Yeast Tip20, a subunit of the Dsl1 complex, is implicated in Golgi-to–endoplasmic reticulum retrograde transport. Differing from Tip20, its mammalian counterpart, RINT-1, is required for endosome-to–trans-Golgi network transport. RINT-1 in coordination with the COG complex regulates SNARE complex assembly at the trans-Golgi network.
Abstract
Docking and fusion of transport vesicles/carriers with the target membrane involve a tethering factor–mediated initial contact followed by soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE)–catalyzed membrane fusion. The multisubunit tethering CATCHR family complexes (Dsl1, COG, exocyst, and GARP complexes) share very low sequence homology among subunits despite likely evolving from a common ancestor and participate in fundamentally different membrane trafficking pathways. Yeast Tip20, as a subunit of the Dsl1 complex, has been implicated in retrograde transport from the Golgi apparatus to the endoplasmic reticulum. Our previous study showed that RINT-1, the mammalian counterpart of yeast Tip20, mediates the association of ZW10 (mammalian Dsl1) with endoplasmic reticulum–localized SNARE proteins. In the present study, we show that RINT-1 is also required for endosome-to–trans-Golgi network trafficking. RINT-1 uncomplexed with ZW10 interacts with the COG complex, another member of the CATCHR family complex, and regulates SNARE complex assembly at the trans-Golgi network. This additional role for RINT-1 may in part reflect adaptation to the demand for more diverse transport routes from endosomes to the trans-Golgi network in mammals compared with those in a unicellular organism, yeast. The present findings highlight a new role of RINT-1 in coordination with the COG complex.
INTRODUCTION
Eukaryotic cells contain an endomembrane system that consists of morphologically and functionally distinct organelles. Communication between organelles is mediated by coated vesicular/tubular carriers that are generated from the donor compartment, traffic to their destinations, lose their coat, and fuse with the acceptor compartment. Docking and fusion of transport carriers with the target membrane involve an initial contact mediated by Rab GTPases and tethering factors, followed by soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE)–catalyzed membrane fusion (Bonifacino and Glick, 2004).
Tethering factors are large proteins or protein complexes that not only facilitate long-range interactions between transport carriers and the acceptor membrane, but also coordinate SNARE complex assembly (Bröcker et al., 2010; Brown and Pfeffer, 2010; Yu and Hughson, 2010). They are classified into two types: homo-oligomeric extended coiled-coil proteins and multisubunit tethering complexes. The CATCHR complexes are a subfamily of the multisubunit tethering complexes consisting of the Dsl1, COG, exocyst, and GARP complexes (Yu and Hughson, 2010). Despite often-subtle sequence homology among subunits, recent structural studies revealed that they share a common structure and suggest that they derive from a common progenitor (Yu and Hughson, 2010).
The Dsl1 complex consists of three subunits (Dsl1, Tip20, and Sec39/Dsl3) and participates in retrograde transport from the Golgi to the endoplasmic reticulum (ER; Schmitt, 2010). X-ray crystallographic analyses revealed that Dsl1 and Tip20 have common α-helical folds (Tripathi et al., 2009), whereas Sec39/Dsl3 lacks the shared fold (Ren et al., 2009). The Dsl1 complex resides on ER membranes and binds to and regulates the assembly of the ER SNAREs Sec20, Ufe1, and Use1/Slt1 (Kraynack et al., 2005; Ren et al., 2009; Diefenbacher et al., 2011; Meiringer et al., 2011). The structural model suggests that the Dsl1 complex forms a 20-nm-tall tower from the ER surface (Ren et al., 2009). Given that Dsl1 interacts with subunits of the COPI, the Dsl1 complex likely serves as an acceptor for Golgi-derived COPI-coated carriers (Andag et al., 2001; Andag and Schmitt, 2003; Reilly et al., 2001). Moreover, the Dsl1 complex may assist uncoating of COPI-coated carriers tethered on ER membranes (Zink et al., 2009).
The mammalian orthologue of the Dsl1 complex is composed of ZW10 (Dsl1), RINT-1 (Tip20), and NAG (Sec39/Dsl3; Aoki et al., 2009). Like the Dsl1 complex (Meiringer et al., 2011), the ZW10 complex can associate with the ER SNAREs syntaxin 18 (Ufe1), BNIP1 (Sec20), p31 (Use1/Slt1), and Sec22b (Sec22; Hatsuzawa et al., 2000; Hirose et al., 2004; Nakajima et al., 2004; Aoki et al., 2009). The protein–protein interactions among Dsl1 complex subunits and between Dsl1 complex and SNARE subunits are also well conserved between yeast and mammals (Sweet and Pelham, 1993; Hirose et al., 2004; Nakajima et al., 2004; Aoki et al., 2009; Ren et al., 2009; Tripathi et al., 2009; Uemura et al., 2009; Meiringer et al., 2011).
RINT-1 was originally discovered as a Rad50-interacting protein and implicated in cell cycle control (Xiao et al., 2001). Depletion of RINT-1 causes partial Golgi fragmentation (Arasaki et al., 2006; Sun et al., 2007), together with defects in mitosis, including centrosome amplification and chromosome loss (Lin et al., 2007). Rint-1 heterozygotes succumb to multiple tumor formation with haploinsufficiency (Lin et al., 2007). RINT-1 also interacts with Rb-related p130 and has been implicated in telomere length control (Kong et al., 2006). These results raise the possibility that RINT-1 plays additional roles by interacting with unidentified partners. Indeed, in yeast Tip20 mutants, crystal-like structures are formed in the nucleus by an unknown mechanism (Spang, 2012).
In the present study, we show that RINT-1 is required for endosome-to–trans-Golgi network (TGN) transport. Immunoprecipitation and binding studies revealed that RINT-1 interacts with TGN SNAREs and Cog1, a subunit of the octameric COG complex (Smith and Lupashin, 2008; Miller and Ungar, 2012), which also belongs to the CATCHR family complex (Yu and Hughson, 2010).
RESULTS
Depletion of RINT-1 causes redistribution of TGN proteins
We and others previously reported that depletion of RINT-1 causes partial Golgi fragmentation with some change in the staining pattern for ER-Golgi intermediate compartment (ERGIC)-53 (Arasaki et al., 2006; Lin et al., 2007; Sun et al., 2007), an ERGIC marker that maintains its steady-state localization by cycling between the ERGIC and ER (Appenzeller-Herzog and Hauri, 2006). RINT-1 depletion, however, does not significantly inhibit brefeldin A– or Sar1p mutant–induced retrograde transport from the Golgi to the ER (Arasaki et al., 2006). To further characterize the contribution of RINT-1 to retrograde transport to the ER, we examined the distribution of other ER-Golgi/ERGIC recycling proteins in RINT-1–depleted cells. When HeLa cells were treated for 72 h with short interfering RNA (siRNA) targeting RINT-1 (RINT-1 (1149)), which was used in our previous study (Arasaki et al., 2006), RINT-1 expression was markedly reduced concomitant with partial dispersal of a cis-Golgi protein, GM130 (Supplemental Figure S1A). RINT-1 depletion by this siRNA, however, did not markedly disrupt the distribution of two ER-Golgi/ERGIC recycling proteins (Rer1 and KDEL receptor; Figure 1A, second row). Merged images show the colocalization of Rer1 and KDEL receptor with a cis-Golgi marker GM130 in RINT-1 (1149)–treated cells. Whereas treatment of cells with another siRNA (RINT-1 (687)) caused almost complete depletion of RINT-1 and substantial dispersal of GM130 (Supplemental Figure S1A), GM130 was found to be still colocalized with Rer1 and KDEL receptor (Figure 1A, third row). Depletion of RINT-1 by either siRNA had no effect on the ER structure (Figure 1B). These results suggest that RINT-1 depletion has a similar effect on the localization of cis-Golgi and recycling proteins (Rer1 and KDEL receptor). On the other hand, depletion of ZW10 mildly disrupted the localization of these recycling proteins, as seen by an increase in diffuse staining throughout the cytosol, with partial fragmentation of GM130-positive Golgi structure (Supplemental Figure S1B and Figure 1A, fourth row). Moreover, NAG depletion markedly disturbed the distribution of the recycling proteins with marginal effect on the distribution of GM130 (Figure 1A, bottom row; Aoki et al., 2009). These results suggest that depletion of RINT-1 may have less effect on the distribution of Rer1 and KDEL receptor than that of NAG and perhaps ZW10, although it affects the distribution of ERGIC-53 (Arasaki et al., 2006)
FIGURE 1:
Effects of depletion of RINT-1 on the distribution of ER-Golgi/ERGIC recycling proteins and the ER structure. (A) HeLa cells were mock transfected (top) or transfected with siRNA RINT-1 (1149) (second row), RINT-1 (687) (third row), ZW10 (102) (fourth row), or NAG (4160) (bottom). After 72 h, the cells were double stained with antibodies against GM130 and Rer1 or KDEL receptor. Merged images are also shown. Bar, 5 μm. (B) HeLa cells were mock transfected or transfected with siRNA RINT-1 (1149) or RINT-1 (687). After 72 h, the cells were single stained with an antibody against calnexin or Hsp47. Bar, 5 μm.
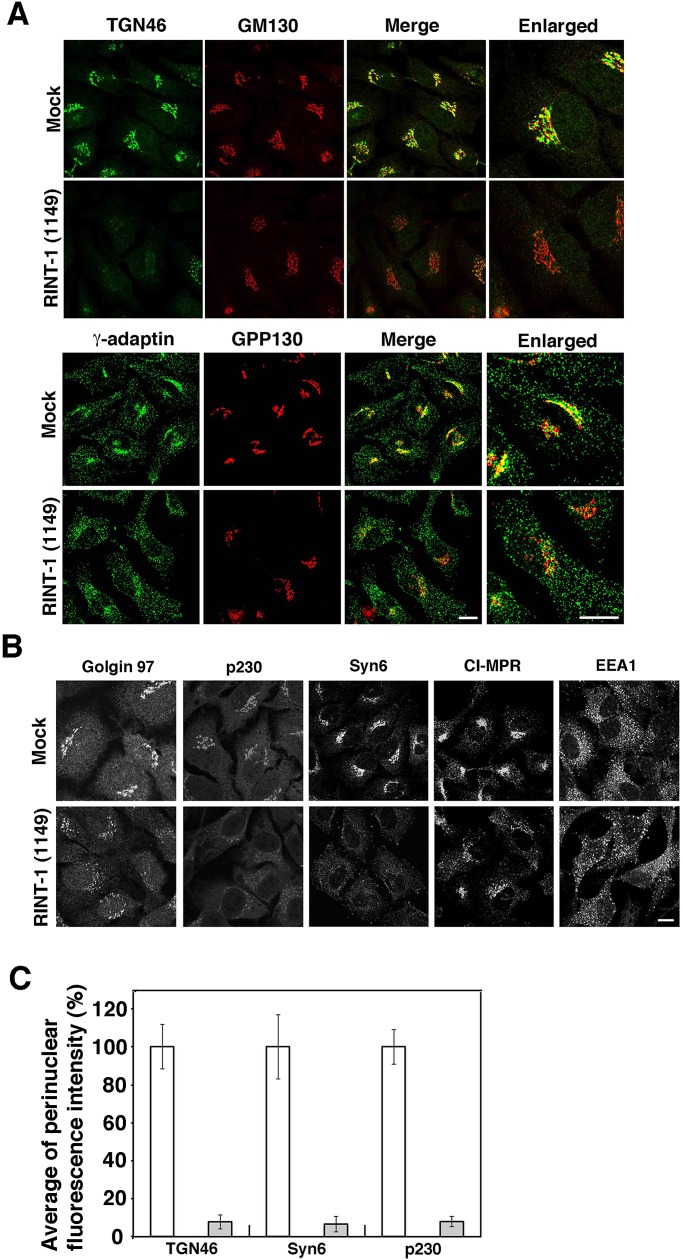
Unexpectedly, RINT-1 depletion severely disrupted the distribution of TGN proteins. RINT-1 depletion caused loss of TGN46 staining and the dispersal of γ-adaptin with relatively minor changes in the localization of cis-Golgi proteins (GM130 and GPP130; Figure 2, A and C). As shown in Figure 2, B and C, depletion of RINT-1 also caused the dispersal of other TGN markers (golgin-97, p230, and syntaxin 6), and a TGN-endosome marker, cation-independent mannose 6-phosphate receptor (CI-MPR). On the other hand, the distribution of an early endosome marker, EEA1, was not significantly altered by RINT-1 depletion, although EEA1-positive structures might be slightly dilated (Figure 2B). When HeLa cells were treated with RINT-1 (687), similar dispersal of TGN markers was observed (Supplemental Figure S2). Merged images clearly showed that TGN markers (TGN46 and γ-adaptin) were more dispersed than cis-Golgi markers (GM130 and GPP130) in RINT-1 (687)–treated cells (Supplemental Figure S2A).
FIGURE 2:
Depletion of RINT-1 disrupts the localization of TGN proteins more than that of cis-Golgi proteins. (A) HeLa cells were mock transfected or transfected with RINT-1 (1149), incubated for 72 h, and double stained with antibodies against TGN46 and GM130 (top two rows) or γ-adaptin and GPP130 (bottom two rows). Merged and enlarged images are also shown. Bars, 5 μm. (B) HeLa cells were treated as described in A and single stained with the indicated antibodies. Bar, 5 μm. (C) Quantitative data. HeLa cells were double stained with antibodies against each TGN protein and a cis-Golgi protein (GM130 or GPP130). Fluorescence intensity for each TGN protein in the perinuclear region was measured using ImageJ (National Institutes of Health, Bethesda, MD). In RINT-1 (1149)–treated cells, the perinuclear Golgi region was deduced from the position of cis-Golgi markers. The average fluorescence intensity in RINT-1 (1149)–treated cells (gray bars) was expressed as percentage of that in mock-treated cells (white bars). Data are the average of three independent experiments (n [cell number] = 30). Error bars represent SD.
To exclude the possibility that dispersal of TGN markers by RINT-1 depletion is due to off-target effects of the siRNAs used, we performed rescue experiments. Because overexpression of RINT-1 per se caused dispersal of TGN markers, as described later, we transfected RINT-1 (1149)–treated cells with an unmutated RINT-1 construct fused to the C-terminus of green fluorescent protein (GFP). We expected that overexpression of GFP–RINT-1 would be prevented by the siRNA, which might allow expression of GFP–RINT-1 at a level adequate for rescuing siRNA-treated cells. As shown in Supplemental Figure S3, perinuclear TGN46 staining was retained in 37% of the GFP–RINT-1–expressing cells treated with RINT-1 (1149). This relatively low rescue efficiency is likely attributable to an overexpression effect. On the other hand, moderate expression of the central and C-terminal regions of RINT-1 (amino acids 265–792) failed to prevent siRNA-induced TGN46 dispersion. These results corroborate that TGN dispersion is a consequence of RINT-1 depletion.
In contrast to RINT-1 depletion, depletion of ZW10, a RINT-1 partner in the ER, did not induce marked changes in the distribution of TGN46 (Supplemental Figure S4, top) or γ-adaptin (bottom).
Depletion of RINT-1 inhibits transport from endosomes to the TGN
Because the localization of TGN proteins is regulated by transport from endosomes to the TGN (Pfeffer, 2011), the most straightforward interpretation of the foregoing results is that RINT-1 is involved in endosome-to-TGN transport. To verify this possibility, we first measured the trafficking of cholera toxin B (CTB). CTB is endocytosed and transported to the ER via endosomes and the Golgi (Sandvig and van Deurs, 2002). To monitor CTB transport, we incubated HeLa cells with fluorescence-labeled CTB at 4ºC for 30 min, followed by incubation at 37ºC for 45 and 90 min. As shown in Figure 3, top, CTB reached the perinuclear, Golgi region at 45 min in mock-treated cells. In RINT-1–depleted cells, on the other hand, CTB did not accumulate at the perinuclear region even after incubation for 90 min, and instead it was almost completely colocalized with an early endosome marker, EEA1, in most cells (Figure 3, middle). Note that depletion of ZW10 had essentially no effect on the transport of CTB (Figure 3, bottom). These results suggest that the endosome-to-TGN trafficking is impaired in RINT-1–depleted cells.
FIGURE 3:
Endosome-to-TGN transport of CTB is impaired in RINT-1–depleted cells. HeLa cells were mock transfected (top) or transfected with RINT-1 (1149) (middle) or ZW10 (102) (bottom). After 72 h, the cells were allowed to bind Alexa Fluor 594–conjugated CTB for 30 min at 4ºC (left) followed by chase at 37ºC for 45 (middle) and 90 min (right). The cells were analyzed by immunofluorescence microscopy after staining with an anti-EEA1 antibody. Bar, 5 μm.
We next examined the effect of RINT-1 depletion on the transport of TGN38 (the rodent orthologue of primate TGN46) from the cell surface to the TGN, using an antibody uptake assay (Reaves et al., 1993). TGN38 constitutively cycles between the TGN and the plasma membrane through early/recycling endosomes (Ghosh et al., 1998). A plasmid encoding FLAG-tagged TGN38 was transfected into cells that had been treated with RINT-1 (1149). At 24 h after transfection, the cells were incubated with an anti-FLAG antibody for 3, 15, and 45 min. In mock-treated cells, the antibody was rapidly internalized into most cells and reached the perinuclear region at 15 min (Supplemental Figure S5, top). In RINT-1-depleted cells, on the other hand, at 15 min most of the antibody incorporated was found to be localized in punctate structures that were diffusely distributed throughout the cytoplasm, and this distribution remained unchanged at 45 min (Supplemental Figure S5, middle). Note that depletion of ZW10 had no marked effect on the internalization of FLAG-TGN38 (Supplemental Figure S5, bottom). These results confirm that endosome-to-TGN trafficking is impaired in RINT-1–depleted cells and indicate distinct roles of RINT-1 and ZW10.
RINT-1, but not ZW10, interacts with the COG complex
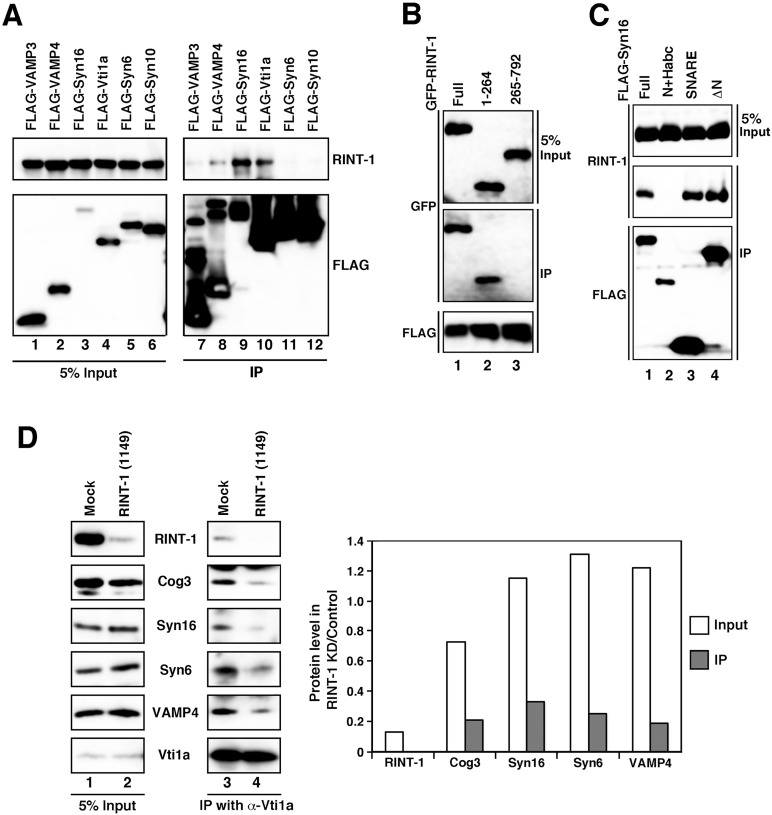
We explored the possibility that RINT-1 interacts with the COG complex because the phenotype of cells depleted of RINT-1 is very similar to that of cells depleted of Cog6 (Laufman et al., 2011). Moreover, a comprehensive analysis of protein–protein interactions in yeast demonstrated that Tip20 (yeast RINT-1) can interact with Cog4 (Uetz et al., 2000). We first performed immunoprecipitation using an anti–RINT-1 antibody. Solubilized membrane fractions from 293T cells were immunoprecipitated with an anti–RINT-1 antibody and analyzed by immunoblotting. As shown in Figure 4A, endogenous Cog3 coprecipitated with an anti–RINT-1 antibody (lane 2). Of note, Vti1a and syntaxin 6 also coprecipitated with RINT-1. On the other hand, no coprecipitation of Cog3, Vti1a, or syntaxin 6 occurred with an anti-ZW10 antibody (lane 3). Cog1 coprecipitated with an anti–RINT-1 antibody with almost equal efficiency to Cog3 (Figure 4B), perhaps suggesting coprecipitation of the COG complex with RINT-1.
FIGURE 4:
Interaction of RINT-1 with Cog1. (A, B) Solubilized membrane fractions of 293T cells were incubated for 1 h with an antibody against RINT-1 (lane 2) or ZW10 (lane 3). Immunocomplexes were precipitated with protein G–Sepharose, subjected to SDS–PAGE, and then analyzed by immunoblotting with the indicated antibodies. Three percent input was also analyzed (lane 1). (C) Each of the FLAG-Cog subunits was coexpressed with GFP–RINT-1 in 293T cells. Cell lysates were immunoprecipitated with anti-FLAG beads, subjected to SDS–PAGE, and then analyzed by immunoblotting with antibodies against GFP (middle) and FLAG (bottom). Five percent input was also analyzed by immunoblotting with an anti-GFP antibody (top). (D) Each of the GFP-tagged RINT-1 constructs was coexpressed with FLAG-Cog1, and cell lysates were immunoprecipitated with anti-FLAG beads, subjected to SDS–PAGE, and analyzed by immunoblotting with antibodies against GFP (middle) and FLAG (bottom). Five percent input was also analyzed by immunoblotting with an anti-GFP antibody (top). (E) GST fused to each of the Cog1 constructs was coexpressed with FLAG–RINT-1, and cell lysates were pulled down with glutathione beads and analyzed by immunoblotting with antibodies against FLAG (middle) and GST (bottom). Five percent input was also analyzed by immunoblotting with an anti-FLAG antibody (top).
Next we sought to determine which COG subunit(s) interact with RINT-1. For this purpose, each of the eight COG subunits (carrying a FLAG tag) was coexpressed with GFP–RINT-1 and immunoprecipitated with anti-FLAG beads. As shown in Figure 4C, GFP–RINT-1 coprecipitated with FLAG-Cog1 (lane 1). Some GFP–RINT-1 coprecipitated with FLAG-Cog8 (lane 8) but not with other subunits (lanes 2–7). To confirm this interaction, we examined the interaction of endogenous RINT-1 with FLAG-Cog1. As shown in Supplemental Figure S6, endogenous RINT-1 also coprecipitated with FLAG-Gog1 (lane 1). In this experiment some RINT-1 also coprecipitated with FLAG-Cog4 and -Cog7. Given that these Cog subunits, when expressed, did not bind to GFP–RINT-1 (Figure 4C), they may indirectly bind to RINT-1. It is possible that expressed FLAG-Cog4 and -Cog7 are incorporated into the COG complex.
Consistent with the result that endogenous Cog1 or Cog3 was not precipitated with an anti-ZW10 antibody (Figure 4, A and B, lane 3), ZW10, NAG, or syntaxin 18 did not coprecipitate with FLAG-Cog1 (Supplemental Figure S6, lane 1). These results suggest that RINT-1 uncomplexed with ZW10 interacts with the COG complex.
Vps51-like domain of Cog1 is responsible for the association with the N-terminal, ZW10-interacting region of RINT-1
The fact that RINT-1 uncomplexed with ZW10 interacts with Cog1 suggested that the Cog1-binding site on RINT-1 overlaps with the binding site for ZW10. To test this, we expressed FLAG-Cog1 together with GFP fused to the N-terminal 264 amino acids of RINT-1, which was previously shown to bind to ZW10 (Arasaki et al., 2006; Supplemental Figure S7A) and conducted immunoprecipitation with anti-FLAG beads. As shown in Figure 4D, GFP–RINT-1 (amino acids 1–264) coprecipitated with FLAG-Cog1 (lane 2, middle), whereas no significant coprecipitation was observed for the fragment containing the central and C-terminal region (amino acids 265–792; lane 3, middle). The latter finding may partly explain why expression of this fragment failed to rescue the RINT-1 depletion effect on TGN protein localization (Supplemental Figure S3).
Next we determined the RINT-1-binding site on Cog1. The SMART program (http://smart.embl-heidelberg.de/) predicts the presence of a Vps51-like domain (amino acids 12–93) in Cog1 (Supplemental Figure S7B). Vps51 is a subunit of the GARP complex, another member of the CATCHR family complexes (Bonifacino and Hierro, 2011). Of interest, the Pfam database (http://pfam.sanger.ac.uk/) indicates that the RINT-1 partner ZW10 is a member of the Vps51 clan. To determine whether the Vps51-like domain is required for the association with RINT-1, we expressed glutathione S-transferase (GST)–Cog1 full-length or GST-Cog1 lacking the N-terminal 93 amino acids (GST–Cog1Δ1-93) together with FLAG–RINT-1, pulled down with glutathione beads and analyzed by immunoblotting with an anti-FLAG antibody. As shown in Figure 4E, FLAG–RINT-1 was pulled down with the full-length Cog1 construct (lane 1, middle) but much less with GST–Cog1Δ1-93 (lane 2). Given that the COILS program (www.ch.embnet.org/software/COILS_form.html) predicts the presence of a putative coiled-coil region (amino acids 27–55) in the Vps51-like domain, we examined the interaction of RINT-1 with the N-terminal Vps51-like domain (Vps51: amino acids 12–93) and the putative coiled-coil region (CC: amino acids 27–55). As shown in Figure 4E, the Vps51-like domain bound to FLAG–RINT-1 (lane 3, middle), whereas no binding was observed between the putative coiled-coil region and FLAG–RINT-1 (lane 4).
We next examined whether GARP subunits interact with RINT-1. The GARP complex consists of Vps51/Ang2, Vps52, Vps53, and Vps54 (Bonifacino and Hierro, 2011). Each GARP subunit as a V5-tagged protein was expressed together with FLAG–RINT-1, and then cell lysates were immunoprecipitated with anti-FLAG beads. Although a very small amount of V5-tagged Vps52 coprecipitated with FLAG–RINT-1 (Supplemental Figure S8, lane 1), no interaction was observed between V5-Vps51/Ang2 and FLAG–RINT-1 (lane 4). These results suggest that the interaction of RINT-1 with Cog1 is specific.
Overexpression of RINT-1 causes redistribution of Cog3
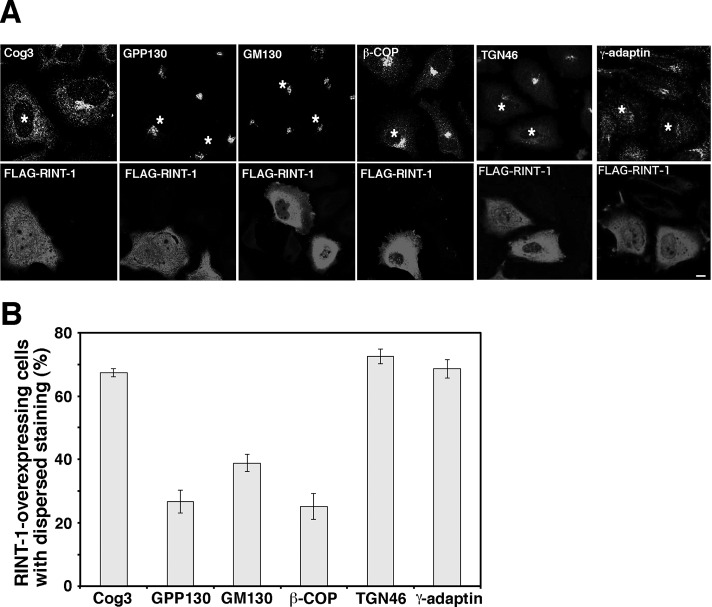
To further characterize the relationship between RINT-1 and the COG complex, we examined the effect of overexpression and depletion of RINT-1 on the localization of the COG complex. Our previous data showed that overexpression of a RINT-1 full-length construct has no marked effect on the cis/medial-Golgi structure, whereas its N-terminal, ZW10-interacting region causes Golgi fragmentation (Arasaki et al., 2006). When FLAG–RINT-1 full-length construct was overexpressed, dispersal of Cog3, as well as that of TGN46 and γ-adaptin, was observed in many cells (Figure 5). Dispersal of TGN46 and γ-adaptin may be a consequence of the disruption of the TGN structure. Consistent with our previous result (Arasaki et al., 2006), on the other hand, cis-Golgi markers (GPP130, GM130, and β-COP) remained in the perinuclear region in a substantial fraction of RINT-1–overexpressing cells. These results confirm a link between RINT-1 and the COG complex, although the precise mechanism underlying the dispersal of Cog3 by the overexpression of FLAG–RINT-1 is unclear.
FIGURE 5:
Overexpression of RINT-1 disrupts the localization of the COG complex but not cis-Golgi proteins. (A) The plasmid encoding FLAG–RINT-1 was transfected into HeLa cells, and at 24 h after transfection, the cells were double stained for indicated proteins (top) and FLAG–RINT-1 (bottom). Asterisks indicate FLAG–RINT-1–overexpressing cells. Bar, 5 μm. (B) Quantitation of the data shown in A. Data are the average of three independent experiments (n [cell number] ≥ 50). Error bars, SEM.
In contrast to overexpression, depletion of RINT-1 did not markedly affect Cog3 localization; it was principally localized in partially fragmented structures (Supplemental Figure S9). Double staining revealed that the fragmented Cog3 staining almost completely overlapped with GM130 (cis-Golgi) staining (Supplemental Figure S9, second row) but was markedly different from ERGIC-53 staining (bottom). Because the GOG complex seems be evenly distributed among cisternae along the cis-to-trans direction (Vasile et al., 2006), this fragmented Cog3 pattern likely reflects its cis/medial-Golgi localized pool. As in the case of TGN proteins (Figure 2), a TGN fraction of Cog3 might be redistributed upon depletion of RINT-1.
Interaction between RINT-1 and TGN SNAREs
In addition to Cog1 and Cog3, endogenous Vti1a and syntaxin 6, both implicated in endosome-to-TGN trafficking (Mallard et al., 2002; Ganley et al., 2008; Laufman et al., 2011), were precipitated with an anti–RINT-1 antibody (Figure 4A, lane 2). Next we determined the binding specificity of RINT-1 with TGN SNARE proteins. In addition to Vti1a and syntaxin 6, we examined the interactions with syntaxin 10, syntaxin 16, and VAMP3/4, which have been implicated in endosome-to-TGN trafficking (Hong 2005, Ganley et al., 2008; Laufman et al., 2011). As shown in Figure 6A, endogenous RINT-1 coprecipitated with FLAG-syntaxin 16 (lane 9) and less efficiently with FLAG-Vti1a (lane 10) but not with FLAG-syntaxin 6 (lane 11) or FLAG-syntaxin 10 (lane 12). Some RINT-1 coprecipitated with FLAG-VAMP4 (lane 8). Given that the expression level of FLAG-syntaxin 16 was much lower than that of FLAG-Vti1a (lane 3 vs. lane 4), these results suggest that syntaxin 16 is the major partner for RINT-1. The precipitation of endogenous syntaxin 6 with an anti–RINT-1 antibody (Figure 4A) may be explained by the idea that syntaxin 6 is connected to RINT-1 through syntaxin 16, Vti1a, and/or the COG complex.
FIGURE 6:
RINT-1 modulates TGN SNARE complex assembly. (A) Lysates of cells expressing each of the FLAG-SNARE constructs were immunoprecipitated with anti-FLAG-beads (lanes 7–12) and analyzed by immunoblotting with antibodies against RINT-1 (top) and FLAG (bottom). Five percent input was also analyzed (lanes 1–6). (B) Interaction between RINT-1 and syntaxin 16. Lysates of cells expressing FLAG–syntaxin 16 and each of the GFP–RINT-1 constructs were immunoprecipitated with anti-FLAG beads and analyzed by immunoblotting with antibodies against GFP (middle) and FLAG (bottom). Five percent input was also analyzed by immunoblotting with an anti-GFP antibody (top). (C) SNARE domain is responsible for the interaction with RINT-1. Lysates of cells expressing each of the FLAG–syntaxin 16 constructs were immunoprecipitated with anti-FLAG beads and analyzed by immunoblotting against RINT-1 (middle) and FLAG (bottom). Five percent input was also analyzed by immunoblotting with an ant-RINT-1 antibody (top) (D) TGN SNARE complex assembly is abrogated in RINT-1–depleted cells. HeLa cells were mock transfected (lanes 1 and 3) or transfected with RINT-1 (1149) (lanes 2 and 4). At 72 h after transfection, cell lysates were immunoprecipitated with an anti-Vti1a antibody and analyzed by immunoblotting with the indicated antibodies (lanes 3 and 4). Five percent input was also analyzed (lanes 1 and 2). The intensities of immunostained bands were quantitated with ImageJ. The quantitative data represent the average of two independent experiments.
We next determined the region of RINT-1 responsible for the interaction with syntaxin 16. As shown in Figure 6B, a GFP-tagged, N-terminal 264–amino acid fragment of RINT-1 coprecipitated with FLAG-syntaxin16 (lane 2, middle), whereas the remaining region was not (lane 3). We then determined the RINT-1–interacting site on syntaxin 16. Syntaxin 16 (isoform B: 325 amino acids) is a Qa-SNARE (Hong, 2005) that contains, from the N-terminus, a Vps45 (a SM protein)-binding site (Dulubova et al., 2002; Carr and Rizo, 2010), a regulatory domain of trihelical bundle called the Habc domain, a SNARE motif, and a transmembrane domain (Supplemental Figure S7C). We therefore constructed three mutants each with FLAG tag, Syn16N+Habc (amino acids 1–207), Syn16SNARE (amino acids 208–325), and Syn16ΔN (amino acids 54–325) and performed immunoprecipitation. Figure 6C shows that the SNARE domain of syntaxin 16 (lane 3, middle), but not the Habc domain-containing region (Syn16N+Habc; lane 2), interacts with RINT-1.
Assembly of SNARE complexes containing syntaxin 16 is disturbed in RINT-1–depleted cells
Tethering factors contribute to the regulation of SNARE complex assembly (Bröcker et al., 2010; Brown and Pfeffer, 2010; Yu and Hughson, 2010). We next examined whether RINT-1 depletion affects TGN SNARE complex assembly. To this end, RINT-1–depleted cells were lysed, immunoprecipitated with an anti-Vti1a antibody, and analyzed by immunoblotting. As shown in Figure 6D, the amounts of syntaxin 16, syntaxin 6, and VAMP4 coprecipitated with Vti1a were decreased by 67–81% compared with the control levels, suggesting that RINT-1 regulates the assembly of TGN SNARE proteins. The amount of Cog3 coprecipitated with Vti1a was also decreased by 80%, supporting the idea that RINT-1 is important for the connection between the TGN SNAREs and the COG complex. The fact that the steady-state level of Cog3 was decreased by 30% upon RINT-1 knockdown might imply that RINT-1 affects the stability of Cog3. The primary effect of RINT-1 depletion, however, is the defect in SNARE complex assembly and the abrogation of the association of Cog3 with syntaxin 16. In contrast to RINT-1 depletion, essentially no change in immunoprecipitation profile was observed when ZW10 was depleted (data not shown).
DISCUSSION
The ZW10/Dsl1 and COG complexes, as well as the GARP and exocyst complexes, belong to the CATCHR family of multisubunit tethering complexes (Yu and Hughson, 2010). These family members were believed to function in distinct membrane trafficking pathways, although the COG and GARP complexes are involved in endosome-to-TGN trafficking (Bonifacino and Hierro, 2011; Miller and Ungar, 2012). Whereas CATCHR family proteins share subtle sequence homology (Whyte and Munro, 2002), emerging structural similarities provide strong evidence of a common evolutionary origin and may reflect a shared mechanism of action (Richardson et al., 2009; Tripathi et al., 2009; Pérez-Victoria et al., 2010a; Vasan et al., 2010). The present study reveals for the first time interplay between members of the CATCHAR family proteins; RINT-1, a component of the ZW10/Dsl1 complex, regulates endosome-to-TGN transport by interacting with Cog1, one of the eight subunits of the COG complex, whose size and sequence are markedly different from those of yeast Cog1 (Chatterton et al., 1999; Ungar et al., 2002). The interaction between RINT-1 and Cog1 relies upon known structural similarities and protein–protein interactions of CATCHR family proteins. Both the N-terminal region of Cog1, which is responsible for binding to RINT-1, and an ER RINT-1 partner ZW10 have a Vps51-like structure. Moreover, RINT-1 bears strong resemblance to Cog4 (Richardson et al., 2009), which can directly interact with Cog1 in the mammalian COG complex (Loh and Hong, 2004).
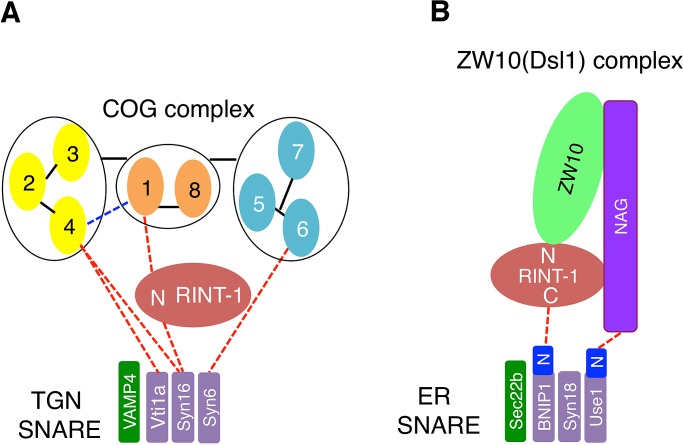
Figure 7 depicts two distinct RINT-1–containing complexes, localized at the TGN (Figure 7A) and the ER (Figure 7B), respectively. There are similar and different protein–protein interactions in these complexes. Given that RINT-1/Tip20 interacts with ZW10/Dsl1 (tether) through its N-terminal region in the ER complex (Kraynack et al., 2005; Arasaki et al., 2006; Diefenbacher et al., 2011; Meiringer et al., 2011), the N-terminal region of RINT-1 is responsible for the binding to Cog1 (tether) in the TGN complex. In terms of SNARE binding, however, RINT-1 uses its N-terminal region for syntaxin 16 (Qa-SNARE) binding, whereas the central and C-terminal regions of RINT-1/Tip20 are involved in the interaction with BNIP1/Tip20 (Qb-SNARE; Tripathi et al., 2009; Supplemental Figure S7A). Moreover, the SNARE domain of syntaxin 16 participates in the interaction with RINT-1 in the TGN complex, whereas the N-terminal regulatory domain of Sec20 is involved in the interaction with Tip20 in the ER complex (Ren et al., 2009).
FIGURE 7:
The interactions of RINT-1 with tethers and SNAREs in the TGN and the ER. (A) At the TGN, RINT-1 serves as a link between Cog1 and syntaxin 16 through its N-terminal region (N). The Vps51-like domain of Cog1 and the SNARE domain of syntaxin 16 are responsible for the interactions with RINT-1. The COG complex model is according to Oka et al. (2005) and Ungar et al. (2005). The interaction between Cog1 and Cog4 (blue dashed line) was reported (Loh and Hong, 2004). (B) At the ER, RINT-1/Tip20 serves as a link between ZW10/Dsl1 and BNIP1/Sec20. The N-terminal regulatory region of Sec20 (shown as BNIP1N) is responsible for the interaction with Tip20 (Ren et al., 2009), and the central and C-terminal regions (shown as C) of RINT-1/Tip20 bind to BNIP1/Sec20 (Tripathi et al., 2009; this study; Supplemental Figure S7A).
In addition to a RINT-1–mediated link, there are links between tethers and SNAREs in the TGN and ER. In the TGN, the SNARE domain of syntaxin 6 interacts with the N-terminal region of Cog6 (Laufman et al., 2011). More recently, it was reported that the SNARE domains of syntaxin 16 and Vti1a bind to the N-terminal region of Cog4, although Vti1a also interacts with the C-terminal fragment of Cog4 (Laufman et al., 2013). These links may provide explanations for the coprecipitation of some Cog3 with Vti1a in the absence of RINT-1 (Figure 6D). Moreover, the GARP complex, consisting of Vps51, Vps52, Vps53, and Vps54 (Bonifacino and Hierro, 2011), can bind to TGN SNAREs. Vps51 (Ang2 in mammals) interacts with the N-terminal Habc region of Tlg2 (syntaxin 6 [Qc-SNARE] in mammals; Siniossoglou and Pelham, 2002; Conibear et al., 2003; Fridmann-Sirkis et al., 2006; Pérez-Victoria et al., 2010b), and the N-terminal coiled-coil domains of Vps53 and Vps54 of the mammalian complex interact with the SNARE motifs of syntaxin 6, syntaxin 16, and VAMP4 (Pérez-Victoria and Bonifacino, 2009). Of interest, the N-terminal regions of COG and GARP subunits seem to be commonly involved not only in the interactions with SNAREs (RINT-1 in the case of Cog1), but also in complex assembly (Ungar et al., 2005; Lees et al., 2010; Bonifacino and Hierro, 2011). In the ER, on the other hand, p31/Use1 (Qc-SNARE) binds to the N-terminal region of NAG, and the extreme N-terminal region of p31/Use1 is responsible for this interaction (Aoki et al., 2009; Ren et al., 2009).
Although RINT-1 plays an important role as a tether for ZW10 to syntaxin 18–containing SNARE complexes, it does not significantly contribute to SNARE complex assembly on the ER membrane (Arasaki et al., 2006). This is in contrast to the critical role of yeast Tip20 in ER SNARE complex assembly (Kraynack et al., 2005). The minor contribution of RINT-1 to ER SNARE complex assembly may be related to a unique mechanism of ER SNARE complex assembly. Association of syntaxin 18 with BNIP1 and p31/Use1 is dramatically induced by the v-SNARE Sec22b (Aoki et al., 2008), which may reduce the requirement of tethers such as RINT-1 in SNARE complex assembly. Under this circumstance, RINT-1 might have acquired, during the course of evolution, an additional function, that is, participation in endosome-to-TGN transport. Of interest, the sites of the RINT-1–syntaxin 16 interaction are different from those of the Tip20/RINT-1–Sec20/BNIP1 interaction. The central and C-terminal regions of Tip20/RINT-1 bind to the N-terminal region of Sec20/BNIP1 on the ER (Ren et al., 2009; Tripathi et al., 2009), whereas RINT-1 on the TGN binds to the SNARE domain of syntaxin 16 through its N-terminal region (Figure 7). Mammals have a greater complexity in their transport pathways from endosomes to the TGN than a unicellular organism, yeast (Bonifacino and Rojas, 2006). In addition to the increase in the number of transport devices (Kloepper et al., 2007), mammals might adopt a preexisting device to fulfill the requirement for diverse transport routes from endosome to the TGN. Moreover, mammals might endow RINT-1 with the ability to interact with Rad50 (Xiao et al., 2001) and Rb-related p130 (Kong et al., 2006).
In conclusion, the present findings highlight a new layer of complexity for the interactions of proteins responsible for the tethering of transport carriers with SNAREs. The CATCHR family COG complex interacts with coiled-coil tethers at the cis-side of the Golgi apparatus (Sohda et al., 2007, 2010) and with the CATCHR family tether RINT-1 at the TGN (this study). Taken together, these data indicate that CATCHR family members function in vesicle tethering by regulating SNARE complex assembly.
MATERIALS AND METHODS
Antibodies and reagents
Monoclonal antibodies against GM130, γ-adaptin, p230, syntaxin 6, EEA1, calnexin, Cog1, and Vti1a were purchased from BD Biosciences PharMingen (San Diego, CA). Polyclonal antibodies against TGN46 and Golgin-97 were obtained from Abcam (Cambridge, MA). Monoclonal and polyclonal antibodies against FLAG were purchased from Sigma-Aldrich (St. Louis, MO). Polyclonal antibodies against syntaxin 16 and VAMP4 were purchased from Synaptic Systems (Göttingen, Germany). Polyclonal antibodies against GPP130, GFP, and GST and monoclonal antibodies against CI-MPR and Hsp47 were obtained from Covance (Princeton, NJ), Invitrogen (Carlsbad, CA), Santa Cruz Biotechnology (Santa Cruz, CA), Thermo Fisher Scientific (Waltham, MA), and Enzo Life Science (Farmingdale, NY), respectively. Polyclonal antibody against Cog3 was prepared as described (Sohda et al., 2007). Polyclonal antibodies against RINT-1, ZW10, NAG, syntaxin 18, and Rer1 were prepared in our laboratory (Hirose et al., 2004; Nakajima et al., 2004, Aoki et al., 2009). Glutathione Sepharose 4B was from GE Healthcare (Piscataway, NJ).
Cell culture
293T cells were grown in DMEM supplemented with 50 IU/ml penicillin, 50 μg/ml streptomycin, and 10% fetal calf serum. HeLa cells were cultured in Eagle's MEM supplemented with the same materials.
Plasmids and transfection
The plasmids encoding FLAG–Cog1-8 were constructed previously (Sohda et al., 2007, 2010). The cDNAs encoding full-length or truncated mutants of RINT-1 were inserted into pEGFP-C3 vector so as to express proteins with an N-terminal GFP tag. The cDNAs encoding full-length syntaxin 16 and its truncated mutants, syntaxin 6, syntaxin 10, VAMP3, VAMP4, and Vti1a were inserted into pFLAG-CMV-6 (Sigma-Aldrich) so as to express proteins with an N-terminal FLAG tag. To express full-length or truncated Cog1 fragments as GST fusion proteins in mammalian cells, the cDNAs encoding Cog1 and its fragments were inserted into the SmaI site of pEBG-Sma vector. The plasmids encoding GARP subunits were a generous gift from J. S. Bonifacino (National Institutes of Health, Bethesda, MD). Transfection was carried out using LipofectAMINE PLUS or LipofectAMINE 2000 (Invitrogen) according to the manufacturer's protocol. At 24 h after transfection, the cells were fixed for immunofluorescence analysis or lysed for immunoprecipitation or pull-down assays.
Immunofluorescence microscopy
For immunofluorescence microscopy, cells were fixed with 4% paraformaldehyde for 20 min at room temperature or ice-cold methanol at −20ºC and observed with an Olympus FluoView 300 or 1000 laser scanning microscope (Olympus, Tokyo, Japan).
Immunoprecipitation
For immunoprecipitation of endogenous RINT-1 or ZW10, ∼90% confluent 293T cells (two 15-cm dishes) were washed twice in phosphate-buffered saline (PBS) and then once in homogenization buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES]–KOH, pH 7.2, 150 mM KCl, 2 mM EDTA, 1 mM dithiothreitol, 10 μg/ml leupeptin, 1 μM pepstatin A, 2 μg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride). The cells were collected, suspended in 1 ml of homogenization buffer, and homogenized with 20 strokes in a Dounce homogenizer. The homogenate was centrifuged at 1000 × g for 10 min, and then the supernatant was centrifuged at 100,000 × g for 30 min to separate the cytosol and membrane fractions. The membrane pellet was solubilized in homogenization buffer containing 1% Triton X-100. Equal volumes of Triton X-100 extracts were incubated for 1 h with 2 μg of a polyclonal antibody against RINT-1 or ZW10. After incubation, 10 μl of protein G–Sepharose (GE Healthcare) was added, and the suspension was gently mixed for 2 h. The beads were thoroughly washed, and the attached proteins were eluted by SDS sample buffer, resolved by SDS–PAGE, and analyzed by immunoblotting.
For immunoprecipitation of expressed proteins, 293T cells expressing FLAG-tagged proteins were lysed in homogenization buffer containing 1% Triton X-100 and centrifuged at 17,000 × g for 10 min. The supernatants were immunoprecipitated with anti–FLAG M2 affinity gels (Sigma-Aldrich). After extensive washing of the beads, the bound proteins were eluted from the gels by adding SDS-sample buffer and analyzed by immunoblotting.
RNA interference
The RNA duplexes used for targeting RINT-1 (1149), ZW10 (102), and NAG (4160) were described previously (Hirose et al., 2004; Arasaki et al., 2006; Aoki et al., 2009). The RNA duplexes used for targeting RINT-1 (687) (5′-aagugauuuugaggaaauu-3′, which corresponds to positions 687–707 relative to the start codon) were purchased from Japan Bioservice (Asaka, Japan). Transfection was performed using Oligofectamine (Invitrogen) according to the manufacturer's protocol.
CTB transport assay
At 48 h after transfection of siRNA, HeLa cells were incubated in OPTI-MEM containing 20 mM HEPES, pH 7.4, and 5 μg/ml Alexa Fluor 594–labeled CTB (Invitrogen) for 30 min at 4ºC. The cells were then washed with PBS and incubated in a complete growth medium at 37ºC. The cells were fixed, immunostained with an anti-EEA1 antibody, and analyzed by confocal microscopy.
Antibody uptake assay
At 48 h after transfection of siRNA, HeLa cells were transfected with a mammalian expression vector encoding FLAG-TGN38. At 24 h later, the cells were incubated at 37ºC in medium containing 1.5 μg/ml anti–FLAG M2 monoclonal antibody for different time periods (3, 15, and 45 min). The cells were extensively washed with PBS, fixed, stained with a Texas red–labeled anti-mouse secondary antibody, and analyzed by confocal microscopy.
Two-hybrid assay
Two-hybrid analysis was carried out essentially according to the manufacturer's protocol using pGBKT7 vector for BNIP1 and pACT2 vector for RINT-1 and its fragments. To detect β-galactosidase activity, filters were incubated at 30ºC for 1 h.
Supplementary Material
Acknowledgments
We thank Suzanne Pfeffer (Stanford University, Stanford, CA) for critical comments and suggestions on the manuscript. We are grateful to Nobuhiro Nakamura (Kyoto Sangyo University, Kyoto, Japan) and Shin-ichiro Yoshimura (Osaka University, Osaka, Japan) for a generous gift of a FLAG-TGN38 vector and Juan S. Bonifacino (National Institutes of Health, Bethesda, MD) for a generous gift of GARP subunit vectors. We also thank Chikano Noda, Kahori Okada, Nanako Oyama, and Mei Taniguchi for technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research 24790425 (to K.A.), 23113726 and 24657141 (to M.T.), and 23570174 (to H.I.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Abbreviations used:
- CI-MPR
cation-independent mannose 6-phosphate receptor
- CTB
cholera toxin B
- ER
endoplasmic reticulum
- ERGIC
ER-Golgi intermediate compartment
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- PBS
phosphate-buffered saline
- siRNA
short interfering RNA
- SNARE
N-ethylmaleimide–sensitive factor attachment protein receptor
- TGN
trans-Golgi network
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-01-0014) on July 24, 2013.
*These authors contributed equally to this work.
REFERENCES
- Andag U, Neumann T, Schmitt HD. The coatomer-interacting protein Dsl1p is required for Golgi-to-endoplasmic reticulum retrieval in yeast. J Biol Chem. 2001;276:39150–39160. doi: 10.1074/jbc.M105833200. [DOI] [PubMed] [Google Scholar]
- Andag U, Schmitt HD. Dsl1p, an essential component of the Golgi-endoplasmic reticulum retrieval system in yeast, uses the same sequence motif to interact with different subunits of the COPI vesicle coat. J Biol Chem. 2003;278:51722–51734. doi: 10.1074/jbc.M308740200. [DOI] [PubMed] [Google Scholar]
- Aoki T, Ichimura S, Itoh A, Kuramoto M, Shinkawa T, Isobe T, Tagaya M. Identification of the neuroblastoma-amplified gene product as a component of the syntaxin 18 complex implicated in Golgi-to-endoplasmic reticulum retrograde transport. Mol Biol Cell. 2009;20:2639–2649. doi: 10.1091/mbc.E08-11-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T, Kojima M, Tani K, Tagaya M. Sec22b-dependent assembly of endoplasmic reticulum Q-SNARE proteins. Biochem J. 2008;410:93–100. doi: 10.1042/BJ20071304. [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Hauri HP. The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J Cell Sci. 2006;119:2173–2183. doi: 10.1242/jcs.03019. [DOI] [PubMed] [Google Scholar]
- Arasaki K, Taniguchi M, Tani K, Tagaya M. RINT-1 regulates the localization and entry of ZW10 to the syntaxin 18 complex. Mol Biol Cell. 2006;17:2780–2788. doi: 10.1091/mbc.E05-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Hierro A. Transport according to GARP: receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol. 2011;21:159–167. doi: 10.1016/j.tcb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- Bröcker C, Engelbrecht-Vandré S, Ungermann C. Multisubunit tethering complexes and their role in membrane fusion. Curr Biol. 2010;20:R943–R952. doi: 10.1016/j.cub.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Brown FC, Pfeffer SR. An update on transport vesicle tethering. Mol Membr Biol. 2010;27:457–461. doi: 10.3109/09687688.2010.501765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr CM, Rizo J. At the junction of SNARE and SM protein function. Curr Opin Cell Biol. 2010;22:488–495. doi: 10.1016/j.ceb.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterton JE, Hirsch D, Schwartz JJ, Bickel PE, Rosenberg RD, Lodish HF, Krieger M. Expression cloning of LDLB, a gene essential for normal Golgi function and assembly of the ldlCp complex. Proc Natl Acad Sci USA. 1999;96:915–920. doi: 10.1073/pnas.96.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Cleck JN, Stevens TH. Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol Biol Cell. 2003;14:1610–1623. doi: 10.1091/mbc.E02-10-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbacher M, Thorsteinsdottir H, Spang A. The Dsl1 tethering complex actively participates in soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptor (SNARE) complex assembly at the endoplasmic reticulum in Saccharomyces cerevisiae. J Biol Chem. 2011;286:25027–25038. doi: 10.1074/jbc.M110.215657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Yamaguchi T, Gao Y, Min SW, Huryeva I, Südhof TC, Rizo J. How Tlg2p/syntaxin 16 “snares” Vps45. EMBO J. 2002;21:3620–3631. doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridmann-Sirkis Y, Kent HM, Lewis MJ, Evans PR, Pelham HR. Structural analysis of the interaction between the SNARE Tlg1 and Vps5. Traffic. 2006;7:182–190. doi: 10.1111/j.1600-0854.2005.00374.x. [DOI] [PubMed] [Google Scholar]
- Ganley IG, Espinosa E, Pfeffer SR. A syntaxin 10-SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. J Cell Biol. 2008;180:159–172. doi: 10.1083/jcb.200707136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh RN, Mallet WG, Soe TT, McGraw TE, Maxfield FR. An endocytosed TGN38 chimeric protein is delivered to the TGN after trafficking through the endocytic recycling compartment in CHO cells. J Cell Biol. 1998;142:923–936. doi: 10.1083/jcb.142.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsuzawa K, Hirose H, Tani K, Yamamoto A, Scheller RH, Tagaya M. Syntaxin 18, a SNAP receptor that functions in the endoplasmic reticulum, intermediate compartment, and cis-Golgi vesicle trafficking. J Biol Chem. 2000;275:13713–13720. doi: 10.1074/jbc.275.18.13713. [DOI] [PubMed] [Google Scholar]
- Hirose H, Arasaki K, Dohmae N, Takio K, Hatsuzawa K, Nagahama M, Tani K, Yamamoto A, Tohyama M, Tagaya M. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 2004;23:1267–1278. doi: 10.1038/sj.emboj.7600135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;174:493–517. [PubMed] [Google Scholar]
- Kloepper TH, Kienle CN, Fasshauer D. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol Biol Cell. 2007;18:3463–3471. doi: 10.1091/mbc.E07-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong LJ, Meloni AR, Nevins JR. The Rb-related p130 protein controls telomere lengthening through an interaction with a Rad50-interacting protein, RINT-1. Mol Cell. 2006;22:63–71. doi: 10.1016/j.molcel.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Kraynack BA, Chan A, Rosenthal E, Essid M, Umansky B, Waters MG, Schmitt HD. Dsl1p, Tip20p, and the novel Dsl3(Sec39) protein are required for the stability of the Q/t-SNARE complex at the endoplasmic reticulum in yeast. Mol Biol Cell. 2005;16:3963–3977. doi: 10.1091/mbc.E05-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufman O, Hong W, Lev S. The COG complex interacts directly with syntaxin 6 and positively regulates endosome-to-TGN retrograde transport. J Cell Biol. 2011;194:459–472. doi: 10.1083/jcb.201102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufman O, Hong W, Lev S. The COG complex interacts with multiple Golgi SNAREs and enhances fusogenic assembly of SNARE complexes. J Cell Sci. 2013;126:1506–1516. doi: 10.1242/jcs.122101. [DOI] [PubMed] [Google Scholar]
- Lees JA, Yip CK, Walz T, Hughson FM. Molecular organization of the COG vesicle tethering complex. Nat Struct Mol Biol. 2010;17:1292–1297. doi: 10.1038/nsmb.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Liu CC, Gao Q, Zhang X, Wu G, Lee WH. RINT-1 serves as a tumor suppressor and maintains Golgi dynamics and centrosome integrity for cell survival. Mol Cell Biol. 2007;27:4905–4916. doi: 10.1128/MCB.02396-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E, Hong W. The binary interacting network of the conserved oligomeric Golgi tethering complex. J Biol Chem. 2004;279:24640–24648. doi: 10.1074/jbc.M400662200. [DOI] [PubMed] [Google Scholar]
- Mallard F, Tang BL, Galli T, Tenza D, Saint-Pol A, Yue X, Antony C, Hong W, Goud B, Johannes L. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J Cell Biol. 2002;156:653–664. doi: 10.1083/jcb.200110081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiringer CT, Rethmeier R, Auffarth K, Wilson J, Perz A, Barlowe C, Schmitt HD, Ungermann C. The Dsl1 protein tethering complex is a resident endoplasmic reticulum complex, which interacts with five soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptors (SNAREs): implications for fusion and fusion regulation. J Biol Chem. 2011;286:25039–25046. doi: 10.1074/jbc.M110.215327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VJ, Ungar D. Re'COG'nition at the Golgi. Traffic. 2012;13:891–897. doi: 10.1111/j.1600-0854.2012.01338.x. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Hirose H, Taniguchi M, Kurashina H, Arasaki K, Nagahama M, Tani K, Yamamoto A, Tagaya M. Involvement of BNIP1 in apoptosis and endoplasmic reticulum membrane fusion. EMBO J. 2004;23:3216–3226. doi: 10.1038/sj.emboj.7600333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Vasile E, Penman M, Novina CD, Dykxhoorn DM, Ungar D, Hughson FM, Krieger M. Genetic analysis of the subunit organization and function of the COG complex: studies of Cog5- and Cog7-deficient mammalian cells. J Biol Chem. 2005;280:32736–32745. doi: 10.1074/jbc.M505558200. [DOI] [PubMed] [Google Scholar]
- Pérez-Victoria FJ, Abascal-Palacios G, Tascón I, Kajava A, Magadán JG, Pioro EP, Bonifacino JS, Hierro A. Structural basis for the wobbler mouse neurodegenerative disorder caused by mutation in the Vps54 subunit of the GARP complex. Proc Natl Acad Sci USA. 2010a;107:12860–12865. doi: 10.1073/pnas.1004756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Victoria FJ, Bonifacino JS. Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-Golgi network. Mol Cell Biol. 2009;29:5251–5263. doi: 10.1128/MCB.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Victoria FJ, Schindler C, Magadán JG, Mardones GA, Delevoye C, Romao M, Raposo G, Bonifacino JS. Ang2/Fat-free is a conserved subunit of the Golgi-associated retrograde protein complex. Mol Biol Cell. 2010b;21:3386–3395. doi: 10.1091/mbc.E10-05-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR. Entry at the trans-face of the Golgi. Cold Spring Harb Perspect Biol. 2011;3:a005272. doi: 10.1101/cshperspect.a005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaves B, Horn M, Banting G. TGN38/41 recycles between the cell surface and the TGN: brefeldin A affects its rate of return to the TGN. Mol Biol Cell. 1993;4:93–105. doi: 10.1091/mbc.4.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly BA, Kraynack BA, VanRheenen SM, Waters MG. Golgi-to-endoplasmic reticulum (ER) retrograde traffic in yeast requires Dsl1p, a component of the ER target site that interacts with a COPI coat subunit. Mol Biol Cell. 2001;12:3783–3796. doi: 10.1091/mbc.12.12.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Yip CK, Tripathi A, Huie D, Jeffrey PD, Walz T, Hughson FM. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 2009;139:1119–1129. doi: 10.1016/j.cell.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BC, Smith RD, Ungar D, Nakamura A, Jeffrey PD, Lupashin VV, Hughson FM. Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene. Proc Natl Acad Sci USA. 2009;106:13329–13334. doi: 10.1073/pnas.0901966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig K, van Deurs B. Membrane traffic exploited by protein toxins. Annu Rev Cell Dev Biol. 2002;18:1–24. doi: 10.1146/annurev.cellbio.18.011502.142107. [DOI] [PubMed] [Google Scholar]
- Schmitt HD. Dsl1p/Zw10: common mechanisms behind tethering vesicles and microtubules. Trends Cell Biol. 2010;20:257–268. doi: 10.1016/j.tcb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HR. Vps51p links the VFT complex to the SNARE Tlg1p. J Biol Chem. 2002;277:48318–48324. doi: 10.1074/jbc.M209428200. [DOI] [PubMed] [Google Scholar]
- Smith RD, Lupashin VV. Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr Res. 2008;343:2024–2031. doi: 10.1016/j.carres.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohda M, Misumi Y, Yamamoto A, Nakamura N, Ogata S, Sakisaka S, Hirose S, Ikehara Y, Oda K. Interaction of Golgin-84 with the COG complex mediates the intra-Golgi retrograde transport. Traffic. 2010;11:1552–1566. doi: 10.1111/j.1600-0854.2010.01123.x. [DOI] [PubMed] [Google Scholar]
- Sohda M, Misumi Y, Yoshimura S, Nakamura N, Fusano T, Ogata S, Sakisaka S, Ikehara Y. The interaction of two tethering factors, p115 and COG complex, is required for Golgi integrity. Traffic. 2007;8:270–284. doi: 10.1111/j.1600-0854.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- Spang A. The DSL1 complex: the smallest but not the least CATCHR. Traffic. 2012;13:908–913. doi: 10.1111/j.1600-0854.2012.01362.x. [DOI] [PubMed] [Google Scholar]
- Sun Y, Shestakova A, Hunt L, Sehgal S, Lupashin V, Storrie B. Rab6 regulates both ZW10/RINT-1 and conserved oligomeric Golgi complex-dependent Golgi trafficking and homeostasis. Mol Biol Cell. 2007;18:4129–4142. doi: 10.1091/mbc.E07-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet DJ, Pelham HR. The TIP1 gene of Saccharomyces cerevisiae encodes an 80 kDa cytoplasmic protein that interacts with the cytoplasmic domain of Sec20p. EMBO J. 1993;12:2831–2840. doi: 10.1002/j.1460-2075.1993.tb05944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Ren Y, Jeffrey PD, Hughson FM. Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex. Nat Struct Mol Biol. 2009;16:114–123. doi: 10.1038/nsmb.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Sato T, Aoki T, Yamamoto A, Okada T, Hirai R, Harada R, Mori K, Tagaya M, Harada A. p31 deficiency influences endoplasmic reticulum tubular morphology and cell survival. Mol Cell Biol. 2009;29:1869–1881. doi: 10.1128/MCB.01089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Ungar D, Oka T, Brittle EE, Vasile E, Lupashin VV, Chatterton JE, Heuser JE, Krieger M, Waters MG. Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J Cell Biol. 2002;157:405–415. doi: 10.1083/jcb.200202016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D, Oka T, Vasile E, Krieger M, Hughson FM. Subunit map of the conserved oligomeric Golgi complex. J Biol Chem. 2005;280:32729–32735. doi: 10.1074/jbc.M504590200. [DOI] [PubMed] [Google Scholar]
- Vasan N, Hutagalung A, Novick P, Reinisch KM. Structure of a C-terminal fragment of its Vps53 subunit suggests similarity of Golgi-associated retrograde protein (GARP) complex to a family of tethering complexes. Proc Natl Acad Sci USA. 2010;107:14176–14181. doi: 10.1073/pnas.1009419107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasile E, Oka T, Ericsson M, Nakamura N, Krieger M. IntraGolgi distribution of the conserved oligomeric Golgi (COG) complex. Exp Cell Res. 2006;312:3132–3141. doi: 10.1016/j.yexcr.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- Xiao J, Liu CC, Chen PL, Lee WH. RINT-1, a novel Rad50-interacting protein, participates in radiation-induced G2/M checkpoint control. J Biol Chem. 2001;276:6105–6111. doi: 10.1074/jbc.M008893200. [DOI] [PubMed] [Google Scholar]
- Yu IM, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol. 2010;26:137–156. doi: 10.1146/annurev.cellbio.042308.113327. [DOI] [PubMed] [Google Scholar]
- Zink S, Wenzel D, Wurm CA, Schmitt HD. A link between ER tethering and COP-I vesicle uncoating. Dev Cell. 2009;17:403–416. doi: 10.1016/j.devcel.2009.07.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.