H2O2-stressed yeast cells increase superoxide radical production, dependent on the mitochondrial respiratory chain. This is protective during H2O2 stress at low levels; however, higher superoxide levels are deleterious. This hormesis may further elucidate the role of reactive oxygen species in oxidative stress and aging.
Abstract
Reactive oxygen species (ROS) consist of potentially toxic, partly reduced oxygen species and free radicals. After H2O2 treatment, yeast cells significantly increase superoxide radical production. Respiratory chain complex III and possibly cytochrome b function are essential for this increase. Disruption of complex III renders cells sensitive to H2O2 but not to the superoxide radical generator menadione. Of interest, the same H2O2-sensitive mutant strains have the lowest superoxide radical levels, and strains with the highest resistance to H2O2 have the highest levels of superoxide radicals. Consistent with this correlation, overexpression of superoxide dismutase increases sensitivity to H2O2, and this phenotype is partially rescued by addition of small concentrations of menadione. Small increases in levels of mitochondrially produced superoxide radicals have a protective effect during H2O2-induced stress, and in response to H2O2, the wild-type strain increases superoxide radical production to activate this defense mechanism. This provides a direct link between complex III as the main source of ROS and its role in defense against ROS. High levels of the superoxide radical are still toxic. These opposing, concentration-dependent roles of the superoxide radical comprise a form of hormesis and show one ROS having a hormetic effect on the toxicity of another.
INTRODUCTION
Most organisms rely on the role of oxygen as a terminal electron acceptor for efficient energy production in the form of ATP. Increased intracellular levels of oxygen, however, are potentially toxic. This toxicity is mainly due to partially reduced forms of O2 (Gille and Sigler, 1995), since the O2 molecule per se has low reactivity (Halliwell and Gutteridge, 1990). The molecules and radicals formed by the incomplete reduction of oxygen are termed reactive oxygen species (ROS; Halliwell and Gutteridge, 1989). ROS commonly formed in vivo include the superoxide radical anion (O2•−), hydrogen peroxide (H2O2), and the hydroxyl radical (OH•), and their formation can result in damage to proteins, lipids, and nucleic acids (Aung-Htut et al., 2012).
The first intermediate in the sequential reduction of oxygen is often O2•−. This is produced by single-electron reduction of oxygen to a large extent by electron carriers of the respiratory chain in the mitochondria (Lambert and Brand, 2004). Systems that produce O2•− also produce H2O2 as a result of disproportionation reactions (Gille and Sigler, 1995). These reactions are among the main sources of H2O2 in vivo and are either nonenzymatic or catalyzed by superoxide dismutases (SODs). In Saccharomyces cerevisiae the SOD1 gene encodes a copper- and zinc-containing enzyme located in the cytoplasm and mitochondrial intermembrane space. SOD2 encodes a manganese-containing enzyme found in the mitochondrial matrix (Bermingham-McDonogh et al., 1988; Gralla and Kosman, 1992; O'Brien et al., 2004). The main threat to the cell is the subsequent transformation of O2•− and H2O2 to stronger oxidants, in particular the hydroxyl radical, which is the most oxidizing radical known to arise in biological systems (Youngman, 1984; Buettner, 1993).
The incomplete reduction of O2 primarily occurs at two sites in the respiratory chain: complex I at NADH dehydrogenase (Turrens and Boveris, 1980) and complex III at ubisemiquinone (Boveris et al., 1976; Cadenas et al., 1977; Turrens et al., 1985). It is widely accepted that radicals derived from the partial reduction of ubiquinone in complex III are the main generators of ROS during respiration. This principally occurs at the semiquinone at the center o (Qo) electron carrier in complex III (Trumpower, 2002; Andreyev et al., 2005). Several studies showed that the complex I and III sites generate mostly O2•− rather than a mixture of partially reduced species (Turrens, 1997).
S. cerevisiae is a useful tool in the study of mitochondrial function because mutations in mitochondrial DNA or deletion of nuclear genes encoding mitochondrial proteins are not lethal. Although there are differences between the electron transport chain (ETC) of mammals and yeast, mostly at complex I, O2•− production at complex III results from similar mechanisms in yeast and mammals (Sun and Trumpower, 2003).
Yeast cells have evolved complex defense mechanisms to protect themselves against ROS. These mechanisms use small molecules such as glutathione and d-erythroascorbate and enzymatic defenses such as SOD, catalase, thioredoxins, and glutathione peroxidases, as reviewed in Temple(2005). These defenses are often inducible and are activated by transcription factors such as those encoded by YAP1 and SKN7, forming part of the environmental stress response (Gasch et al., 2000). Other active responses the cell can mount to ROS damage include adaptation, in which exposure to a non-LD of ROS confers resistance to a subsequent and normally LD, and cell cycle delay (Collinson and Dawes, 1992; Flattery-O'Brien et al., 1993; Alic et al., 2001, 2004). These responses have also been linked to upstream transcription factors and signaling pathways such as those encoded by SWI6, YAP1, SKN7, MPK1, ROX1, and MGA2 (Alic et al., 2003; Beckhouse et al., 2008; Fong et al., 2008; Ng et al., 2008; Kelley and Ideker, 2009).
The importance of mitochondrial function during oxidative stress has been demonstrated in many studies. ρ0 petite strains, which contain no mitochondrial DNA, are sensitive to oxidative stress caused by H2O2 and O2•− when compared with their isogenic wild type (WT; Collinson and Dawes, 1992; Jamieson, 1992; Flattery-O'Brien et al., 1993; Lee et al., 2001). Furthermore, strains with deletions in nuclear genes encoding components of the respiratory chain and WT strains treated with respiratory inhibitors are sensitive to H2O2 (Grant et al., 1997). This has been highlighted in a global genome-wide study in which nearly half the H2O2-sensitive mutants were deficient in electron transport chain function (Thorpe et al., 2004). Of interest, this was specific to H2O2-induced stress and not the other oxidative stress conditions examined, suggesting that complex III may have a specific and central role in survival during H2O2 stress.
ROS have been implicated in aging in yeast, with replicatively aged cells showing markers of oxidative stress (Laun et al., 2001). H2O2 activates a caspase-like enzyme in yeast that regulates apoptosis, and oxidative stress is intimately involved in the activation and progression of apoptosis in yeast (Madeo et al., 1997, 1999, 2002). This is consistent with the widely held view that ROS and oxidative stress are always harmful to cells. Recent results regarding the role of ROS in chronological aging, however, have offered apparently conflicting views on their contribution. It has been proposed that superoxide radicals mediate chronological aging, whereas it has been suggested that H2O2 has a positive effect, increasing chronological lifespan (Mesquita et al., 2010; Weinberger et al., 2010; Lewinska et al., 2011; Pan et al., 2011). To fully understand this, more research into the complex roles of different ROS in the cell is needed to elucidate their exact contribution to cell viability in different conditions.
We therefore sought to determine the role of the mitochondrial electron transport chain and the ROS it produces (O2•−) in the tolerance of H2O2-induced oxidative stress in S. cerevisiae. The results of these experiments show that mitochondrially produced O2•− is protective against H2O2, providing the first link between complex III as the main source of ROS in the cell and protection against ROS.
RESULTS
H2O2 stress increases superoxide radical levels in a mitochondrial-dependent manner
Mitochondrial function is crucial for the survival of yeast treated with H2O2 despite mitochondria being a main source of O2•− production. To investigate the relationship between mitochondrial function and O2•− levels in cells during H2O2-induced oxidative stress, we used electron paramagnetic resonance spectroscopy (EPR) spin trapping using the trap 5-(diethoxyphosphoryl)-5-methyl-1-pyrroline N-oxide (DEPMPO), a method that gives very specific signals with O2•− (Frejaville et al., 1995).
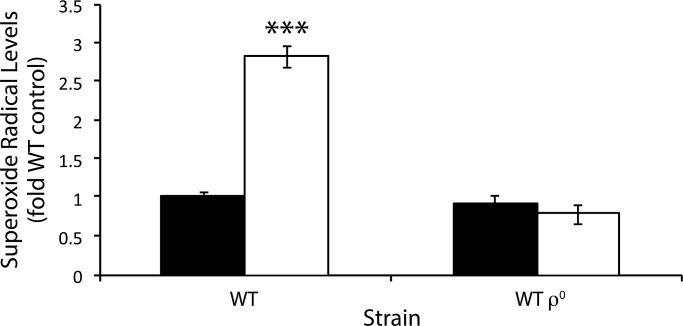
Cells were grown in yeast extract/peptone/dextrose (YEPD) liquid to late exponential phase (OD600 of 4). Half were treated with 4 mM H2O2 for 60 min, and cells in both sample and control cultures were washed and incubated with DEPMPO. EPR spectra were obtained on the culture supernatant. Treatment of the WT with H2O2 increased superoxide levels to nearly three times that of the untreated control (Figure 1). This was in contrast to the isogenic WT ρ0 strain, in which H2O2 treatment resulted in no increase in O2•− levels, demonstrating that yeast cells need respiration to increase O2•− levels in response to H2O2 stress. The mitochondrial genome contains only genes that encode proteins that are part of the ETC or transcripts that are required for their synthesis. Consequently, the complexes in the ETC may be the mitochondrial function that is involved in O2•− production in response to H2O2.
FIGURE 1:
Superoxide radical production in the wild type and a petite mutant during oxidative stress. O2•− was assayed in vivo by EPR using the spin trap DEPMPO and is expressed relative to the unstressed WT cells. Cells were grown in YEPD liquid to late exponential phase (OD600 of 4), half were treated with 4 mM H2O2 for 60 min, and then cells were washed and incubated with DEPMPO for 20 min. The supernatant was then taken for EPR analysis. Black bars represent the control; white bars represent treatment with H2O2. H2O2 was compared with the control, ***p < 0.001.
The superoxide radical levels observed during H2O2-induced oxidative stress depend on cytochrome b
The ETC in mitochondria is a major source of endogenous ROS in cells, and since the increase in O2•− levels in the WT treated with H2O2 depended on functional mitochondria, we sought to further investigate their role.
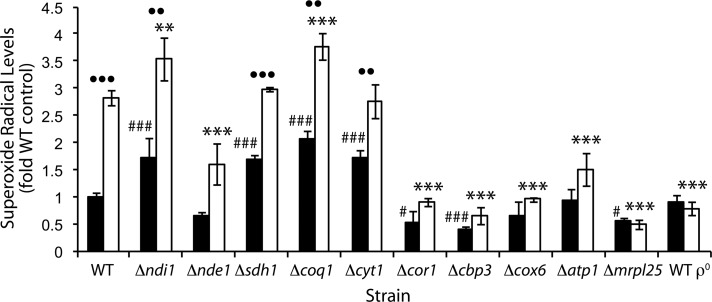
Representative strains covering deficiencies in all complexes in the ETC were chosen, and their O2•− levels were determined in the absence of added H2O2. In general, strains lacking genes encoding for upstream components of the electron transport chain (NADH dehydrogenase, complex II, and ubiquinone) had increased levels of O2•− when compared with the WT, whereas strains lacking downstream components (complexes III–V) had reduced levels (Figure 2). Deletion of NDI1 and SDH1 resulted in a significant increase in O2•−. This may have occurred due to the electron carriers in the ETC being in a more reduced state, since lack of NADH and succinate dehydrogenase activity may result in reduced flux of electron flow into the ETC. The strain deleted for COQ1, which encodes the protein responsible for the first step of ubiquinone biosynthesis, also showed high O2•− levels. The increased O2•− levels may be a consequence of the absence of antioxidant activity of ubiquinol, increased superoxide production, or a combination of both. Because the coq1∆ strain has no ubiquinol to carry electrons to complex III, complex II and the internal and external NADH dehydrogenases may be the sites of O2•− production.
FIGURE 2:
Superoxide radical production in electron transport chain mutants during oxidative stress. O2•− was assayed in vivo by EPR using the spin trap DEPMPO and is expressed relative to the unstressed WT. Cells were grown in YEPD liquid to late exponential phase (OD600 of 4), half were treated with 4 mM H2O2 for 60 min, and then cells were washed and incubated with DEPMPO for 20 min. The supernatant was then taken for EPR analysis. Black bars represent the untreated control sample; white bars represent treatment with H2O2. In the statistical analysis, mutant strains in control conditions were compared with the WT in control conditions (number sign or hash symbol), H2O2 treated strains were compared with the H2O2-treated WT (asterisks), and for each strain, H2O2 treatment was compared with no treatment (dots).
The O2•− levels in the strain deleted for ATP1 were not significantly different from those in the WT. The atp1Δ mutant contains the full complement of genes encoding electron-carrying respiratory complexes but cannot use the membrane potential created to produce ATP. This reinforces the view that the ETC function responsible for O2•− production comes from the transfer of electrons from NADH and succinate to O2 rather than the production of ATP.
Complex III was essential for normal O2•− production in S. cerevisiae, since removal of either function resulted in the lowest levels of O2•− (Figure 2). Two of the three deletion strains that are specifically affected in complex III assembly or function (cor1∆ and cbp3∆) had significantly reduced levels of O2•−. In contrast, the strain deleted for CYT1, which encodes cytochrome c1, had significantly increased levels of O2•−. The low O2•− levels in the cor1∆ and cbp3∆ strains cannot be explained by the abolition of respiration, since the cyt1∆ strain, which is also respiratory deficient, had increased levels of O2•−.
Of interest, the cor1∆ and cbp3∆ mutants that had low O2•− levels are deficient in cytochrome b function or assembly (Tzagoloff et al., 1986; Gruschke et al., 2011), whereas the cyt1∆ strain that did not have reduced superoxide levels is not known to be deficient in cytochrome b. Consistent with this, the mrpl25Δ strain, which cannot express the mitochondrially encoded cytochrome b gene (COB), also had reduced levels of O2•−. This strongly indicates that O2•− production in these strains may depend on cytochrome b.
The superoxide levels in the wild type and the foregoing mutants were also determined after exposure to 4 mM H2O2 for 60 min. For some mutants this treatment did lead to loss of longer-term viability (in some mutants to as low as 4% survival vs. 57% survival for the wild type) as determined by ability of cells to replicate (by estimation of colony-forming units) but not to loss of viability in the time span used to measure superoxide production, as determined by membrane integrity measurements based on propidium iodide staining (see Supplemental Table S1). During H2O2-induced oxidative stress the cor1∆, cbp3∆, cox6 ∆, atp1∆, mrpl25∆, and WT ρ0 strains all showed a significant reduction in O2•− levels compared with the WT in the same conditions (Figure 2). These strains are affected in the function of complexes III–V. Strains with NDI1, SDH1, COQ1, and CYT1 deleted had either increased or no significant difference in their O2•− levels compared with the WT. Therefore H2O2 stress resulted in the same general pattern as observed in the absence of stress (Figure 2), that is, higher O2•− levels in strains deleted for upstream components of the ETC and lower O2•− levels in strains deleted for downstream components.
The WT and strains deleted for NDI1, SDH1, COQ1, and CYT1 all had significant increases in O2•− levels after H2O2 treatment when compared with unstressed conditions for each strain (Figure 2). All other strains did not significantly increase their O2•− levels after H2O2 stress from their initial low levels. Because the strains with low superoxide levels could not significantly increase them in response to H2O2, it appears that the mechanisms responsible for unstressed O2•− levels, H2O2-treated O2•− levels, and increased O2•− levels in response to H2O2 stress all rely on complex III and possibly cytochrome b.
SOD gene expression does not account for differences in superoxide radical levels
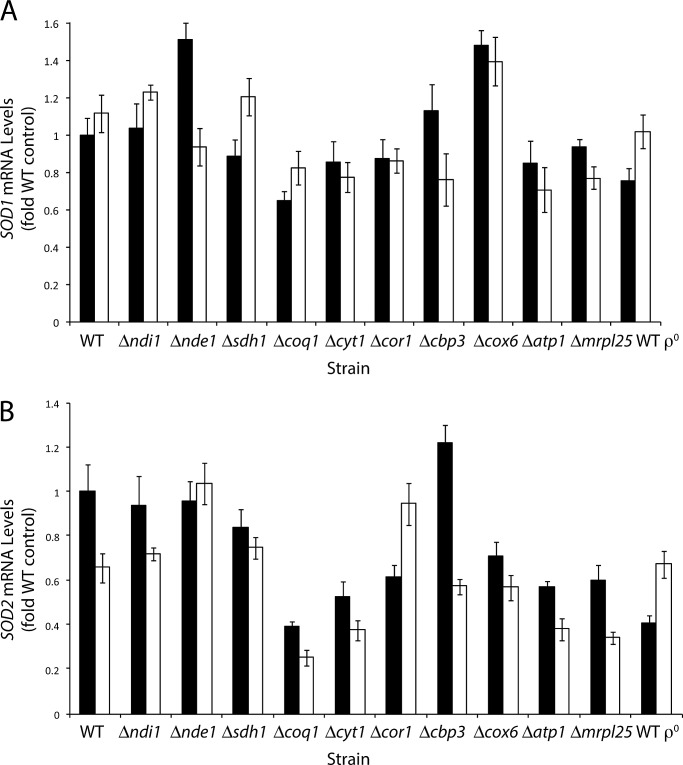
One factor that may influence the O2•− levels measured in these mutant strains is the detoxification of O2•− by SOD. To determine whether changes in the amount of SOD due to increased transcription were responsible for the differences in O2•− levels in some mutants, we determined the transcript levels of the SOD1 and SOD2 genes (Figure 3). Differences in the extent of SOD1 mRNA induction between mutant strains in both test conditions were relatively small. SOD1 mRNA changes ranged from 0.65-fold for the unstressed coq1Δ strain to 1.5-fold for the unstressed nde1Δ strain. This strongly suggests that the SOD1 mRNA levels were not having an effect on O2•− levels observed in Figure 2. A similar result was found in the case of SOD2 mRNA, for which we observed small changes (Figure 3B). There was no consistent pattern showing that SOD1 or SOD2 mRNA levels were responsible for the differences in O2•− levels observed in the test strains. Often the opposite was observed, in which strains with lower levels of O2•− also had lower levels of SOD mRNA. Examples of this included the cbp3Δ, cor1Δ, and mrpl25Δ mutants, which had lower O2•− levels than the WT in the presence of H2O2 while also having slightly lower SOD1 mRNA levels than the WT.
FIGURE 3:
CuZnSOD and MnSOD mRNA transcript levels in electron transport chain mutants during oxidative stress. Cells were grown in YEPD to OD600 of 4 and then split into one control sample and one sample treated with 4 mM H2O2. After 15 min, total cellular RNA was extracted. Purified mRNA was reverse transcribed and quantitative PCR analysis was then performed on the genes (A) SOD1 (CuZnSOD) and (B) SOD2 (MnSOD). The relative amounts of the SOD1 and SOD2 transcripts were then normalized to the control ACT1 signal. All mRNA levels are expressed as the multiple relative to the unstressed WT strain. Black bars represent the control; white bars represent treatment with H2O2.
Strains deficient in complex III and IV function are sensitive to H2O2 but not to superoxide radicals
It would be expected that the increase in O2•− in the WT and some of the other strains after the addition of H2O2 would result in increased oxidative stress and damage to the cell. This is due to the generation of species such as the hydroxyl radical via Fenton chemistry. To investigate this further, we assayed the sensitivities of the ETC mutants to H2O2 and the O2•− generator menadione (Figure 4).
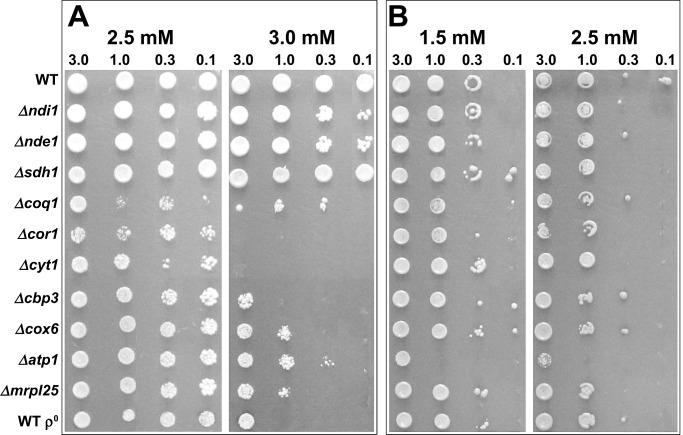
FIGURE 4:
Sensitivity of electron transport chain mutants to oxidative stress. Strains were grown in liquid YEPD for 2 d and diluted in YEP to OD600 of 3, 1, 0.3, and 0.1. Then 5 μl was spotted onto SC agar containing (A) H2O2 and (B) menadione. The plates were incubated at 30°C for 2 d.
The mutants with different degrees of H2O2 sensitivity could be placed into three distinct functional groups: those with extreme sensitivity, high sensitivity, and little or no sensitivity. The strains most sensitive to H2O2 (Figure 4A) were those deleted for complex III subunits (cor1Δ, cyt1Δ) and those deficient in CoQ biosynthesis (coq1Δ). Sensitivity of ubiquinone-deficient mutants to oxidative stress may be due to loss of respiratory activity and the antioxidant function of the CoQ pool (Schultz and Clarke, 1999; James et al., 2004). The next group of strains also showed a high degree of H2O2 sensitivity but not to the levels seen in the first group. The functions disrupted in these strains were complex III assembly (cbp3Δ), complex IV (cox6Δ), complex V (atp1Δ), and mitochondrial genome expression (mrpl25Δ, and WT ρ0).
The strains in the group showing little or no H2O2 sensitivity were deleted for NADH dehydrogenase (ndi1Δ or nde1Δ) or succinate dehydrogenase (sdh1Δ) activity (Figure 4A). When electron transport is slowed (e.g., decreased supply of electrons into the electron transport chain), electron carriers are mostly reduced. This situation promotes the one-electron reduction of O2, which increases ROS production (Andreyev et al., 2005), although this does not appear to have any detrimental effect on the cell during H2O2-induced stress.
In all strains in which electron flow is completely abolished (coq1Δ, strains with mutations in complexes III and IV, and mrpl25Δ and WT ρ0), an extreme or high degree of H2O2 sensitivity was observed (Figure 4A). Therefore the ETC is essential for WT H2O2 tolerance. The different degrees of sensitivity of these mutants—with complex III mutants the most sensitive—indicate that additional factors may be affecting sensitivity. In the presence of menadione, the atp1Δ strain showed the highest degree of sensitivity, whereas all other strains showed either little or no difference in sensitivity compared with the WT (Figure 4B). The differences in O2•− levels observed in Figure 2 did not seem to affect tolerance of these strains to menadione.
Survival of H2O2 treatment correlates with higher levels of superoxide radicals
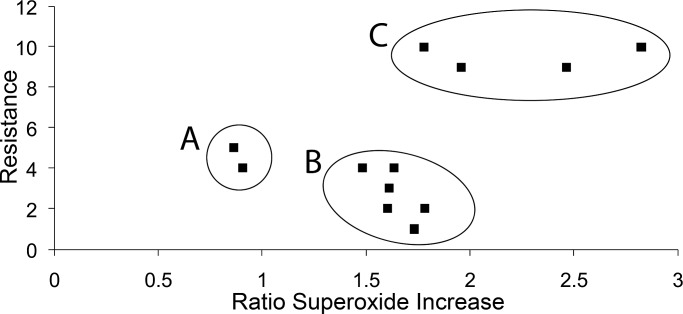
To further investigate the relationship between levels of O2•− and resistance to oxidative stress, we compared O2•− levels with H2O2 sensitivity of each mutant. For H2O2 sensitivity, a numerical score was assigned based on growth in the spot test in Figure 4A, with the WT scoring 10 for maximum resistance. In Figure 5, H2O2 sensitivity (data from Figure 4A) is compared with the ratio of O2•− levels during H2O2 to control conditions for each strain. There was clearly no linear correlation, but the mutants fell into three distinct functional classes. Group A contained the strains deleted for MRPL25 and the WT ρ0, and both mutations result in deficient expression of the entire mitochondrial genome. Because they form a distinct group, the mechanisms by which these strains are sensitive to H2O2 may therefore be different from that of the other ETC deletion strains studied. Group B contained all of the other strains that were sensitive to H2O2, and these were deleted for genes required for the biosynthesis of ubiquinone and for function and assembly of complexes III–V of the ETC. Group C contained the WT and strains deficient in entry of NADH into the ETC from either side of the inner mitochondrial membrane and succinate dehydrogenase function. This group had the highest resistance to H2O2 and included strains among those with the highest O2•− levels.
FIGURE 5:
The relationship between H2O2 resistance and increase in superoxide radical levels. H2O2 resistance was plotted against O2•− level data to determine any correlation. H2O2 resistance data from Figure 4 were assigned numerical scores and plotted against the ratio of O2•− levels during H2O2 stress to unstressed conditions, which was calculated for each strain (data from Figure 2). Areas A–C denote the three areas where the plotted data appeared to cluster and are discussed in the text.
The relationship between H2O2 sensitivity and O2•− levels during H2O2 stress was examined. The strains were placed into one of four classes based on their relative O2•− levels and H2O2 sensitivity, as shown in Table 1. The WT, ndi1Δ, and sdh1Δ strains had the highest O2•− levels and the lowest H2O2 sensitivity. The cor1Δ, cbp3Δ, cox6Δ, mrpl25Δ, WT ρ0, and atp1Δ strains were the opposite, with all having low or very low O2•− levels and high or very high H2O2 sensitivity (Table 1). These nine strains may provide evidence that there is a correlation between H2O2 sensitivity and O2•− levels during H2O2 stress. When O2•− levels are higher during H2O2 stress, resistance to H2O2 stress appears to be increased. Therefore it is the overall level of O2•− that may influence H2O2 sensitivity and not the relative increase in O2•− in response to H2O2. The coq1Δ strain was very sensitive to H2O2 but also had very high levels of O2•−. This may be due to loss of the antioxidant function of the coenzyme Q pool (Schultz and Clarke, 1999; James et al., 2004). Similarly, the cyt1Δ strain was very sensitive to H2O2 but also had high levels of O2•−. This may be due to the possible presence of cytochrome b in this strain, which is not present in most other low-O2•− strains. Therefore the coq1Δ and cyt1Δ strains may deviate from this correlation because of these additional unique factors.
TABLE 1:
Relationship between superoxide radical level and H2O2 sensitivity.
| Very low superoxide | Low superoxide | High superoxide | Very high superoxide | |
|---|---|---|---|---|
| Very low sensitivity | nde1Δ | WT, ndi1Δ, sdh1Δ | ||
| Low sensitivity | ||||
| High sensitivity | cbp3Δ, cox6Δ, mrpl25Δ, WT ρ0 | atp1Δ | ||
| Very high sensitivity | cor1Δ | cyt1Δ | coq1Δ |
Reducing superoxide levels in the wild type confirms its protective role during H2O2 stress
Because low O2•− levels appeared to correlate with increased H2O2 sensitivity, we tested the hypothesis that a decrease in O2•− levels from overexpressing SOD would increase H2O2 sensitivity. Overexpression vectors pRS425 and pRS426 containing the SOD1 and SOD2 genes, respectively, were transformed into BY4743, and the resulting strains were then treated with H2O2.
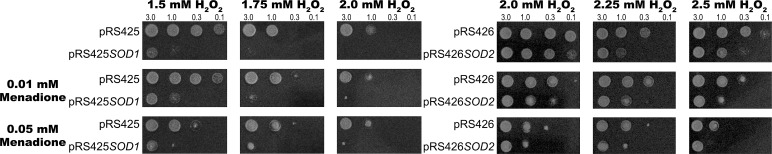
Overexpression of SOD increased the sensitivity of the WT to H2O2 (Figure 6). This was more pronounced in the case of overexpression of SOD1 (Figure 6). Overexpression of SOD2 also made the cell more sensitive to H2O2 but to a lesser extent than with SOD1 overexpression. This was not surprising, considering that SOD1 encodes the CuZnSOD, which in yeast accounts for 90–99% of total SOD activity (Longo et al., 1999; Sturtz and Culotta, 2002). We hypothesized that the increase in H2O2 sensitivity observed in the SOD-overexpressing strain could be due to either decreased O2•− levels (as the substrate of increased SOD) or increased H2O2 levels (as the product of increased SOD). To distinguish between these two possibilities, we repeated the experiment in the presence of non-LDs of the O2•− generator menadione. If increased H2O2 was the cause of sensitivity in the SOD-overexpressing strains, the overexpressed SOD would detoxify the menadione-generated O2•−, forming even higher levels of H2O2, which would further increase sensitivity. Addition of menadione, however, did not increase the sensitivity of the SOD1-overexpressing strain, confirming that increased production of H2O2 was unlikely to be the cause of sensitivity. Not only did the addition of menadione cause no increase in H2O2 sensitivity in the SOD1-overexpressing strain, it actually increased its resistance to H2O2, as seen in the growth of the test strain compared with that containing the vector-only control (pRS425) under the same conditions (Figure 6). This indicates that overexpression of SOD1 confers H2O2 sensitivity by reducing O2•− levels. Therefore O2•− levels appear to have a protective effect during oxidative stress induced by H2O2.
FIGURE 6:
The effect of superoxide radical levels on H2O2 tolerance. Strains were grown in liquid SD dropout medium to OD600 of 1 and suspended in SD minus glucose to OD600 of 3, 1, 0.3, and 0.1. Then 5 μl was spotted onto SD dropout agar containing H2O2 and menadione (where specified). Top, H2O2 only; middle, H2O2 of the same concentrations plus 0.01 mM menadione; bottom, H2O2 of the same concentrations plus 0.05 mM menadione. The plates were incubated at 30°C for 2 d.
The partial rescue of the H2O2-sensitive phenotype by adding menadione was accompanied by decreased growth of the control, whereas growth of the SOD1-overexpressing strain was mostly unchanged. This loss of yield of the control in increasing menadione concentrations shows that when superoxide levels get too high, a negative effect results. O2•− levels are therefore protective at low concentrations but toxic at higher concentrations.
DISCUSSION
The increased O2•− levels observed in the H2O2-treated WT cells are unlikely to result directly from metabolism of H2O2 since the systems detoxifying H2O2 do not directly produce O2•−. Moreover, the differences in O2•− levels in the various strains did not appear to be due to differences in detoxification of O2•− since the expression levels of SOD genes did not correlate with the different O2•− levels observed. Therefore differences in superoxide levels observed in the WT appear to be due to a change in O2•− production, and the dependence on functional mitochondria supports this. Studies using dihydroethidium to detect O2•− are consistent with these results in the case of the ρ0 (Reddi and Culotta, 2013) and mrpl25Δ, where the MRPL25 gene was involved in replicative aging, also named AFO1 (Heeren et al., 2009). The O2•− observed in our study may not be due to the NADPH oxidase encoded by YNO1 since O2•− production by this enzyme is independent of mitochondrial function (Rinnerthaler et al., 2012).
The abolition of complex III and/or complex IV function significantly reduced O2•− levels below those seen in the WT, presumably due to loss of complex III function, since complex III is recognized as the major in vivo source of O2•− (Andreyev et al., 2005). Specifically, the O2•− levels observed depended on cytochrome b function. This is consistent with accepted theory that a ubisemiquinone intermediate of the Q-cycle mechanism in complex III is the main source of electrons donated to O2 to form O2•−. An increased lifetime of the center N ubisemiquinone (Qo ), which is located at cytochrome b, is the main cause of higher levels of mitochondrial O2•− production (Trumpower, 1990; Hunte et al., 2003; Crofts, 2004).
Previously ROS was believed to have only a deleterious role in yeast, apart from activating cellular defenses to combat them and the damage they cause. More recently it was proposed that ROS might have a positive signaling role during chronological aging (Mesquita et al., 2010; Weinberger et al., 2010; Lewinska et al., 2011; Pan et al., 2011). Here we show that O2•− has a protective signaling role in yeast during H2O2-induced oxidative stress. We propose the mechanism for O2•− production and provide an explanation of hormesis to address the seemingly conflicting positive and negative effects of the O2•− and ROS. Specifically, in response to H2O2, the cell increases O2•− production in a complex III–dependent manner. Preventing or diminishing this response by gene deletion or increased O2•− detoxification renders cells more sensitive to H2O2.
The mitochondrial electron transport chain is crucial for cell viability during H2O2 stress, with complex III implicated as having critical importance (Grant et al., 1997; Thorpe et al., 2004). One possible explanation for the importance of the ETC during H2O2 stress could be a requirement for energy (Grant et al., 1997). Because there may be little or no proton-motive force in the H2O2-sensitive strains, they may therefore be unable to make ATP at complex V. Furthermore, the atp1Δ strain, which has an intact electron transport chain but is unable to synthesize ATP, was sensitive to H2O2. A previous large-scale screening study, however, highlighted only strains lacking respiratory chain functions to be H2O2 sensitive and not ones lacking other mitochondrial energy-generating reactions such as the trichloroacetic acid cycle (Thorpe et al., 2004). Moreover, these studies were carried out in cells primarily generating energy by fermentation rather than respiration. The metabolic adaptation to loss of mitochondrial function induced by the retrograde response also offers no explanation for H2O2 sensitivity since the ρ0 strain showed little difference in sensitivity or O2•− levels compared with other mitochondrial mutants.
The specific involvement of the electron transport chain in H2O2 tolerance may therefore be due to other factors, such as ROS production. Here we found that increased O2•− levels resulted in cells becoming more resistant to H2O2. This correlation with O2•− levels is interesting because elevated levels of the radical in the presence of H2O2 would be expected, on the basis of Fenton chemistry, to increase oxidative stress by generating the hydroxyl radical and therefore render the cells more sensitive to H2O2. Other studies support our finding; for example, grande strains have been proposed to produce more ROS than petite strains (Guidot et al., 1993; Longo et al., 1999), but they are more resistant to oxidative stress elicited by H2O2 and menadione than petites (Collinson and Dawes, 1992; Jamieson, 1992; Flattery-O'Brien et al., 1993). Moreover, a ρ0 mutant that showed significant viability loss during chronological aging produced low amounts of ROS (Trancikova et al., 2004).
How can lower levels of O2•− production result in increased sensitivity? The answer may lie in O2•− having a protective effect. The normal response to H2O2-induced stress in WT cells was to increase O2•− production, and when this was inhibited by either increased SOD levels or inhibition of complex III function, sensitivity to H2O2 increased. The protective effect of increased O2•− is distinct from the adaptive response, since pretreatment with menadione does not protect against LD of H2O2, and adaptation occurs in strains lacking mitochondrial function (Collinson and Dawes, 1992; Jamieson, 1992). Therefore adaptation and O2•− signaling are two distinct examples of the many overlapping defenses against H2O2, and full activation of H2O2 defenses may only happen in response to both H2O2 and O2•−. Consequently, one function of the electron transport chain during H2O2 stress may be to produce increased levels of O2•−, which then has a signaling role capable of activating antioxidant defenses.
Although our results show that O2•− produced in the mitochondria has a protective effect at low concentrations, high concentrations were still detrimental. The importance of O2•− concentration means that SODs may have an important role during H2O2 stress beyond simply detoxifying O2•−. The levels of O2•− must also be tightly regulated to promote their protective signaling effect. Supporting this, it was proposed that in vivo SOD activity can either increase or decrease ROS damage, depending on the conditions (Lushchak et al., 2005). Furthermore, SOD overexpression has been shown to extend chronological lifespan but reduce replicative lifespan (Harris et al., 2003; Fabrizio et al., 2004). This concentration-dependent dual role of O2•−, in which at low concentration it can have a positive effect and at higher concentration a negative effect, is a form of mitochondrial hormesis. The concept of mitochondrial hormesis has been proposed as an explanation for the role of ROS in aging in higher organisms and humans (Linnane et al., 2007; Yang and Hekimi, 2010; Ristow and Schmeisser, 2011). In yeast, O2•− has been proposed to provide an adaptive signal extending chronological lifespan, and the levels of H2O2 have been proposed to have a hormetic effect on chronological aging (Mesquita et al., 2010; Pan et al., 2011). Here we show the hormetic effect of O2•− on tolerance of another ROS, H2O2.
O2•− production in yeast and mammalian complex III appears to occur via a similar mechanism (Sun and Trumpower, 2003), and yeast is an important model for research into mitochondrial function and oxidative stress. This hormetic signaling role of O2•− has implications for the role of ROS and mitochondria in oxidative stress, aging, and apoptosis in yeast and higher organisms and consequently a possible role in human diseases such as cancer (Singh, 2004).
MATERIALS AND METHODS
Strains and plasmids
S. cerevisiae strains used were in the genetic background of BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) and BY4742 (MATα; his3Δ1; leu2Δ0; lysΔ0; ura3Δ0), with BY4743 (MATa/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; MET15/met15Δ0; lysΔ0/LYS2; ura3Δ0/ura3Δ0) the diploid product of these two strains (Brachmann et al., 1998). The term wild type is used to refer to BY4743. Diploid deletion strains, homozygous at the deleted locus, constructed as part of International Saccharomyces Gene Deletion Project (Winzeler et al., 1999), were obtained from the European Saccharomyces cerevisiae Archive for Functional Analysis, Frankfurt, Germany. To confirm that strains deleted for nuclear genes were grande, haploid strains were mated with a haploid strain lacking mitochondrial DNA, and the resultant diploid was able to grow on a respiratory carbon source. The BY4743 ρ0 petite (WT ρ0) was generated by growth of the parent strain on YEPD agar containing 20 mg/l ethidium bromide. The plasmids used in the study were the pRS42X series (Christianson et al., 1992). pRS425SOD1 and pRS426SOD2 were obtained from John Park (Department of Biochemistry, Medical College of Wisconsin, Milwaukee, WI; Park et al., 1998).
Media and chemicals
Cells were grown in YEPD (2% [wt/vol] d-glucose, 2% [wt/vol] bactopeptone, and 1% yeast extract), SC (2% [wt/vol] d-glucose, 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 0.074% complete supplement mixture from Difco [Franklin Lakes, NJ]), and SD minimal medium (2% [wt/vol] d-glucose, 0.17% yeast nitrogen base, 0.5% ammonium sulfate). SD medium was supplemented with appropriate auxotrophic requirements to the concentrations prescribed for SC medium (Adams et al., 1998). Leucine auxotrophs were also supplemented with isoleucine and valine. Agar plates were solidified with 2% (wt/vol) agar (type 1 agar for SD and SC; type 3 agar for YEPD). Dropout plates lacking only the selected nutrient were used to select transformants with acquired prototrophic genes. S. cerevisiae was grown at 30°C with shaking and aeration when in liquid media. DEPMPO was purchased from Sapphire Bioscience (Waterloo, Australia). Diethylenetriaminepentaacetic acid (DTPA), ethidium bromide, and menadione sodium bisulfate were obtained from Sigma-Aldrich (Castle Hill, Australia).
Sensitivity to oxidative stress
Sensitivity to a chronic exposure of oxidative stress was performed as spot tests on agar plates. Liquid cultures were grown to the growth phase specified, centrifuged, and resuspended in 0.17% yeast nitrogen base and 0.5% ammonium sulfate to an OD600 of 3.0, 1.0, 0.3, and 0.1. A 5-μl amount of each dilution was spotted onto SC or SC-dropout agar plates if strains contained a plasmid, with oxidants of differing concentrations, and incubated at 30°C for 2 d. All agar was cooled to 50ºC before addition of the oxidant and then immediately allowed to set.
Viability estimation
Cells of the wild type and mutants were grown in YEPD to OD600 of 4, and each culture was split into one control sample and one sample treated with 4 mM H2O2 for 60 min. Samples of treated and untreated cultures were diluted appropriately for plate counts on YEPD plates and also taken for propidium iodide staining to determine cell viability as measured by retention of membrane permeability. For the propidium iodide (PI) staining, cells were harvested by centrifugation, resuspended in 1 ml phosphate-buffered saline (PBS), and stained with propidium iodide (10 µg/ml) for 20 min in the dark and washed twice with PBS, and the level of PI staining was analyzed by flow cytometry. Stained cells were analyzed with a FACS Canto II (BD Biosciences, San Diego, CA) instrument equipped with a laser that excites at 488 nm, and emission was detected at 617 nm using a 556-nm long-pass filter and a 585/42-nm broad-pass filter.
Measurement of superoxide radicals
Intracellular superoxide radicals were estimated based on the method of Heeren et al. (2004). Before the experiment, 100 μl of methanol was added to each 50-mg tube of DEPMPO, and all tubes to be used in the experiment were pooled together. DEPMPO aliquots were transferred to individual Eppendorf tubes so that the final concentration in 350 μl of buffer was 100 mM. Cells were grown in YEPD to the specified OD600/growth phase, and a sample was taken to determine viable cells per milliliter. If pretreatment was involved (H2O2-induced oxidative stress), this was applied at the specified concentration for 1 h. Cultures were centrifuged (4 min, 2000 rpm) and washed twice in 10 mM potassium phosphate buffer, pH 7.0. Triplicate samples of 2.25 × 108 cells were resuspended in 350 μl of 10 mM potassium phosphate buffer, pH 7.0, and 0.1 mM DTPA. Cell concentration calculations were based on OD600 derived from a standard curve obtained from the WT and checked 2 d later from viable colony counts. The 350 μl of cells in PI/DTPA buffer was transferred to an Eppendorf tube containing DEPMPO and incubated for 20 min at room temperature with shaking, with lid open to allow aeration, and protected from light. After the 20-min incubation, samples were centrifuged and the supernatant taken for subsequent EPR analysis. The supernatant was immediately frozen in liquid N2 and stored protected from light until immediately before EPR analysis.
EPR spectra were recorded at room temperature using a Bruker EMX spectrometer with 100-kHz modulation equipped with a cylindrical (ER4103TM) cavity. Samples were contained in a flattened aqueous sample cell (Wilmad WG-813-SQ), and recording of the spectra was initiated within 2 min of the start of the reaction. Hyperfine coupling constants were measured directly from the field scan and confirmed by spectral simulation with the program WINSIM. Typical EPR spectrometer settings were as follows: gain, 2 × 106; modulation amplitude, 0.1 mT; time constant, 163 s; scan time, 83 s; resolution, 1024 points; center field, 348.5 mT; field scan, 12.0 mT; power, 20.068 mW; and frequency, 9.658 GHz; with eight scans averaged. The relative concentrations of the O2− adducts to DEPMPO were determined from measurement of peak-to-peak line heights of the EPR absorption lines specific to this adduct, relative to the WT control spectra.
RNA extraction and quantitative real time-PCR
Total cell RNA was extracted with TRIzol reagent (Life Technologies, Carlsbad, CA) and purified using the RNeasy Mini Kit (Qiagen, Valencia, CA), with optional on-column DNase digestion. Extracted RNA was quantified using the Bioanalyzer (Agilent Technologies, Santa Clara, CA) and a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, MA). The absence of genomic DNA in samples was confirmed using non–reverse-transcribed RNA as a template in real-time PCR assays. RNA was then reverse transcribed into cDNA using the iScript Select cDNA Synthesis Kit (Bio-Rad, Hercules, CA).
Oligonucleotides were designed using Primer3 (Rozen and Skaletsky, 2000). The primers used to amplify a 144–base pair section of ACT1 were CTGCCGGTATTGACCAAACT (forward) and CGGTGATTTCCTTTTGCATT (reverse). A 129–base pair section of SOD1 was amplified using CACATGGTGCTCCAACTGAC (forward) and CAACGGAGGTAGGACCGATA (reverse). A 131–base pair sequence of SOD2 was amplified using AACCAGGATACCGTCACAGG (forward) and TTCCAGTTGACCACATTCCA (reverse). PCRs were performed in quadruplicate samples using the iTaq SYBR Green Supermix With ROX and analyzed on a Chromo4 Real-Time PCR Detection System (Bio-Rad), with a no-template control included in each assay. The thermocycling program consisted of 95°C for 150 s, followed by 40 cycles of 20 s at 95, 58, 72°C. Melting-curve data were collected to verify PCR specificity and absence of contamination and primer dimers. PCR efficiency was determined using the dilution series method, and data were analyzed using Opticon Monitor Software (Bio-Rad). Expression relative to ACT1 was determined with efficiency correction (Pfaffl, 2001).
Statistics
Values in graphs are means ± SD. Student's t test was used for the statistical analysis of each strain in control conditions compared with H2O2 treatment. A p < 0.01 was deemed indicative of a statistically significant difference for these tests. Analysis of mutants strains compared with the WT was performed using a one-way analysis of variance followed by Dunnett's test for multiple comparisons. A p < 0.05 was deemed indicative of a statistically significant difference for these tests. *p < 0.05, **p < 0.01, and ***p < 0.001.
Supplementary Material
Acknowledgments
We thank Klause Stolze, Hans Nohl, Shixiong Tan, Richard Morris, and William Burhans for helpful discussions. This work was supported by grants from the Australian Research Council and by Australian Post Graduate Awards to G.W.T. and M.R. We are grateful to the Austrian Science Fund Fonds zur Förderung der wissenschaftlichen Forschung (Vienna, Austria) for Grant S9302-B05 to M.B.
Abbreviations used:
- DEPMPO
5-(diethoxyphosphoryl)-5-methyl-1-pyrroline N-oxide
- EPR
electron paramagnetic resonance spectroscopy
- ETC
electron transport chain
- O2•−
superoxide radical anion
- ROS
reactive oxygen species
- SOD
superoxide dismutase
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-01-0052) on July 17, 2013.
REFERENCES
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- Alic N, Felder T, Temple MD, Gloeckner C, Higgins VJ, Briza P, Dawes IW. Genome-wide transcriptional responses to a lipid hydroperoxide: adaptation occurs without induction of oxidant defenses. Free Radic Biol Med. 2004;37:23–35. doi: 10.1016/j.freeradbiomed.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Alic N, Higgins VJ, Dawes IW. Identification of a Saccharomyces cerevisiae gene that is required for G1 arrest in response to the lipid oxidation product linoleic acid hydroperoxide. Mol Biol Cell. 2001;12:1801–1810. doi: 10.1091/mbc.12.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alic N, Higgins VJ, Pichova A, Breitenbach M, Dawes IW. Lipid hydroperoxides activate the mitogen-activated protein kinase Mpk1p in Saccharomyces cerevisiae. J Biol Chem. 2003;278:41849–41855. doi: 10.1074/jbc.M307760200. [DOI] [PubMed] [Google Scholar]
- Andreyev AY, Kushnareva YE, Starkov AA. Mitochondrial metabolism of reactive oxygen species. Biochemistry (Mosc) 2005;70:200–214. doi: 10.1007/s10541-005-0102-7. [DOI] [PubMed] [Google Scholar]
- Aung-Htut MT, Ayer A, Breitenbach M, Dawes IW. Oxidative stresses and ageing. In: Breitenbach M, Jazwinski SM, Laun P, editors. Aging Research in Yeast. Dordrecht, Netherlands: Springer; 2012. pp. 13–54. [Google Scholar]
- Beckhouse AG, Grant CM, Rogers PJ, Dawes IW, Higgins VJ. The adaptive response of anaerobically grown Saccharomyces cerevisiae to hydrogen peroxide is mediated by the Yap1 and Skn7 transcription factors. FEMS Yeast Res. 2008;8:1214–1222. doi: 10.1111/j.1567-1364.2008.00439.x. [DOI] [PubMed] [Google Scholar]
- Bermingham-McDonogh O, Gralla EB, Valentine JS. The copper, zinc-superoxide dismutase gene of Saccharomyces cerevisiae: cloning, sequencing, and biological activity. Proc Natl Acad Sci USA. 1988;85:4789–4793. doi: 10.1073/pnas.85.13.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveris A, Cadenas E, Stoppani AO. Role of ubiquinone in the mitochondrial generation of hydrogen peroxide. Biochem J. 1976;156:435–444. doi: 10.1042/bj1560435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Buettner GR. The pecking order of free radicals and antioxidants: lipid peroxidation, alpha-tocopherol, and ascorbate. Arch Biochem Biophys. 1993;300:535–543. doi: 10.1006/abbi.1993.1074. [DOI] [PubMed] [Google Scholar]
- Cadenas E, Boveris A, Ragan CI, Stoppani AO. Production of superoxide radicals and hydrogen peroxide by NADH-ubiquinone reductase and ubiquinol-cytochrome c reductase from beef-heart mitochondria. Arch Biochem Biophys. 1977;180:248–257. doi: 10.1016/0003-9861(77)90035-2. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Collinson LP, Dawes IW. Inducibility of the response of yeast cells to peroxide stress. J Gen Microbiol. 1992;138:329–335. doi: 10.1099/00221287-138-2-329. [DOI] [PubMed] [Google Scholar]
- Crofts AR. The cytochrome bc1 complex: function in the context of structure. Annu Rev Physiol. 2004;66:689–733. doi: 10.1146/annurev.physiol.66.032102.150251. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pletcher SD, Minois N, Vaupel JW, Longo VD. Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS Lett. 2004;557:136–142. doi: 10.1016/s0014-5793(03)01462-5. [DOI] [PubMed] [Google Scholar]
- Flattery-O'Brien J, Collinson LP, Dawes IW. Saccharomyces cerevisiae has an inducible response to menadione which differs from that to hydrogen peroxide. J Gen Microbiol. 1993;139:501–507. doi: 10.1099/00221287-139-3-501. [DOI] [PubMed] [Google Scholar]
- Fong CS, Temple MD, Alic N, Chiu J, Durchdewald M, Thorpe GW, Higgins VJ, Dawes IW. Oxidant-induced cell-cycle delay in Saccharomyces cerevisiae: the involvement of the SWI6 transcription factor. FEMS Yeast Res. 2008;8:386–399. doi: 10.1111/j.1567-1364.2007.00349.x. [DOI] [PubMed] [Google Scholar]
- Frejaville C, Karoui H, Tuccio B, Le Moigne F, Culcasi M, Pietri S, Lauricella R, Tordo P. 5-(Diethoxyphosphoryl)-5-methyl-1-pyrroline N-oxide: a new efficient phosphorylated nitrone for the in vitro and in vivo spin trapping of oxygen-centered radicals. J Med Chem. 1995;38:258–265. doi: 10.1021/jm00002a007. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille G, Sigler K. Oxidative stress and living cells. Folia Microbiol (Praha) 1995;40:131–152. doi: 10.1007/BF02815413. [DOI] [PubMed] [Google Scholar]
- Gralla EB, Kosman DJ. Molecular genetics of superoxide dismutases in yeasts and related fungi. Adv Genet. 1992;30:251–319. doi: 10.1016/s0065-2660(08)60322-3. [DOI] [PubMed] [Google Scholar]
- Grant CM, MacIver FH, Dawes IW. Mitochondrial function is required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. FEBS Lett. 1997;410:219–222. doi: 10.1016/s0014-5793(97)00592-9. [DOI] [PubMed] [Google Scholar]
- Gruschke S, Kehrein K, Rompler K, Grone K, Israel L, Imhof A, Herrmann JM, Ott M. Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J Cell Biol. 2011;193:1101–1114. doi: 10.1083/jcb.201103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidot DM, McCord JM, Wright RM, Repine JE. Absence of electron transport (Rho 0 state) restores growth of a manganese-superoxide dismutase-deficient Saccharomyces cerevisiae in hyperoxia. Evidence for electron transport as a major source of superoxide generation in vivo. J Biol Chem. 1993;268:26699–26703. [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. London: Clarendon Press; 1989. [Google Scholar]
- Halliwell B, Gutteridge JM. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- Harris N, Costa V, MacLean M, Mollapour M, Moradas-Ferreira P, Piper PW. MnSOD overexpression extends the yeast chronological (G0) life span but acts independently of Sir2p histone deacetylase to shorten the replicative life span of dividing cells. Free Radic Biol Med. 2003;34:1599–1606. doi: 10.1016/s0891-5849(03)00210-7. [DOI] [PubMed] [Google Scholar]
- Heeren G, et al. The mitochondrial ribosomal protein of the large subunit, Afo1p, determines cellular longevity through mitochondrial back-signaling via TOR1. Aging (Albany NY) 2009;1:622–636. doi: 10.18632/aging.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeren G, Jarolim S, Laun P, Rinnerthaler M, Stolze K, Perrone GG, Kohlwein SD, Nohl H, Dawes IW, Breitenbach M. The role of respiration, reactive oxygen species and oxidative stress in mother cell-specific ageing of yeast strains defective in the RAS signaling pathway. FEMS Yeast Res. 2004;5:157–167. doi: 10.1016/j.femsyr.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Hunte C, Palsdottir H, Trumpower BL. Protonmotive pathways and mechanisms in the cytochrome bc1 complex. FEBS Lett. 2003;545:39–46. doi: 10.1016/s0014-5793(03)00391-0. [DOI] [PubMed] [Google Scholar]
- James AM, Smith RA, Murphy MP. Antioxidant and prooxidant properties of mitochondrial coenzyme Q. Arch Biochem Biophys. 2004;423:47–56. doi: 10.1016/j.abb.2003.12.025. [DOI] [PubMed] [Google Scholar]
- Jamieson DJ. Saccharomyces cerevisiae has distinct adaptive responses to both hydrogen peroxide and menadione. J Bacteriol. 1992;174:6678–6681. doi: 10.1128/jb.174.20.6678-6681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R, Ideker T. Genome-wide fitness and expression profiling implicate Mga2 in adaptation to hydrogen peroxide. PLoS Genet. 2009;5:e1000488. doi: 10.1371/journal.pgen.1000488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AJ, Brand MD. Inhibitors of the quinone-binding site allow rapid superoxide production from mitochondrial NADH:ubiquinone oxidoreductase (complex I) J Biol Chem. 2004;279:39414–39420. doi: 10.1074/jbc.M406576200. [DOI] [PubMed] [Google Scholar]
- Laun P, Pichova A, Madeo F, Fuchs J, Ellinger A, Kohlwein S, Dawes I, Frohlich KU, Breitenbach M. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol Microbiol. 2001;39:1166–1173. [PubMed] [Google Scholar]
- Lee JC, Straffon MJ, Jang TY, Higgins VJ, Grant CM, Dawes IW. The essential and ancillary role of glutathione in Saccharomyces cerevisiae analysed using a grande gsh1 disruptant strain. FEMS Yeast Res. 2001;1:57–65. doi: 10.1111/j.1567-1364.2001.tb00013.x. [DOI] [PubMed] [Google Scholar]
- Lewinska A, Macierzynska E, Grzelak A, Bartosz G. A genetic analysis of nitric oxide-mediated signaling during chronological aging in the yeast. Biogerontology. 2011;12:309–320. doi: 10.1007/s10522-011-9329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnane AW, Kios M, Vitetta L. The essential requirement for superoxide radical and nitric oxide formation for normal physiological function and healthy aging. Mitochondrion. 2007;7:1–5. doi: 10.1016/j.mito.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Longo VD, Liou LL, Valentine JS, Gralla EB. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch Biochem Biophys. 1999;365:131–142. doi: 10.1006/abbi.1999.1158. [DOI] [PubMed] [Google Scholar]
- Lushchak V, Semchyshyn H, Mandryk S, Lushchak O. Possible role of superoxide dismutases in the yeast Saccharomyces cerevisiae under respiratory conditions. Arch Biochem Biophys. 2005;441:35–40. doi: 10.1016/j.abb.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Madeo F, et al. A caspase-related protease regulates apoptosis in yeast. Mol Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Frohlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, Frohlich KU. Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita A, Weinberger M, Silva A, Sampaio-Marques B, Almeida B, Leao C, Costa V, Rodrigues F, Burhans WC, Ludovico P. Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proc Natl Acad Sci USA. 2010;107:15123–15128. doi: 10.1073/pnas.1004432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CH, Tan SX, Perrone GG, Thorpe GW, Higgins VJ, Dawes IW. Adaptation to hydrogen peroxide in Saccharomyces cerevisiae: the role of NADPH-generating systems and the SKN7 transcription factor. Free Radic Biol Med. 2008;44:1131–1145. doi: 10.1016/j.freeradbiomed.2007.12.008. [DOI] [PubMed] [Google Scholar]
- O'Brien KM, Dirmeier R, Engle M, Poyton RO. Mitochondrial protein oxidation in yeast mutants lacking manganese-(MnSOD) or copper- and zinc-containing superoxide dismutase (CuZnSOD): evidence that MnSOD and CuZnSOD have both unique and overlapping functions in protecting mitochondrial proteins from oxidative damage. J Biol Chem. 2004;279:51817–51827. doi: 10.1074/jbc.M405958200. [DOI] [PubMed] [Google Scholar]
- Pan Y, Schroder EA, Ocampo A, Barrientos A, Shadel GS. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JI, Grant CM, Davies MJ, Dawes IW. The cytoplasmic Cu,Zn superoxide dismutase of Saccharomyces cerevisiae is required for resistance to freeze-thaw stress. Generation of free radicals during freezing and thawing. J Biol Chem. 1998;273:22921–22928. doi: 10.1074/jbc.273.36.22921. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddi AR, Culotta VC. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell. 2013;152:224–235. doi: 10.1016/j.cell.2012.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinnerthaler M, et al. Yno1p/Aim14p, a NADPH-oxidase ortholog, controls extramitochondrial reactive oxygen species generation, apoptosis, and actin cable formation in yeast. Proc Natl Acad Sci USA. 2012;109:8658–8663. doi: 10.1073/pnas.1201629109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Schmeisser S. Extending life span by increasing oxidative stress. Free Radic Biol Med. 2011;51:327–336. doi: 10.1016/j.freeradbiomed.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Schultz JR, Clarke CF. Characterization of Saccharomyces cerevisiae ubiquinone-deficient mutants. Biofactors. 1999;9:121–129. doi: 10.1002/biof.5520090206. [DOI] [PubMed] [Google Scholar]
- Singh KK. Mitochondrial dysfunction is a common phenotype in aging and cancer. Ann NY Acad Sci. 2004;1019:260–264. doi: 10.1196/annals.1297.043. [DOI] [PubMed] [Google Scholar]
- Sturtz LA, Culotta VC. Superoxide dismutase null mutants of baker's yeast, Saccharomyces cerevisiae. Methods Enzymol. 2002;349:167–172. doi: 10.1016/s0076-6879(02)49332-9. [DOI] [PubMed] [Google Scholar]
- Sun J, Trumpower BL. Superoxide anion generation by the cytochrome bc1 complex. Arch Biochem Biophys. 2003;419:198–206. doi: 10.1016/j.abb.2003.08.028. [DOI] [PubMed] [Google Scholar]
- Temple MD, Perrone GG, Dawes IW. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 2005;15:319–326. doi: 10.1016/j.tcb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc Natl Acad Sci USA. 2004;101:6564–6569. doi: 10.1073/pnas.0305888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trancikova A, Weisova P, Kissova I, Zeman I, Kolarov J. Production of reactive oxygen species and loss of viability in yeast mitochondrial mutants: protective effect of Bcl-xL. FEMS Yeast Res. 2004;5:149–156. doi: 10.1016/j.femsyr.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Trumpower BL. The protonmotive Q cycle. Energy transduction by coupling of proton translocation to electron transfer by the cytochrome bc1 complex. J Biol Chem. 1990;265:11409–11412. [PubMed] [Google Scholar]
- Trumpower BL. A concerted, alternating sites mechanism of ubiquinol oxidation by the dimeric cytochrome bc1 complex. Biochim Biophys Acta. 2002;10:166–173. doi: 10.1016/s0005-2728(02)00273-6. [DOI] [PubMed] [Google Scholar]
- Turrens JF. Superoxide production by the mitochondrial respiratory chain. Biosci Rep. 1997;17:3–8. doi: 10.1023/a:1027374931887. [DOI] [PubMed] [Google Scholar]
- Turrens JF, Alexandre A, Lehninger AL. Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch Biochem Biophys. 1985;237:408–414. doi: 10.1016/0003-9861(85)90293-0. [DOI] [PubMed] [Google Scholar]
- Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J. 1980;191:421–427. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Wu MA, Crivellone M. Assembly of the mitochondrial membrane system. Characterization of COR1, the structural gene for the 44-kilodalton core protein of yeast coenzyme QH2-cytochrome c reductase. J Biol Chem. 1986;261:17163–17169. [PubMed] [Google Scholar]
- Weinberger M, Mesquita A, Caroll T, Marks L, Yang H, Zhang Z, Ludovico P, Burhans WC. Growth signaling promotes chronological aging in budding yeast by inducing superoxide anions that inhibit quiescence. Aging (Albany NY) 2010;2:709–726. doi: 10.18632/aging.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 2010;8:e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman RJ. Oxygen activation: is the hydroxyl radical always biologically relevant. Trends Biochem Sci. 1984;9:280–283. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.