Higher-order structure of chromatin is essential for chromosome segregation and repetitive DNA stability. Monopolin recruits condensin to organize centromere DNA irrespective of the number of kinetochore–microtubule attachments. In addition, the role of monopolin in stabilizing repeat tracts observed in budding yeast is conserved in Candida albicans.
Abstract
The establishment and maintenance of higher-order structure at centromeres is essential for accurate chromosome segregation. The monopolin complex is thought to cross-link multiple kinetochore complexes to prevent merotelic attachments that result in chromosome missegregation. This model is based on structural analysis and the requirement that monopolin execute mitotic and meiotic chromosome segregation in Schizosaccharomyces pombe, which has more than one kinetochore–microtubule attachment/centromere, and co-orient sister chromatids in meiosis I in Saccharomyces cerevisiae. Recent data from S. pombe suggest an alternative possibility: that the recruitment of condensin is the primary function of monopolin. Here we test these models using the yeast Candida albicans. C. albicans cells lacking monopolin exhibit defects in chromosome segregation, increased distance between centromeres, and decreased stability of several types of repeat DNA. Of note, changing kinetochore–microtubule copy number from one to more than one kinetochore–microtubule/centromere does not alter the requirement for monopolin. Furthermore, monopolin recruits condensin to C. albicans centromeres, and overexpression of condensin suppresses chromosome segregation defects in strains lacking monopolin. We propose that the key function of monopolin is to recruit condensin in order to promote the assembly of higher-order structure at centromere and repetitive DNA.
INTRODUCTION
Centromere DNA is organized into higher-order chromatin, and this organization is critical for proper chromosome segregation. Numerous protein complexes, including cohesin, condensin, and monopolin, contribute to centromere organization (reviewed in Poon and Mekhail, 2011). Cohesin binds to regions flanking centromere DNA and promotes chromatin condensation and accurate chromosome segregation (Eckert et al., 2007). Condensin binding is enriched near centromeres in both Saccharomyces cerevisiae and Schizosaccharomyces pombe (D'Ambrosio et al., 2008; Tada et al., 2011).
The monopolin complex is important for meiotic and mitotic chromosome segregation in multiple fungal species. It was originally identified in S. cerevisiae and found to be essential for co-orienting kinetochores on sister chromatids in meiosis I (Toth et al., 2000). The S. cerevisiae meiotic monopolin complex includes the monopolin core complex (Csm1/Lrs4), Hrr25, and Mam1 (Rabitsch et al., 2003; Petronczki et al., 2006). Monopolin and condensin cooperate to establish centromere structure in meiosis I (Brito et al., 2010b). The monopolin core complex also localizes to kinetochores and enhances plasmid segregation accuracy during mitosis (Brito et al., 2010a). S. cerevisiae CSM1 and LRS4 exhibit synthetic genetic interactions with genes critical for mitotic chromosome segregation, further supporting a mitotic, as well as a meiotic, role for the monopolin core complex (Pan et al., 2006). In S. pombe, homologues of the monopolin core complex (Pcs1/Mde4) function during both mitosis and meiosis II to prevent merotelic attachments (Gregan et al., 2007). Cells lacking either the monopolin core complex or proteins required to establish centromeric heterochromatin have high frequencies of merotelic attachments (Rumpf et al., 2010). The functions of monopolin and condensin are closely linked in S. pombe. Both complexes promote kinetochore orientation, faithful sister chromosome segregation, and ribosomal DNA (rDNA) separation during mitosis (Nakazawa et al., 2008; Tada et al., 2011). Furthermore, monopolin recruits condensin to the centromere, and tethering of condensin to the centromere rescues many phenotypes of a monopolin-deletion mutant (Tada et al., 2011). Together these results highlight the importance of the monopolin core complex, and its recruitment of condensin, in accurate chromosome segregation.
The chromosome segregation phenotype of monopolin mutants is related to centromere structure. S. cerevisiae has small, point centromeres with a single kinetochore microtubule attachment per centromere (Winey et al., 1995; Joglekar et al., 2008). S. pombe has larger, regional centromeres, each with more than one kinetochore-microtubule attachment (Ding et al., 1993; Joglekar et al., 2008). S. cerevisiae monopolin mutants have stronger phenotypes in meiosis than in mitosis, whereas the monopolin complex in S. pombe is required for both mitotic and meiotic chromosome segregation. These observations led to the hypothesis that monopolin is primarily required to coordinate the segregation of centromeres with multiple kinetochore–microtubule attachments (Rabitsch et al., 2003).
Monopolin also functions at repetitive DNA other than centromeres. Repetitive DNA regions are packaged into higher-order chromatin structures that repress sister chromatid exchange and other mechanisms of recombination. In S. cerevisiae vegetative cells, the monopolin core complex, also known as cohibin, is important for rDNA stability and telomere maintenance (Huang et al., 2006; Mekhail et al., 2008; Chan et al., 2011). Tof2, a nucleolar component important for rDNA silencing, associates with Csm1 and Lrs4 to enhance condensin binding to rDNA repeats. In addition, LRS4 deletion results in subtelomeric DNA instability (Chan et al., 2011), and the CSM1 deletion strain exhibits increased telomere length (Askree et al., 2004). The role of the monopolin core complex at repetitive DNA, both rDNA and telomeres, has only been tested in S. cerevisiae and has not been explored in other organisms.
Several models have been proposed for the mechanisms of monopolin function. One model, based on structural data, is that it functions as a cross-linker. The S. cerevisiae Csm1/Lrs4 core complex has a V-shaped structure, with the globular domain of Csm1 containing a conserved patch that interacts with kinetochore proteins and the rDNA-associated protein Tof2 in vitro (Corbett et al., 2010). This V-shaped structure suggests that the monopolin core complex may function as a molecular clamp to cross-link kinetochore components and proteins bound to rDNA repeats. However, the proposal that monopolin is required for the alignment of multiple kinetochore–microtubule attachments has not been directly tested. An alternative model, that the primary function of monopolin is to recruit condensin, which in turn provides higher-order structure to DNA within centromere regions, was proposed recently based on experiments in S. pombe demonstrating that tethering of condensin bypasses the requirement for monopolin in chromosome segregation (Dudas et al., 2011; Tada et al., 2011).
We exploited the flexibility of kinetochore size in the yeast Candida albicans to test the requirement for the monopolin complex as a function of one or more than one kinetochore–microtubule/centromere. C. albicans has small, regional centromeres that span ∼4 kb and are inherited epigenetically (Baum et al., 2006). Normally, C. albicans centromeres associate with one kinetochore–microtubule attachment (Joglekar et al., 2008). On overexpression of CSE4, the C. albicans gene encoding the homologue of the centromere-specific histone H3 CENP-A, the majority of centromeres bind more than one kinetochore–microtubule complex (Burrack et al., 2011). The C. albicans genome also has a highly repetitive region found in one or two copies on all but one chromosome. Each of these major repeat sequences (MRSs) is composed of tandem repeats that span 10–50 kb and undergo shifts in repeat length, presumably due to recombination between tandem repeats and/or unequal sister chromatid recombination (Chibana and Magee, 2009).
Here we find that deletion or repression of monopolin in C. albicans causes defects in chromosome segregation, an increase in metaphase sister centromere separation, and increased recombination at rDNA, telomeres, and the MRSs. If monopolin was required specifically to cross-link chromosomes with multiple microtubules, then we would expect an increased requirement for monopolin in C. albicans cells having more than one kinetochore–microtubule/centromere. This was not the case: increasing the kinetochore–microtubule copy number did not affect the requirement for monopolin. Instead, the results support a model in which the primary role of the monopolin complex is to recruit condensin and organize DNA at regional centromeres. We also demonstrate that the monopolin core complex member Csm1 has critical roles in maintaining the integrity of a range of repetitive DNA tracts, including rDNA, telomeres, and MRSs, indicating that the role of monopolin in the maintenance of repetitive DNA tracts, which was previously observed in S. cerevisiae, is more widely conserved in fungi.
RESULTS
C. albicans Csm1, a monopolin core complex homologue, localizes to centromeres and kinetochores in actively dividing C. albicans cells
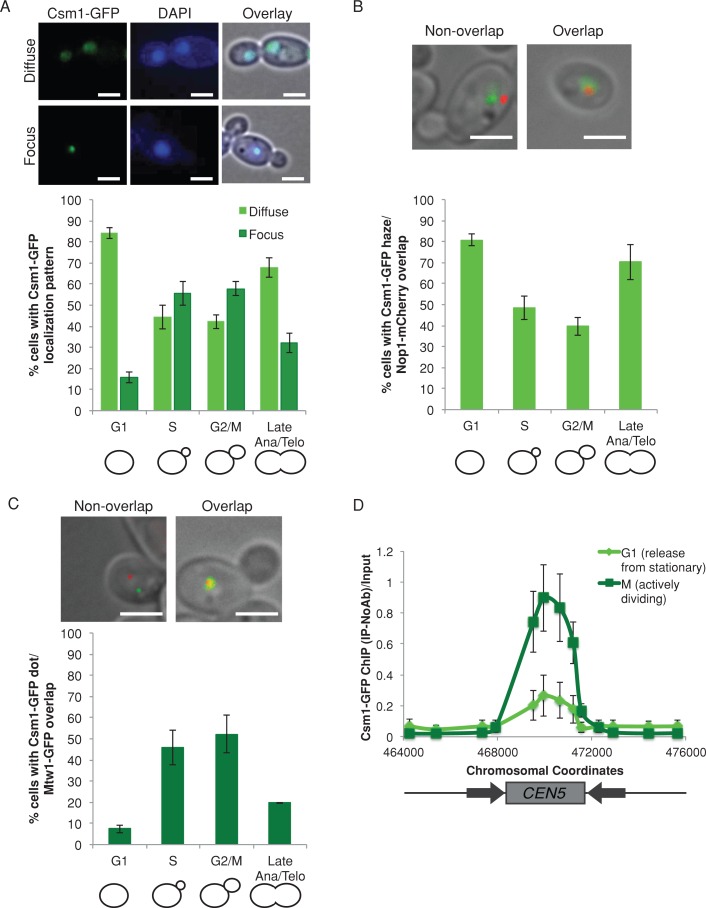
CSM1 (ORF19.7663) is the C. albicans gene most similar to the S. cerevisiae CSM1 monopolin complex subunit gene. We constructed a Csm1–green fluorescent protein (GFP) fusion protein to track the localization pattern of Csm1 in cells also stained with 4′,6-diamidino-2-phenylindole (DAPI) to demarcate the nucleus. Csm1-GFP localized both as a diffuse pattern coincident with the nucleus (Figure 1A, top) and a localized focus within the nucleus (Figure 1A, bottom). The localization was cell cycle dependent. The diffuse localization was seen in 84% of unbudded (G1) cells and 68% of cells with segregated nuclei (late anaphase/telophase); the focal localization was more prevalent in budded cells, with 56% of small-budded cells (S) and 58% of medium-budded cells (G2/M) having Csm1-GFP foci (Figure 1A). Thus the Csm1 localization pattern is consistent with monopolin binding throughout the nuclear DNA in unbudded cells and with more specific monopolin binding to kinetochores in actively dividing cells.
FIGURE 1:
Csm1 colocalizes with kinetochores in actively dividing cells. (A) Csm1-GFP (green) exhibited both diffuse (top) and focal (bottom) localization patterns within DAPI-stained nuclei (blue). Cells were imaged by fluorescence microscopy at 1000× total magnification. The percentage of cells in each cell cycle stage with each localization pattern was quantified. Data shown are mean ± SEM of three biological replicates. Scale bar, 5 μm. (B) Csm1-GFP (green) and Nop1-mCherry (red) were coexpressed and imaged by fluorescence microscopy at 1000× total magnification. The percentage of cells in each cell cycle stage exhibiting colocalization of the Csm1-GFP diffuse region with Nop1-mCherry was quantified. Data shown are mean ± SEM of three biological replicates. Scale bar, 5 μm. (C) Csm1-GFP (green) and Mtw1-mCherry (red) were coexpressed and imaged by fluorescence microscopy at 1000× total magnification. The percentage of cells in each cell cycle stage exhibiting colocalization of the Csm1-GFP focus with Mtw1-mCherry was quantified. Data shown are mean ± SEM of three biological replicates. Scale bar, 5 μm. (D) Anti-GFP ChIP analyzed with primers amplifying CEN5 for a CSM1-GFP strain released from stationary phase into YPA-glucose for 15 min (G1, light green) and 105 min (M, dark green). Data shown are mean ± SEM of three biological replicates.
We used colocalization of Csm1-GFP with other proteins tagged with mCherry to more carefully map the intracellular localization of Csm1. Because Csm1 colocalizes with the nucleolus in S. cerevisiae and S. pombe (Rabitsch et al., 2003), we examined the localization of Csm1 relative to the nucleolar marker Nop1-mCherry (Lavoie et al., 2008). Of interest, Csm1-GFP colocalized with Nop1-mCherry in 91% of cells with diffuse Csm1-GFP localization (Figure 1B). The diffuse Csm1-GFP signal usually extended beyond the Nop1-mCherry signal (Figure 1B, top right) and was similar to the DAPI localization patterns (Figure 1A). Therefore Csm1-GFP appears to be distributed throughout the nucleus, including the nucleolus in C. albicans.
During mitosis, monopolin binds to kinetochores in both S. cerevisiae (Brito et al., 2010a) and S. pombe (Rabitsch et al., 2003). Accordingly, we quantified the localization of Csm1-GFP foci relative to kinetochore protein Mtw1-mCherry. A majority (64%) of cells with a Csm1-GFP focus colocalized with an Mtw1-mCherry focus. Of note, approximately half of the S-phase and G2/M cells exhibited a Csm1-GFP focus that colocalized with the Mtw1-mCherry focus (46% of S-phase [small-budded] cells and 52% of G2/M [medium-budded] cells; Figure 1C).
We next asked whether Csm1 associated with centromere DNA during mitosis using chromatin immunoprecipitation (ChIP). In G1 phase (15 min after release from stationary phase, >90% unbudded cells), only weak binding of Csm1-GFP to centromere DNA was detected by ChIP (Figure 1D, light green). In contrast, in S and G2/M phases (105 min after release from stationary phase, >80% budded cells) we detected strong binding of Csm1-GFP to centromere DNA (Figure 1D, dark green). Csm1-GFP binding was restricted to the centromere core region, which is bound by inner and outer kinetochore proteins (Burrack et al., 2011). Therefore Csm1 associates with the core region of centromere DNA in C. albicans, and this is especially evident in actively dividing S- and G2/M-phase cells.
Csm1 is required for accurate chromosome segregation and cell cycle progression
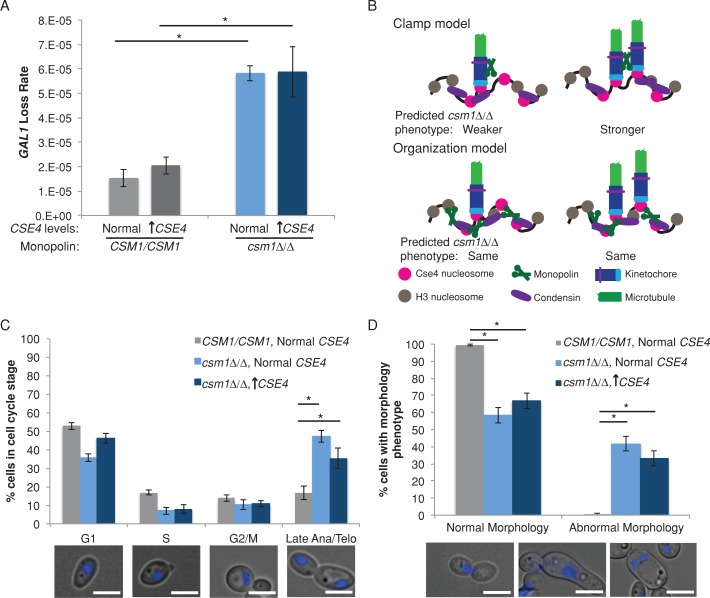
Having established that C. albicans Csm1 is present at kinetochores and centromere DNA in actively dividing cells, we asked whether the monopolin core complex is required for chromosome segregation or for cell cycle progression in wild-type C. albicans, which have a single kinetochore–microtubule. We followed marker loss using two counterselectable markers: loss of GAL1 was detected by resistance to 2-deoxygalactose (2-DOG), and loss of URA3 was detected by resistance to 5-fluoroorotic acid (5-FOA). Deletion of both copies of CSM1 resulted in increased loss of the heterozygous GAL1 marker on Chr1 (Figure 2A), suggesting an increased rate of chromosome loss. Similar results were obtained with conditional repression of CSM1, using the MET3 promoter to drive CSM1 expression in a strain with the other copy deleted. Growth of this strain under inducing conditions (media lacking methionine and cysteine) permitted expression of CSM1, whereas expression was dramatically reduced under repressing conditions (media with high levels of methionine and cysteine; Supplemental Figure S1A). Conditional depletion of CSM1 resulted in increased marker loss, detected with GAL1 on Chr1 and with the URA3 marker gene inserted on Chr3 (Supplemental Figure S1B). In addition, we measured chromosome segregation defects by observing segregation patterns of TetR-GFP bound to TetO sequences inserted immediately adjacent to CEN7. Abnormal segregation patterns, such as no TetR-GFP signal or >2 TetR-GFP foci/cell, were increased 11-fold in csm1Δ/Δ cells compared with control cells (Supplemental Figure S2).
FIGURE 2:
Requirement for CSM1 is not dependent on the number of kinetochore proteins and spindle microtubules in C. albicans. (A) Fluctuation analysis of loss of GAL1 in control and csm1Δ/Δ CSE4/PCK1p-CSE4 strains during growth in repressing conditions (YPA-glucose, normal expression) and activating conditions (YPA-succinate, overexpression). Loss of GAL1 was quantified by plating cells on nonselective media and on media containing 2-DOG. Colony counts were used to calculate the rate of loss per cell division. Results are mean ± SEM of the rates calculated from at least three experiments, each with eight cultures per condition. Significance was determined by analysis of variance (ANOVA) and Tukey posttest. *p < 0.05. (B) Schematic of models for monopolin function. (C) Control strains were grown in SDC-glucose, and csm1Δ/Δ CSE4/PCK1p-CSE4 strains were grown in repressing conditions (SDC-glucose, normal expression) and activating conditions (SDC-succinate, overexpression) for 6 h. Cells were stained with DAPI and imaged at 1000× total magnification with a DAPI filter set. The percentage of cells in each cell cycle stage was determined (bottom, representative cells at each cell cycle stage). Results are mean ± SEM of three biological replicates. Significance was determined by ANOVA and Tukey posttest. *p < 0.01. Scale bar, 5 μm. (D) Cells were grown and imaged as in B. The percentage of cells with abnormal nuclear segregation and/or morphology was determined (bottom, representative abnormal cells of three types: left, lagging DNA segregation; middle, highly elongated, pseudohyphal-like cells; right, multiple nuclei in a single cell). Results are mean ± SEM of three biological replicates. Significance was determined by ANOVA and Tukey posttest. *p < 0.01. Scale bar, 5 μm.
Marker loss was due to increase in whole-chromosome loss rather than increase in recombination events, as determined by single nucleotide polymorphism–restriction fragment length polymorphism (SNP-RFLP) analysis of a subset of strains. Whole-chromosome loss of heterozygosity occurred in 40 of 55 (73%) of csm1Δ/Δ 2-DOGR isolates compared with 5–10% whole-chromosome loss of heterozygosity previously observed in wild-type strains (Forche et al., 2011). Similarly, 17 of 24 (71%) of 2-DOGR isolates (Chr1) and 12 of 24 (50%) of 5-FOAR isolates (Chr3) underwent whole-chromosome loss of heterozygosity when CSM1 was repressed, which is significantly more than in cells in which CSM1 was expressed, in which 0 of 24 (0%) of 2-DOGR (Chr1) isolates and 2 of 23 (9%) of 5-FOAR isolates (Chr3) had whole-chromosome loss. Thus strains lacking monopolin, as well as strains that expressed very low amounts of monopolin, underwent whole-chromosome loss of heterozygosity, presumably via chromosome missegregation.
The number of kinetochore–microtubule attachments does not alter the requirement for monopolin
C. albicans chromosomes normally have 1 kinetochore–microtubule attachment/centromere. In C. albicans, Csm1 was required for accurate mitotic chromosome segregation; therefore this suggests that the monopolin complex function facilitates mitotic chromosome segregation even when there is only 1 kinetochore–microtubule/centromere. We tested the hypothesis by manipulating the number of assembled kinetochore complexes and kinetochore–microtubules using a conditional promoter system previously developed to increase C. albicans kinetochore copy number (Burrack et al., 2011). If Csm1 contributes primarily to the cross-linking of kinetochore–microtubules, we would expect to see a more severe chromosome segregation defect in strains overexpressing CSE4, which results in increased number of kinetochore–microtubules per centromere (Figure 2B). Growth of the cells in succinate activated the PCK1 promoter driving CSE4, resulting in high levels of CSE4 expression (Supplemental Figure S3A), increased binding of Cse4 (CaCENP-A) protein to centromere DNA (Supplemental Figure S3B), increased recruitment of the kinetochore protein Mtw1 to centromeres, and increased number of spindle microtubules per centromere (Supplemental Figure S3C). Approximately 60% of centromeres have two kinetochore–microtubule attachments when CSE4 is overexpressed (Supplemental Figure 3C; Burrack et al., 2011). As a control, growth in glucose (repressing the PCK1 promoter upstream of one copy of CSE4) yielded levels of CSE4 similar to unmodified controls and did not change the association of other kinetochore proteins with centromere DNA (Burrack et al., 2011).
Of importance, marker loss rates were similar in csm1Δ/Δ strains overexpressing CSE4 and csm1Δ/Δ deletion strains with normal levels of Cse4. Thus increased kinetochore–microtubule copy number did not affect the chromosome loss defect in cells lacking the monopolin complex in C. albicans (Figure 2A). This result is consistent with the idea that coordination of multiple kinetochore–microtubules per centromere is not a critical function of the monopolin core complex.
In addition to marker loss, we tracked nuclear segregation and cell cycle progression in DAPI-stained csm1Δ/Δ strains. As expected of cells with defects in chromosome segregation, deletion of the monopolin complex was associated with a significant increase in the fraction of large-budded cells with elongated nuclei stretched along the mother–bud axis, indicating that monopolin mutants are delayed in anaphase (Figure 2C). In addition, cells with abnormal morphologies, including cells with missegregated nuclei (Figure 2D, left), elongated pseudohyphal-like cells (Figure 2D, middle), and multinucleate cells (Figure 2D, right), were more prevalent in csm1Δ/Δ strains (Figure 2D). As before, we did not detect a difference in the phenotypes of csm1Δ/Δ cells with CSE4 overexpression relative to cells with normal CSE4 levels (Figure 2, C and D). Thus neither chromosome segregation phenotypes nor cell cycle delay phenotypes were different in monopolin mutants with a single or multiple kinetochore–microtubules.
Loss of monopolin leads to increased sister centromere separation
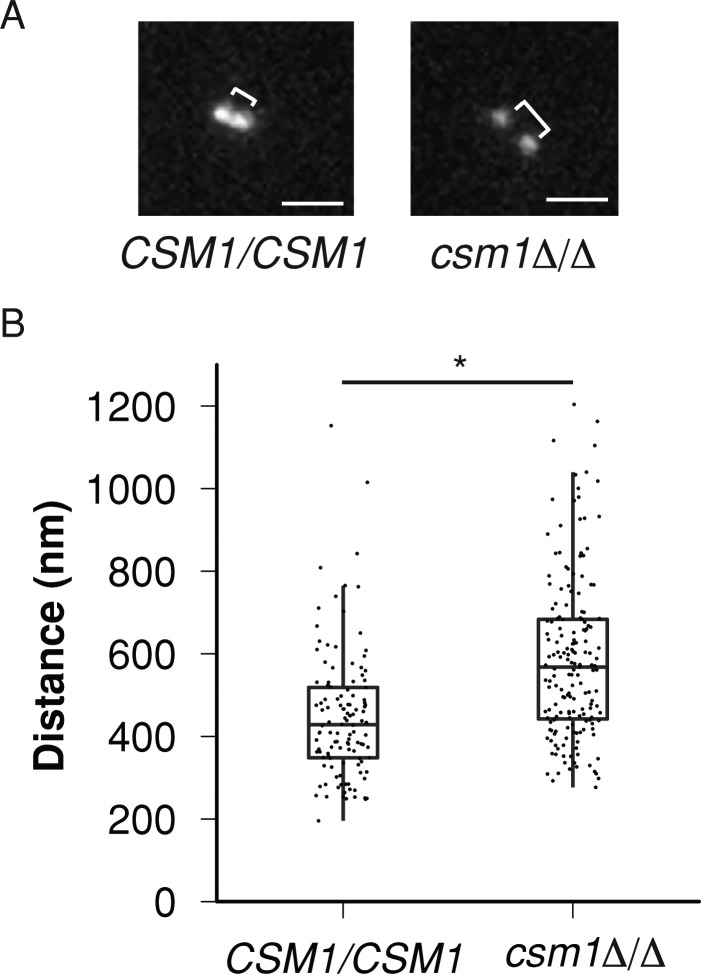
The metaphase separation distance between sister centromeres has been characterized as an indicator of chromosome stiffness, with increased separation distance suggestive of decreased chromosome stiffness (Ribeiro et al., 2009). To assess whether monopolin could affect chromosome stiffness during mitosis, we measured the separation distance between sister centromeres in wild-type and csm1Δ/Δ strains. We did this by inserting TetO sequences immediately adjacent to CEN7 in wild-type and csm1Δ/Δ strains and then expressing a TetR-GFP fusion protein from an intergenic region. We imaged metaphase cells (Figure 3A) and measured the distance between TetR-GFP spots on sister chromosomes, which approximates the distance between sister centromeres. The sister centromere separation distance was ∼30% longer in csm1Δ/Δ than in wild-type cells (587 ± 13 nm vs. 448 ± 12 nm, mean ± SEM, p << 0.001; Figure 3B). This suggests that deletion of CSM1 caused a decrease in chromosome stiffness during mitosis.
FIGURE 3:
Monopolin is required for proper metaphase sister centromere separation distance. (A) Control and csm1Δ/Δ TetO-CEN7/TetR-GFP cells were grown in SDC-glucose until cells reached log phase. Log-phase cells were adhered to concanavalin-coated glass coverslips and imaged in SDC media at 30°C. Distances between two TetR-GFP spots in metaphase (brackets) were then measured. Representative images are shown. Scale bar, 1 μm. (B) The distance between sister centromeres was determined by Gaussian fitting to the diffraction-limited TetR-GFP spots. Significance of centromere distance differences between wild-type and csm1Δ/Δ metaphase cells was determined using a Welch's two-sample, two-tailed t test conducted in R. *p << 0.001.
Monopolin recruits condensin to centromeres and kinetochores
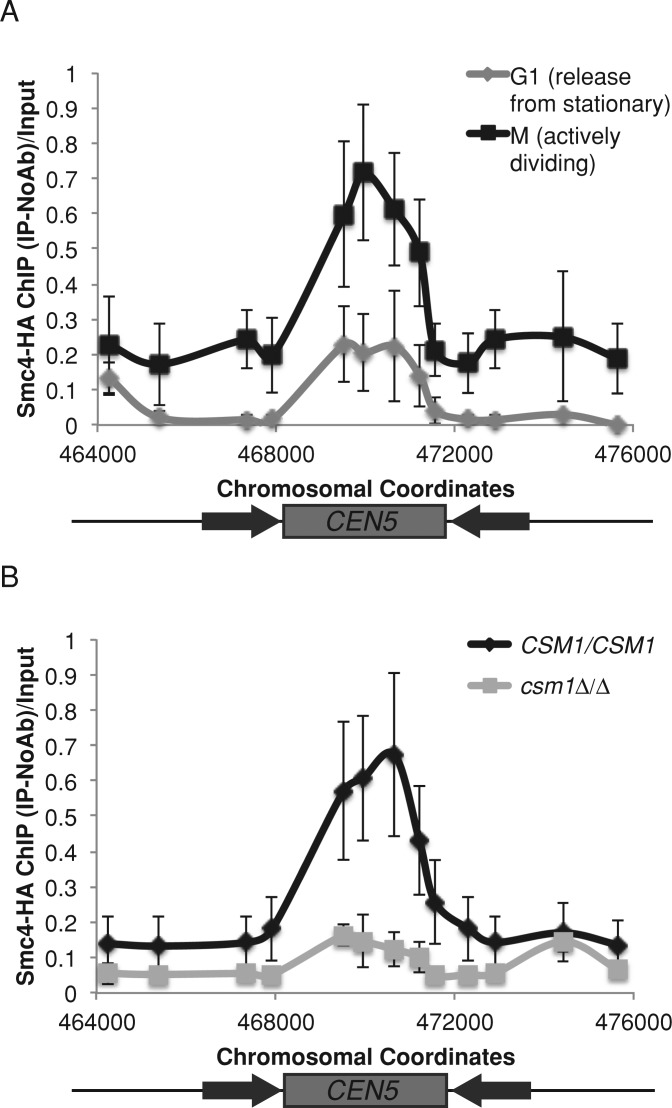
Condensin is believed to be an important contributor to chromosome stiffness (Ribeiro et al., 2009; Stephens et al., 2011). One explanation for decreased chromosome stiffness in the csm1Δ/Δ mutant is that condensin recruitment to the chromosomes might be impaired. Thus we asked whether monopolin recruits condensin in C. albicans by tracking condensin using ChIP of a hemagglutinin (HA) tag on Smc4, an ATPase subunit of the condensin complex. The DNA binding pattern and timing of Smc4-HA binding to centromere sequences mirrored the binding of Csm1-GFP (Figures 4A and 1D). Smc4-HA bound to the core centromere region, and the extent of Smc4-HA binding was higher in actively dividing S-phase and G2/M cells (105 min after release from stationary phase) than in G1 cells (15 min after release from stationary phase; Figure 4A). Of importance, Smc4-HA binding to centromeres in actively dividing cells was dependent on Csm1: in a csm1Δ/Δ strain, binding of Smc4-HA to the centromere central core was reduced (Figure 4B).
FIGURE 4:
Monopolin recruits condensin to centromere regions. (A) Anti-HA ChIP analyzed with primers amplifying CEN5 for an SMC4-HA strain released from stationary phase into YPA-glucose for 15 min (G1, light gray) and 105 min (M, black). Data shown are mean ± SEM of three biological replicates. (B) Anti-HA ChIP analyzed with primers amplifying CEN5 for SMC4-HA (black) and SMC4-HA csm1Δ/Δ (light gray) strains grown in YPA-glucose for 4 h. Data shown are mean ± SEM of three biological replicates.
In S. cerevisiae and human condensin mutants, Cse4/CENP-A incorporation is decreased at centromeres (Yong-Gonzalez et al., 2007; Samoshkin et al., 2009). In S. pombe monopolin mutants that have reduced condensin levels at the kinetochores, however, binding of Cnp1 (the S. pombe homologue of CENP-A) is not altered (Tada et al., 2011). To determine whether decreased Cse4 incorporation at centromeres contributed to the phenotypes of the monopolin mutants in C. albicans, we measured centromere binding of Cse4 in control and monopolin-deletion strains. Deletion of monopolin had no detectable effect on Cse4 incorporation at C. albicans centromeres (Supplemental Figure S4), consistent with observations in S. pombe.
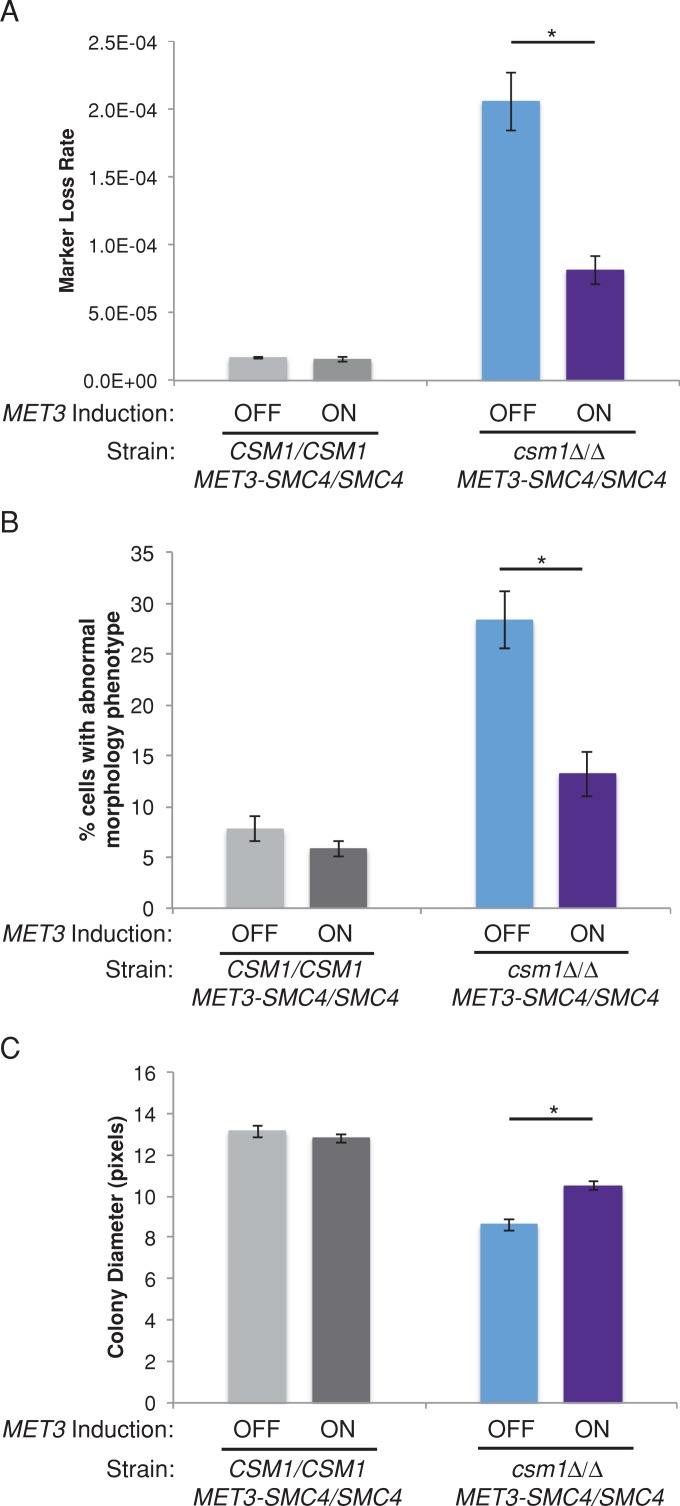
In S. pombe, targeting of condensin to centromeres via a fusion protein rescued monopolin-deletion phenotypes including growth defects and high rates of lagging chromosomes (Tada et al., 2011). Therefore we hypothesized that if a primary function of monopolin is to recruit condensin to centromeres, then overexpression of condensin may rescue the observed chromosome segregation defects. We constructed conditional CSM1/CSM1 and csm1Δ/Δ strains in which one copy of SMC4 was placed under control of the MET3 promoter. Growth of these strains under inducing conditions (media lacking methionine and cysteine) resulted in overexpression of SMC4 relative to noninducing conditions (media with high levels of methionine and cysteine) or a control strain (Supplemental Figure S5). Remarkably, overexpression of SMC4 significantly suppressed chromosome segregation defects in the monopolin-deletion strain: excess Smc4 reduced the rate of GAL1 marker loss relative to a strain with lower Smc4 levels (Figure 5A). When CSM1 was present, chromosome loss rates were not affected by changes in SMC4 expression. In addition, SMC4 overexpression reduced the proportion of csm1Δ/Δ cells that exhibited nuclear segregation defects and abnormal cell morphologies, measured by microscopy of DAPI-stained cells (Figure 5B). Furthermore, growth of csm1Δ/Δ cells, as measured by colony size, was increased in cells overexpressing SMC4 (Figure 5C). Overexpression of a single subunit of condensin was sufficient to counteract many of the defects in chromosome segregation and cell cycle progression characteristic of monopolin mutants, whereas cells with an intact monopolin complex did not exhibit sensitivity to changes in SMC4 expression. Thus an important function of monopolin is to promote assembly of higher-order chromosome structure in cooperation with, or via recruitment of, condensin.
FIGURE 5:
Overexpression of SMC4 suppresses the monopolin-deletion phenotype. (A) Fluctuation analysis of loss of GAL1 in CSM1/CSM1 and csm1Δ/Δ MET3p-SMC4/SMC4 strains during growth in repressing conditions (SDC+Met+Cys, normal expression) and activating conditions (SDC-Met-Cys, overexpression). Loss of GAL1 was quantified by plating cells on nonselective media and media containing 2-DOG to select for loss of GAL1. Colony counts were used to calculate the rate of loss per cell division. Results are mean ± SEM of rates calculated from at least three experiments, each with eight cultures per condition. Significance between inducing and noninducing conditions was determined by two-tailed paired t test. *p < 0.05. (B) CSM1/CSM1 and csm1Δ/Δ MET3p-SMC4/SMC4 strains were grown in repressing conditions (SDC+Met+Cys, normal expression) and activating conditions (SDC-Met-Cys, overexpression) for 6 h. Cells were stained with DAPI and imaged at 1000× total magnification with a DAPI filter set. The percentage of cells with abnormal nuclear segregation and/or morphology was determined as in Figure 2. Results are mean ± SEM of three biological replicates. Significance between inducing and noninducing conditions was determined by two-tailed paired t test. *p < 0.05. (C) Colony size was used to quantify growth of CSM1/CSM1 and csm1Δ/Δ MET3p-SMC4/SMC4 strains. Strains were grown on SDC+Met+Cys to repress the MET3 promoter (normal expression) and SDC-Met-Cys to activate the MET3 promoter (overexpression). Colony sizes were measured after 48 h incubation. Data shown are mean ± SEM of three experiments. Significance between inducing and noninducing conditions was determined by two-tailed paired t test. *p < 0.05.
Monopolin is essential for maintenance of repetitive DNA tracts in the genome
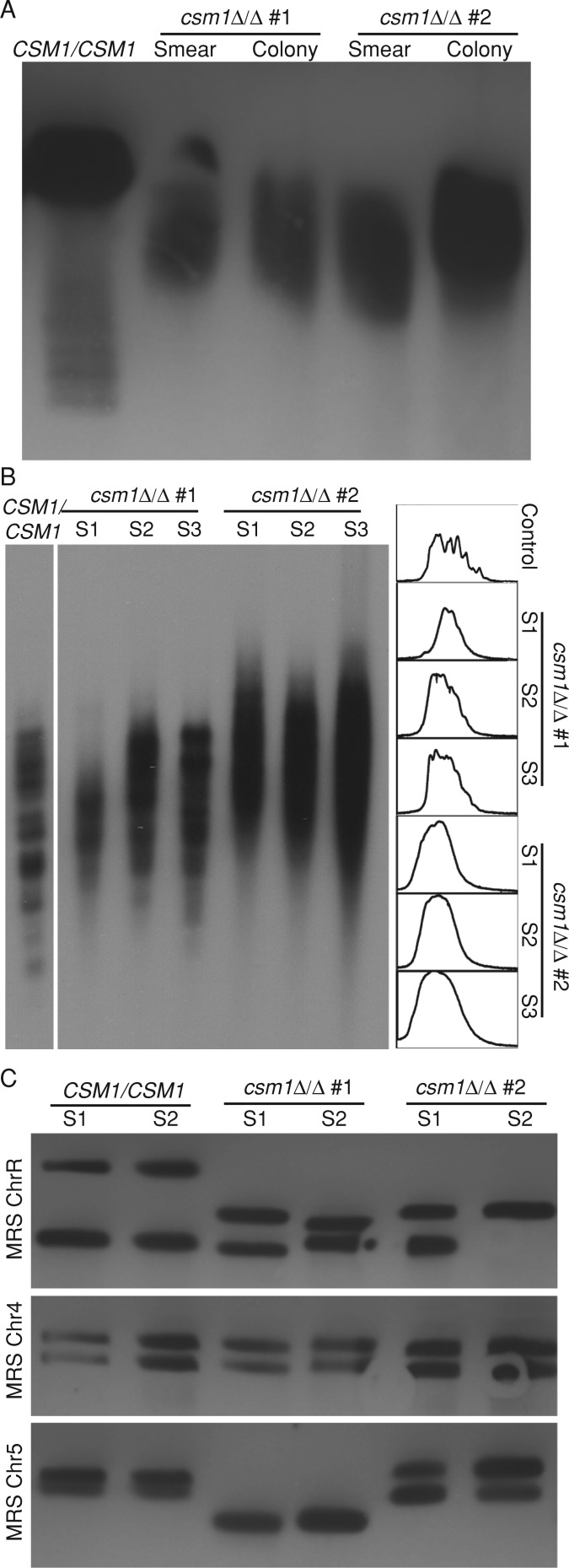
Monopolin is required for the maintenance of rDNA and telomere repeats in S. cerevisiae (Johzuka and Horiuchi, 2009), where it binds to the NTS1 region of rDNA (Huang et al., 2006). The function of monopolin at repetitive DNA in other organisms, however, has not been studied. To ask whether monopolin is also required to maintain repeats in C. albicans, we compared rDNA repeat tract length in csm1Δ/Δ monopolin-deletion strains relative to control CSM1 strains. Fragments containing the entire rDNA tract were digested with KpnI, separated by clamped homogeneous electric field (CHEF) gel electrophoresis, and detected by Southern blot. Two independently constructed csm1Δ/Δ strains both had shorter rDNA tracts relative to control strains (Figure 6A). Of importance, the rDNA was shorter and more variable within a population of csm1Δ/Δ cells than rDNA observed in wild-type cells, regardless of whether the culture was initiated from a mixed population or a single colony. This suggests that rDNA length changes more frequently in monopolin mutants.
FIGURE 6:
Monopolin is required for repeat tract length stability. (A) CHEF Southern blot of KpnI-digested genomic DNA plugs prepared from control and csm1Δ/Δ deletion strains was probed with a DIG probe targeting RDN18. (B) Southern blot of NlaIII- and AluI-digested genomic DNA prepared from control and csm1Δ/Δ deletion strains was probed with a DIG probe targeting telomeric repeat sequences. The intensity pattern of each lane from top to bottom was quantified (right). (C) CHEF Southern blot of XhoI-digested genomic DNA plugs prepared from control and csm1Δ/Δ deletion strains was probed with a DIG probe targeting chromosome-specific regions flanking the MRS on ChrR, Chr4, and Chr5.
Because Csm1-GFP colocalizes with the nucleolus, especially in unbudded G1 cells (Figure 1B), we next asked whether monopolin or condensin interacts with rDNA sequences. Both Csm1-GFP and Smc4-HA bound to the NTS1 region of rDNA as detected by ChIP of chromatin prepared from unbudded G1 cells (Supplemental Figure S6). This indicates that, at least during the early part of the cell cycle, monopolin and condensin associate with the rDNA repeats. These results are consistent with the idea that monopolin is nucleolar and its role in maintaining rDNA length is conserved from S. cerevisiae to C. albicans.
We next measured telomere stability by Southern blot with a probe specific to telomere repeat sequences in C. albicans. Genomic DNA from two independently constructed csm1Δ/Δ strains and a control CSM1 strain was digested, separated by gel electrophoresis, and analyzed by Southern blot with probes to the C. albicans telomere repeat (McEachern and Blackburn, 1994). Telomere length in C. albicans differs between chromosomes, such that telomeres from control cells appear as several discrete bands. In csm1Δ/Δ deletion strains, telomere length was increased and was also more variable, appearing more heterogeneous both visually (Figure 6B, left) and on densitometry scans of the gel (Figure 6B, right), suggesting altered regulation of recombination at telomeres (Basenko et al., 2011).
In addition to rDNA and telomeres, C. albicans has another class of nested repeats, which presents a unique opportunity to examine the role of the monopolin complex at different types of repetitive DNA sequences. One or two MRS tracts of 20–150 kb are present on seven of the eight C. albicans chromosomes in derivatives of SC5314, the progenitor of all C. albicans strains used in this study (Lephart et al., 2005). This variability in MRS length is derived from changes in the copy number of nested repeats within the MRS. Changes in MRS tract length, including the appearance of heterogeneity in MRS length between homologous chromosomes, as well as homozygosis of the two alleles, have been detected occasionally in wild-type strains (Chibana and Magee, 2009). We analyzed MRS stability by following the ChrR, Chr4, and Chr5 MRS tracts using CHEF Southern blots and probes specific to the unique sequences that flank each MRS tract over ∼100 generations of successive culturing. Of interest, csm1Δ/Δ monopolin-deletion strains had an increased frequency of repeat tract length changes on at least one of the six alleles (22 of 48 samples) compared with control cells (4 of 42 samples; Figure 6C). Thus monopolin affects the stability of repetitive tract lengths at the rDNA repeats, at the telomeres, and also at the internal, long tandem MRS repeats. Taken together, these results imply that monopolin is important for the maintenance of all types of repetitive DNA and is not specific to only rDNA or telomere repeats.
Monopolin has a more dramatic effect on the segregation of chromosomes carrying rDNA repeats
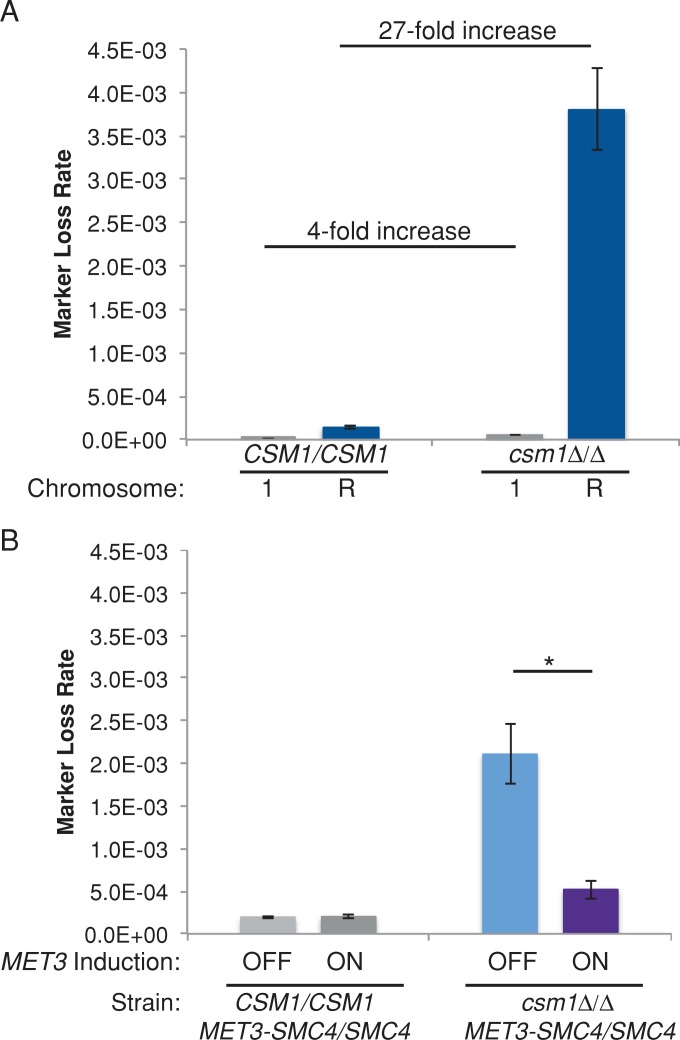
We considered the possibility that excess repeat tract recombination caused lagging chromosomes due to failure to resolve the recombination junctions. To ask whether, in monopolin mutants, the mechanisms that cause increased repeat tract recombination also impair chromosome segregation, we measured marker loss rates using URA3 inserted on ChrR, which contains all of the rDNA repeats in the C. albicans genome. Of note, the csm1Δ/Δ strain exhibited a 27-fold increase in the loss rate (5-FOAR) relative to control strains when URA3 had been inserted on ChrR (Figure 7A). This is in contrast to the fourfold increase in the loss of GAL1 on Chr1 (non–rDNA-containing chromosome) in a csm1Δ/Δ strain relative to control CSM1 strains (Figure 7A). The increase in marker loss on ChrR was primarily due to recombination rather than whole-chromosome loss, as measured by SNP-RFLP analysis: only 1 of 55 (2%) of 5-FOAR isolates had undergone whole-chromosome loss of heterozygosity.
FIGURE 7:
Monopolin deletion results in high levels of loss of heterozygosity on ChrR. (A) Fluctuation analysis of loss of GAL1 (Chr1) or URA3 (ChrR) in control and csm1Δ/Δ strains. Loss of GAL1 or URA3 was quantified by plating cells on nonselective media and media containing 2-DOG to select for loss of GAL1 or 5-FOA to select for loss of URA3. Colony counts were used to calculate the rate of loss per cell division. Results are mean ± SEM of rates calculated from at least three experiments, each with eight cultures per condition. (B) Fluctuation analysis of loss of URA3 (ChrR) in CSM1/CSM1 and csm1Δ/Δ MET3p-SMC4/SMC4 strains during growth in repressing conditions (SDC+Met+Cys, normal expression of MET3p-SMC4) and activating conditions (SDC-Met-Cys, overexpression of MET3p-SMC4). Loss of URA3 was quantified by plating cells on nonselective media and media containing 5-FOA to select for loss of URA3. Colony counts were used to calculate the rate of loss per cell division. Results are mean ± SEM of rates calculated from at least three experiments, each with eight cultures per condition. Significance between inducing and noninducing conditions was determined by two-tailed paired t test. *p < 0.05.
Furthermore, the increased loss of heterozygosity of ChrR (containing the rDNA repeat tracts) in a csm1Δ/Δ strain was suppressed approximately sixfold in a strain overexpressing condensin (Figure 7B). Therefore it appears that the role of monopolin in suppressing excessive recombination involves cooperation with, or recruitment of, condensin. Of interest, we also observed defects in nucleolar segregation in csm1Δ/Δ strains (Supplemental Figure S7), suggesting that the cell cycle defects observed in monopolin-deletion strains may be due to difficulty in resolving rDNA recombination.
To ask whether marker loss rates were increased for the chromosome carrying the rDNA in other yeasts, we inserted URA3 into intergenic regions on S. cerevisiae ChrXII (containing the rDNA repeat tract) and ChrX (non–rDNA-containing) and compared the loss rates in the two strains. The ChrXII URA3 loss rates were 10-fold higher in csm1Δ/Δ strains than in control strains, whereas the ChrX URA3 loss rates were similar in csm1Δ/Δ and control CSM1 strains (Supplemental Figure S8). Consistent with these observations, a large-scale screen for loss of heterozygosity in diploid S. cerevisiae also detected increased rates of marker loss on ChrXII (containing the rDNA repeat tract) in monopolin-deletion strains, whereas marker loss rates on other chromosomes were very similar to those in controls. Approximately 50% of the increase in marker loss on ChrXII was due to reciprocal recombination (Andersen et al., 2008). Thus, as in C. albicans, monopolin reduces the frequency of recombination specifically on the rDNA-containing chromosome in S. cerevisiae.
DISCUSSION
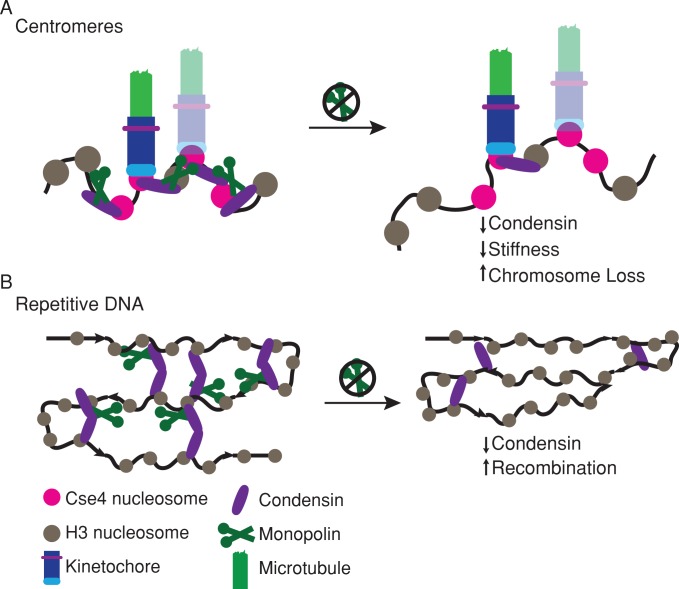
The monopolin complex structure suggests that it may function as a molecular clamp to cross-link kinetochore components and rDNA repeats (Corbett et al., 2010). An alternative model however, posits that the primary function of monopolin is to recruit condensin (Tada et al., 2011). Here we tested the effect of increasing the number of kinetochore–microtubule attachments on the requirement for the monopolin core complex in mitosis. We found that monopolin mutants have a strong phenotype even during mitosis with a single kinetochore–microtubule attachment and that increasing kinetochore copy number does not alter the requirement for the monopolin complex (Figure 2). Deletion of monopolin decreased apparent chromosome stiffness during mitosis (Figure 3) and reduced the recruitment of condensin to centromere regions (Figure 4). These results support a model in which monopolin recruits condensin to organize centromere DNA into the appropriate higher-order structure, irrespective of the number of kinetochore–microtubule attachments (Figure 8A). In addition, monopolin is required for the stability of different types of repetitive DNA sequences by preventing high levels of recombination (Figure 8B).
FIGURE 8:
Monopolin organizes DNA at centromeres and repetitive sequences. (A) At centromeres, monopolin (dark green) functions through the recruitment of condensin (purple) to organize centromere DNA. (B) Monopolin functions at repetitive DNA sites throughout the C. albicans genome, including rDNA repeats, telomere repeats, and the major repeat sequence, to maintain repeat stability.
In other yeasts, monopolin complex members interact physically with condensin in two-hybrid assays (Wysocka et al., 2004; Johzuka and Horiuchi, 2009; Tada et al., 2011). Furthermore, overexpression of a single condensin subunit bypassed chromosome segregation defects associated with knockdown of the telomere-associated Ccq1 protein, a protein that cooperates with condensin at S. cerevisiae telomeres (Motwani et al., 2010). On the basis of the reduced amount of condensin at centromere DNA in csm1Δ/Δ mutants (Figure 4), we infer that monopolin is required to recruit condensin in C. albicans. Of importance, overexpression of C. albicans condensin also suppressed the chromosome segregation and cell cycle progression defects associated with loss of monopolin (Figure 5). Taken together, the genetic and phenotypic results here are consistent with the idea that monopolin recruits condensin to the centromere to maintain proper chromosome segregation dynamics.
Condensin proteins are components of the chromatin spring necessary for chromosome segregation (Stephens et al., 2011) and are hypothesized to contribute to the stiffness of the chromatin spring (Haase et al., 2012; Stephens et al., 2013). Increased distance between centromeres during metaphase is believed to reflect reduced stiffness of centromeric chromatin or loss of sister centromere cohesion (Goshima et al., 1999). This occurs in csm1∆/∆ strains (Figure 3) and is consistent with the elongation of S. pombe spindles in monopolin mutants at the metaphase-to-anaphase transition (Choi et al., 2009). Reduced stiffness correlates with decreased chromosome segregation accuracy. This is believed to be due to reduced structural integrity at the centromere, which in turn can arise from multiple types of defects. For example, reduced stiffness could cause defects in the positioning of multiple centromeric nucleosomes, kinetochore assembly, or the efficiency of checkpoint or error correction proteins, such as Aurora B.
The increased mitotic requirement for the monopolin complex in S. pombe compared with S. cerevisiae was proposed to be due to the presence of more than one kinetochore–microtubule attachment/centromere (Gregan et al., 2007; Brito et al., 2010a). This model, however, is inconsistent with the data we obtained in C. albicans mitosis. Although we cannot exclude the possibility that monopolin functions as a clamp in some situations, such as meiosis (which has not been observed to occur in C. albicans; Butler et al., 2009), our data are more consistent with a model in which monopolin organizes DNA at centromeres and repetitive DNA by recruiting condensin.
We posit that the increased requirement for monopolin during mitosis in C. albicans and S. pombe relative to S. cerevisiae is due to the need for correct positioning of multiple centromeric nucleosomes within the centromeric core and/or organization of longer centromere DNA sequences, perhaps including their flanking repeats. Consistent with this, the severity of the mitotic phenotype in monopolin mutants increases with increased centromere DNA length: S. cerevisiae has short (∼<200 base pair) point centromeres, C. albicans has regional centromeres of intermediate length (∼3- to 5-kb core), and S. pombe has longer regional centromeres (∼10-kb core). Although the exact number of Cse4/CENP-A nucleosomes per centromere remains controversial (Coffman et al., 2011; Lawrimore et al., 2011; Henikoff and Henikoff, 2012; Aravamudhan et al., 2013), the idea that there are more CENP-A nucleosomes per centromere in C. albicans and S. pombe than in S. cerevisiae is quite clear (Joglekar et al., 2008). Another difference between centromeres in these three organisms is the degree of repetitive DNA. S. cerevisiae centromeres are located within nonrepetitive DNA, whereas S. pombe centromeres are flanked by long tracts of tandem repeats (up to ∼45 kb/arm; Allshire and Karpen, 2008). C. albicans centromeres again have an intermediate structure: they are flanked by short (120 base pairs to 2.2 kb) direct and/or inverted repeats (Supplemental Figure S7). Given the role of monopolin at other repeat sequences (Figure 6; Huang et al., 2006; Johzuka and Horiuchi, 2009), its role at centromeres could be to organize repeats into a higher-order chromatin structure necessary for centromere function during mitosis.
Monopolin relocalization from a diffuse distribution in G1 cells to a sharp focus that colocalizes with Mtw1 specifically in actively dividing cells suggests that monopolin is actively recruited to kinetochores. This localization pattern is consistent with work in other yeasts. In S. cerevisiae, condensin localizes to the nucleolus throughout the cell cycle and to kinetochores beginning at S phase and continuing through anaphase (Bachellier-Bassi et al., 2008). In S. pombe, condensin localizes to the nucleus throughout the cell cycle and is enriched in binding to centromeric chromatin during mitosis when Aurora B is active (Tada et al., 2011). We assume that the diffuse nuclear and nucleolar monopolin should be available to bind rDNA, telomeres, and MRS repeats; indeed, monopolin and condensin bound to rDNA repeats by ChIP, although binding levels were lower than at centromeres. Differences in the affinity of monopolin for centromeres (where it forms a bright, focused spot) and rDNA (where binding appears more diffuse) could account for the reduced ChIP signal. Monopolin may bind to only a subset of the repeat sequences, and the ChIP signal may be diluted by unbound rDNA repeat copies. Consistent with this, in S. pombe monopolin (Pcs1/Mde4) bound more tightly to centromere DNA than to nucleolar chromatin (Gregan et al., 2007). Thus the localization of monopolin and condensin is cell cycle regulated to promote accurate chromosome segregation and maintenance of repetitive sequences.
The monopolin core complex is critical for maintenance of the length of rDNA repeats, telomeres, and the MRS. In S. cerevisiae, the monopolin core complex is important for rDNA stability and telomere maintenance (Huang et al., 2006; Mekhail et al., 2008; Chan et al., 2011). Our observation that deletion of the monopolin complex also resulted in repetitive DNA instability in C. albicans implies that the function of monopolin at repetitive DNA is evolutionarily conserved beyond the Saccharomyces clade. Furthermore, the role of monopolin in maintenance of the MRS, a repetitive sequence not found in S. cerevisiae, supports the idea that monopolin has a general role at repetitive DNA sequences and not just at rDNA and telomeres.
We found that the monopolin organization of repetitive DNA is important to prevent recombination. Marker loss rates were significantly higher for markers on the rDNA-containing chromosome in C. albicans (Figure 7) and also in S. cerevisiae (Supplemental Figure S8; Andersen et al., 2008) than for markers on other chromosomes, and in both organisms, the marker loss was due to increased recombination rather than chromosome loss. Nonetheless, defects in nucleolar segregation were also evident in the csm1Δ/Δ mutant, suggesting that segregation of rDNA and the rDNA-containing chromosome was also impaired (Supplemental Figure S7). This differs from what was seen in S. pombe, in which all three chromosomes are approximately equally likely to lag when monopolin is disrupted (Gregan et al., 2007). Condensin mutants also do not show rDNA-specific chromosome segregation defects in S. pombe (Tada et al., 2011) or vertebrate cells (Vagnarelli et al., 2006). The much longer repeat regions flanking each S. pombe centromere may have a more dramatic effect on segregation of each chromosome, thus reducing the relative effect of rDNA seen in S. cerevisiae and C. albicans. Furthermore, we were unable to detect significant changes in rDNA length in S. pombe Δpcs1 and Δmde4 strains. One possibility is that differences in chromatin structure, such as the presence of canonical heterochromatin, reduce the requirement for the monopolin complex in the maintenance of rDNA repeats in S. pombe.
In C. albicans, recombination at the very long (∼2 Mb) rDNA repeats impaired chromosome segregation, whereas recombination at the shorter (∼50–100 kb) MRS sequences did not have a detectable chromosome segregation effect. To ask whether the MRS affected chromosome loss in a monopolin-dependent manner, we compared loss rates for Chr1 (a chromosome that contains an MRS) and Chr3 (a chromosome that does not contain an MRS) in conditional CSM1 depletion strains and controls. Monopolin depletion resulted in segregation defects for both chromosomes, and the severity of the defect was similar for the MRS-containing and the non–MRS-containing chromosome (Supplemental Figure S1). This result supports the idea that monopolin is important for kinetochore function and that the role of monopolin in maintenance of shorter repeat sequences, such as the MRS and telomeres, does not make a major contribution to chromosome stability.
Taken together, our results support a model in which monopolin organizes centromeric DNA to promote accurate chromosome segregation and proper centromere stiffness via the recruitment of condensin to regional centromeres regardless of the number of kinetochore–microtubule attachments (Figure 8A). In addition, monopolin is required for the maintenance of repetitive DNA sequences in C. albicans (Figure 8B) by preventing excess recombination, and this has implications for genome stability as well.
MATERIALS AND METHODS
Strain construction
Strains are listed in Supplemental Table S1. Lithium acetate transformation of linearized plasmids or PCR products with at least 70 base pairs of homology to the targeted gene was used for strain construction. Briefly, strains to be transformed were inoculated in YPA (yeast extract–peptone–adenine)-glucose and grown at 30°C for 16–18 h. Cultures were then diluted 1:166 in YPA-glucose and grown at 30C for 3–4 h. Cells were washed with water, and then TELiAc (10 mM Tris, pH 7.5, 1 mM EDTA, 100 mM lithium acetate [LiAc]) and incubated in TELiAc with transformation DNA and 50 μg of sheared salmon sperm DNA (Ambion, Austin, TX) for 30 min. Four volumes of PLATE mix (40% polyethylene glycol, 10 mM Tris, pH 7.5, 1 mM EDTA, 100 mM LiAc) were then added, and the transformation mix was incubated for 16–18 h at 20–24°C. Transformations were heat shocked at 42°C for 1 h and then plated on selective media, with the exception of NAT1 marker transformations, which were recovered on nonselective media for 6 h before replica plating to selective media containing 400 μg/ml nourseothricin (Werner BioAgents, Jena, Germany). Strains were checked by PCR of genomic DNA.
For the construction of the UAU knockouts of CSM1 the URA3-ARG4-URA3 (UAU) construct was amplified via PCR from plasmid pBME101 (Enloe et al., 2000) using primers with flanking 5′ and 3′ ends containing 70–base pair homology to the targeted genes of interest. Insertion of the UAU cassette at the correct location was confirmed by PCR. Recombination events resulting in Arg+Ura+ cells were selected for by streaking colonies of confirmed transformants onto SDC (synthetic dextrose complete)-Arg-Ura. Recombination and absence of native CSM1 gene were verified with PCR and by Southern blot. To construct the PCK1p-CSE4 strains, either the URA3-PCK1p-CSE4 plasmid (Sanyal and Carbon, 2002) was linearized with EcoRV or the HIS1-PCK1p-CSE4 plasmid was linearized with HindIII. Integration into the CSE4 upstream region was confirmed by PCR. Nop1-GFP, Csm1-GFP, Mtw1-GFP, and Tub1-GFP strains were tagged at the C-terminus with PCR-mediated transformation. Primers containing at least 70–base pair homology to the targeted gene were used to amplify the GFP and marker from the C-terminal GFP-tagging plasmids with URA3, HIS1, or NAT1 markers (Gerami-Nejad et al., 2001, 2012). Nop1-mCherry and Mtw1-mCherry strains were tagged at the C-terminus with PCR-mediated transformation. Primers containing at least 70–base pair homology to the targeted gene were used to amplify the mCherry and marker from the C-terminal mCherry-tagging plasmids (Gerami-Nejad et al., 2012). Smc4-HA strains were tagged at the C-terminus with PCR-mediated transformation. Primers containing at least 70–base pair homology to the targeted gene were used to amplify the HA tag and marker from the C-terminal HA-tagging plasmids (Gerami-Nejad et al., 2009). For MET3p conditional CSM1 strains, one copy of CSM1 was deleted by amplifying HIS1 with primers containing homology to the regions immediately 5′ and 3′ to the gene of interest. The other copy of CSM1 was placed under control of the MET3p by amplifying the NAT1 marker and MET3p region only from a NAT1-MET3p-GFP plasmid (similar to the URA3-MET3p-GFP plasmid described in Gerami-Nejad et al., 2004). For the MET3p conditional SMC4 overexpression strains, one copy of SMC4 was left intact and the other copy was placed under control of the MET3p by amplifying the NAT1 marker and MET3p region only from a NAT1-MET3p-GFP plasmid.
To construct TetO-CEN7/TetR-GFP strains, TetR under the control of the SNU114 promoter was fused with GFP into the plasmid pDIS3 (Gerami-Nejad et al., 2013) by in vivo recombination in S. cerevisiae. The plasmid was recovered, digested with SfiI to isolate the targeting sequences and insert, and transformed into strain SN76. Insertion of the TetR-GFP-NAT1 cassette into the intergenic region between ORF19.1963 and ORF19.1961 in the C. albicans genome was confirmed by PCR. URA3 and surrounding plasmid sequences were inserted adjacent to CEN7. pSR8 (Rohner et al., 2008) was modified for use in C. albicans by swapping out the S. cerevisiae HIS3 gene and replacing it with C. albicans HIS1. This plasmid was linearized with AscI and transformed into the URA3-CEN7/TetR-GFP strain. Transformants were selected on media lacking histidine and then replica plated to media lacking uridine to confirm loss of the URA3 marker flanked by the targeting sequences. Insertions were also confirmed by PCR. Approximately 3 kb of TetO sequences were inserted adjacent to CEN7. CSM1 deletion by UAU was performed in this strain as described.
S. cerevisiae strains BY4743 and csm1Δ/Δ were obtained from the S. cerevisiae diploid deletion collection (www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html). URA3 was targeted to intergenic regions on ChrX and ChrXII by PCR-mediated transformation. Primers containing at least 40–base pair homology to the targeted region were used to amplify the URA3 marker from pRS316 (Sikorski and Hieter, 1989).
Microscopy
For localization studies, strains were inoculated into SDC-glucose and grown at 30°C for 16–18 h. Cultures were then washed and diluted 1:100 into SDC-glucose. For strains with PCK1 conditional promoters, cultures were washed and diluted into repressing conditions (SDC-glucose) and activating conditions (SDC-succinate) and grown at 30°C for 6 h. For conditional MET3 strains, cultures were washed and diluted 1:50 into SDC-glucose without methionine and cysteine (-Met-Cys) or SDC-glucose with added methionine and cysteine (+Met+Cys) and grown at 30°C for 6 h. DNA was stained with 15 μg/ml DAPI in phosphate-buffered saline (PBS) for 10 min before imaging. Cells were imaged with a Nikon Plan Apo 100×/1.4 numerical aperture (NA) objective on a Nikon E600 microscope (Nikon, Melville, NY) with differential interference contrast, DAPI, GFP, and Texas red filter sets as appropriate, using constant exposure times and scaling for each strain. Images were acquired using MetaMorph software (Molecular Devices, Sunnyvale, CA). Quantification of Mtw1-GFP and Tub1-GFP in csm1Δ/Δ PCK1p-CSE4 strains was performed as previously described (Burrack et al., 2011).
To compare centromere separation distances during metaphase between wild-type and csm1Δ/Δ cells, we imaged TetO-CEN7/TetR-GFP cells using a Nikon Eclipse Ti microscope equipped with a Nikon CFI Apochromat 100×/1.49 NA oil objective and an iXon3 EMCCD camera (Andor, Belfast, United Kingdom). Log-phase cells were adhered to concanavalin-coated glass coverslips and imaged in SDC media at 30°C. We chose metaphase cells by including only cells with two distinct centromere spots, indicative of a bipolar spindle, and also by excluding cells in which those spots were separated by >1300 nm, which we chose as a conservative upper limit to the amount of separation possible in a metaphase cell. We measured the distance between centromeres, using a custom Matlab script, with nanometer precision by taking advantage of the fact that the location of a fluorescent particle can be precisely determined by fitting a Gaussian distribution to the captured point-spread function of the particle (Jaqaman et al., 2008). To conduct this fitting in each image, we rotated the image so that the centromeres were aligned on the horizontal axis. Next fluorescence was integrated in a 15-pixel column for each location along the horizontal vector on which the centromeres were aligned. The resulting one-dimensional vector of fluorescence intensities were fitted with a two-mixture Gaussian mixture model using the Matlab function gmdistribution.fit(), which returned the locations of each centromere. We compared the distances between centromeres in wild-type and csm1Δ/Δ metaphase cells using a Welch's two-sample, two-tailed t test conducted in R.
ChIP of Cse4, Csm1-GFP, and Smc4-HA
ChIP was performed essentially as described in Ketel et al. (2009), with the exception that an additional fixation step using 10 mM dimethyl adipimidate (Pierce Biotechnology, Rockford, IL) in PBS for 45 min was added before fixation in 1% formaldehyde. ChIP was performed using rabbit anti-Cse4 (CaCENP-A) antibodies (Ketel et al., 2009), mouse anti-GFP antibodies (Roche Applied Science, Indianapolis. IN), and mouse anti-HA antibodies. DNA pull-down efficiency was measured by quantitative PCR using the Universal Probe Library (Roche Applied Science) with a LightCycler 480 PCR machine (Roche Applied Science) according to the manufacturer's instructions. Enrichment was calculated as relative quantification of (+Ab/Input) − (−Ab/Input) using the second-derivative maximum to determine CT values and corrections for primer efficiency values with the LightCycler 480 software (Roche Applied Science).
Southern blot
To confirm the loss of CSM1 from csm1Δ/Δ UAU deletion strains, genomic DNA was digested with SpeI for 18–20 h at 37°C. The digested genomic DNA was run on a 0.8% agarose gel, transferred to a nylon membrane (GE Osmonics, Minnetonka, MN), and probed with digoxigenin (DIG) probes binding the wild-type copy of CSM1 as described in Ketel et al. (2009).
We measured rDNA length, MRS length, and telomere length by inoculating cultures in 2 ml of YPA-glucose with strains directly cultured from storage at −80°C and with strains passaged on YPA-glucose every day for 10 d. The cultures were grown for 16–18 h at 30°C.
To measure rDNA length and MRS length, we prepared plugs for CHEF gel electrophoresis. A total of 0.5 ml of a stationary-phase culture in YPA-glucose was harvested and washed once with 1 ml of 50 mM EDTA, pH 8.0, and resuspended in 20 μl of EDTA containing 10 mg/ml zymolyase. Three hundred microliters of 1% low-melting-point agarose in 50 mM EDTA, pH 8.0, at 50°C was added and gently mixed. The mixture was then transferred into a sample mold. On solidification, the agarose blocks were incubated for 24 h at 37°C in 3 ml of LET (0.5 M EDTA, 0.01 M Tris-HCl, pH 7.5). The LET was replaced with 0.4 ml of NDS (0.5 M EDTA, 0.01 M Tris-HCl, pH 7.5, 1% N-lauroyl sarcosine, 2 mg/ml proteinase K) and incubated for 48 h at 50°C. The plugs were washed once with 50 mM EDTA, pH 8.0, and stored at 4°C in 50 mM EDTA, pH 8.0. Plugs were cut to size, digested with KpnI (rDNA) or XhoI (MRS) for 18–20 h, and then separated using pulse-field gel electrophoresis as described in Lephart et al. (2005). The conditions for separation were as follows: 1% Megabase agarose gel, 3–8 s, 6 V/cm, 120° included angle for 14 h, and then 8–30 s, 5 V/cm, 120° included angle for 6 h. DNA was then transferred to a nylon membrane (GE Osmonics), probed with DIG probes binding RDN18 (rDNA) and chromosome-specific regions flanking MRS DNA (MRS), and hybridized as described in Ketel et al. (2009).
To measure telomere length, we prepared genomic DNA and digested the DNA with NlaIII and AluI for 2 h at 37°C (Hsu et al., 2007). The digested genomic DNA was run on a 0.8% agarose gel, transferred to a nylon membrane (GE Osmonics), and probed with DIG probes binding the telomere repeat sequence. DIG probes were constructed via PCR of a plasmid containing telomere repeats (pBSF) from Michael McEachern (Department of Genetics, University of Georgia, Athens, GA) and hybridized as described in Ketel et al. (2009). Lanes were analyzed using ImageJ (National Institutes of Health, Bethesda, MD).
Fluctuation analyses of chromosome loss rates
Fluctuation analysis of loss rates was performed as described (Spell and Jinks-Robertson, 2004) using the method of the median (Lea and Coulson, 1949). Briefly, strains were streaked for single colonies and grown on SDC-Uri for 2 d at 30°C. Per strain, eight independent colonies were inoculated into 1 ml of liquid nonselective medium (YPA-glucose, YPA-succinate, SDC-Met-Cys, or SDC+Met+Cys) and grown overnight at 30°C with shaking. Cultures were harvested by centrifugation and washed once in 1 ml of sterile water. Dilutions were plated onto nonselective YPA-glucose for total cell counts and selective media (SD+FOA for URA3 loss or SC+2-DOG for GAL1 loss; Gold Biotechnology, St. Louis, MO). Plates were incubated at 30°C for 2–3 d, and colony counts were used to calculate the rate of FOAR/cell division and 2-DOGR/cell division (Spell and Jinks-Robertson, 2004).
Reverse transcriptase quantitative PCR
Strains were inoculated into YPA-glucose and grown at 30°C for 16–18 h. Cultures were then diluted 1:100 into repressing conditions (YPA-glucose for PCK1p; SDC+Met+Cys for MET3p) and activating conditions (YPA-succinate for PCK1p; SDC-Met-Cys for MET3p) and grown at 30°C for 6 h. RNA was prepared using the MasterPure yeast RNA purification kit (Epicentre Biotechnologies, Madison, WI) according to the manufacturer's instructions. RNA was treated with DNase (Epicentre Biotechnologies) to remove contaminating genomic DNA. cDNA was prepared using the ProtoScript M-MuLV First Strand cDNA Synthesis Kit (New England BioLabs, Ipswich, MA) according to the manufacturer's instructions with oligo dT primers. cDNA was measured by quantitative PCR using the Universal Probe Library (Roche Applied Science) with a LightCycler 480 PCR machine (Roche Applied Science) according to the manufacturer's instructions. Expression was calculated as the amount of CSE4 cDNA, CSM1 cDNA, or SMC4 cDNA relative to the amount of TEF1 cDNA in the same sample using the second-derivative maximum to determine CT values and corrections for primer efficiency values with the LightCycler 480 software (Roche Applied Science).
Supplementary Material
Acknowledgments
We acknowledge Maryam Gerami-Nejad for assistance with strain construction, Damien Tank for technical assistance, Tamar Lahav and Darren Abbey for helpful analysis of C. albicans centromere repeat sequence arrangements, and Meleah Hickman for valuable comments on the manuscript. We also thank David Kirkpatrick for strains from the S. cerevisiae diploid deletion collection, Michael McEachern for the telomere repeat plasmid, Quinn Mitrovich and Sandy Johnson for the TetR construct, and Susan Gasser for the TetO repeat construct. This work was supported by Ruth L. Kirschstein National Research Service Award Fellowship F32 AI800742, a 2011 Williston Postdoctoral Fellowship, and Grant PF-12-108-01-CCG from the American Cancer Society to L.S.B. and National Institutes of Health/National Institute of Allergy and Infectious Diseases Grant AI075096 to J.B. M.K.G. is supported by the Pew Charitable Trusts through the Pew Scholars Program in the Biomedical Sciences and by National Institutes of Health Grant GM100122.
Abbreviations used:
- CHEF
clamped homogeneous electric field
- ChIP
chromatin immunoprecipitation
- 2-DOG
2-deoxygalactose
- 5-FOA
5-fluoroorotic acid
- MRS
major repeat sequence
- rDNA
ribosomal DNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0229) on July 24, 2013.
REFERENCES
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks. Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen MP, Nelson ZW, Hetrick ED, Gottschling DE. A genetic screen for increased loss of heterozygosity in Saccharomyces cerevisiae. Genetics. 2008;179:1179–1195. doi: 10.1534/genetics.108.089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravamudhan P, Felzer-Kim I, Joglekar AP. The budding yeast point centromere associates with two Cse4 molecules during mitosis. Curr Biol. 2013;23:770–774. doi: 10.1016/j.cub.2013.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellier-Bassi S, Gadal O, Bourout G, Nehrbass U. Cell cycle-dependent kinetochore localization of condensin complex in Saccharomyces cerevisiae. J Struct Biol. 2008;162:248–259. doi: 10.1016/j.jsb.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Basenko E, Topcu Z, McEachern MJ. Recombination can either help maintain very short telomeres or generate longer telomeres in yeast cells with weak telomerase activity. Eukaryot Cell. 2011;10:1131–1142. doi: 10.1128/EC.05079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, Sanyal K, Mishra PK, Thaler N, Carbon J. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc Natl Acad Sci USA. 2006;103:14877–14882. doi: 10.1073/pnas.0606958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito IL, Monje-Casas F, Amon A. The Lrs4-Csm1 monopolin complex associates with kinetochores during anaphase and is required for accurate chromosome segregation. Cell Cycle. 2010a;9:3611–3618. doi: 10.4161/cc.9.17.12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito IL, Yu HG, Amon A. Condensins promote coorientation of sister chromatids during meiosis I in budding yeast. Genetics. 2010b;185:55–64. doi: 10.1534/genetics.110.115139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack LS, Applen SE, Berman J. The requirement for the Dam1 complex is dependent upon the number of kinetochore proteins and microtubules. Curr Biol. 2011;21:889–896. doi: 10.1016/j.cub.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JN, Poon BP, Salvi J, Olsen JB, Emili A, Mekhail K. Perinuclear cohibin complexes maintain replicative life span via roles at distinct silent chromatin domains. Dev Cell. 2011;20:867–879. doi: 10.1016/j.devcel.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Chibana H, Magee PT. The enigma of the major repeat sequence of Candida albicans. Future Microbiol. 2009;4:171–179. doi: 10.2217/17460913.4.2.171. [DOI] [PubMed] [Google Scholar]
- Choi SH, Peli-Gulli MP, McLeod I, Sarkeshik A, Yates JR, 3rd, Simanis V, McCollum D. Phosphorylation state defines discrete roles for monopolin in chromosome attachment and spindle elongation. Curr Biol. 2009;19:985–995. doi: 10.1016/j.cub.2009.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Wu P, Parthun MR, Wu JQ. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Yip CK, Ee LS, Walz T, Amon A, Harrison SC. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell. 2010;142:556–567. doi: 10.1016/j.cell.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas A, Polakova S, Gregan J. Chromosome segregation: monopolin attracts condensin. Curr Biol. 2011;21:R634–R636. doi: 10.1016/j.cub.2011.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert CA, Gravdahl DJ, Megee PC. The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 2007;21:278–291. doi: 10.1101/gad.1498707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enloe B, Diamond A, Mitchell AP. A single-transformation gene function test in diploid Candida albicans. J Bacteriol. 2000;182:5730–5736. doi: 10.1128/jb.182.20.5730-5736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A, Abbey D, Pisithkul T, Weinzierl MA, Ringstrom T, Bruck D, Petersen K, Berman J. Stress alters rates and types of loss of heterozygosity in Candida albicans. MBio. 2011;2:e00129–e00111. doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerami-Nejad M, Berman J, Gale CA. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast. 2001;18:859–864. doi: 10.1002/yea.738. [DOI] [PubMed] [Google Scholar]
- Gerami-Nejad M, Dulmage K, Berman J. Additional cassettes for epitope and fluorescent fusion proteins in Candida albicans. Yeast. 2009;26:399–406. doi: 10.1002/yea.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerami-Nejad M, Forche A, McClellan M, Berman J. Analysis of protein function in clinical C. albicans isolates. Yeast. 2012;29:303–309. doi: 10.1002/yea.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerami-Nejad M, Hausauer D, McClellan M, Berman J, Gale C. Cassettes for the PCR-mediated construction of regulatable alleles in Candida albicans. Yeast. 2004;21:429–436. doi: 10.1002/yea.1080. [DOI] [PubMed] [Google Scholar]
- Gerami-Nejad M, Zacchi LF, McClellan M, Matter K, Berman J. Shuttle vectors for facile gap repair cloning and integration into a neutral locus in Candida albicans. Microbiology. 2013;159:565–579. doi: 10.1099/mic.0.064097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J, Riedel CG, Pidoux AL, Katou Y, Rumpf C, Schleiffer A, Kearsey SE, Shirahige K, Allshire RC, Nasmyth K. The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr Biol. 2007;17:1190–1200. doi: 10.1016/j.cub.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J, Stephens A, Verdaasdonk J, Yeh E, Bloom K. Bub1 kinase and Sgo1 modulate pericentric chromatin in response to altered microtubule dynamics. Curr Biol. 2012;22:471–481. doi: 10.1016/j.cub.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. “Point” centromeres of Saccharomyces harbor single CenH3 nucleosomes. Genetics. 2012;190:1575–1577. doi: 10.1534/genetics.111.137711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M, Yu EY, Singh SM, Lue NF. Mutual dependence of Candida albicans Est1p and Est3p in telomerase assembly and activation. Eukaryot Cell. 2007;6:1330–1338. doi: 10.1128/EC.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K, Horiuchi T. The cis element and factors required for condensin recruitment to chromosomes. Mol Cell. 2009;34:26–35. doi: 10.1016/j.molcel.2009.02.021. [DOI] [PubMed] [Google Scholar]
- Ketel C, Wang HS, McClellan M, Bouchonville K, Selmecki A, Lahav T, Gerami-Nejad M, Berman J. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 2009;5:e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H, Sellam A, Askew C, Nantel A, Whiteway M. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics. 2008;9:578. doi: 10.1186/1471-2164-9-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D, Coulson C. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Lephart PR, Chibana H, Magee PT. Effect of the major repeat sequence on chromosome loss in Candida albicans. Eukaryot Cell. 2005;4:733–741. doi: 10.1128/EC.4.4.733-741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc Natl Acad Sci USA. 1994;91:3453–3457. doi: 10.1073/pnas.91.8.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Seebacher J, Gygi SP, Moazed D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 2008;456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motwani T, Doris R, Holmes SG, Flory MR. Ccq1p and the condensin proteins Cut3p and Cut14p prevent telomere entanglements in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell. 2010;9:1612–1621. doi: 10.1128/EC.00339-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa N, Nakamura T, Kokubu A, Ebe M, Nagao K, Yanagida M. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J Cell Biol. 2008;180:1115–1131. doi: 10.1083/jcb.200708170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Poon BP, Mekhail K. Cohesin and related coiled-coil domain-containing complexes physically and functionally connect the dots across the genome. Cell Cycle. 2011;10:2669–2682. doi: 10.4161/cc.10.16.17113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- Ribeiro SA, et al. Condensin regulates the stiffness of vertebrate centromeres. Mol Biol Cell. 2009;20:2371–2380. doi: 10.1091/mbc.E08-11-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner S, Gasser SM, Meister P. Modules for cloning-free chromatin tagging in Saccharomyces cerevisiae. Yeast. 2008;25:235–239. doi: 10.1002/yea.1580. [DOI] [PubMed] [Google Scholar]
- Rumpf C, Cipak L, Schleiffer A, Pidoux A, Mechtler K, Tolic-Norrelykke IM, Gregan J. Laser microsurgery provides evidence for merotelic kinetochore attachments in fission yeast cells lacking Pcs1 or Clr4. Cell Cycle. 2010;9:3997–4004. doi: 10.4161/cc.9.19.13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samoshkin A, Arnaoutov A, Jansen LE, Ouspenski I, Dye L, Karpova T, McNally J, Dasso M, Cleveland DW, Strunnikov A. Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation. PLoS One. 2009;4:e6831. doi: 10.1371/journal.pone.0006831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal K, Carbon J. The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc Natl Acad Sci USA. 2002;99:12969–12974. doi: 10.1073/pnas.162488299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell RM, Jinks-Robertson S. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2004;262:3–12. doi: 10.1385/1-59259-761-0:003. [DOI] [PubMed] [Google Scholar]
- Stephens AD, Haase J, Vicci L, Taylor RM, 2nd, Bloom K. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol. 2011;193:1167–1180. doi: 10.1083/jcb.201103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, et al. Pericentric chromatin loops function as a nonlinear spring in mitotic force balance. J Cell Biol. 2013;200:757–772. doi: 10.1083/jcb.201208163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada K, Susumu H, Sakuno T, Watanabe Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011;474:477–483. doi: 10.1038/nature10179. [DOI] [PubMed] [Google Scholar]
- Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- Vagnarelli P, Hudson DF, Ribeiro SA, Trinkle-Mulcahy L, Spence JM, Lai F, Farr CJ, Lamond AI, Earnshaw WC. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat Cell Biol. 2006;8:1133–1142. doi: 10.1038/ncb1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka M, Rytka J, Kurlandzka A. Saccharomyces cerevisiae CSM1 gene encoding a protein influencing chromosome segregation in meiosis I interacts with elements of the DNA replication complex. Exp Cell Res. 2004;294:592–602. doi: 10.1016/j.yexcr.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Yong-Gonzalez V, Wang BD, Butylin P, Ouspenski I, Strunnikov A. Condensin function at centromere chromatin facilitates proper kinetochore tension and ensures correct mitotic segregation of sister chromatids. Genes Cells. 2007;12:1075–1090. doi: 10.1111/j.1365-2443.2007.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.