation of GRK2 (Chen et al., 2008) and GRK2 is localized to centrosomes (Figures 1 and 2), we hypothesized that GRK2 might contribute to the EGFR pathway that mediates centrosome separation. To test this possibility, we synchronized control, GRK2, and GRK5 shRNA HeLa cells in G1/S by treating the cells with hydroxyurea for 48 h and then incubating the cells with or without EGF. Activation of the EGFR was comparable in all three lines as assessed by EGF-promoted phosphorylation of ERK1/2 and MEK1 after a 5-min treatment with EGF (Figure 3A). This result is in agreement with previous studies showing that GRK2 expression does not change EGFR activity (Freedman et al., 2002). Centrosome separation was determined by counting cells that displayed duplicated centrosomes greater than one centrosome diameter apart, and this was then expressed as a percentage of the total cells. Control and GRK5 shRNA cells demonstrated similar approximately twofold increase in the number of cells with centrosomal separation after 30-min EGF treatment, whereas EGF had no effect in the GRK2 shRNA cells (Figure 3B). To assess whether the EGF-promoted centrosome separation was due to rapid cell cycle progression, we collected cells for cell cycle analysis after a 30-min EGF treatment. No significant difference in cells in G1, S, or G2 phase was observed with or without EGF treatment (Supplemental Figure S2). Centrosome separation was rapid and could be observed in 40–50% of the cells after 45–60 min of EGF treatment (Figure 3C), values comparable to those in previous reports (Sherline and Mascardo, 1982b). To determine whether the catalytic activity of GRK2 was required for EGF-mediated centrosome separation, we expressed wild-type bovine GRK2 or catalytically inactive bovine GRK2-K220R in GRK2 shRNA HeLa cells (Figure 3D). EGF-promoted centrosomal separation was effectively rescued by overexpression of wild-type GRK2 but not by GRK2-K220R, demonstrating that the catalytic activity of GRK2 is important in this process (Figure 3E). We also evaluated whether GRK2 remains localized to centrosomes after EGF-promoted separation and found that GRK2 levels are not significantly different in separated centrosomes compared with unseparated centrosomes (Figure 3F). Thus it does not appear that EGFR activation significantly changes the localization of GRK2 in centrosomes.
FIGURE 3:
Knockdown of GRK2 expression leads to inhibition of EGF-promoted centrosome separation in HeLa cells. (A) Knockdown of GRK2 expression and activation of EGFR, as indicated by ERK1/2-T202 and MEK1/2-S217 phosphorylation, after 5-min treatment with 100 ng/ml EGF (n = 3). Calnexin (CLX) was used as a loading control. (B) Percentage of cells with separated duplicated centrosomes in S phase after vehicle (serum-free media) or 30-min treatment with 100 ng/ml EGF in control, GRK2, and GRK5 shRNA cells. *p < 0.05 (n = 6). (C) Separation of duplicated centrosomes in control or GRK2 shRNA cells after 0–1 h of 100 ng/ml EGF treatment. *p < 0.05 (n = 4). (D) Expression of transiently transfected wild-type bovine GRK2 or GRK2-K220R in GRK2 shRNA cells. (E) Percentage of cells with separated duplicated centrosomes after vehicle or 30-min treatment with 100 ng/ml EGF in GRK2 shRNA cells transiently transfected with empty pcDNA3 vector, wild-type GRK2, or GRK2-K220R. *p < 0.05 (n = 4). F. Centrosomal GRK2 in unseparated or separated duplicated centrosomes, as detected by anti-GRK2/3 antibody, after 100 ng/ml EGF treatment for 30 min.
To further quantify these effects, we measured the distance between centrosomes within centrosomal pairs. Although there was significant increase in the number of cells with separated centrosomes (from 0–2 to ≥6 μm) after a 30-min treatment with EGF, centrosome separation was blocked in the GRK2 shRNA cells (Supplemental Figure S3, A–C). To make sure that our results were not due to cellular changes induced by the stable knockdown of GRK2, we also evaluated HeLa cells with transient knockdown of GRK2 or GRK5 using specific small interfering RNA (siRNA) pools that we previously validated (Busillo et al., 2010). These siRNAs effectively and specifically knocked down expression of either GRK2 (Supplemental Figure S3D) or GRK5 (data not shown), and loss of GRK2 completely attenuated EGF-promoted centrosome separation (Supplemental Figure S3E).
To determine whether GRK2 also plays a role in EGF-promoted centrosome separation in nontransformed cells, we evaluated this process in retinal pigment epithelial 1 (RPE1) cells. GRK2 is expressed in these cells and can be effectively knocked down by siRNA treatment (Figure 4A). Endogenous GRK2 appears to be localized in centrosomes in RPE1 cells, as assessed using a polyclonal GRK2-specific antibody (Figure 4B) or a monoclonal GRK2/3 antibody (data not shown). Similar to HeLa cells, knockdown of GRK2 expression in RPE1 cells had no effect on EGF-promoted activation of ERK1/2 phosphorylation (Figure 4C) but completely attenuated EGF-promoted centrosome separation (Figure 4D). Of interest, RPE1 cells demonstrated less centrosomal separation than that observed in HeLa cells (Figure 3B). Thus GRK2 appears to play an essential role in EGF-promoted centrosomal separation in both transformed and nontransformed cells.
FIGURE 4:
GRK2 is localized to centrosomes in RPE1 cells. (A) Blotting for GRK2, using the anti-GRK2 3A10 antibody, in RPE1 cells transfected with control or GRK2 siRNA (n = 3). (B) Centrosomal localization of GRK2 in RPE1 cells using the GRK2 polyclonal antibody. (C) Expression and activation, as indicated by ERK1/2-T202 phosphorylation, of EGFR in RPE1 cells after vehicle or 5-min treatment with 100 ng/ml EGF (n = 3). Calnexin (CLX) was used as a loading control. (D) Percentage of S phase–synchronized cells with separated duplicated centrosomes after vehicle (serum-free media) or 30-min treatment with 100 ng/ml EGF in control or GRK2 siRNA–transfected RPE1 cells. *p < 0.05 (n = 3).
To better define the link between EGFR and GRK2 in centrosome separation, we compared the ability of wild-type GRK2 and a mutant GRK2 (GRK2-YF) to rescue EGF-promoted centrosome separation in GRK2 shRNA cells. GRK2-YF has mutations in three tyrosine residues (Tyr-13, 86, and 92) that were previously shown to be phosphorylated by activated EGFR and result in GRK2 activation (Chen et al., 2008). Whereas expression of wild-type GRK2 effectively rescued EGF-promoted centrosome separation, GRK2-YF was completely ineffective in rescuing centrosome separation (Figure 5, A and B). These data provide strong evidence that EGFR phosphorylation of tyrosine residues in GRK2 serves as an essential link between EGFR and GRK2 in the EGF-promoted centrosomal separation pathway.
FIGURE 5:
EGF-promoted centrosome separation is mediated by tyrosine phosphorylation of GRK2. (A) Expression of wild-type (wt) GRK2 and GRK2-YF in GRK2 shRNA HeLa cells, detected using anti-GRK2 3A10 antibody. (B) Separation of duplicated centrosomes after vehicle or 100 ng/ml EGF treatment for 30 min transfected with vector, wt GRK2, or GRK2-YF (n = 3). *p < 0.01 (n = 3).
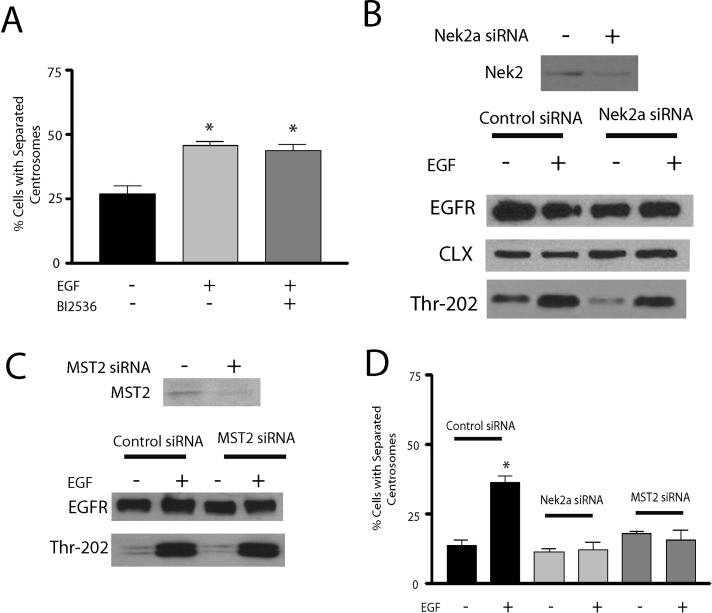
Centrosome separation during the cell cycle is mediated by Plk1, which controls splitting of duplicated centrosomes (centrosome disjunction) in late G2 by phosphorylating and activating Mst2, which in turn phosphorylates and activates Nek2A (Mardin et al., 2011). Therefore we tested whether this Plk1-Mst2-Nek2A pathway is also involved in EGF-promoted centrosome separation. Initial studies revealed that inhibition of Plk1 using the Plk1 inhibitor BI 2536 had no effect on EGF-induced centrosome separation in HeLa cells (Figure 6A) or on the basal level of centrosome separation (data not shown). BI 2536 is a potent Plk1 inhibitor, with Ki ≈ 0.5 nM (Steegmaier et al., 2007), and was previously shown to effectively inhibit centrosome disjunction (Mardin et al., 2011) and nucleophosmin phosphorylation (So et al., 2012). To investigate the potential involvement of Nek2A, Nek2A levels were knocked down by siRNA treatment (Figure 6B). Whereas knockdown of Nek2A had no significant effect on EGFR activation (Figure 6B), it completely attenuated EGF-promoted centrosome separation (Figure 6D). Because the role of Nek2A in centrosomal separation occurs through a process involving members of the Hippo pathway, particularly Mst2 (Mardin et al., 2011), we next evaluated a potential role for Mst2 in this pathway. Similar to Nek2A, knockdown of Mst2 had no effect on EGFR activation (Figure 6C), but it effectively attenuated EGF-promoted centrosome separation (Figure 6D). Taken together, these results reveal an essential role for GRK2, Mst2, and Nek2A in EGF-promoted centrosome separation.
FIGURE 6:
EGF-activated duplicated centrosome separation is mediated by Mst2 and Nek2A in HeLa cells. (A) Separation of duplicated centrosomes after vehicle or 100 ng/ml EGF treatment with or without 60 nM BI 2536 treatment. *p < 0.05 (n = 4). (B) siRNA-mediated knockdown of Nek2A expression and activation of EGFR, as indicated by ERK1/2-T202 phosphorylation, after vehicle or 5-min treatment with 100 ng/ml EGF (n = 3). Calnexin (CLX) was used as a loading control. (C) siRNA-mediated knockdown of Mst2 expression and activation of EGFR, as indicated by ERK1/2-T202 phosphorylation, after vehicle or 100 ng/ml EGF treatment for 5 min (n = 3). D. Percentage of separated duplicated centrosomes, in cells transfected with control, Mst2 siRNA, or Nek2A siRNA, after vehicle or 100 ng/ml EGF treatment for 30 min. *p < 0.05 (n = 3–4).
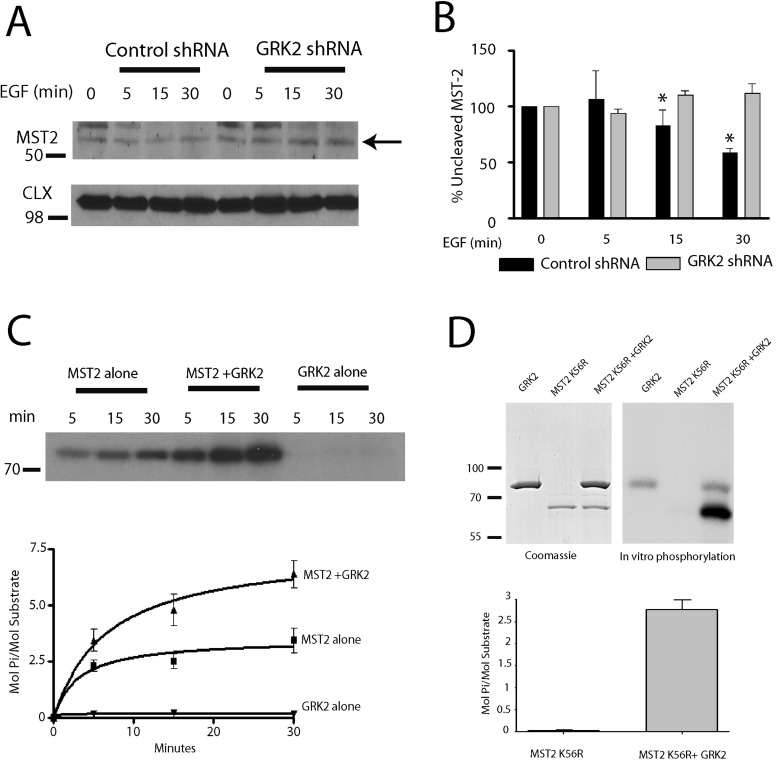
A hallmark of Mst2 activation involves the proteolytic cleavage of full-length Mst2 (∼55 kDa) to produce an active ∼34-kDa form (Lee et al., 2001). To determine whether Mst2 is activated by EGF treatment, we assessed full-length Mst2 levels over a 0- to 30-min treatment with EGF in control and GRK2 shRNA cells. This resulted in ∼40% reduction in full-length Mst2 after 30-min EGF treatment in control cells, whereas-full length Mst2 levels were unchanged in GRK2 shRNA cells (Figure 7, A and B). Thus GRK2 appears to mediate EGF-promoted cleavage and activation of Mst2. To test whether Mst2 is directly phosphorylated by GRK2, we performed in vitro phosphorylation reactions using purified glutathione S-transferase (GST)–Mst2 and GRK2. Although a significant level of Mst2 autophosphorylation occurred over a 30-min incubation, Mst2 phosphorylation was substantially increased in the presence of GRK2 (Figure 7C). Additional analysis of these data revealed that Mst2 is an effective substrate for GRK2, with a stoichiometry of ∼3–4 mol phosphate/mol Mst2 above the level of Mst2 autophosphorylation (Figure 7C). To substantiate these findings, we also evaluated the ability of GRK2 to phosphorylate purified Mst2-K56R, a kinase-dead mutant of Mst2. There was no autophosphorylation of Mst2-K56R, whereas GRK2 phosphorylated Mst2-K56R to a stoichiometry of ∼3 mol/mol (Figure 7D). These studies demonstrate that GRK2 can directly phosphorylate Mst2 and suggest that this phosphorylation results in Mst2 activation.
FIGURE 7:
Mst2 is activated by the EGFR-GRK2 pathway in HeLa cells. (A, B) Decrease in full-length Mst2 from 0–30 min of EGF treatment in control shRNA cells. *p < 0.05 (n = 4). (C) Time course of in vitro phosphorylation of GST-Mst2 by GRK2 (n = 3) and quantitation of Mst2 phosphorylation in the presence or absence of GRK2. (D) In vitro phosphorylation of purified Mst2-K56R by GRK2 for 60 min and quantitation of Mst2-K56R phosphorylation in the presence or absence of GRK2.
To identify the GRK2 phosphorylation sites on Mst2, we analyzed tryptic digests of nonphosphorylated and GRK2-phosphorylated Mst2-K56R using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Seven sites of phosphorylation were identified, three major sites (Ser-18, Thr-174, and Ser-316) and four minor sites (Thr-180, Thr-252, Ser-284, and Thr-292; Table 1). Two of the major sites of GRK2 phosphorylation (Ser-18 and Ser-316) were previously identified as sites for Plk1 phosphorylation of Mst2 (Mardin et al., 2011). These studies demonstrate that GRK2 can directly phosphorylate Mst2 and there is significant overlap between the sites phosphorylated by GRK2 and Plk1.
TABLE 1:
Identified tryptic phosphopeptides from Mst2-K56R.
| Relative abundanceb | ||
|---|---|---|
| Sequencea | Nonphoshoc | GRK2-phosphod |
| 13KLSEDpSLTK21 | — | ++ |
| 162LADFGVAGQLTDpTMAK177 | — | ++ |
| 178RNpTVIGTPF186 | — | + |
| 243KPELWSDDFpTDFVK256 | — | + |
| 280NAKPVpSILR288 | — | + |
| 289DLIpTEAMEIK298 e | — | + |
| 308ELEEEEENpSDEDELDSHTMVK328 | + | ++ |
aPrimary sequence of phosphopeptides identified on the basis of MS/MS spectra; pS, phosphoserine; pT, phosphothreonine.
bRelative abundance of the identified phosphopeptide was estimated from the MS/MS spectra counting method. —, not present; +, low counts; ++ high counts.
cNonphosphorylated kinase-dead mutant Mst2-K56R purified from Sf9 cells.
dPhosphorylated in the presence of GRK2 kinase-dead mutant Mst2-K56R.
eOwing to no coverage of this area in the MS/MS spectra of Mst2-K56R, the abundance of this phosphopeptide was estimated from the MS/MS spectra of tryptic proteolytic digests of GST-Mst2 WT. No phosphorylation of this peptide was found in the autophosphorylated form of GST-Mst2 WT.
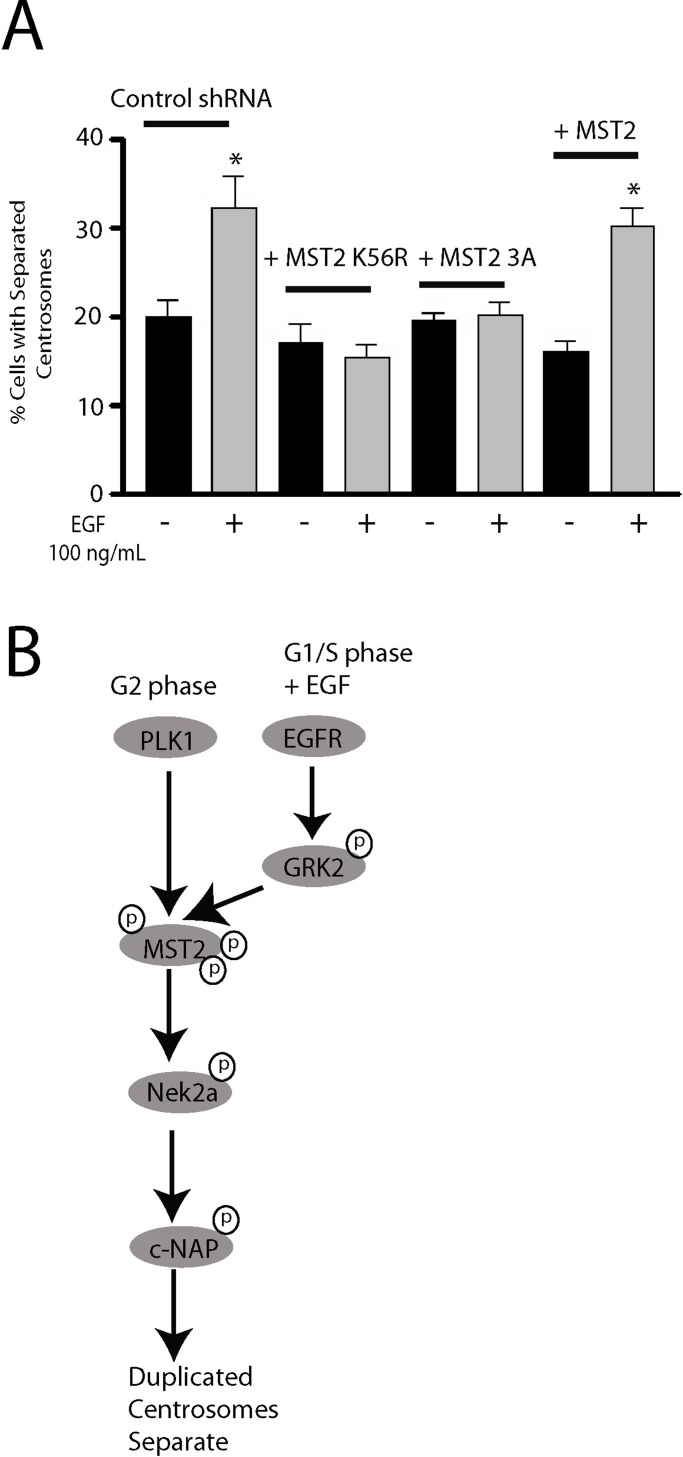
To further characterize the role of Mst2 in the EGF pathway, we expressed catalytically inactive Mst2-K56R in HeLa cells. This mutant functioned as an effective dominant-negative mutant and abolished EGF-mediated centrosome separation (Figure 8A). Because our mass spectrometry analysis identified Ser-18 and 316 as two of the major GRK2 phosphorylation sites on Mst2 and previous studies identified Ser-15, 18, and 316 as putative Plk1 phosphorylation sites involved in centrosome separation (Mardin et al., 2011), we also evaluated an Mst2 mutant defective in Plk1 phosphorylation in this pathway. Expression of an Mst2-3A (S15A, S18A, S316A) mutant in control cells effectively suppressed EGF-mediated centrosome separation, whereas expression of wild-type Mst2 had no effect (Figure 8A). Taken together, these studies support a role for GRK2 phosphorylation of Mst2 residues Ser-18 and Ser-316 in EGF-promoted centrosome separation.
FIGURE 8:
GRK2 phosphorylation of Mst2 mediates EGF-mediated centrosomal separation. (A) Decrease in separation of duplicated centrosomes mediated by overexpression of Mst2-K56R (n = 3) or Mst2-3A (n = 5) with or without 30-min treatment with EGF (100 ng/ml) in HeLa cells. *p < 0.05. (B) Model for centrosome separation mediated by EGFR in G1/S phase and Plk1 in G2 phase.
DISCUSSION
In addition to their more commonly known role in phosphorylating activated GPCRs, GRKs also appear to have many additional functional roles (Gurevich et al., 2012). In this article, we demonstrate that GRK2 is localized to centrosomes and mediates EGF-promoted separation of duplicated centrosomes. The ability of GRK2 to regulate this pathway is both Nek2A and Mst2 dependent and appears to involve direct phosphorylation of Mst2 by GRK2, resulting in Mst2 activation.
The pathway involved in centrosomal separation that occurs during the cell cycle has been extensively characterized (Mardin and Schiebel, 2012). Centrosomes duplicate once per cell cycle and normally start to separate at G2/M. Duplicated centrosomes are held together by two structural proteins that function as a linker or tether, C-Nap1 and rootletin. The kinase Nek2A phosphorylates C-Nap1 and rootletin in late G2, which displaces these proteins from the centrosome and allows centrosome separation. Upstream control of Nek2A activity is mediated by two components of the Hippo pathway, the protein kinase Mst2 and the scaffolding protein hSav1 (Mardin et al., 2010). Aurora A and Plk1 also play a critical role in this process, with Aurora A functioning to activate Plk1, which in turn phosphorylates Mst2. Mst2 phosphorylation results in reduced interaction of PP1γ within an Mst2-Nek2A-PP1γ complex, leading to enhanced ability of Nek2A to phosphorylate C-Nap1 (Mardin et al., 2010, 2011).
The role of mitogens such as EGF in centrosomal separation was first described in 1982 (Sherline and Mascardo, 1982a). Additional studies proposed a role for calmodulin-stimulated contraction of microfilaments attached to centrosomes in this process (Sherline and Mascardo, 1982b), whereas a link between mitogen-promoted centrosome separation and DNA synthesis was also proposed (Sherline and Mascardo, 1984). These early studies clearly demonstrated that mitogens such as EGF stimulate centrosome separation and help to coordinately link centrosome separation with DNA synthesis. In the present study, we started to further dissect the mechanism of this process and identified an essential role for GRK2, Mst2, and Nek2A in this pathway.
While the manuscript of this article was under review, Mardin et al. (2013) published an article demonstrating that EGF-mediated centrosome separation was Mst2 and Nek2A dependent and also involved hSav1, rootletin, and C-Nap1. Although the link between EGFR and Mst2 was not clearly established in their study, they also found that the pathway was dependent on Akt activation (Mardin et al., 2013). Of interest, previous studies established a link between Akt and Mst2 and showed that Akt can directly phosphorylate Mst2 on Thr-117 and Thr-384, although this phosphorylation inhibits Mst2 activity (Kim et al., 2010; Romano et al., 2010). Thus the mechanistic link between Akt activation and EGF-promoted centrosome separation is unclear. Our work provides additional mechanistic input on this pathway and reveals that GRK2 can directly phosphorylate Mst2 and likely regulate Mst2 activation. The combination of EGFR and GRK2 activation also results in partial cleavage of Mst2 into its more active conformation, a process that may be mediated by caspase-3 (Deng et al., 2003). Although it is unclear how caspase-3 might be activated under these conditions, previous studies showed that Mst1 can function as an upstream activator of caspase-3 (Lee et al., 2001). GRK2 has also been reported to mediate the G2/M phase of the cell cycle (Penela et al., 2010). Given that we found that GRK2 does not affect centrosome separation during the cell cycle, however, it appears likely that GRK2 has a distinct functional role at different stages of the cell cycle.
LC-MS/MS analysis of GRK2-phosphorylated Mst2 revealed Ser-18 and Ser-316 as the likely two major phosphorylation sites involved in centrosome separation. These two sites were among the three sites previously identified to be important for linking Mst2 phosphorylation by Plk1 with centrosome separation in the G2 phase (Mardin et al., 2011). Ser-18 and Ser-316 are surrounded by acidic residues and are thus in the preferred environment for both Plk1-mediated (Dephoure et al., 2008) and GRK2-mediated (Onorato et al., 1991) phosphorylation. We propose that GRK2 activation by EGFR leads to GRK2 phosphorylation of Mst2 at these sites, which, in turn, regulates the Mst2-Nek2A-PP1γ complex (Figure 8B). We cannot rule out a potential role for Thr-174 phosphorylation, however, since this site was also identified to be a major phosphorylation site for GRK2 in our in vitro analysis.
An important component in this pathway is the link between EGFR and GRK2. Previous studies showed that EGFR activation results in association of GRK2 with the EGFR and subsequent phosphorylation of GRK2 at three tyrosine residues (Tyr-13, -86, and -92), resulting in activation of GRK2 (Chen et al., 2008). It is worth noting that Src also phosphorylates GRK2 on the same three tyrosines and results in GRK2 activation (Sarnago et al., 1999). Here we show that these tyrosine residues are, in fact, important for EGF-promoted centrosome separation, as a GRK2 mutant with these residues mutated to Phe was not able to rescue centrosomal separation in GRK2-null cells. In addition to direct phosphorylation of GRK2 by EGFR, it is also possible that the EGFR might activate GRK2 through GPCR transactivation. GPCR-EGFR cross-talk has been well documented (Leserer et al., 2000) and might even be transmitted to GPCRs localized in centrosomes (Gillies et al., 2009). Although the source of EGFR signaling to centrosomal GRK2 could stem from plasma membrane–localized, endocytosed, or nuclear EGFRs (Brand et al., 2011), EGFR activation ultimately leads to activation of GRK2 with centrosomal GRK2 being optimally localized to regulate the Mst2-Nek2A-PP1γ complex.
Although we did not address the functional role of EGFR-promoted centrosome separation, recent studies suggested that activating EGFR during S phase might allow cells to continue into G2/M and have beneficial effects (Mardin et al., 2013). For example, EGF-treated cells allowed to progress into G2/M demonstrate decreased mitotic defects such as lagging chromosomes, chromosome bridges, and chromosome missegregation (Mardin et al., 2013). Furthermore, EGF activation also allows cells to bypass the requirement for the Eg5 pathway in mitosis, revealing a way for cancer cells to bypass potential cancer therapeutics targeting this pathway (Mardin et al., 2013). The fact that GRK2 is involved in a process that could modify mitotic spindle fidelity supports the potential role of GRK2 in the development of cancer. For example, GRK2 levels are elevated in human granulosa cell tumors (King et al., 2003) and thyroid cancer (Metaye et al., 2002). Because EGFR signaling is associated with many cancers (Herbst, 2004), abnormal EGFR-GRK2 signaling could lead to unintended defects especially when activated in G1 or S phase. Potentially, cells with abnormal GRK2 levels may be more prone to genomic instability.
Overall these studies provide additional insight into the myriad of functions regulated by GRKs and provide a clear link between EGF receptors, GRK2, and regulation of the cellular machinery that controls centrosome separation. These results also provide insight into the specificity of these kinases since GRK5 is also localized in centrosomes, where it has a role in regulating microtubule nucleation (Michal et al., 2012). Although it is unclear whether GPCRs have a role in regulating GRK function in centrosomes, several GPCR signaling components, including GPCRs (Gillies et al., 2009), Gi (Cho and Kehrl, 2007), and β-arrestins (Shankar et al., 2010), are found in centrosomes. Further characterizing the potential links between GPCRs and centrosomal function should provide important insight into the regulation of cell function. Moreover, targeting the EGFR-GRK2 pathway may be important in helping to treat cancers mediated by aberrant EGFR signaling.
MATERIALS AND METHODS
Materials
Nocodazole, cytochalasin B, hydroxyurea, and thymidine were purchased from Sigma-Aldrich (St. Louis, MO). Calyculin A was from Cell Signaling Technology (Danvers, MA). Monoclonal antibodies used were anti-Nek2 (1:2000; BD Biosciences, San Jose, CA), anti–γ-tubulin (1:1000; Sigma-Aldrich), anti-GRK2/3 (1:100 for immunofluorescence, 1:2000 for immunoblotting; Millipore, Billerica, MA), anti-GRK2 3A10 (1:2000 for immunoblotting), anti–α-tubulin (1:1000; Sigma-Aldrich), and anti-EGFR (1:1000; Covance, Princeton, NJ). Polyclonal antibodies used were anti–γ-tubulin (1:1000; Sigma-Aldrich), anti–α-tubulin (1:1000; Sigma-Aldrich), anti-pericentrin (1:2000; Abcam, Cambridge, MA), anti-GRK2 (1:100 for immunofluorescence, 1:1000 for immunoblotting; Santa Cruz Biotechnology, Santa Cruz, CA), anti-Mst2 (1:2000; Cell Signaling Technology), and anti–ERK-pT202/Y204 and antixpMEK1/2-pS217/S221 (1:2000; Cell Signaling Technology). Secondary antibodies for immunofluorescence were from Invitrogen (Carlsbad, CA) and included Alexa Fluor 488 goat anti-mouse, Alexa Fluor 594 goat anti-mouse, Alexa Fluor 488–conjugated goat anti-rabbit, Alexa Fluor 594–conjugated goat anti-rabbit, and Alexa Fluor 633–conjugated goat anti-rabbit, all used at 1:100 dilution. Secondary antibodies for immunoblotting were from Vector Laboratories (Burlingame, CA).
Cell culture
HeLa and HEK293 cells were cultured in DMEM (Mediatech, Manassas, VA) supplemented with 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and 10% fetal bovine serum (FBS; GIBCO, Invitrogen). MDA-MB-231 cells were grown in MEM (Mediatech) supplemented with 10 mM HEPES and 10% FBS. RPE1 cells were cultured in DMEM:F12 medium (Mediatech) with 10% FBS. HeLa cells with stable GRK knockdown were grown in DMEM supplemented with 10 mM HEPES, 10% FBS, and 2 μg/ml puromycin. HeLa cells stably expressing GFP-centrin were a generous gift from Timothy Yen (Fox Chase Cancer Center, Philadelphia, PA) and were cultured in DMEM supplemented with 10 mM HEPES, 10% FBS, and 800 μg/ml G418. The GFP-centrin signal of the daughter centriole was ∼40% that of the mother centriole.
For cell synchronization, double-thymidine block was used for G1/S-phase synchronization, and hydroxyurea treatment was used for S-phase synchronization and to promote centrosome duplication. For double-thymidine block, HeLa cells were plated at 25% confluency, blocked in media containing 2 mM thymidine for 16 h, released for 9–12 h into fresh medium, and then blocked a second time with 2 mM thymidine for 16 h. Cells were then released into fresh medium and the appropriate time points collected. For hydroxyurea block, HeLa cells were plated at 70% confluency and blocked in media containing 2 mM hydroxyurea for 48 h. Samples were prepared, and standard immunoblotting procedures were followed. Flow cytometry was similar to that described previously (Michal et al., 2012).
DNA constructs, siRNAs, and shRNAs
HA-Mst2 and Flag-Mst2-K56R were obtained from Addgene (Cambridge, MA). HA-Mst2-3A (S15A, S18A, and S316A) was a generous gift of Elmar Schiebel (University of Heidelberg, Heidelberg, Germany).
To create stable cell lines, shRNA constructs specific to GRK2 (5′-GATCAAGAAGTACGAGAAGCT-3′) or GRK5 (5′-ACGAGATGATAGAAACAGAAT-3′) along with a scrambled control shRNA (Sigma-Aldrich) were transfected into HeLa cells using Lipofectamine 2000 (Invitrogen). Cells were then selected in 2 μg/ml puromycin to obtain a stably expressing population. For GRK2 siRNA transfection, four independent GRK2 siRNAs (5′-GGGACGUGUUCCAGAAAUU-3′, 5′-GCUCGCAUCCCUUCUCGAA-3′, 5′-GGAAUCAAGUUACUGGACA-3′, and 5′-GCGAUAAGUUCACACGGUU-3′) were pooled, and 60 pmol of the pool was used per transfection. For GRK5 siRNA transfection, four independent GRK5 siRNAs (5′-CCAACACGGUCUUGCUGAA-3′, 5′-GGGAGAACCAUUCCACGAA-3′, 5′-CAAACCAUGUCAGCUCGAA-3′, and 5′-GAUUAUGGCCACAUUAGGA-3′) were pooled and 60 pmol of the pool was used per transfection. For Mst2 and Nek2A, 5′-GGAUAGUUUUUCAAAUAGG-3′ was used to knock down Mst2 expression (Matallanas et al., 2007), and 5′-TTCTGAGAGTCAGCTCACA-3′ was used for Nek2A (Fletcher et al., 2005). Control siRNA–scrambled sequences were purchased from Thermo Fisher Scientific (Waltham, MA).
Fractionation and immunoblotting
For immunoblotting, cells were pelleted by centrifugation, lysed in Triton lysis buffer (0.2 mM HEPES, pH 7.4, 1% Triton X-100, 0.5 M NaCl, 10 mM EDTA, containing protease inhibitors and PhosSTOP phosphatase inhibitor tablets [Roche, Indianapolis, IN]) for 20 min on ice and then centrifuged at 18,000 × g for 30 min at 4°C. Samples were loaded onto 10% SDS–PAGE gels, transferred to nitrocellulose membranes, and blocked in 5% nonfat milk in Tris-buffered saline (TBS; 20 mM Tris-HCl, pH 7.5, 150 mM NaCl) with 0.1% Tween-20. Blots were incubated with primary antibody overnight at 4°C, washed in TBS with 0.1% Tween-20, and incubated with appropriate secondary antibody for 1 h at room temperature. Blots were then washed extensively and developed with either West PICO or DURA Chemiluminescence Kit (Pierce, Rockford, IL). For Mst2 immunoblotting, cells were pretreated for 5 min with 1 μM calyculin A in phosphate-buffered saline (PBS) and then lysed as described. Protein concentrations were determined by Bradford assay. Centrosome preparations were made following the procedure of Bornens and Moudjou (1999).
Immunofluorescence microscopy
For all immunofluorescence studies, cells were split onto poly-l-lysine–coated coverslips 24–72 h before fixation/staining. For all antibodies, except γ-tubulin when used alone, cells were first preextracted with 1% Triton X-100 in PBS for 30 s. For samples used for centrosomal separation and duplication analyses, only methanol fixation was used, followed by blocking with 1% bovine serum albumin/PBS overnight. To costain for endogenous GRK2 (3A10 monoclonal antibody) and γ-tubulin (polyclonal antibody), cells were preextracted with 1% Triton X-100 in PBS for 30 s, fixed in 4% paraformaldehyde in PBS for 10 min, and permeabilized with methanol for 10 min. To costain for GRK2, using a polyclonal GRK2 antibody or GRK2/3 monoclonal antibody, and γ-tubulin or pericentrin, cells were preextracted with 1% Triton X-100 in PBS, fixed in methanol for 20 min at −20°C, and then rehydrated in PBS for 10 min at room temperature. For all GRK2 antibodies, after fixation, slides were incubated with quench buffer (PBS, 2.5% nonfat milk, 150 mM sodium acetate) and block buffer (PBS, 0.1% Tween-20, 2.5% nonfat milk) at room temperature and the appropriate primary antibodies overnight at 4°C. Slides were then washed with PBS, incubated with the designated secondary antibodies for 1 h at room temperature, and washed with PBS with 0.1% Tween-20. DNA was stained with 4′,6-diamidino-2-phenylindole (Molecular Probes, Eugene, OR) and slides mounted with Pro-Long Anti-Fade (Molecular Probes). Images were taken using either a Zeiss LSM 510 META confocal microscope with a Plan-Apo 63× 11.4 oil immersion lens (Carl Zeiss, Jena, Germany) or a Nikon Eclipse E800 fluorescence microscope (Nikon, Melville, NY). All intensity and area analysis was performed using MetaMorph software (Molecular Devices, Sunnyvale, CA), Image-Pro Plus (Media Cybernetics, Rockville, MD), or ImageJ (National Institutes of Health, Bethesda, MD).
Microtubule nucleation assay
HeLa cells with stable GRK2 knockdown were treated for 1 h with 10 μg/ml nocodazole to completely depolymerize microtubules. To assess microtubule regrowth, cells were washed twice with warm media to remove nocodazole and incubated at 37°C for 5 min. Cells were then fixed and stained with anti-rabbit pericentrin and anti-mouse α-tubulin antibodies. Images were taken of at least 80 cells from four independent experiments using a Nikon Eclipse E800 fluorescence microscope, and each aster was treated independently. Only asters nucleated from either single or paired duplicated centrosomes were analyzed. Aster areas (μm2) were obtained by drawing a region around the aster and measuring the area of this region. Images were quantified using QED camera software (Media Cybernetics) and processed with Image-Pro Plus.
In vitro kinase assay
GRK2 was purified as described (Kunapuli et al., 1994), and purified GST-Mst2 was from Sigma-Aldrich. Purified Mst2-K56R prepared from Sf9 cells was a generous gift from Elmar Schiebel (University of Heidelberg, Heidelberg, Germany). Protein phosphorylation assays were similar to previous protocols (Pronin et al., 1997). For reactions involving GST-Mst2, ∼50 nM purified GST-Mst2 was incubated with or without 50 nM purified GRK2 for 5–30 min at 30°C in a buffer containing 20 mM Tris-HCl, pH 7.5, 2 mM EDTA, 7.5 mM MgCl2, 100 μM ATP. and [γ-32P]ATP (250–1000 cpm/pmol ATP). Reactions were stopped with SDS sample buffer and electrophoresed on 10% SDS–polyacrylamide gel. Proteins were visualized by autoradiography, and phosphorylation stoichiometries were determined by excising bands from the dried gels and counting in a scintillation counter.
A similar protocol was used with purified Mst2-K56R, and although initial studies revealed that Mst2 is a substrate for GRK2, the stoichiometry of phosphate incorporation was only ∼0.8 mol phosphate/mol Mst2. LC-MS/MS analysis identified multiple sites of basal phosphorylation on Mst2-K56R, suggesting that Mst2-K56R is a substrate of insect cell kinases when expressed in Sf9 cells. To remove these phosphates, the protein was treated with lambda phosphatase according to the manufacturer's protocol (New England BioLabs, Ipswich, MA). Briefly, 1.2 μM Mst2-K56R was incubated with 1600 U of recombinant lambda phosphatase in lambda phosphatase buffer (50 mM HEPES, pH 7.5, 100 mM NaCl, 2 mM dithiothreitol, 0.01% Brij35) with 1 mM MnCl2 at 30°C for 30 min, and the reaction was stopped by adding 1 μl of PhosSTOP solution (Roche) to 160 μl of reaction mixture. Dephosphorylated Mst2-K56R was then isolated by incubation with 20 μl of nickel-nitriloacetic acid Sepharose beads for 1 h at 4°C while rocking, followed by elution and overnight dialysis against 20 mM Tris, pH 7.5, 2 mM MgCl2, and 50 mM NaCl. The dephosphorylated Mst2-K56R was then incubated with 1 μM GRK2 for 60–90 min at 30°C using the buffer conditions described for GST-Mst2 phosphorylation. GRK2 phosphorylated Mst2-K56R was then subjected to LC-MS/MS analysis to identify the specific sites of GRK2 phosphorylation. It is important to note that the phosphatase treatment removed all basal phosphates on Mst2-K56R except for a low level of phosphate on Ser-316. Incubation of Mst2-K56R with GRK2, however, significantly enhanced phosphorylation of Ser-316, based on comparison of MS/MS spectra counts before and after treatment with GRK2 (Table 1).
Tandem mass spectrometry
LC-MS/MS analyses were performed at the Wistar Institute Proteomics Facility (Philadelphia, PA). Gel bands containing Mst2 proteins were excised, digested with trypsin, and analyzed using an LTQ-Orbitrap XL mass spectrometer as described previously (Beer et al., 2011; Fong et al., 2011). Peptide sequences were interpreted from MS/MS spectra by searching against a human database containing Mst2 and common contaminant sequences using Bioworks, version 3.3.1 SP1 (Thermo Fisher Scientific). All database searches allowed for up to two missed cleavages, a static modification of Cys with iodoacetamide, and the following dynamic modifications: Met oxidation, Asn deamidation, and Ser, Thr, and Tyr phosphorylation. Outputs from Bioworks searches were filtered using mass tolerance of 10 ppm and DCn > 0.05, and identified phosphorylated peptides were manually verified.
Supplementary Material
Acknowledgments
This work was supported in part by a fellowship from the Canadian Heart and Stroke Foundation (to C.H.S.) and National Institutes of Health Grants GM44944 and CA129626 (to J.L.B.). We thank Tim Yen and Phil Wedegaertner for helpful comments on the manuscript, Hsin-Yao Tang for helpful discussion of the mass spectrometry data, Elmar Schiebel for providing Mst2 expression constructs and purified Mst2-K56R, and Federico Mayor, Jr., for providing a GRK2-YF expression construct.
Abbreviations used:
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- GPCR
G protein–coupled receptor
- GRK
G protein–coupled receptor kinase
- GST
glutathione S-transferase
- PBS
phosphate-buffered saline
- Mst2
mammalian STE20-like kinase 2
- Plk1
polo-like kinase 1
- RPE1
retinal pigment epithelial 1
- TBS
Tris-buffered saline
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-01-0013) on July 31, 2013.
*Present address: Department of Pathology, Yale University School of Medicine, New Haven, CT 06520.
REFERENCES
- Barker BL, Benovic JL. G protein-coupled receptor kinase 5 phosphorylation of hip regulates internalization of the chemokine receptor CXCR4. Biochemistry. 2012;50:6933–6941. doi: 10.1021/bi2005202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthet G, et al. Beta-arrestin1 phosphorylation by GRK5 regulates G protein-independent 5-HT4 receptor signalling. EMBO J. 2009;28:2706–2718. doi: 10.1038/emboj.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer LA, Tang HY, Sriswasdi S, Barnhart KT, Speicher DW. Systematic discovery of ectopic pregnancy serum biomarkers using 3-D protein profiling coupled with label-free quantitation. J Proteome Res. 2011;10:1126–1138. doi: 10.1021/pr1008866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- Bornens M, Moudjou M. Studying the composition and function of centrosomes in vertebrates. Methods Cell Biol. 1999;61:13–34. doi: 10.1016/s0091-679x(08)61973-1. [DOI] [PubMed] [Google Scholar]
- Brand TM, Iida M, Li C, Wheeler DL. The nuclear epidermal growth factor receptor signaling network and its role in cancer. Discov Med. 2011;12:419–432. [PMC free article] [PubMed] [Google Scholar]
- Busillo JM, Armando S, Sengupta R, Meucci O, Bouvier M, Benovic JL. Site-specific phosphorylation of CXCR4 is dynamically regulated by multiple kinases and results in differential modulation of CXCR4 signaling. J Biol Chem. 2010;285:7805–7817. doi: 10.1074/jbc.M109.091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bychkov ER, Ahmed MR, Gurevich VV, Benovic JL, Gurevich EV. Reduced expression of G protein-coupled receptor kinases in schizophrenia but not in schizoaffective disorder. Neurobiol Dis. 2011;44:248–258. doi: 10.1016/j.nbd.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bychkov ER, Gurevich VV, Joyce JN, Benovic JL, Gurevich EV. Arrestins and two receptor kinases are upregulated in Parkinson's disease with dementia. Neurobiol Aging. 2008;29:379–396. doi: 10.1016/j.neurobiolaging.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant SH, Pitcher JA. G protein-coupled receptor kinase 2-mediated phosphorylation of ezrin is required for G protein-coupled receptor-dependent reorganization of the actin cytoskeleton. Mol Biol Cell. 2005;16:3088–3099. doi: 10.1091/mbc.E04-10-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman CV, Som T, Kim CM, Benovic JL. Binding and phosphorylation of tubulin by G protein-coupled receptor kinases. J Biol Chem. 1998;273:20308–20316. doi: 10.1074/jbc.273.32.20308. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhu H, Yuan M, Fu J, Zhou Y, Ma L. G-protein-coupled receptor kinase 5 phosphorylates p53 and inhibits DNA damage-induced apoptosis. J Biol Chem. 2010;285:12823–12830. doi: 10.1074/jbc.M109.094243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Long H, Wu Z, Jiang X, Ma L. EGF transregulates opioid receptors through EGFR-mediated GRK2 phosphorylation and activation. Mol Biol Cell. 2008;19:2973–2983. doi: 10.1091/mbc.E07-10-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kehrl JH. Localization of Gi proteins in the centrosomes and at the midbody: implication for their role in cell division. J Cell Biol. 2007;178:245–255. doi: 10.1083/jcb.200604114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Pang A, Wang JH. Regulation of mammalian STE20-like kinase 2 (MST2) by protein phosphorylation/dephosphorylation and proteolysis. J Biol Chem. 2003;278:11760–11767. doi: 10.1074/jbc.M211085200. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher L, Cerniglia GJ, Yen TJ, Muschel RJ. Live cell imaging reveals distinct roles in cell cycle regulation for Nek2A and Nek2B. Biochim Biophys Acta. 2005;1744:89–92. doi: 10.1016/j.bbamcr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Fong KP, Tang HY, Brown AC, Kieba IR, Speicher DW, Boesze-Battaglia K, Lally ET. Aggregatibacter actinomycetemcomitans leukotoxin is post-translationally modified by addition of either saturated or hydroxylated fatty acyl chains. Mol Oral Microbiol. 2011;26:262–276. doi: 10.1111/j.2041-1014.2011.00617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman NJ, Kim LK, Murray JP, Exum ST, Brian L, Wu JH, Peppel K. Phosphorylation of the platelet-derived growth factor receptor-beta and epidermal growth factor receptor by G protein-coupled receptor kinase-2. Mechanisms for selectivity of desensitization. J Biol Chem. 2002;277:48261–48269. doi: 10.1074/jbc.M204431200. [DOI] [PubMed] [Google Scholar]
- Fusco A, Santulli G, Sorriento D, Cipolletta E, Garbi C, Dorn GW, 2nd, Trimarco B, Feliciello A, Iaccarino G. Mitochondrial localization unveils a novel role for GRK2 in organelle biogenesis. Cell Signal. 2012;24:468–475. doi: 10.1016/j.cellsig.2011.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sevilla JA, Alvaro-Bartolome M, Diez-Alarcia R, Ramos-Miguel A, Puigdemont D, Perez V, Alvarez E, Meana JJ. Reduced platelet G protein-coupled receptor kinase 2 in major depressive disorder: antidepressant treatment-induced upregulation of GRK2 protein discriminates between responder and non-responder patients. Eur Neuropsychopharmacol. 2010;20:721–730. doi: 10.1016/j.euroneuro.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Gillies L, Lee SC, Long JS, Ktistakis N, Pyne NJ, Pyne S. The sphingosine 1-phosphate receptor 5 and sphingosine kinases 1 and 2 are localised in centrosomes: possible role in regulating cell division. Cell Signal. 2009;21:675–684. doi: 10.1016/j.cellsig.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Gros R, Benovic JL, Tan CM, Feldman RD. G-protein-coupled receptor kinase activity is increased in hypertension. J Clin Invest. 1997;99:2087–2093. doi: 10.1172/JCI119381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich EV, Tesmer JJ, Mushegian A, Gurevich VV. G protein-coupled receptor kinases: more than just kinases and not only for GPCRs. Pharmacol Ther. 2012;133:40–69. doi: 10.1016/j.pharmthera.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RA, Spurney RF, Premont RT, Rahman N, Blitzer JT, Pitcher JA, Lefkowitz RJ. G protein-coupled receptor kinase 6A phosphorylates the Na+/H+ exchanger regulator factor via a PDZ domain-mediated interaction. J Biol Chem. 1999;274:24328–24334. doi: 10.1074/jbc.274.34.24328. [DOI] [PubMed] [Google Scholar]
- Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59:21–26. doi: 10.1016/j.ijrobp.2003.11.041. [DOI] [PubMed] [Google Scholar]
- Huang ZM, Gold JI, Koch WJ. G protein-coupled receptor kinases in normal and failing myocardium. Front Biosci. 2012;17:3047–3060. doi: 10.2741/3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LR, Scott MG, Pitcher JA. G protein-coupled receptor kinase 5 contains a DNA-binding nuclear localization sequence. Mol Cell Biol. 2004;24:10169–10179. doi: 10.1128/MCB.24.23.10169-10179.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Shu S, Coppola MD, Kaneko S, Yuan ZQ, Cheng JQ. Regulation of proapoptotic mammalian ste20-like kinase MST2 by the IGF1-Akt pathway. PLoS One. 2010;5:e9616. doi: 10.1371/journal.pone.0009616. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kim JI, Chakraborty P, Wang Z, Daaka Y. G-protein coupled receptor kinase 5 regulates prostate tumor growth. J Urol. 2012;187:322–329. doi: 10.1016/j.juro.2011.09.049. [DOI] [PubMed] [Google Scholar]
- King DW, Steinmetz R, Wagoner HA, Hannon TS, Chen LY, Eugster EA, Pescovitz OH. Differential expression of GRK isoforms in nonmalignant and malignant human granulosa cells. Endocrine. 2003;22:135–142. doi: 10.1385/ENDO:22:2:135. [DOI] [PubMed] [Google Scholar]
- Kunapuli P, Onorato JJ, Hosey MM, Benovic JL. Expression, purification, and characterization of the G protein-coupled receptor kinase GRK5. J Biol Chem. 1994;269:1099–1105. [PubMed] [Google Scholar]
- Lee KK, Ohyama T, Yajima N, Tsubuki S, Yonehara S. MST, a physiological caspase substrate, highly sensitizes apoptosis both upstream and downstream of caspase activation. J Biol Chem. 2001;276:19276–19285. doi: 10.1074/jbc.M005109200. [DOI] [PubMed] [Google Scholar]
- Leserer M, Gschwind A, Ullrich A. Epidermal growth factor receptor signal transactivation. IUBMB Life. 2000;49:405–409. doi: 10.1080/152165400410254. [DOI] [PubMed] [Google Scholar]
- Onorato J, Palczewski K, Regan JW, Caron MG, Lefkowitz RJ, Benovic JL. Role of acidic amino acids in peptide substrates of the beta-adrenergic receptor kinase and rhodopsin kinase. Biochemistry. 1991;30:5118–5125. doi: 10.1021/bi00235a002. [DOI] [PubMed] [Google Scholar]
- Mardin BR, Lange C, Baxter JE, Hardy T, Scholz SR, Fry AM, Schiebel E. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat Cell Biol. 2010;12:1166–1176. doi: 10.1038/ncb2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardin BR, Agircan FG, Lange C, Schiebel E. Plk1 controls the Nek2A-PP1gamma antagonism in centrosome disjunction. Curr Biol. 2011;21:1145–1151. doi: 10.1016/j.cub.2011.05.047. [DOI] [PubMed] [Google Scholar]
- Mardin BR, Schiebel E. Breaking the ties that bind: new advances in centrosome biology. J Cell Biol. 2012;197:11–18. doi: 10.1083/jcb.201108006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardin BR, Isokane M, Cosenza MR, Kramer A, Ellenberg J, Fry AM, Schiebel E. EGF-induced centrosome separation promotes mitotic progression and cell survival. Dev Cell. 2013;25:229–240. doi: 10.1016/j.devcel.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini JS, Raake P, Vinge LE, DeGeorge BR, Jr, Chuprun JK, Harris DM, Gao E, Eckhart AD, Pitcher JA, Koch WJ. Uncovering G protein-coupled receptor kinase-5 as a histone deacetylase kinase in the nucleus of cardiomyocytes. Proc Natl Acad Sci USA. 2008;105:12457–12462. doi: 10.1073/pnas.0803153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, Baccarini M, Vass JK, Kolch W, O'Neill E. RASSF1A elicits apoptosis through an MAST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell. 2007;27:962–975. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi J, et al. Expression of G protein-coupled receptor kinase 4 is associated with breast cancer tumourigenesis. J Pathol. 2008;216:317–327. doi: 10.1002/path.2414. [DOI] [PubMed] [Google Scholar]
- Metaye T, Menet E, Guilhot J, Kraimps JL. Expression and activity of g protein-coupled receptor kinases in differentiated thyroid carcinoma. J Clin Endocrinol Metab. 2002;87:3279–3286. doi: 10.1210/jcem.87.7.8618. [DOI] [PubMed] [Google Scholar]
- Michal AM, So CH, Beeharry N, Shankar H, Mashayekhi R, Yen TJ, Benovic JL. G Protein-coupled receptor kinase 5 is localized to centrosomes and regulates cell cycle progression. J Biol Chem. 2012;287:6928–6940. doi: 10.1074/jbc.M111.298034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CA, Milano SK, Benovic JL. Regulation of receptor trafficking by GRKs and arrestins. Annu Rev Physiol. 2007;69:451–482. doi: 10.1146/annurev.physiol.69.022405.154712. [DOI] [PubMed] [Google Scholar]
- Parameswaran N, Pao CS, Leonhard KS, Kang DS, Kratz M, Ley SC, Benovic JL. Arrestin-2 and G protein-coupled receptor kinase 5 interact with NFkappaB1 p105 and negatively regulate lipopolysaccharide-stimulated ERK1/2 activation in macrophages. J Biol Chem. 2006;281:34159–34170. doi: 10.1074/jbc.M605376200. [DOI] [PubMed] [Google Scholar]
- Patial S, Luo J, Porter KJ, Benovic JL, Parameswaran N. G-protein-coupled-receptor kinases mediate TNFalpha-induced NFkappaB signalling via direct interaction with and phosphorylation of IkappaBalpha. Biochem J. 2009;425:169–178. doi: 10.1042/BJ20090908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penela P, Rivas V, Salcedo A, Mayor F., Jr G protein-coupled receptor kinase 2 (GRK2) modulation and cell cycle progression. Proc Natl Acad Sci USA. 2010;107:1118–1123. doi: 10.1073/pnas.0905778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peregrin S, Jurado-Pueyo M, Campos PM, Sanz-Moreno V, Ruiz-Gomez A, Crespo P, Mayor F, Jr, Murga C. Phosphorylation of p38 by GRK2 at the docking groove unveils a novel mechanism for inactivating p38MAPK. Curr Biol. 2006;16:2042–2047. doi: 10.1016/j.cub.2006.08.083. [DOI] [PubMed] [Google Scholar]
- Pitcher JA, Hall RA, Daaka Y, Zhang J, Ferguson SS, Hester S, Miller S, Caron MG, Lefkowitz RJ, Barak LS. The G protein-coupled receptor kinase 2 is a microtubule-associated protein kinase that phosphorylates tubulin. J Biol Chem. 1998;273:12316–12324. doi: 10.1074/jbc.273.20.12316. [DOI] [PubMed] [Google Scholar]
- Pronin AN, Morris AJ, Surguchov A, Benovic JL. Synucleins are a novel class of substrates for G protein-coupled receptor kinases. J Biol Chem. 2000;275:26515–26522. doi: 10.1074/jbc.M003542200. [DOI] [PubMed] [Google Scholar]
- Pronin AN, Satpaev DK, Slepak VZ, Benovic JL. Regulation of G protein-coupled receptor kinases by calmodulin and localization of the calmodulin binding domain. J Biol Chem. 1997;272:18273–18280. doi: 10.1074/jbc.272.29.18273. [DOI] [PubMed] [Google Scholar]
- Romano D, Matallanas D, Weitsman G, Preisinger C, Ng T, Kolch W. Proapoptotic kinase MST2 coordinates signaling crosstalk between RASSF1A, Raf-1 and Akt. Cancer Res. 2010;70:1195–1203. doi: 10.1158/0008-5472.CAN-09-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gomez A, Humrich J, Murga C, Quitterer U, Lohse MJ, Mayor F., Jr Phosphorylation of phosducin and phosducin-like protein by G protein-coupled receptor kinase 2. J Biol Chem. 2000;275:29724–29730. doi: 10.1074/jbc.M001864200. [DOI] [PubMed] [Google Scholar]
- Sarnago S, Elorza A, Mayor F., Jr Agonist-dependent phosphorylation of G protein-coupled receptor kinase 2 (GRK2) by Src tyrosine kinase. J Biol Chem. 1999;274:34411–34416. doi: 10.1074/jbc.274.48.34411. [DOI] [PubMed] [Google Scholar]
- Shankar H, Michal A, Kern RC, Kang DS, Gurevich VV, Benovic JL. Non-visual arrestins are constitutively associated with the centrosome and regulate centrosome function. J Biol Chem. 2010;285:8316–8329. doi: 10.1074/jbc.M109.062521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherline P, Mascardo RN. Epidermal growth factor induces rapid centrosomal separation in HeLa and 3T3 cells. J Cell Biol. 1982a;93:507–512. doi: 10.1083/jcb.93.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherline P, Mascardo R. Epidermal growth factor-induced centrosomal separation: mechanism and relationship to mitogenesis. J Cell Biol. 1982b;95:316–322. doi: 10.1083/jcb.95.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherline P, Mascardo RN. Co-ordinate control of centrosomal separation and DNA synthesis by growth regulators. Exp Cell Res. 1984;153:109–120. doi: 10.1016/0014-4827(84)90453-1. [DOI] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D. Timing of centrosome separation is important for accurate chromosome segregation. Mol Biol Cell. 2012;23:401–411. doi: 10.1091/mbc.E11-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, et al. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. EMBO J. 2011;30:2233–2245. doi: 10.1038/emboj.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So CH, Michal AM, Mashayekhi R, Benovic JL. G protein-coupled receptor kinase 5 phosphorylates nucleophosmin and regulates cell sensitivity to polo-like kinase 1 inhibition. J Biol Chem. 2012;287:17088–17099. doi: 10.1074/jbc.M112.353854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- Suo Z, Cox AA, Bartelli N, Rasul I, Festoff BW, Premont RT, Arendash GW. GRK5 deficiency leads to early Alzheimer-like pathology and working memory impairment. Neurobiol Aging. 2007;28:1873–1888. doi: 10.1016/j.neurobiolaging.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Tiedemann RE, et al. Kinome-wide RNAi studies in human multiple myeloma identify vulnerable kinase targets, including a lymphoid-restricted kinase, GRK6. Blood. 2010;115:1594–1604. doi: 10.1182/blood-2009-09-243980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer M, Bohm M, Elce JS, Erdmann E, Lohse MJ. Altered expression of beta-adrenergic receptor kinase and beta 1-adrenergic receptors in the failing human heart. Circulation. 1993;87:454–463. doi: 10.1161/01.cir.87.2.454. [DOI] [PubMed] [Google Scholar]
- Woerner BM, Luo J, Brown KR, Jackson E, Dahiya SM, Mischel P, Benovic JL, Piwnica-Worms D, Rubin JB. Suppression of G-protein-coupled receptor kinase 3 expression is a feature of classical GBM that is required for maximal growth. Mol Cancer Res. 2012;10:156–166. doi: 10.1158/1541-7786.MCR-11-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi XP, Gerdes AM, Li F. Myocyte redistribution of GRK2 and GRK5 in hypertensive, heart-failure-prone rats. Hypertension. 2002;39:1058–1063. doi: 10.1161/01.hyp.0000019130.09167.3b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.