The authors investigated the regulation of the Drosophila actin-microtubule cross-linker Short stop (Shot) and found that Shot undergoes an intramolecular conformational change that regulates its cross-linking activity. This intramolecular interaction depends on Shot's NH2-terminal actin-binding domain and EF-hand-GAS2 domain.
Abstract
Actin and microtubule dynamics must be precisely coordinated during cell migration, mitosis, and morphogenesis—much of this coordination is mediated by proteins that physically bridge the two cytoskeletal networks. We have investigated the regulation of the Drosophila actin-microtubule cross-linker Short stop (Shot), a member of the spectraplakin family. Our data suggest that Shot's cytoskeletal cross-linking activity is regulated by an intramolecular inhibitory mechanism. In its inactive conformation, Shot adopts a “closed” conformation through interactions between its NH2-terminal actin-binding domain and COOH-terminal EF-hand-GAS2 domain. This inactive conformation is targeted to the growing microtubule plus end by EB1. On activation, Shot binds along the microtubule through its COOH-terminal GAS2 domain and binds to actin with its NH2-terminal tandem CH domains. We propose that this mechanism allows Shot to rapidly cross-link dynamic microtubules in response to localized activating signals at the cell cortex.
INTRODUCTION
Although often studied as isolated networks, actin filaments and microtubules are functionally intertwined during cellular processes such as polarization, directed migration, and asymmetric cell division (Waterman-Storer et al., 2000; Schaefer et al., 2002; Rodriguez et al., 2003; Wittmann et al., 2003; Drabek et al., 2006; Gupta et al., 2010). This type of cross-talk often requires mechanical coupling between the two cytoskeletal networks by proteins or protein complexes that act as physical cross-bridges (Salmon et al., 2002; Rothenberg et al., 2003; Wittmann et al., 2003; Miller et al., 2004; Rosales-Nieves et al., 2006; Lee and Suter, 2008; Applewhite et al., 2010). The spectraplakins are one such family of giant cytoskeletal cross-linking proteins that have been highly conserved across most animal lineages (Lee et al., 2000; Röper et al., 2002; Röper and Brown, 2003; Jefferson et al., 2004; Sonnenberg and Liem, 2007; Kim et al., 2011; Suozzi et al., 2012). Wherever studied genetically, spectraplakins are found to be essential genes that play multiple roles in development. For example, Drosophila has a single spectraplakin, Short stop (Shot), that is required for cell–cell and cell–matrix adhesion. Shot mutants exhibit defects in axon outgrowth, trachea development, and muscle attachment to the cuticle (Strumpf and Volk, 1998; Lee et al., 2000; Lee and Kolodziej, 2002a, b; Röper and Brown, 2003; Subramanian et al., 2003; Bottenberg et al., 2009). Mammals possess two spectraplakin genes, best studied in mouse, in which the orthologues are called dystonin/BPAG1 and ACF7/MACF1 (Bernier et al., 1996; Gong et al., 2001; Jefferson et al., 2007). BPAG1 acts as a cytoskeletal cross-linker in epidermal cells and neurons; mutants with tissue-specific loss of the protein exhibit skin-blistering phenotypes and neurodegeneration (Jefferson et al., 2004; Sonnenberg and Liem, 2007; Edvardson et al., 2012). Studies conducted with cells derived from ACF7 knockout mice revealed that ACF7 regulates microtubule dynamics and plays important roles in cell migration (Kodama et al., 2003; Wu et al., 2008, 2011). Collectively these studies have revealed essential roles for spectraplakin-mediated cytoskeletal cross-linking in tissue morphogenesis and cell migration.
Spectraplakins are characterized by a complex, multidomain architecture that is alternatively spliced to generate a diversity of isoforms (Röper et al., 2002; Sonnenberg and Liem, 2007; Suozzi et al., 2012). Spectraplakins typically possess an NH2-terminal actin-binding domain (ABD) composed of two calponin homology repeats, a central rod-like domain comprising plakin- and spectrin-like repeats, two EF-hand motifs, a GAS2-like domain that binds to microtubules, and a 300-residue COOH-terminal domain (CTD) that binds to the microtubule plus end–binding protein EB1 (Lee et al., 2000; Wu et al., 2008; Honnappa et al., 2009; Applewhite et al., 2010; Alves-Silva et al., 2012). Thus spectraplakins are thought to be elongated, bivalent, rod-like proteins with actin- and microtubule-binding sites at each end.
Given the many important roles that spectraplakins play throughout development, investigating the spatiotemporal regulation of these proteins is key to understanding their function at the molecular level. We previously published a structure–function study of Drosophila Shot that led us to propose a model in which its actin-microtubule cross-linking activity is regulated by an intramolecular inhibition mechanism (Applewhite et al., 2010). In the current report, we used a combination of biochemical interaction studies and advanced imaging techniques to test our hypothesis. Our data indicate that Shot is able to adopt a conformation in which the ABD contacts the EF-hand-GAS2 domains. While in this autoinhibited conformation, Shot does not interact with actin or the microtubule lattice, but does interact with EB1 to localize to the plus ends of growing microtubules. On reaching the actin-rich zones located at the cell periphery, the ABD and EF-hand-GAS2 domains dissociate and are able to bridge nearby actin filaments and microtubules, respectively. These data provide the first mechanistic detail of how spectraplakin-mediated cytoskeletal cross-linking is regulated at the molecular level.
RESULTS
The NH2-terminal ABD of Shot directly interacts with the EF-hand/GAS2 domain
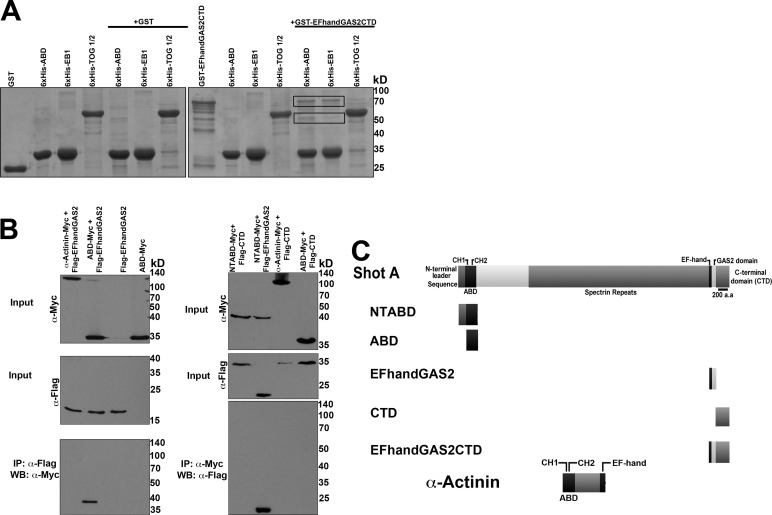
Our previous work showed that Shot localizes either to the microtubule plus end or to the microtubule lattice and that the lattice-associated pool was actively engaged as a cross-linker to actin (Applewhite et al., 2010). Our hypothesis is that actin-microtubule cross-linking is inhibited by a direct intramolecular contact between the actin- and microtubule-binding domains of the protein. To test this idea, we purified 6xHis- and glutathione S-transferase (GST)-tagged fusions of the ABD and EF-hand-GAS2-CTD from Shot and tested their ability to interact by Ni-NTA resin pull-down assays (Figure 1A). We observed that 6xHis-ABD bound to GST-EF-hand-GAS2-CTD, but not to GST alone. As a positive control, we also showed that 6xHis-EB1 also bound to GST-EF-hand-GAS2-CTD. We noted that recombinant GST-EF-hand-GAS2-CTD always exhibited some degradation products; 6xHis-EB1 selectively bound to the higher-molecular-weight fragments, consistent with our previous observation that the EB1-binding sites are present in the CTD of Shot (Slep et al., 2005; Applewhite et al., 2010). 6xHis-ABD, however, interacted with all the observed truncated fragments, consistent with an interaction with the EF-hand-GAS2 domains (Figure 1A, black boxes). As a negative control for specificity of the interaction, we found that the first two tumor overexpressed gene (TOG) domains of the microtubule tip-tracking protein Mini spindles (6xHis-TOG1-2) did not interact with GST-EF-hand-GAS2-CTD. To test whether the ABD and EF-hand-GAS2 domains could also interact in vivo, we cotransfected S2 cells with epitope-tagged versions of ABD-myc and a more refined Flag-EF-hand-GAS2 construct and immunoprecipitated with antibodies to each epitope (Figure 1B). Immunoblotting revealed that the ABD and EF-hand-GAS2 fusion coimmunoprecipitated, while the ABD did not coimmunoprecipitate with the CTD alone. These data indicate that the ABD and EF-hand-GAS2 domains are able to directly interact.
FIGURE 1:
Shot's NH2-terminal ABD interacts with its COOH-terminal EF-hand-GAS2 domain. (A) Coomassie-stained polyacrylamide gel of purified 6xHis-tagged NH2-terminal and COOH-terminal halves of Shot. Left, GST alone failed to interact with 6xHis-tagged ABD (Shot's tandem calponin homology domains), EB1, or TOG domains 1 and 2 from Mini spindles (control). Right, GST-EF-handGAS2CTD did interact with 6xHis-ABD and 6xHis-EB1 (black boxes), but did not interact with TOG1/2. (B) Western blot analysis of S2 cell lysate collected from cells expressing NH2-terminal Flag- tagged and COOH-terminal Myc-tagged fragments of Shot. NTABD is the actin-binding domain plus an NH2-terminal leader sequence (residues 1–142); ABD-Myc lacks this NH2-terminal leader sequence. CTD is Shot's COOH-terminal domain lacking both EF-hand motifs and the GAS2 domain. Only Shot's ABD and EF-hand-GAS2 domain coimmunoprecipitated. Also included are Myc-tagged Drosophila α-actinin, which contains both NH2-terminal CH domains, and a COOH-terminal EF-hand motif (control). (C) Diagram of the Shot fragments used in A and B.
Shot exhibits an intramolecular head-to-tail interaction when localized to microtubule plus ends
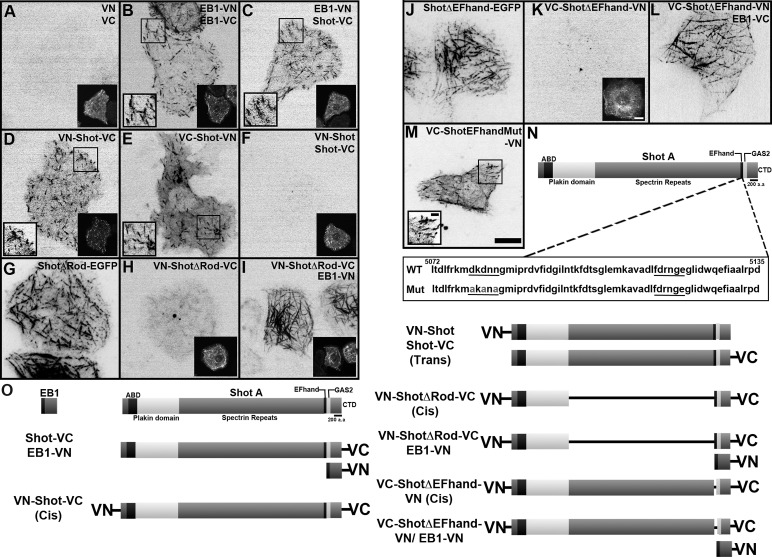
To examine the conformation of Shot within living cells, we designed a series of bimolecular fluorescent complementation (BiFC) probes to serve as a readout for interactions between the ABD and EF-hand-GAS2 domains (Figure 2). The basis of this technique is that two nonfluorescent fragments of the yellow fluorescent protein Venus are fused to candidate proteins; if the chimeric proteins directly interact, they reconstitute full-length Venus and become fluorescent (Kerppola, 2008, 2009). Coexpressed complementary halves of split Venus probe alone (VN and VC) did not fluoresce in S2 cells; however, coexpression of two EB1 constructs, EB1-VN and EB1-VC, heterodimerized and reconstituted fluorescence at growing microtubule plus ends (Figure 2, A and B). To further validate the specificity of this approach, we differentially tagged EB1 and Shot with VC and VN, as both proteins interact via their COOH-termini (Figure 2C). Expression of EB1-VN and Shot-VC reconstituted fluorescence and the proteins localized to microtubules; however, as expected, expression of EB1-VC and VN-Shot failed to produce fluorescence above background levels (unpublished data). We next fused VN and VC to the Shot coding sequence to determine whether its NH2- and COOH-termini could interact in cis (Figure 2D and Supplemental Movie S1). Both VN-Shot-VC and VC-Shot-VN reconstituted fluorescence in S2 cells and localized exclusively to microtubule plus ends, indicating that both ends of the protein were in close proximity. To determine whether Venus reconstitution occurred as the result of inter- or intramolecular interaction, we coexpressed VN-Shot and Shot-VC (Figure 2F) but failed to observe fluorescence, indicating that Shot does not form an intermolecular interaction between NH2- and COOH-terminal domains in trans. These data suggest that Shot is able to adopt a head-to-tail, folded conformation when localized to the microtubule plus end.
FIGURE 2:
BiFC assay indicates an intramolecular interaction dependent on Shot's Rod domain. S2 cells transfected with BiFC probes. Regions defined by a black box are shown at higher magnification (where indicated). To identify transfectants, we also transfected cells with EB1-mRFP, which is shown at lower magnification (where indicated), as it is our experience that there is high cotransfection efficiency when constructs share the same promoter. (A) NH2- and COOH-terminal halves (VN and VC) of split Venus do not reconstitute fluorescence. (B) Coexpression of EB1 tagged with VN or VC as positive control for the reconstitution of Venus. (C) Coexpression of EB1-VN and Shot-VC also reconstitutes fluorescence, as these two proteins interact via their COOH-termini. (D and E) Expression of a single molecule of Shot tagged at both its NH2- and COOH-termini with VN and VC in a cis orientation reconstitutes fluorescence. (F) Coexpression of Shot tagged at its NH2-terminus with VN and Shot tagged at its COOH-terminus with VC in a trans orientation fails to reconstitute fluorescence. (G) ShotΔRod-EGFP. (H) Expression of VN-ShotΔRod-VC in a cis orientation fails to reconstitute fluorescence. (I) Coexpression of VN-ShotΔRod-VC and EB1-VN. (J) ShotΔEF-hand-EGFP. (K) VC-ShotΔEF-hand-VN fails to reconstitute fluorescence. (L) Coexpression of VC-ShotΔEF-hand-VN and EB1-VC. (M) VC-Shot-EF-handMut-VN (Mut, EF-hand motif mutant, D5080A, D5082A, D5084A) reconstitutes fluorescence. (N) Diagram of Shot's domain organization. Highlighted by the box are the amino acids that correspond to the EF-hand motif. Underlined are the putative calcium-coordinating residues. Residues mutated in VC-Shot-EF-handMut-VN (D5080A, D5082A, N5084A) are shown in gray. (O) Diagram of the BiFC probes used in this study. Each transfection condition was repeated two to five times, with five to ten time-lapsed images obtained per condition. Scale bars: 2 μm in high-magnification images; 10 μm in low-magnification images.
The central rod domain of shot is required for interactions between the ABD and EF-hand-GAS2 domains
Given Shot's size, we reasoned that its rod domain must be flexible enough to allow the long-distance interactions between the NH2- and COOH-termini we observed using the BiFC assay. The triple-helical, coiled-coil motifs of spectrin endow it with a high degree of flexibility; thus we hypothesized that the spectrin-like rod domain of Shot confers a similar degree of flexibility and that deletion of this domain would eliminate intramolecular inhibition (Kusunoki et al., 2004; Määttä et al., 2004; Bhasin et al., 2005). To test this, we used the BiFC assay to determine whether the NH2- and COOH-termini of Shot could interact if the rod domain were deleted (Figure 2H and Movie S2). When transfected into S2 cells, VN-ShotΔRod-VC (lacking residues 1205–4599) was unable to reconstitute Venus fluorescence above background levels. However, when we coexpressed VN-ShotΔRod-VC with EB1-VN (Figure 2I), we did observe reconstitution of fluorescence along the lattice of microtubules, indicating that the lack of fluorescence from VN-ShotΔRod-VC alone was not due to misfolding of the protein, as it was still competent to bind EB1. These results indicate that the spectrin-like domain is necessary for interactions between the NH2- and COOH-termini and for normal dynamics of the molecule. They also suggest that, upon binding to the microtubule lattice, ShotΔRod acts to mislocalize EB1 when bound via reconstituted split Venus.
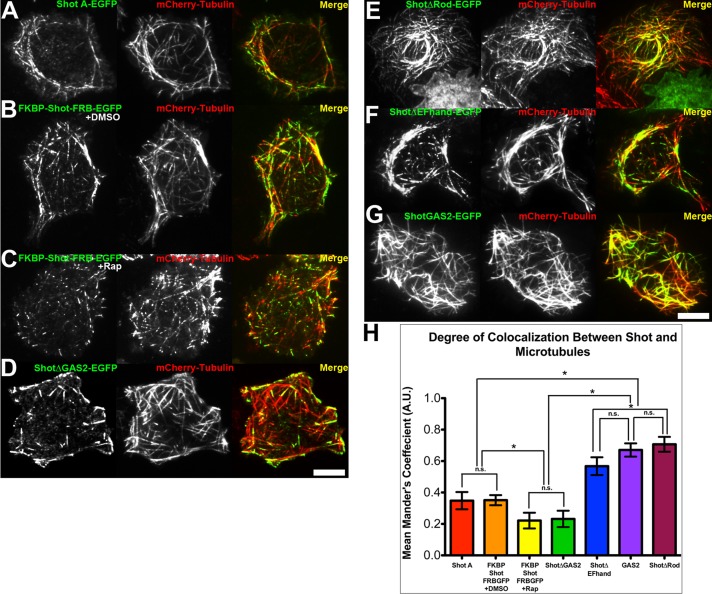
Next we measured the degree of colocalization between Shot–green fluorescent protein (Shot-GFP) and mCherry-tubulin in living cells, using Mander's coefficient (M; a scale of zero to one, where zero indicates no colocalization; Bolte and Cordelières, 2006; Figure 3). In our experiments, Shot-GFP exhibited an intermediate colocalization with microtubules (M = 0.34; Figure 3, A and H) that reflected its dual localization to the microtubule lattice and plus end. As a reference, Shot∆GAS2-EGFP, a construct that localizes exclusively to microtubule plus ends, exhibited a significantly lower value (M = 0.23; Figure 3, D and H), and ShotGAS2-GFP, a construct that binds exclusively to the lattice, produced a significantly higher value (M = 0.66; Figure 3, G and H). To test the contribution of the central rod domain to Shot localization, we expressed ShotΔRod-EGFP in S2 cells and examined its interactions with microtubules (Figure 3E). We found that ShotΔRod-EGFP predominantly associated with the microtubule lattice in the central cytoplasm, as well as in the cell periphery. Consistent with this localization pattern, ShotΔRod-EGFP exhibited a significantly higher degree of colocalization with microtubules (M = 0.70; Figure 3H) than full-length Shot A-GFP and was statistically indistinguishable from that of ShotGAS2-EGFP. These data suggested that deletion of the central rod domain disrupted regulation of ShotΔRod-EGFP and shifted it to a lattice-bound conformation.
FIGURE 3:
Colocalization between Shot constructs and microtubules. S2 cells cotransfected with (A) Shot A-EGFP, (B) FKBP-Shot-FRB-EGFP treated with DMSO, (C) FKBP-Shot-FRB-EGFP treated with 500 nM rapamycin, (D) Shot∆GAS2-EGFP, (E) Shot∆Rod-EGFP, (F) Shot∆EF-hand-EGFP, and (G) ShotGAS2-EGFP (green) and mCherry-tubulin (red). Scale bar: 10 μm. (H) Graph representing the mean Mander's coefficient quantifying fraction of overlap/colocalization between microtubules and EGFP-tagged Shot constructs. Error bars represent 95% confidence intervals. Statistical analysis was determined by one-way analysis of variance test with Tukey's multiple-comparison post hoc analysis performed on ∼30–40 different cells per condition. Statistical significance is denoted by asterisks (*, p < 0.05), while statistically insignificant comparisons between groups are denoted by n.s.
The EF-hand motifs contribute to regulation of Shot
The two EF-hand motifs that lie immediately upstream of the GAS2 domain are essential for axon extension, a process that requires actin-microtubule cross-linking by Shot (Lee and Kolodziej, 2002b). To test the hypothesis that the EF-hand motifs contribute to intramolecular inhibition of Shot, we constructed a full-length Shot BiFC probe lacking the EF-hand motifs (VC-Shot∆EF-hand-VN; lacking residues 5072–5135) and tested whether this construct could reconstitute fluorescence (Figure 2, J–L). We did not observe fluorescence above background levels when we expressed this construct in S2 cells (Figure 2K). However, when we coexpressed VC-Shot∆EF-hand-VN with EB1-VC, we did observe fluorescence along the lattice of microtubules in a manner similar to that seen in Shot∆EF-hand-EGFP, which contains a similar deletion (residues 5011–5152; Figure 2, J and L). This result suggests that without the EF-hand motifs, Shot remains in the “open” lattice-bound conformation.
EF-hand motifs are most often found in tandem, whereby they function as calcium-dependent molecular switches (Yap et al., 1999; Atkinson et al., 2001; Gifford et al., 2007). The presence of tandem EF-hand motifs in Shot raises the intriguing possibility that intramolecular inhibition could be regulated by calcium. In fact, a recent study of a mammalian homologue of Shot, BPAG1n4, demonstrated that increased calcium led to a switch in how the COOH-terminal domain of this protein interacted with microtubules (Kapur et al., 2012). To test the hypothesis that the EF-hand motifs of Shot regulate its activity by binding to calcium, we constructed a full-length Shot BiFC probe carrying mutations (D5080A, D5082A, N5084A) predicted to abolish calcium binding (Figure 2, M and N, and Movie S3). When expressed in S2 cells, this probe did reconstitute fluorescence at the microtubule plus end, which was similar to results seen with the full-length VC-Shot-VN BiFC probe (Figure 2, D and E). The coordinating residues of the second EF-hand motif appear to diverge from other canonical calcium-binding EF-hands, and we did not test their contribution. To further test the potential role of calcium in Shot's regulation, we visualized Shot A-EGFP dynamics in S2 cells following perfusion with the calcium ionophore A23187; however, this treatment did not lead to any major changes to Shot's localization or dynamics (unpublished data). We also tested whether addition of free calcium altered the degree of interaction between recombinant 6xHis-ABD and GST-EF-hand-GAS2-CTD, but did not observe this to be the case (unpublished data). These results indicate that the EF-hand motifs present at the Shot COOH-terminus are unlikely to contribute to Shot's conformational state by acting as calcium sensors.
We next compared the dynamics and localization of Shot∆EF-hand-EGFP with those of other previously characterized Shot constructs. Consistent with the BiFC results, Shot∆EF-hand-EGFP localized primarily to the microtubule lattice and colocalized with microtubules (M = 0.56; Figure 3, F and H) to an extent that was statistically indistinguishable from that of ShotGAS2-EGFP. These data further support the model that the EF-hand motifs are critical to maintaining the “closed” tip-tracking conformation of Shot.
The folded conformation of shot is cis-inhibited and unable to act as an actin-microtubule cross-linker
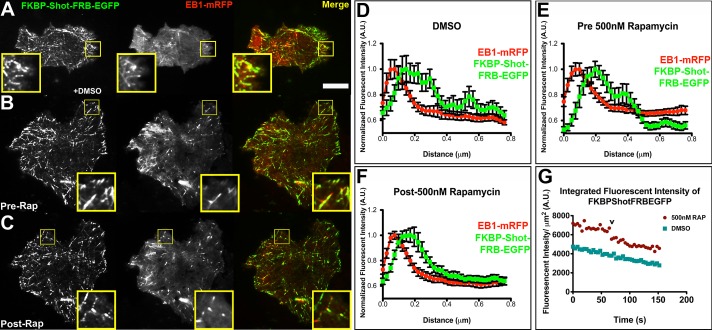
To experimentally control the conformation of Shot, we used the rapamycin-induced FKBP/FRB dimerization system to engineer FKBP-Shot-FRB-EGFP (Banaszynski et al., 2005, 2006; Slep and Vale, 2007; Figure 4). With the coding sequence of Shot flanked by FKBP and FRB, we predicted that addition of rapamycin would force the molecule into a closed and potentially inactive conformation. When FKBP-Shot-FRB-EGFP was transfected into S2 cells, its dynamics were indistinguishable from Shot A-GFP; it tracked with microtubule plus ends in the central regions of the cells and localized to the microtubule lattice in the cell periphery (Figure 4, A and B). On perfusion with rapamycin, however, FKBP-Shot-FRB-EGFP interaction with microtubules was dramatically altered (Figure 4C and Movies S4 and S5). We observed a reduction in microtubule lattice association of FKBP-Shot-FRB-EGFP association compared with dimethyl sulfoxide (DMSO) controls and documented this change by measuring the lengths of microtubule lattice decorated by FKBP-Shot-FRB-EGFP from the plus end (Supplemental Figure S1). We also measured this change by comparing the distribution of FKBP-Shot-FRB-EGFP with that of EB1, using fluorescence intensity line scans, and observed a decrease in the extent of lattice binding (Figure 4, D–F). In addition we calculated the Mander's coefficient to quantify the degree of colocalization between FKBP-Shot-FRB-EGFP and mCherry-tubulin in S2 cells treated with DMSO or 500 nM rapamycin (Figure 3, B, C, and H). Consistent with our live-cell imaging results, the Mander's coefficient for FKBP-Shot-FRB-EGFP and mCherry-tubulin was similar to that of Shot A-EGFP in cells treated with DMSO (M = 0.35); however, upon treatment with rapamycin, the degree to which Shot and mCherry-tubulin colocalized decreased (M = 0.22). The Mander's coefficient for rapamycin-treated cells was statistically indistinguishable from that of Shot∆GAS2-EGFP (Figure 3, D, C, and H). These data indicate that intramolecular association between Shot's NH2- and COOH-termini is sufficient to target it to microtubule plus ends.
FIGURE 4:
An induced conformational change is sufficient to target Shot to microtubule plus ends. S2 cells coexpressing FKBP-Shot-FRB-EGFP (green) and EB1-mRFP (red) following perfusion of DMSO (A), before perfusion of 500 nM rapamycin (B), and after perfusion of 500 nM rapamycin (C). Highlighted by the yellow boxes are portions of the cell periphery shown at higher magnification. (D–F) Quantitative line scans of 12–15 microtubules found in the cell periphery (∼1–5 μm from the cell edge) following perfusion of DMSO (D), just prior to perfusion of 500 nM rapamycin (E), and following perfusion of 500 nM rapamycin (F) from the cells pictured in (A–C); error bars represent SE. The quantification shown is from a single time-lapsed movie; however, the experiment was repeated at least five times with similar results. (G) Graphical representation of the integrated fluorescence intensity per cell area vs. time from the cells pictured in (A) and (B). The “v” denotes the time point when the rapamycin was perfused during the imaging. Image acquisition was sequential, with the “green” image acquired first. Scale bars: 10 μm in high-magnification images; 2 μm in low-magnification images.
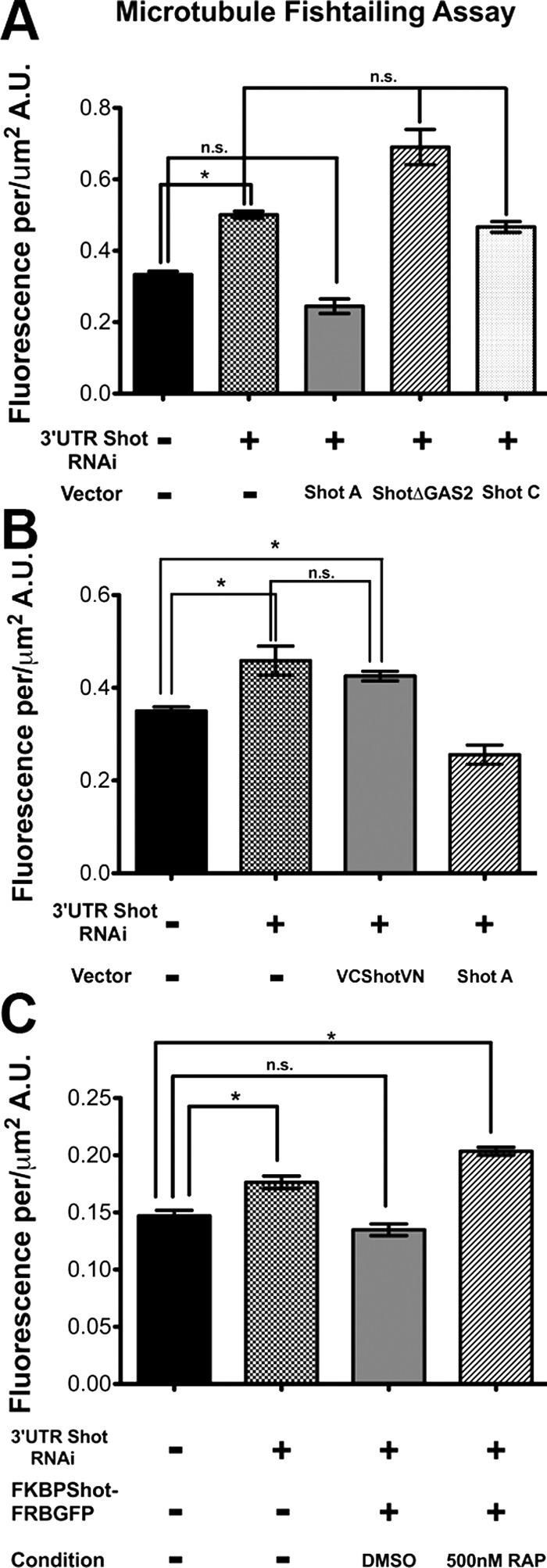
Our previous data suggested that Shot functions as a cross-linker when it is bound to the microtubule lattice (Applewhite et al., 2010). Shot depletion causes microtubules to exhibit abnormal lateral, “fishtail” movements in S2 cells that can be measured from time-lapse images of fluorescently-tagged tubulin (Figure 5). In this assay an increase in fluorescence correlates with increased microtubule displacement or “fishtailing.” Microtubule fishtailing was rescued in S2 cells depleted of endogenous Shot (3′UTR Shot RNA interference [RNAi]) by transfection with full-length Shot isoform A, but not with a version of Shot with the GAS2 domain deleted (ΔGAS2) nor with the Shot isoform C, which lacks a functional actin-binding domain (Figure 5A). These domains are also required for axon extension during Drosophila neurogenesis, a process that depends on Shot's cross-linking activity (Lee and Kolodziej, 2002b). Reconstitution of split Venus is an irreversible reaction and, therefore, is predicted to “trap” the NH2- and COOH-termini of Shot in its folded conformation. Because neither VN-Shot-VC nor VC-Shot-VN exhibited robust localization to the microtubule lattice (Figure 2, D and E), we hypothesized that these constructs would be unable to act as cytoskeletal cross-linkers. We tested the ability of VC-Shot-VN to rescue cells depleted of endogenous Shot with RNAi and found that this construct did not suppress microtubule fishtailing movements (Figure 5B). Comparison of the amount of microtubule lateral movements revealed that VC-Shot-VN–expressing cells were statistically indistinguishable from cells treated with 3′UTR Shot RNAi alone. Furthermore, we found that expression of FKBP-Shot-FRB-EGFP could rescue fishtailing, but that FKBP-Shot-FRB-EGFP treated with rapamycin could not (Figure 5C). These data support our hypothesis that, when Shot adopts a “closed” conformation at microtubule plus ends, it is unable to bind to the microtubule lattice and to function as an actin-microtubule cross-linker.
FIGURE 5:
Actin-microtubule cross-linking is required to stabilize the microtubule cytoskeleton. (A–C) Quantification of microtubule fishtailing using image subtraction analysis at 15-s intervals, as described by Applewhite et al. (2010). For each condition, 5 to 10 cells were analyzed. Asterisks indicate statistical significance as compared with control samples (*, p < 0.01, Student's t test); error bars represent SE. Statistically insignificant comparisons are denoted by n.s.
DISCUSSION
A major outstanding question that arises from our work is: What is the regulatory input that causes Shot to undergo this conformational change and actively cross-link actin filaments to microtubules? Two recent studies have addressed this issue and have presented two distinct (although not necessarily exclusive) activation signals. Wu et al. (2011) demonstrated that microtubule association of mouse ACF7/MACF1 is regulated by phosphorylation by GSK3β. In this system, phosphorylation of two GSK3β consensus sites in its COOH-terminus uncoupled ACF7 from microtubules, thus abrogating actin-microtubule cross-linking. Shot possesses putative Zeste-white 3 (Zw3; the fly GSK3β homologue) consensus sequences in the CTD; therefore, we looked for a regulatory role for this pathway by overexpressing constitutively active mCherry-Zw3 in S2 cells or inhibited the kinase by treatment with LiCl or Zw3 RNAi. None of these treatments affected the localization or dynamics of endogenous Shot, full-length Shot-GFP, or CTD-GFP (Figures S2 and S3).
In a second study, Kapur et al. (2012) proposed a different intramolecular inhibitory mechanism for human BPAG1n4 in which the EF-hand motif binds to the GAS2 domain to inhibit microtubule lattice association, and this inhibition is relieved by free calcium. The authors also showed that interactions between BPAG1n4 and EB1 were also inhibited in the presence of calcium. To test whether calcium affected Shot dynamics in our system, we manipulated cytosolic calcium levels in S2 cells by treatment with ionophore, thapsigargin, or BAPTA (1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid) but did not observe significant changes in Shot-GFP dynamics (unpublished data). We did confirm that the EF-hand motifs contribute to Shot regulation; however, our data do not support a role for calcium binding as the regulatory trigger. Thus the signal that relieves Shot autoinhibition in Drosophila remains an open area of investigation.
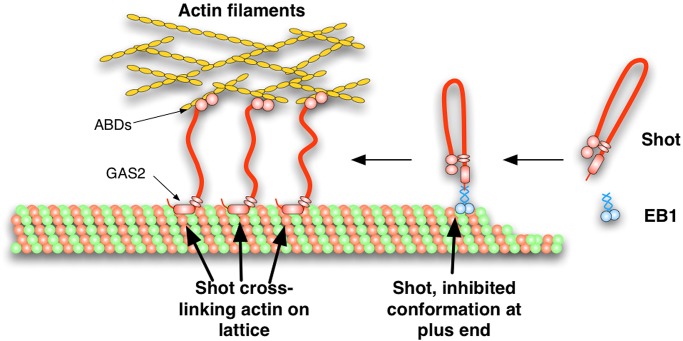
In this article, we demonstrate that Shot is able to adopt a folded conformation through an interaction between its NH2- and COOH-termini. We propose a model in which Shot interconverts between an extended conformation that is able to make cross-bridges between microtubules and actin filaments via the ABD and EF-hand-GAS2 domains, and a folded conformation that is targeted to the microtubule plus end by interaction with EB1 (Figure 6). A central component of this model is that the central rod domain of Shot, composed of 17 tandem spectrin repeats, affords the molecule sufficient flexibility for the NH2- and COOH-termini to interact. It will be interesting to examine the contributions of these individual repeats, which represent potentially flexible elements of the protein, and the plakin repeat domain to intramolecular inhibition of Shot. Given the high degree of conservation between Shot and spectraplakins in other species, as well as the published observations that mammalian homologues exhibit similar transitions between microtubule tip- and lattice-associated states, we speculate that this mechanism of regulation is likely conserved among members of this protein superfamily.
FIGURE 6:
A model for the autoinhibition of Shot's actin-microtubule cross-linking. Shot is targeted to the growing ends of microtubules through EB1 and is maintained in an autoinhibited conformation through and intramolecular interaction between Shot's ABD and EF-hand/GAS2 domains. On relief of this autoinhibition, Shot's conformation is “open,” allowing it to bind to the sides of microtubules through its GAS2 domain and to actin through its ABD, thereby cross-linking the two networks.
While our BiFC data are consistent with our biochemical data in supporting an intramolecular inhibition model, we were unable to directly observe the dynamics of this conformational change due to the irreversibility of this assay (Kerppola, 2008, 2009). Further experiments involving the use of Förster resonance energy transfer (FRET) may be required to capture the spatiotemporal regulation of this process. Additionally, techniques that can examine the structure of purified Shot, such as atomic force microscopy or electron microscopy, will provide a more conclusive test of our model (Lansbergen et al., 2004).
Autoinhibition is a common theme for protein regulation and has been described for a number of cytoskeletal proteins, such as conventional kinesin, CLIP-170, moesin, and the Diaphanous family of proteins (Alberts, 2001; Li and Higgs, 2003; Lansbergen et al., 2004; Wong et al., 2009; Wong and Rice, 2010; Lee et al., 2010; Kaan et al., 2011; Ben-Aissa et al., 2012). We speculate that this regulatory mechanism has evolved in spectraplakins so that they may utilize microtubule dynamic instability to search for localized upstream activation signals by a “search and capture” mechanism. For example, inhibited Shot interacts with microtubule plus ends via EB1; if the microtubule undergoes catastrophe in the absence of Shot-activating signal, Shot is released back into the soluble pool. If the growing microtubule encounters a Shot activation signal (e.g., a region of high kinase activity, second messenger molecule, or integrin activation), then Shot adopts the open conformation and cross-links the microtubule to the local actin network. Microtubules tethered in this manner may then be stabilized, perhaps by interaction with CLASP/Orbit, or they may continue to grow using actin structures as polymerization guides (Drabek et al., 2006).
MATERIALS AND METHODS
Cell culture and RNAi
Drosophila S2 cell culture and RNAi were performed as described by Rogers and Rogers (2008). Briefly, S2 cells (Drosophila Genomics Resource Center, Bloomington, IN) were cultured in SF900II medium supplemented with 100× antibiotic–antimycotic (Invitrogen, Carlsbad, CA). RNAi was administered in six-well plates by treating cells (∼50% confluent) with 10 μg double-stranded RNA (dsRNA) in 1 ml of medium each day for 7 d.
DNA constructs and transfection
The pUAST-ShotA-EGFP, pUAST-ShotC-EGFP, pUAST-ShotΔGAS2-EGFP, and pUAST-ShotΔRod1-EGFP plasmids were first described by Lee and Kolodziej (2002b). For expression in S2 cells, pUAST-plasmids were cotransfected with a pMT-Gal4 plasmid. For BiFC-tagged constructs, the NH2 half (VN, residues 1–173) and COOH half (VC, residues 155–239) of Venus were generated by standard PCR procedures and cloned into pMT vectors (Invitrogen). We included a five–amino acid linker region (RSIAT) on both ends. Full-length Shot PCR products were generated by standard procedures using KOD Xtreme polymerase (EMD Millipore, Billerica, MA). pMT-mCherry-Zw3 was constructed using standard PCR procedures and Ser-9 was mutated to alanine by site-directed mutagenesis. Expression of pMT vectors was achieved with ∼25–50 μM final concentration of copper sulfate (Thermo Fisher Scientific, Waltman, MA). pMT Shot fragments used in immunoprecipitation experiments were cloned using standard PCR procedures. Expression was achieved with 250–500 μM final concentration of copper sulfate.
Protein purification and in vitro binding assays
Full-length EB1, GST, and fragments of Mini spindles and Shot were expressed and harvested from BL21 Escherichia coli. For production of 6xHis-tagged fusion proteins, the NH2-terminal ABD of Shot A isoform (residues 143–398), full-length EB1, and the first two TOG domains from Mini spindles (residues 1–512) were cloned into pET28a and purified on Ni-NTA resin (Qiagen, Valencia, CA). GST-tagged fusion of the EF-hand-GAS2-CTD fragment of Shot (residues 5001–5409) was cloned into pGEX6P2 and purified using glutathione fast-flow resin (GE Healthcare, Pittsburgh, PA).
In vitro binding assays were performed by incubating Ni-NTA resin with ∼10 mg 6xHis fusion prey proteins in 20 mM Tris (pH 8) and 150 mM NaCl and mixing with ∼10 μg GST fusion protein bait proteins for 30 min. The beads were washed three times and then boiled in SDS–PAGE sample buffer, separated by SDS–PAGE, and visualized with Coomassie Blue stain.
Immunoprecipitation/Western blotting
Transfected S2 cells were induced to express the various epitope-tagged constructs with 500 μM CuSO4 for 24 h. Cells were then lysed in cell lysis buffer (CLB; 50 mM Tris, 150 mM NaCl, 0.5 mM EDTA, 1 mM dithiothreitol, 0.5% Triton X-100, and 2.5 mM phenylmethylsulfonyl fluoride) and precleared, and lysates were diluted twofold in CLB. Lysates were mixed with anti-Myc (Sigma-Aldrich, St. Louis, MO) or anti-FLAG (Sigma-Aldrich) for 2 h at 4°C, and then protein A-Sepharose beads or protein G-Sepharose beads (Sigma-Aldrich) were incubated with the antibody–lysate mixture for 2 h at 4°C. The beads were washed three times with 0.5 ml of CLB and boiled in SDS–PAGE sample buffer, and then proteins were analyzed by SDS–PAGE immunoblotted with anti-FLAG Clone M2 (Sigma-Aldrich) or anti-Myc 9E10 (provided by the Developmental Hybridoma Bank, Department of Biology, University of Iowa, Iowa City, IA). Western blots were developed with horseradish peroxidase–tagged secondary antibodies and ECL reagents (Thermo Fisher Scientific). In addition to the antibodies listed above, Western blots were also probed with anti-DM1α (Sigma-Aldrich), anti-dsRed (Clontech, Mountain View, CA), and anti-Armadillo (Developmental Hybridoma Bank).
Immunofluorescence and live-cell imaging
Immunofluorescence was performed on S2 cells plated on 0.5 Rogers et al. (2002). Antibodies used this study include the anti-Rod (Short-stop; Developmental Hybridoma Bank), diluted 1:300 in phosphate-buffered solution and 0.1% Triton X-100 (PBST), and anti-EB1 (Rogers et al., 2002), diluted 1:400 in PBST. Secondary antibodies (Cy2 and Rhodamine Red; Jackson ImmunoResearch, West Grove, PA) were used at a final dilution of 1:100 in PBST. For live-cell imaging, S2 cells were plated on 0.5 mg/m Con A–treated coverslips attached to drilled 35-mm tissue culture dishes with UV-curable adhesive (Norland Products, Cranbury, NJ) in Schneider's Drosophila medium supplemented with 10% fetal bovine serum and 100× antibiotic–antimycotic (Invitrogen). Cells were allowed to attach for at least 1 h before imaging. When treated with rapamycin (EMD Millipore), cells were perfused with 250–500 nM final concentration. Time-lapse imaging was performed on two microscopes. The first was a Hawk-VT multi-point array scanning confocal (VisiTech International, Sunderland, UK) mounted on an inverted microscope (Eclipse TE300; Nikon, Tokyo, Japan) equipped with a 100×/1.4 numerical aperture objective lens driven by VisiTech Vox software. The second was a laser total internal reflection fluorescence (TIRF) system (Nikon) mounted on an inverted microscope (Ti; Nikon) equipped with a 100×/1.49 objective lens driven by Nikon Elements software. Images were captured with an Orca-R2 camera (Hamamatsu, Hamamatsu, Japan) or an Andor-Clara Interline camera (Andor Technology, Belfast, UK). All images were processed for brightness and contrast and prepared for publication using Photoshop (CS version 8.0; Adobe Systems, Mountain View, CA).
Colocalization analysis
The mean fraction of overlap between tubulin and EGFP-tagged Shot was calculated using Mander's coefficient (described in Bolte and Cordelières, 2006) and the JACoP plug-in in ImageJ (National Institutes of Health [NIH], Bethesda, MD). Prior to analysis, images were thresholded. The Mander's coefficient varies from zero to one, with zero representing nonoverlapping images and one representing complete overlap/colocalization. All images used for analysis were acquired from live cells by TIRF microscopy using identical microscope settings.
To exclude the possibility that overexpression artifacts affected the degree of colocalization between the EGFP-tagged Shot constructs and mCherry-tubulin, we compared the Mander's coefficient with the fluorescence intensity per cell area (Figure S4). The correlation between the Mander's coefficient and the fluorescence per cell area in cells expressing Shot A-EGFP, FKBP-Shot-FRB-EGFP (DMSO treated), and ShotGAS2-EGFP was statistically significant, indicating that the extent of colocalization increased with increasing expression levels of these constructs. In contrast, correlation between the Mander's coefficient and the fluorescence intensity per cell area was not statistically significant for ShotΔEFhand-EGFP, ShotΔGAS2-EGFP, and ShotΔRod-EGFP, indicating that microtubule localization of these constructs was independent of expression level and did not reflect an artifact of overexpression.
Supplementary Material
Acknowledgments
We thank members of the Peifer, Rogers, and Slep lab for their helpful comments during the preparation of this article. This work was supported in part by National Cancer Institute grant K01-CA163972-01 (to D.A.A.) and NIH grant R01GM081645 (to S.L.R.).
Abbreviations used:
- +TIPs
microtubule plus end–interacting proteins
- 3′UTR
3′ untranslated region of mRNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-11-0798) on July 24, 2013.
REFERENCES
- Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276:2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- Alves-Silva J, et al. Spectraplakins promote microtubule-mediated axonal growth by functioning as structural microtubule-associated proteins and EB1-dependent +TIPs (tip interacting proteins) J Neurosci. 2012;32:9143–9158. doi: 10.1523/JNEUROSCI.0416-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applewhite DA, Grode KD, Keller D, Zadeh AD, Zadeh A, Slep KC, Rogers SL. The spectraplakin Short stop is an actin-microtubule cross-linker that contributes to organization of the microtubule network. Mol Biol Cell. 2010;21:1714–1724. doi: 10.1091/mbc.E10-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson RA, Joseph C, Kelly G, Muskett FW, Frenkiel TA, Nietlispach D, Pastore A. Ca2+-independent binding of an EF-hand domain to a novel motif in the α-actinin-titin complex. Nat Struct Biol. 2001;8:853–857. doi: 10.1038/nsb1001-853. [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Chen L-C, Maynard-Smith LA, Ooi AGL, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP⋅rapamycin⋅FRB ternary complex. J Am Chem Soc. 2005;127:4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- Ben-Aissa K, Patino-Lopez G, Belkina NV, Maniti O, Rosales T, Hao J-J, Kruhlak MJ, Knutson JR, Picart C, Shaw S. Activation of moesin, a protein that links actin cytoskeleton to the plasma membrane, occurs by phosphatidylinositol 4,5-bisphosphate (PIP2) binding sequentially to two sites and releasing an autoinhibitory linker. J Biol Chem. 2012;287:16311–16323. doi: 10.1074/jbc.M111.304881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier G, Mathieu M, De Repentigny Y, Vidal SM, Kothary R. Cloning and characterization of mouse ACF7, a novel member of the dystonin subfamily of actin binding proteins. Genomics. 1996;38:19–29. doi: 10.1006/geno.1996.0587. [DOI] [PubMed] [Google Scholar]
- Bhasin N, Law R, Liao G, Safer D, Ellmer J, Discher BM, Sweeney HL, Discher DE. Molecular extensibility of mini-dystrophins and a dystrophin rod construct. J Mol Biol. 2005;352:795–806. doi: 10.1016/j.jmb.2005.07.064. [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Bottenberg W, Sánchez-Soriano N, Alves-Silva J, Hahn I, Mende M, Prokop A. Context-specific requirements of functional domains of the Spectraplakin Short stop in vivo. Mech Dev. 2009;126:489–502. doi: 10.1016/j.mod.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Drabek K, et al. Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Curr Biol. 2006;16:2259–2264. doi: 10.1016/j.cub.2006.09.065. [DOI] [PubMed] [Google Scholar]
- Edvardson S, Cinnamon Y, Jalas C, Shaag A, Maayan C, Axelrod FB, Elpeleg O. Hereditary sensory autonomic neuropathy caused by a mutation in dystonin. Ann Neurol. 2012;71:569–572. doi: 10.1002/ana.23524. [DOI] [PubMed] [Google Scholar]
- Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- Gong TW, Besirli CG, Lomax MI. MACF1 gene structure: a hybrid of plectin and dystrophin. Mamm Genome. 2001;12:852–861. doi: 10.1007/s00335-001-3037-3. [DOI] [PubMed] [Google Scholar]
- Gupta T, Marlow FL, Ferriola D, Mackiewicz K, Dapprich J, Monos D, Mullins MC. Microtubule actin crosslinking factor 1 regulates the Balbiani body and animal-vegetal polarity of the zebrafish oocyte. PLoS Genet. 2010;6:e1001073. doi: 10.1371/journal.pgen.1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Jefferson JJ, Ciatto C, Shapiro L, Liem RKH. Structural analysis of the plakin domain of bullous pemphigoid antigen1 (BPAG1) suggests that plakins are members of the spectrin superfamily. J Mol Biol. 2007;366:244–257. doi: 10.1016/j.jmb.2006.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson JJ, Leung CL, Liem RKH. Plakins: goliaths that link cell junctions and the cytoskeleton. Nat Rev Mol Cell Biol. 2004;5:542–553. doi: 10.1038/nrm1425. [DOI] [PubMed] [Google Scholar]
- Kaan HYK, Hackney DD, Kozielski F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 2011;333:883–885. doi: 10.1126/science.1204824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur M, Wang W, Maloney MT, Millan I, Lundin VF, Tran T-A, Yang Y. Calcium tips the balance: a microtubule plus end to lattice binding switch operates in the carboxyl terminus of BPAG1n4. EMBO Rep. 2012;13:1021–1029. doi: 10.1038/embor.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem Soc Rev. 2009;38:2876–2886. doi: 10.1039/b909638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-S, Murakami R, Quintin S, Mori M, Ohkura K, Tamai KK, Labouesse M, Sakamoto H, Nishiwaki K. VAB-10 spectraplakin acts in cell and nuclear migration in Caenorhabditis elegans. Development. 2011;138:4013–4023. doi: 10.1242/dev.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E. ACF7: an essential integrator of microtubule dynamics. Cell. 2003;115:343–354. doi: 10.1016/s0092-8674(03)00813-4. [DOI] [PubMed] [Google Scholar]
- Kusunoki H, Minasov G, Macdonald RI, Mondragón A. Independent movement, dimerization and stability of tandem repeats of chicken brain α-spectrin. J Mol Biol. 2004;344:495–511. doi: 10.1016/j.jmb.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Lansbergen G, et al. Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization. J Cell Biol. 2004;166:1003–1014. doi: 10.1083/jcb.200402082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AC, Suter DM. Quantitative analysis of microtubule dynamics during adhesion-mediated growth cone guidance. Dev Neurobiol. 2008;68:1363–1377. doi: 10.1002/dneu.20662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-S, et al. Phosphorylation controls autoinhibition of cytoplasmic linker protein-170. Mol Biol Cell. 2010;21:2661–2673. doi: 10.1091/mbc.E09-12-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Harris KL, Whitington PM, Kolodziej PA. Short stop is allelic to kakapo, and encodes rod-like cytoskeletal-associated proteins required for axon extension. J Neurosci. 2000;20:1096–1108. doi: 10.1523/JNEUROSCI.20-03-01096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kolodziej PA. The plakin Short Stop and the RhoA GTPase are required for E-cadherin-dependent apical surface remodeling during tracheal tube fusion. Development. 2002a;129:1509–1520. doi: 10.1242/dev.129.6.1509. [DOI] [PubMed] [Google Scholar]
- Lee S, Kolodziej PA. Short Stop provides an essential link between F-actin and microtubules during axon extension. Development. 2002b;129:1195–1204. doi: 10.1242/dev.129.5.1195. [DOI] [PubMed] [Google Scholar]
- Li F, Higgs HN. The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- Määttä A, Hutchison CJ, Watson MD. Spectraplakins and nesprins, giant spectrin repeat proteins participating in the organization of the cytoskeleton and the nuclear envelope. Symp Soc Exp Biol. 2004;2004:265–277. [PubMed] [Google Scholar]
- Miller AL, Wang Y, Mooseker MS, Koleske AJ. The Abl-related gene (Arg) requires its F-actin-microtubule cross-linking activity to regulate lamellipodial dynamics during fibroblast adhesion. J Cell Biol. 2004;165:407–419. doi: 10.1083/jcb.200308055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat Protoc. 2008;3:606–611. doi: 10.1038/nprot.2008.18. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röper K, Brown NH. Maintaining epithelial integrity: a function for gigantic spectraplakin isoforms in adherens junctions. J Cell Biol. 2003;162:1305–1315. doi: 10.1083/jcb.200307089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röper K, Gregory SL, Brown NH. The “spectraplakins”: cytoskeletal giants with characteristics of both spectrin and plakin families. J Cell Sci. 2002;115:4215–4225. doi: 10.1242/jcs.00157. [DOI] [PubMed] [Google Scholar]
- Rosales-Nieves AE, Johndrow JE, Keller LC, Magie CR, Pinto-Santini DM, Parkhurst SM. Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat Cell Biol. 2006;8:367–376. doi: 10.1038/ncb1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg ME, Rogers SL, Vale RD, Jan LY, Jan Y-N. Drosophila pod-1 crosslinks both actin and microtubules and controls the targeting of axons. Neuron. 2003;39:779–791. doi: 10.1016/s0896-6273(03)00508-7. [DOI] [PubMed] [Google Scholar]
- Salmon WC, Adams MC, Waterman-Storer CM. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J Cell Biol. 2002;158:31–37. doi: 10.1083/jcb.200203022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AW, Kabir N, Forscher P. Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J Cell Biol. 2002;158:139–152. doi: 10.1083/jcb.200203038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep KC, Rogers SL, Elliott SL, Ohkura H, Kolodziej PA, Vale RD. Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J Cell Biol. 2005;168:587–598. doi: 10.1083/jcb.200410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg A, Liem RKH. Plakins in development and disease. Exp Cell Res. 2007;313:2189–2203. doi: 10.1016/j.yexcr.2007.03.039. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Volk T. Kakapo, a novel cytoskeletal-associated protein is essential for the restricted localization of the neuregulin-like factor, Vein, at the muscle-tendon junction site. J Cell Biol. 1998;143:1259–1270. doi: 10.1083/jcb.143.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Prokop A, Yamamoto M, Sugimura K, Uemura T, Betschinger J, Knoblich JA, Volk T. Shortstop recruits EB1/APC1 and promotes microtubule assembly at the muscle-tendon junction. Curr Biol. 2003;13:1086–1095. doi: 10.1016/s0960-9822(03)00416-0. [DOI] [PubMed] [Google Scholar]
- Suozzi KC, Wu X, Fuchs E. Spectraplakins: master orchestrators of cytoskeletal dynamics. J Cell Biol. 2012;197:465–475. doi: 10.1083/jcb.201112034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer C, Duey DY, Weber KL, Keech J, Cheney RE, Salmon ED, Bement WM. Microtubules remodel actomyosin networks in Xenopus egg extracts via two mechanisms of F-actin transport. J Cell Biol. 2000;150:361–376. doi: 10.1083/jcb.150.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of leading edge microtubule and actin dynamics downstream of Rac1. J Cell Biol. 2003;161:845–851. doi: 10.1083/jcb.200303082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YL, Dietrich KA, Naber N, Cooke R, Rice SE. The kinesin-1 tail conformationally restricts the nucleotide pocket. Biophys J. 2009;96:2799–2807. doi: 10.1016/j.bpj.2008.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YL, Rice SE. Kinesin's light chains inhibit the head- and microtubule-binding activity of its tail. Proc Natl Acad Sci USA. 2010;107:11781–11786. doi: 10.1073/pnas.1005854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Kodama A, Fuchs E. ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity. Cell. 2008;135:137–148. doi: 10.1016/j.cell.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Shen Q-T, Oristian DS, Lu CP, Zheng Q, Wang H-W, Fuchs E. Skin stem cells orchestrate directional migration by regulating microtubule-ACF7 connections through GSK3β. Cell. 2011;144:341–352. doi: 10.1016/j.cell.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Ames JB, Swindells MB, Ikura M. Diversity of conformational states and changes within the EF-hand protein superfamily. Proteins. 1999;37:499–507. doi: 10.1002/(sici)1097-0134(19991115)37:3<499::aid-prot17>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.