ERES function is required for assembly of the autophagy machinery immediately downstream of the Atg1 kinase complex and is associated with formation of autophagosomes at every stage of the process. ERES are core components of the autophagy machinery for the biogenesis of autophagosomes.
Abstract
Autophagy is a central homeostasis and stress response pathway conserved in all eukaryotes. One hallmark of autophagy is the de novo formation of autophagosomes. These double-membrane vesicular structures form around and deliver cargo for degradation by the vacuole/lysosome. Where and how autophagosomes form are outstanding questions. Here we show, using proteomic, cytological, and functional analyses, that autophagosomes are spatially, physically, and functionally linked to endoplasmic reticulum exit sites (ERES), which are specialized regions of the endoplasmic reticulum where COPII transport vesicles are generated. Our data demonstrate that ERES are core autophagosomal biogenesis components whose function is required for the hierarchical assembly of the autophagy machinery immediately downstream of the Atg1 kinase complex at phagophore assembly sites.
INTRODUCTION
Autophagy is a degradative process in eukaryotic cells that is critical for homeostasis and adaptive responses to cellular stresses such as nutrient starvation. Defects in autophagy are implicated in the pathophysiology of aging and a growing number of diseases, including neurodegeneration, metabolic disorders, and cancer (Ravikumar et al., 2010b; Rubinsztein et al., 2011; Choi et al., 2013). A key feature of autophagy is the de novo formation of double-membrane vesicular structures termed autophagosomes (He and Klionsky, 2009). Autophagosome formation is coupled to the encapsulation of cargo, which includes long-lived proteins, protein aggregates, organelles, and invasive pathogens (Knodler and Celli, 2011; Weidberg et al., 2011). On completion of autophagosome formation, cargo is delivered to vacuoles/lysosomes for degradation (Huang and Klionsky, 2007).
Autophagosome formation is characterized by distinct biochemical and morphological phases, including initiation, phagophore nucleation and expansion, autophagosome completion, and autophagosome fusion with vacuoles/lysosomes and degradation. Initiation of autophagy is tightly controlled by nutrient-sensing pathways, including the conserved target of rapamycin (TOR) kinase, protein kinase A, and AMP-activated kinase, which converge at the Atg1/Ulk1 kinase complex (Kamada et al., 2000, 2010; Budovskaya et al., 2005; Stephan et al., 2009; Kim et al., 2011). Atg1/Ulk1 kinase activation initiates the hierarchical and dynamic assembly of the autophagy machinery at sites of autophagosome formation, termed phagophore assembly sites (PAS; Suzuki et al., 2007; Cheong et al., 2008; Kawamata et al., 2008; Itakura and Mizushima, 2010). The cooperative action of the autophagy machinery, including Atg1 kinase complex, phosphatidylinositol-3 kinase complex I (PdtIns-3 kinase I), the Atg2-18 complex, and Atg9, catalyzes the nucleation of a membranous structure termed the phagophore (Kihara et al., 2001; Obara et al., 2006, 2008; Mari et al., 2010). Recruitment of additional autophagy factors, including Atg8 and the Atg5-Atg12/Atg16 complex, facilitates the incorporation of additional membrane, with the result that the phagophore expands into a cup-shaped structure and grows into a mature autophagosome.
The factors that define the site of autophagosomal biogenesis are not completely understood. This question is critical, given that the site of autophagosome biogenesis is linked to the membrane source for nascent phagophore formation and growth and to the selection of autophagic cargo. Atg9 is a conserved integral membrane protein that is a key player in defining the membrane identity of autophagosomes. Atg9-labeled compartments are vesicular-tubular membrane structures that are believed to mediate the net transfer of membranes to PAS (Mari et al., 2010; Yamamoto et al., 2012). In addition to Atg9 membrane compartments, previous reports spatially linked the PAS to the Golgi apparatus, endoplasmic reticulum (ER), mitochondria, ER–mitochondria contact sites, and plasma membrane, suggesting that these organelles also serve as a membrane source (Furuno et al., 1990; Axe et al., 2008; Hayashi-Nishino et al., 2009; Hailey et al., 2010; Ravikumar et al., 2010a; Van der Vaart et al., 2010; Hamasaki et al., 2013). In yeast, the process of autophagy also occurs in close proximity to vacuolar membranes, but this tight coupling is not observed in mammalian cells (Suzuki et al., 2001). Here we address where autophagosome biogenesis occurs in cells and show that in yeast and mammalian cells, it is spatially linked to specialized regions of the ER, termed ER exit sites (ERES), which function in COPII vesicle formation. Our analysis indicates that ERES are tethered to autophagosomes and function at an early stage of autophagosome formation. Thus our findings support a model in which ERES work in concert with the Atg9 membrane compartment to facilitate the assembly of the autophagy machinery and potentially provide membranes for phagophore nucleation, maturation, and growth.
RESULTS
Common and direct physical interactions of the autophagy and COPII machinery
The site of autophagosome biogenesis is linked to the membrane source for nascent phagophores and the selection of autophagic cargo. Several sites of membrane sources for autophagosomal biogenesis have been reported in various species under diverse growth conditions, raising the possibility that site selection is a plastic process. To address this, we asked where autophagosomes form, using a forward proteomic approach in Saccharomyces cerevisiae, which explores the physical interaction space of the core autophagy factors: Atg1, 2, 6, 8, 9, 11, 13, 14, 17, 18, 23, 27, 29, and 31 and Vps34 and 15. Specifically, we created functional epitope-tagged versions of these core components by inserting cassettes encoding triple green fluorescent protein (GFP) tags at the 3′ end of the respective genomic open reading frame (ORF) or, in the case of ATG8, a double GFP 5′-fusion into the 5′ end of the ORF, resulting in a functional N-terminal fusion protein (2xGFP-ATG8; Table 1). To focus on general autophagy, we constructed all tagged autophagy components in a strain lacking Atg19 (Δatg19), which is defective in the yeast-specific cytoplasm-to-vacuole (Cvt) pathway (Scott et al., 2001). We refer to this strain background as wild type.
TABLE 1:
Yeast strains used in this study.
| Name | Genotype |
|---|---|
| W303 | ade2-1 leu2-3 his3-11, 15 trp1-1 ura3-1 can1-100 |
| MG100 | W303 Δatg19::prATG8-2xGFP-ATG8-NATMX6 |
| MG101 | W303 Δatg19::prATG8-2xGFP-ATG8-NATMX6 Δatg7::KanMX6 |
| MG102 | W303 Δatg19::HIS3MX6 |
| MG103-116 | MG102 ATG1, 2, 6, 9, 11, 13, 14, 17, 18, 23, 27, 29, or 31-3xGFP-CaURA |
| MG117 | MG102 VPS34-3xGFP-CaURA |
| MG118 | MG102 VPS15-3xGFP-CaURA |
| MG119 | MG102 SEC13-GFP-CaURA |
| MG120 | MG102 SEC23-GFP-CaURA |
| MG121 | MG102 SEC24-GFP-CaURA |
| MG122 | W303 Δatg19::prATG8-2xGFP-ATG8-NATMX6 pRS304-Cherry-HDEL |
| MG123 | W303 Δatg19::prATG8-2xGFP-ATG8-NATMX6 pXY142-mito-dsRED |
| MG124 | W303 Δatg19::prATG8-2xGFP-ATG8-NATMX6-dsRed-HDEL pRS305-pPGK1-3xtagBFP-PHO8 |
| MG125 | MG102 SEC13-3xGFP-CaURA pRS304-Cherry-ATG8 |
| MG126 | MG102 SEC16-GFP-CaURA pRS304-Cherry-ATG8 |
| MG127 | MG102 SEC23-GFP-CaURA pRS304-Cherry-ATG8 |
| MG128 | MG102 SEC24-GFP-CaURA pRS304-Cherry-ATG8 |
| MG129 | MG102 SEC31-3xGFP-CaURA pRS304-Cherry-ATG8 |
| MG130 | MG124 Δvam7::NATMX6 |
| MG131 | MG125 sec12-4-NATMX6 |
| MG132-137 | MG102 ATG9, 1, 13, 14, 2, or 5-3xGFP-CaURA pRS304-Cherry-ATG8 |
| MG138-143 | MG132-137 sec12-4-NATMX6 |
| rsy252 | ura3-52 his4-619 |
| rsy263 | ura3-52 his4-619 sec12-4 |
| MG144 | ura3-52 his4-619 pRS306-2xGFP-ATG8 |
| MG145 | ura3-52 his4-619 sec12-4 pRS306-2xGFP-ATG8 |
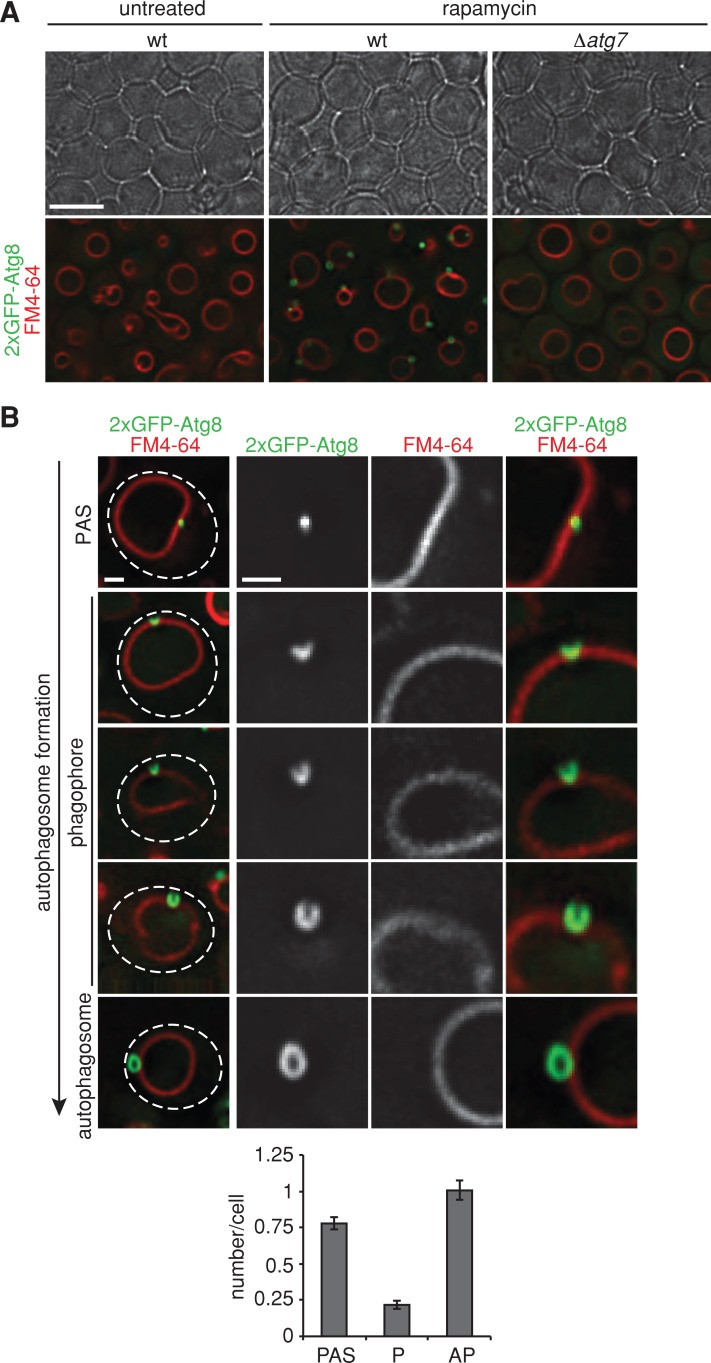
We induced autophagy using the TORC1-specific inhibitor rapamycin and harvested cells for proteomic analysis after 1 h of treatment. Under this condition, all morphological stages of autophagosome biogenesis were observed in wild-type cells by fluorescence microscopy, using the autophagosome membrane marker 2xGFP-Atg8 (Figure 1; Kirisako et al., 1999). Morphologically distinguishable intermediates include PAS, phagophores, and autophagosomes, and each class was on average significantly represented in the cell population (Figure 1B). In addition, the final step in the autophagy pathway—autophagosomal fusion to vacuoles—was also apparent in the population as free GFP generated by the vacuolar proteases (compare to Figure 6A later in the article). Autophagy factors were purified under this condition from detergent-solubilized, lysed cells using anti-GFP antibodies, and purified samples were analyzed by liquid chromatography with tandem mass spectrometry (LC-MS/MS). To capture transient interactions and further probe the spatial environment, we chemically cross-linked samples before immunopurification.
FIGURE 1:
Monitoring autophagosome formation in vivo. (A) Wild-type or Δatg7 cells expressing 2xGFP-ATG8 were grown to log phase in the presence of the vital dye FM4-64 (1 μM) for visualization of vacuolar membranes and treated with rapamycin (400 ng/ml) for 1 h. Cells were imaged using transmission (top) and fluorescence (bottom) microscopy. Single focal planes of representative cells are shown. Scale bar, 7 μm. (B) Wild-type cells expressing 2xGFP-ATG8 were treated as in A and analyzed by fluorescence microscopy. Numbers of PAS, phagophores (P), and autophagosomes (AP) per wild-type cell are shown as mean ± SD of three independent experiments (n = 180 cells). Dashed lines indicate cell boundaries. Scale bars, 1 μm.
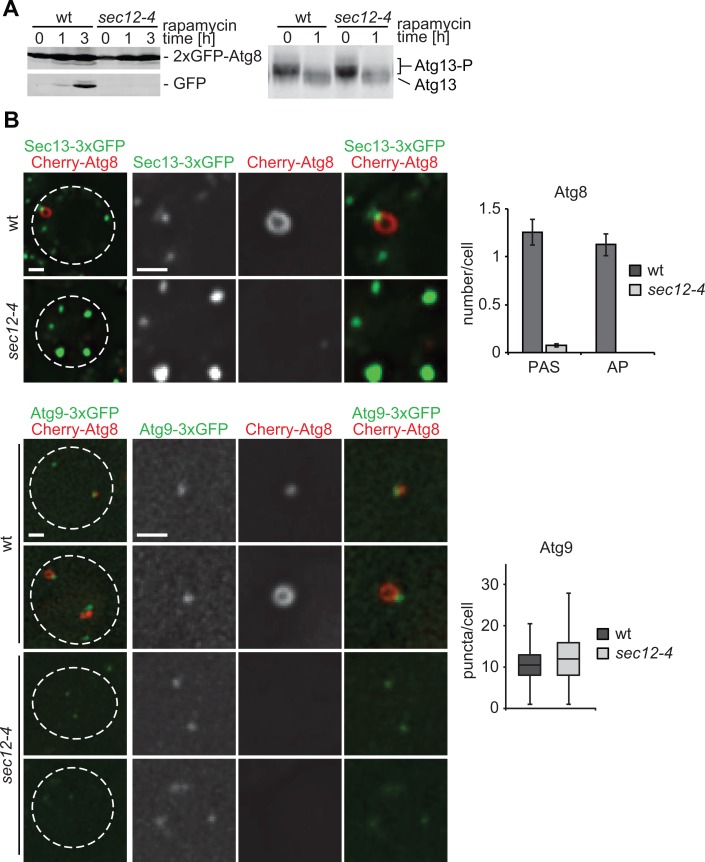
FIGURE 6:
Function of ER exit sites is required for early stages of autophagosome biogenesis. (A) Wild-type or sec12-4 cells expressing 2xGFP-ATG8 were grown to log phase at permissive temperature (23°C). Cells were treated with rapamycin (400 ng/ml) and shifted to nonpermissive temperature (37°C) for up to 3 h. Cell samples harvested at indicated time points were examined by whole-cell extraction and Western blot analysis using α-GFP (left) or α-Atg13 (right) antibodies. (B) Wild type or sec12-4 cells coexpressing Cherry-ATG8 and SEC13-3xGFP or ATG9-3xGFP, respectively, were grown to log phase at permissive temperature (23°C) and treated for 1 h as described in A. Cells were analyzed by fluorescence microscopy. Quantifications of PAS and autophagosomes (AP) or Atg9 puncta per cell are shown as mean ± SD of three independent experiments (n = 120 cells). Single focal planes of representative cells are shown. Dashed lines indicate cell boundaries. Scale bar, 1 μm.
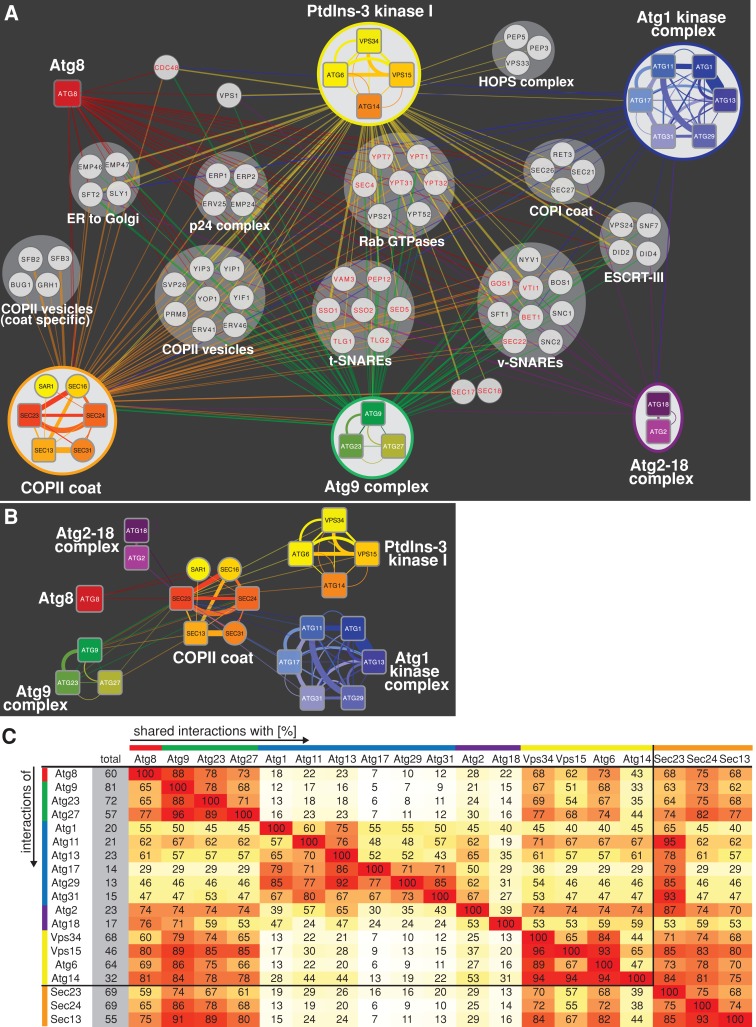
We identified 2705 physical protein interactors with the 16 autophagy components, representing 729 proteins from at least one unique peptide, with probability scores ≥70%, which were absent from untagged control samples processed in parallel. As validation of our proteomic analysis, we detected significant reciprocal interactions between core autophagy components, which represent previously characterized distinct physical complexes of the autophagy machinery: Atg1 kinase complex (Atg1, 11, 13, 17, 29, and 31), Atg2-18 complex (Atg2 and 18), Atg9 complex (Atg9, 23, and 27), and PtdIns-3 kinase I (Vps34 and 15, Atg6 and 14) (Figure 2A and Supplemental Table S1; Cebollero and Reggiori, 2009; Nakatogawa et al., 2009).
FIGURE 2:
Common interactions of the autophagy and COPII machineries. (A) The physical protein interaction network of autophagy and COPII coat complexes within the functional cluster “vesicle-mediated transport” was generated using total peptide counts. Strains expressing individual, genomically GFP-tagged autophagy and COPII coat components (rounded rectangles) were grown to log phase and treated with rapamycin as described in Figure 1, and physical protein interactions were determined by LC-MS/MS after cell lysis, chemical cross-linking, and immunopurification. Physical protein complexes were determined based on reciprocal and common interactions among complex subunits. For network simplification, interactions of bait proteins of the same complex are shown as single color-coded edges between complex and prey proteins (circles) based on the sum of total peptides. Factors previously implicate in autophagy are labeled in red. (B) Direct physical interactions between the autophagy components and coat components of COPII vesicles. Physical interactions of individual autophagy proteins and COPII coat proteins based on total peptide counts are shown. (C) Pairwise analysis of shared interactions of individual components of the autophagy and COPII machinery within the functional cluster “vesicle-mediated transport.” The number of shared interactions of two bait proteins was counted and expressed as percentage (shades of yellow to red) of total (gray) interactions. Bait proteins are organized and color coded according to physical complexes as shown in A.
To identify functional similarities among the 729 interactors, we grouped them according to their functional annotations using the DAVID bioinformatic resource (Huang da et al., 2009a, b). This analysis revealed a large functional cluster for “vesicle-mediated transport” consisting of 128 proteins (Figure 2A and Supplemental Table S1). This group represents ∼18% of total interactors, indicating an enrichment of this functional cluster compared with its representation within all verified yeast ORFs (∼7%; 375 “vesicle-mediated transport” ORFs/5050 total ORFs), consistent with autophagy network analysis in mammalian cells (Behrends et al., 2010). Within this functional cluster, ∼74% of identified proteins interacted with at least 2 and on average 4.9 autophagy components (25th percentile, 1.75 interactions; median, 4 interactions; 75th percentile, 7 interactions; highest number, 15 interactions), indicating a high degree of connectivity between components in this functional class and the autophagy machinery. Several proteins within the “vesicle-mediated transport” functional group, including Rab GTPases such as Ypt1, vesicle soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs), and target SNAREs, were previously implicated to function in autophagy, supporting the functional relevance of the observed physical interactions (Figure 2A, labeled in red; Darsow et al., 1997; Ishihara et al., 2001; Geng et al., 2010; Krick et al., 2010; Lynch-Day et al., 2010; Ohashi and Munro, 2010; Nair et al., 2011).
A prominent feature of the “vesicle-mediated transport” interactor group is the density of interactions between the autophagy machinery and COPII vesicle transport components, which include COPII coat components (Sar1, Sec16, Sec23), the p24 complex, COPII vesicle components, and ER-to-Golgi transport components (Figure 2, A and B, and Supplemental Table S1; Dancourt and Barlowe, 2010). To explore this connection, we analyzed the physical interactions of structural components of the inner and outer COPII coat, Sec23/Sec24 and Sec13, respectively, by purification of functional tagged versions from rapamycin-treated cells (Figure 2A and Supplemental Table S1). Using the same criteria as for the analysis of autophagy components, we identified direct reciprocal interactions between Sec23/Sec24 and Atg9, as well as interactions of Sec13 with Atg9 and Atg27 (Figure 2B). In addition, inclusion of the COPII interactors in the “vesicle-mediated transport” functional group revealed 794 interactions, representing 134 proteins (Supplemental Table S1), with 75% of these identified proteins interacting with at least 2 and on an average 6 of autophagy and COPII coat components (25th percentile, 2 interactions; median, 4 interactions; 75th percentile, 10 interactions; highest number, 18 interactions). These data indicate a high number of common interactors of the COPII and autophagy machinery. As a further test, we performed a pairwise analysis of shared interactions of autophagy and COPII components (Figure 2C). As expected, we observed that the core autophagy machinery shares a significant percentage of interactions with each other, with the degree of shared interactions reflecting the organization of the autophagy machinery into complexes (Figure 2C). Significantly, we also observed a high percentage of shared interactions between autophagy and COPII components, in particular between COPII components and the PtdIns-3 kinase complex I and the Atg9 complex subunits, which are believed to work together to mark and provide the membrane source for phagophore nucleation and expansion, as well as Atg8, implicated in membrane remodeling and substrate recognition (Figure 2C). Thus the direct interactions observed between the autophagy and COPII components and the high degree of shared interactions suggest a role for the COPII machinery in the biogenesis of autophagosomes.
Autophagosomes form in contact with both the ER and vacuoles
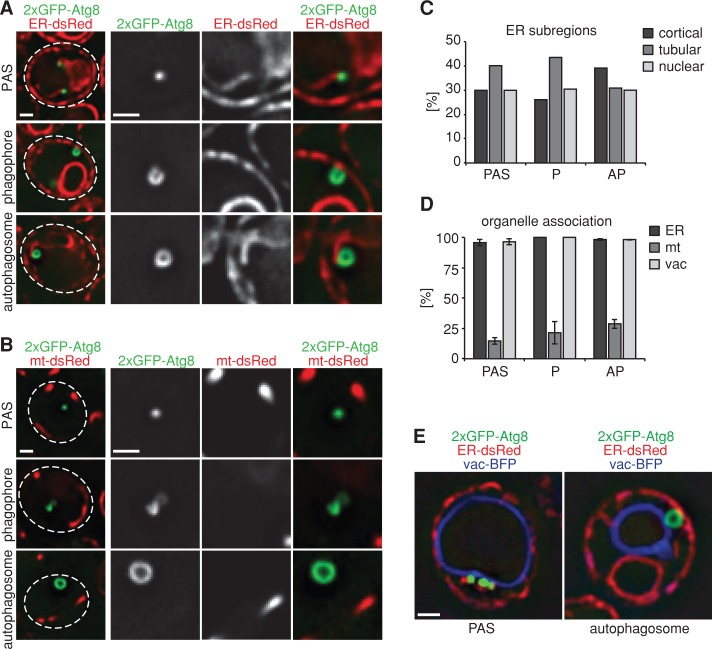
To test for a possible role of the COPII machinery in autophagosome biogenesis, we first examined the spatial relationship of each step of autophagosome biogenesis with the vacuole and the ER in vivo (Figures 1 and 3). Consistent with previous reports, after the induction of autophagy, we observed that the autophagosomal structures PAS, phagophores, and autophagosomes form in close proximity to the cytosolic side of vacuolar membranes (Figures 1 and 3, D and E; Suzuki et al., 2001). Coexpression of an ER-targeted dsRed reporter with 2xGFP-Atg8 revealed that all three structures are also quantitatively spatially linked to the ER, including cortical, tubular, and nuclear ER (Figure 3, A and C–E). In addition, nascent phagophores were associated with the ER in a nonrandom orientation, with their edges/openings always facing the proximal ER region (Figure 3, A, C, and D). The quantitative association of PAS with the vacuole and the ER suggests that sites of vacuole–ER contact are linked to the initiation of autophagosome formation. Consistent with this, in cells expressing fluorescent markers for both ER (ER-targeted dsRed) and vacuolar membranes (vac-BFP), we observed PAS at sites of vacuole–ER contacts or at regions where the vacuole and ER were in close proximity (Figure 3E). In addition, autophagosomes often bridged the distance between the vacuole and the ER (Figure 3E), suggesting that the bridging distance between ER and vacuole at sites of autophagosome formation adapts to the dimensions of the growing phagophore. We also examined the relationship between autophagosomal structures and mitochondria, as observations in mammalian cells suggest that autophagosomal biogenesis occurs on mitochondria or at ER–mitochondria contact sites (Hailey et al., 2010; Hamasaki et al., 2013). In cells expressing a mitochondrial matrix–targeted dsRed reporter, we detected only a minor fraction of PAS, phagophores, and autophagosomes in close proximity to mitochondria, suggesting that mitochondria or ER–mitochondria contacts do not play a general role in autophagy (Figure 3, B and D). Taken together, our data demonstrate that autophagosome formation occurs proximal to both the vacuole and the ER. The association of autophagosomal structures with the ER is consistent with observations in mammalian cells (Axe et al., 2008; Hayashi-Nishino et al., 2009). Thus the spatial organization of autophagosome biogenesis is an evolutionarily conserved feature critical to the process of autophagosome biogenesis.
FIGURE 3:
Autophagosomes are simultaneously tethered to the vacuole and ER during formation. (A) Wild type cells coexpressing 2xGFP-ATG8 and ER-dsRed were grown to log phase and treated with rapamycin (400 ng/ml) for 1 h and cells were imaged by fluorescence microscopy. Single focal planes of representative cells are shown. (B) Wild-type cells coexpressing 2xGFP-ATG8 and mt-dsRed were treated and analyzed as in A. (C) Relative association of PAS, phagophores, and autophagosomes with cortical, tubular, and nuclear subregions of the ER. (D) Association of PAS, phagophores, and autophagosomes with ER, mitochondria, and vacuoles. Data are shown as mean ± SD of three independent experiments. In total, ≥200 PAS or autophagosomes and ≥23 phagophores were analyzed in C and D. (E) Wild-type cells coexpressing 2xGFP-ATG8, ER-dsRed, and vac-BFP were treated as in A. Cells were imaged using three-dimensional structure illumination microscopy (Nikon 3D-SIM). Dashed lines indicate cell boundaries. P, phagophore; AP, autophagosome. Scale bars, 1 μm (A, B, E).
Autophagosomes form at ER exit sites
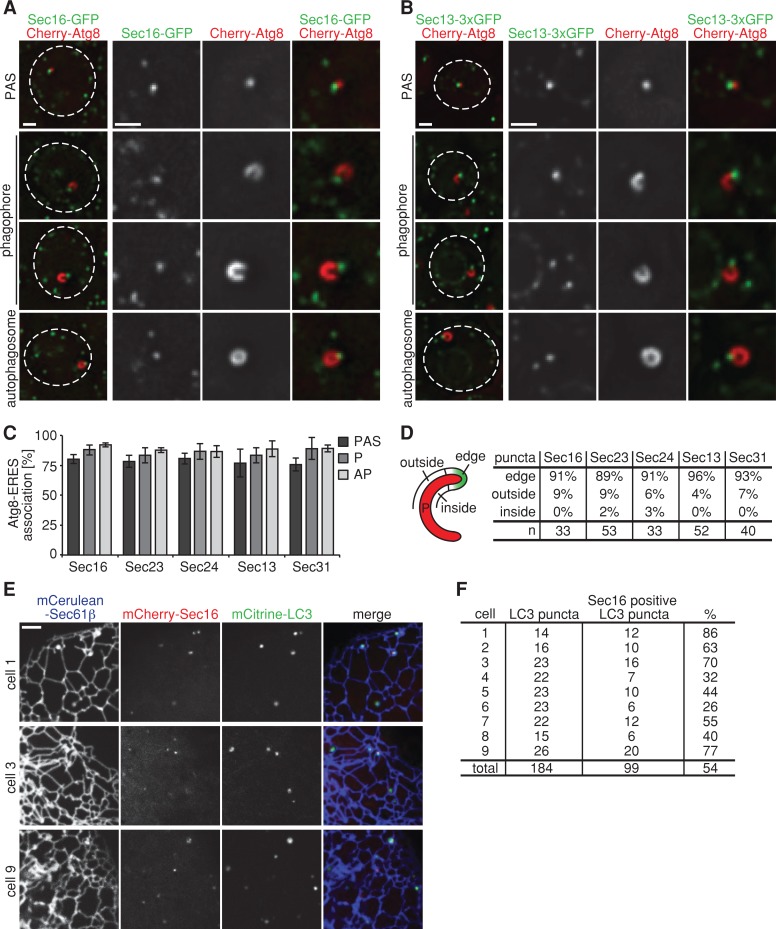
On the basis of the physical links between the COPII and autophagy machinery and the quantitative association of autophagosomal structures with the ER, we tested whether autophagosome formation is spatially linked to the specialized regions of the ER, so-called ER exit sites, where COPII vesicles form (Jensen and Schekman, 2011). To visualize ERES, we expressed the ERES-specific marker Sec16, the inner COPII coat proteins Sec23 or Sec24, or the outer COPII coat proteins Sec13 or Sec31 as functional GFP fusions. In agreement with previous reports, each ERES component localized to multiple puncta distributed throughout the cell (Figure 4, A and B, and Supplemental Figure S1; Castillon et al., 2009; Levi et al., 2010; Shindiapina and Barlowe, 2010; Okamoto et al., 2012). For all five ERES markers used, we observed that ∼80% of Cherry-Atg8–labeled PAS, phagophores, and mature autophagosomes formed in close proximity to ERES in rapamycin-treated cells (Figure 4, A–C, and Supplemental Figure S1). In addition, phagophores associated with ERES in a nonrandom orientation: in a vast majority of the cases (∼90%), a single ERES was found at the edge of the phagophore—a site of highly positive membrane curvature (Figure 4D). In contrast, we rarely observed ERES localized at regions of negative curvature, that is, the inside of the phagophore (≤ 3%; Figure 4D). In a small percentage of phagophores (4–9%), however, ERES was associated with the outside of the phagophore, but in these instances, a second ERES was also detected at the edge (Figure 4D). Together with the observed nonrandom phagophore–ER association, these data strongly support a model in which phagophore expansion proceeds in a directed manner at regions of high ER membrane curvature, where an associated ERES is localized.
FIGURE 4:
Formation of autophagosomes at ER exit sites is a conserved feature of autophagy. (A, B) Wild-type cells coexpressing Cherry-ATG8 and SEC16-GFP (A) or SEC13-3xGFP (B), respectively, were grown to log phase and treated with rapamycin (400 ng/ml) for 1 h. Cells were analyzed by fluorescence microscopy. Single focal planes of representative cells are shown. Dashed lines indicate cell boundaries. (C) Association of PAS, phagophores, and autophagosomes with ERES, labeled by fluorescent protein fusions of indicated components. Data are shown as mean ± SD of three independent experiments. For each ERES marker ≥150 PAS, ≥35 phagophores (P), and ≥180 autophagosomes (AP) were analyzed. (D) Association of ERES, labeled by fluorescent protein fusions of indicated components, with regions of the phagophore as defined in the schematic. n, number of ERES–phagophore structures analyzed. (E) Cos-7 cells transiently expressing mCitrine-LC3, mCherry-Sec16, and mCerulean-Sec61β were treated with rapamycin (218 ng/ml) for 4–6 h and analyzed by fluorescence microscopy. Peripheral sections of cells 1, 3, and 9 as quantified in F are shown. (F) Quantification of mCitrine-LC3 and mCherry-Sec16 colocalization from nine representative cells treated as described in E. Scale bars, 1 μm (A, B), 3 μm (E).
We asked whether the spatial link of autophagosomal structures to ERES is evolutionarily conserved by examining the localization of the transiently expressed mammalian Atg8 orthologue, mCitrine-LC3, and the mammalian ERES marker mCherry-Sec16 in Cos-7 cells whose ER was labeled with mCerulean-Sec61β. After induction of autophagy by rapamycin treatment, a majority of LC3-labeled puncta colocalized with Sec16-labeled ERES, and, in a given Cos-7 cell, multiple ERES were spatially linked to LC3-labeled puncta (Figure 4, E and F). These observations indicate that autophagosome biogenesis at ERES is a conserved feature of autophagy.
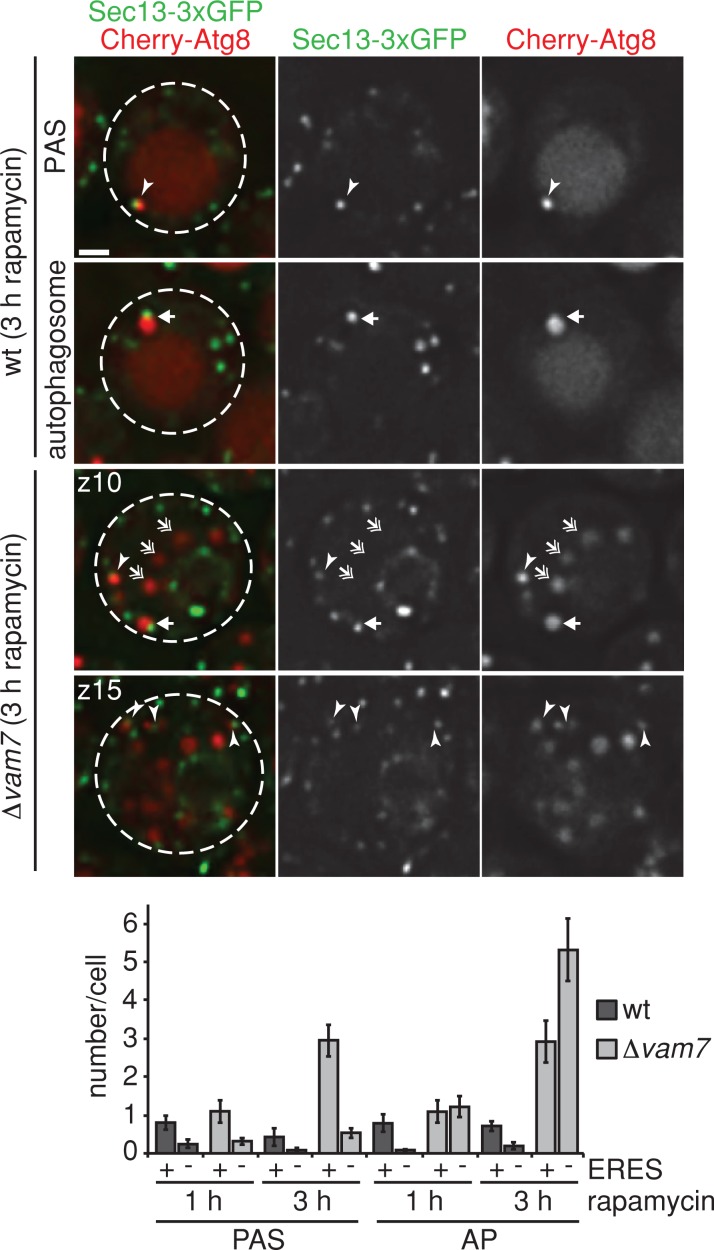
In contrast to mammalian cells, relatively few ERES (on average up to two) were associated with a PAS and an autophagosome in a given wild-type yeast cell after 1–3 h of rapamycin treatment (Figure 5). The basis for this difference is believed to be a consequence of the limited capacity of yeast cells to form only one PAS. We explored this model by monitoring autophagosome biogenesis in Δvam7 yeast cells, which are defective for autophagosome vacuole fusion (Sato et al., 1998). In Δvam7 cells, we observed a significant increase per cell in both PAS-ERES– and autophagosome-ERES–linked sites (on average up to six sites) compared with wild-type cells after 3 h of rapamycin treatment (Figure 5). We also observed an increase in the number of motile autophagosomes, which were not physically linked to ERES, in the cytosol of Δvam7 cells (53 and 65% vs. 13 and 20% in wild-type cells after 1 and 3 h, respectively; Figure 5). These data indicate that yeast cells possess the capacity to initiate autophagosomal biogenesis at multiple sites. In addition, the quantitative differences observed between wild-type yeast and mammalian cells in the association of autophagosomal structures with ERES likely reflects the fact that in yeast, all autophagosomes are in proximity to both the vacuole and ERES and after release from ERES rapidly fuse with the vacuolar membrane. As a consequence, non–ERES-associated autophagosomes are rarely detected in wild-type yeast cells. Thus our data indicate that as a general feature, autophagosomal biogenesis is initiated at multiple ERES in a cell, and ERES remains spatially linked or tethered to autophagosomal structures during autophagosome maturation. Our data indicate that the release of autophagosomes from ERES results in untethered motile autophagosomes that subsequently fuse with the vacuole/lysosome. During phagophore expansion, phagophores are positioned in a way that brings regions of high membrane curvature on the phagophores (edges) and ERES in close proximity to each other, an environment conducive for membrane remodeling and fusion events required for phagophore maturation.
FIGURE 5:
Autophagosome biogenesis is tethered to multiple ER exit sites in yeast cells in the absence of autophagosomal–vacuolar fusion. Wild-type or Δvam7 cells coexpressing Cherry-ATG8 and SEC13-3xGFP were grown to log phase and treated with rapamycin (400 ng/ml) for 1–3 h. Cells were analyzed by fluorescence microscopy at indicated time points. Single or indicated focal planes (z) of representative cells are shown. Examples for ERES–PAS (arrowheads) and ERES–autophagosome (arrows) association, as well as autophagosomes not associated with ERES (double-headed arrows), are indicated. For quantification, ≥90 cells were analyzed for PAS and autophagosome (AP) association with Sec13-3xGFP–marked ERES for each time point and strain as shown in the bar graph. Data are shown as mean ± SD of three independent experiments. Dashed lines indicate cell boundaries. Scale bar, 1 μm.
Assembly of the autophagy machinery requires ERES function
To test our model that ERES, potentially via COPII vesicles, serve as a proximal membrane source and/or platform for the nucleation and/or expansion of phagophores, we used a temperature-sensitive allele of the essential SEC12 gene, sec12-4, allowing for conditional inhibition of ERES function (Kaiser and Schekman, 1990; Barlowe and Schekman, 1993). We compared autophagic flux in wild-type and sec12-4 cells expressing 2xGFP-ATG8 after induction of autophagy by rapamycin treatment and exposure to nonpermissive temperature. Autophagic flux was assayed by monitoring the vacuolar conversion of autophagosome-associated 2xGFP-Atg8 to free GFP by Western blot analysis, which is a semiquantitative readout for the rate of formation and degradation of autophagosomes (Shintani and Klionsky, 2004). Consistent with previous reports, under nonpermissive conditions, autophagic flux was blocked in sec12-4 as compared with wild-type cells, demonstrating an essential function for ERES during autophagy (Figure 6A, left; Ishihara et al., 2001; Hamasaki et al., 2003). To test whether ERES functions in autophagy by modulating the regulatory signaling that controls autophagy initiation, we examined the activation of the Atg1 kinase complex by monitoring Atg13 phosphorylation patterns. Inhibition of TORC1 leads to Atg13 dephosphorylation at multiple sites, resulting in an increase in Atg1 kinase activity, and is required and sufficient for autophagy induction (Abeliovich et al., 2000; Kamada et al., 2000, 2010). On rapamycin treatment at nonpermissive temperature, sec12-4 cells displayed changes in Atg13 phosphorylation that were indistinguishable from wild-type cells, indicating that Atg1 kinase complex activation is not defective in sec12-4 cells (Figure 6A, right). Thus ERES likely function in the mechanics of autophagosome formation, a model consistent with our cytological data demonstrating the persistent physical association of ERES during autophagosomal maturation.
To find the stage at which ERES functions during autophagosome biogenesis, we performed cytological analysis of cells expressing Cherry-Atg8 and the ERES marker Sec13-3xGFP in sec12-4 cells. On induction of autophagy by rapamycin treatment under nonpermissive conditions, we observed aberrant ERES morphology in sec12-4 cells, with fewer brighter Sec13-3xGFP puncta compared with wild-type cells, consistent with inhibition of ERES function (Figure 6B; Castillon et al., 2009; Levi et al., 2010). Under these conditions, sec12-4 cells also displayed significant reduction in the number of Cherry-Atg8–labeled puncta compared with wild-type cells, indicating impaired PAS formation (Figure 6B). Furthermore, no autophagosomes were detected in sec12-4 cells (Figure 6B). These observations are in apparent contrast to previous reports describing the accumulation of aberrant Atg8-marked structures in the absence of Sec12 function (Hamasaki et al., 2003; Reggiori et al., 2004). This difference is likely due to effects on the distinct autophagy-related Cvt pathway, which is absent in our experiments. Our observations demonstrate that the early stages of autophagosome biogenesis, critical for PAS formation, depend on ERES function.
Phagophore nucleation at the PAS is an early autophagy event linked to the recruitment of Atg9-marked membranes. Atg9-labeled compartments are vesicular-tubular membrane structures whose biogenesis depends on the secretory pathway, including ERES function (Mari et al., 2010). Thus we tested whether the observed block in autophagosome formation in sec12-4 was a consequence of loss of the Atg9 compartment. We examined Atg9 compartments and autophagosome formation in wild-type and sec12-4 strains expressing genomic ATG9-3xGFP and Cherry-ATG8 markers, respectively. After autophagy induction, we observed a similar number of Atg9 puncta/compartments in wild-type and sec12-4 cells under nonpermissive conditions, excluding the possibility that the block in autophagy caused by loss of ERES function results from Atg9 compartment depletion.
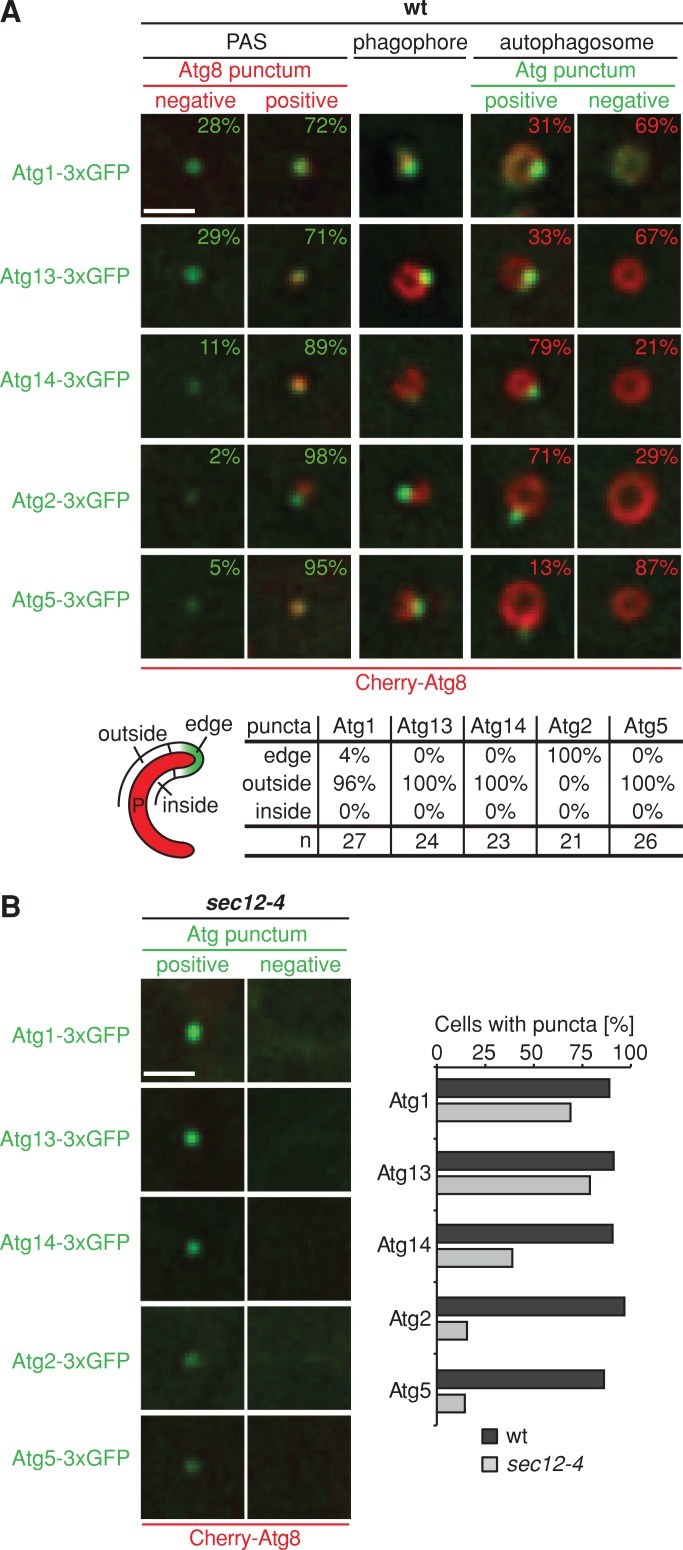
We tested the idea that ERES functions to promote the assembly of the autophagy machinery. First, we compared the association of ERES and core autophagy components with autophagic structures during autophagosome biogenesis. We performed cytological analysis of cells expressing genomic Cherry-ATG8 and ATG1-3xGFP or ATG13-3xGFP (Atg1 kinase complex), ATG14-3xGFP (PtdIns-3 kinase I), ATG2-3xGFP (Atg2-18 complex), or ATG5-3xGFP (Atg5-12/16 complex) after rapamycin treatment. All components were strongly associated with PAS, as characterized by punctate Cherry-Atg8–labeled structures (Atg8 positive, Figure 7A). In addition, we observed a significant number of Atg1- and Atg13-labeled puncta (∼30%) that were not labeled by Cherry-Atg8 (Atg8 negative), suggesting that the Atg1 kinase complex precedes Atg8 in the recruitment to PAS (Figure 7A). Similarly, all components analyzed were quantitatively associated with phagophores (Figure 7A). An analysis of the pattern of phagophore labeling indicated that whereas a majority of the autophagy components formed punctate structures localized to the outside of the phagophores, Atg2-3xGFP puncta were exclusively detected at the phagophore edge in a manner similar to ERES (Figure 7A; compare to Figure 4, A–D), consistent with Suzuki et al. (2013). Consistently, we observed quantitative association and colocalization of Atg2 with ERES in wild-type cells coexpressing Atg2-3xGFP and Sec13-Cherry under autophagy induction conditions (Supplemental Figure S2). All autophagy components tested labeled punctate structures associated with mature autophagosomes but with significant quantitative differences, which likely reflect the dynamic and temporally ordered dissociation of the autophagy machinery in the order of Atg5 followed by Atg1 and Atg13 and, at a late stage, Atg2 and Atg14 (Figure 7A). Taken together, our data indicate that ERES components colocalize with Atg2 at the edge of the phagophore and dissociate from mature autophagosomes at a late stage similar to Atg2 and Atg14 (Supplemental Figures S2 and Figures 4, A–C, and 7A).
FIGURE 7:
Hierarchical assembly of the autophagy machinery depends on ERES function. (A, B) Wild-type or sec12-4 cells expressing Cherry-ATG8 and ATG1-, ATG13-, ATG14-, ATG2-, or ATG5-3xGFP were grown to log phase at permissive temperature (23°C), treated with rapamycin (400 ng/ml), and shifted to nonpermissive temperature (37°C) for 1 h and analyzed by fluorescence microscopy. Single focal planes of sections of representative cells are shown. (A) Association of indicated Atgs with PAS or autophagosomes marked by Cherry-Atg8 is shown in percentage (n ≥ 100). Associations of Atgs, labeled by fluorescent protein fusions of indicated components, with regions of the phagophore as defined in the schematic are shown. n, number of Atg-phagophore structures analyzed. (B) sec12-4 cells without Cherry-Atg8–marked structures were analyzed for punctate structures formed by indicated autophagy components. Left, single focal planes of sections of example cells. The bar graph shows the number of total wild-type cells displaying punctate structures of indicated Atgs compared with the number of sec12-4 cells showing punctate Atgs not marked by Cherry-Atg8 (n ≥ 100). Scale bars, 1 μm.
We next examined directly whether loss of ERES function affects the hierarchical assembly of core autophagy components by comparing the localization of Cherry-ATG8 and ATG1-, ATG13-, ATG14-, ATG2-, or ATG5-3xGFP in rapamycin-treated wild-type and sec12-4 cells under nonpermissive conditions. Whereas all components formed punctate structures associated with Atg8-positive autophagosomal structures in wild-type cells, sec12-4 cells lacked Atg8-marked structures but possessed Atg8-negative Atg1- or Atg13-marked puncta, similar to wild-type cells (Figure 7, A and B). In contrast, Atg14, Atg2, and Atg5 puncta were strongly reduced in sec12-4 cells compared with wild type (Figure 7B). Thus our data indicate that ERES are core autophagic components that play a direct and essential role immediately downstream of the recruitment of the Atg1 kinase complex in the assembly of the autophagy machinery required for phagophore nucleation and/or initial phagophore expansion.
DISCUSSION
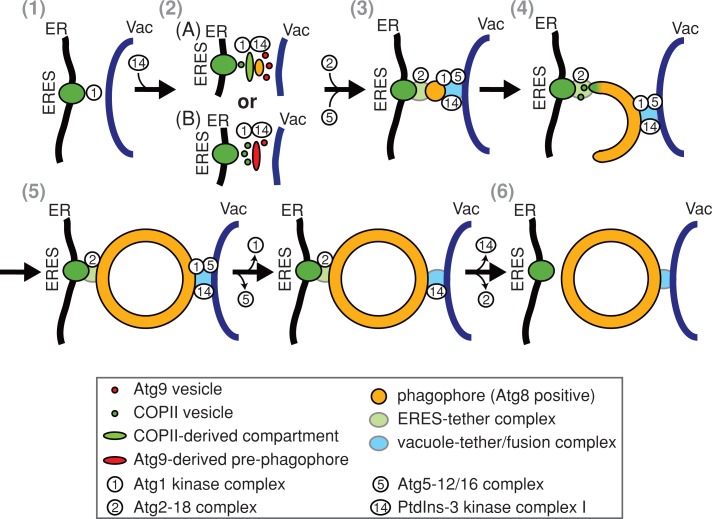
Our work shows that localization of autophagosome biogenesis at ERES is an evolutionarily conserved feature of autophagy. This finding is consistent with work of Suzuki et al. (2013), who also reported ERES in proximity to phagophores in yeast. They artificially enlarged a known cargo protein complex by overexpression of prApe1 to allow for analysis of phagophores by light microscopy. These particular conditions, however, did not permit mature autophagosomes to form and consequently limited the analysis to nonproductive phagophore intermediates. Despite these technical differences, analysis of our native system largely confirmed distinct associations of autophagy components with forming phagophores and a close association of ERES and Atg2. Our system, however, allowed us to analyze the role of ERES during the entire process of autophagosome biogenesis and, in combination with our proteomics and functional analysis, identify a fundamentally different role for ERES during autophagosome formation than proposed by Suzuki and colleagues. ERES associate with autophagic membranes at every stage of the autophagosomal biogenesis process, and ERES function is required directly downstream of the Atg1 kinase complex for the assembly of the autophagy machinery. In addition, our proteomic analyses demonstrate that autophagy and ERES components share many functionally related protein interactors. Thus our analysis identifies ERES as core autophagy components, which play an essential role in phagophore nucleation and likely also in phagophore growth. The Atg1/Ulk kinase complex constitutes the most upstream component in the hierarchical assembly of the autophagy machinery in yeast and mammals (Suzuki et al., 2007; Itakura and Mizushima, 2010), which recruits and provides a structural framework to tether Atg9-marked membranes to PAS by direct physical interaction (Sekito et al., 2009; Ragusa et al., 2012). On the basis of these and our observations, we postulate that ERES work in concert with the Atg1 kinase complex and the Atg9 membrane compartment to facilitate the recruitment of PdtIns-3 kinase I, the Atg2-18 complex, and additional downstream autophagy components for phagophore nucleation and growth. In this capacity, ERES may generate COPII-derived membrane compartments, equivalent to ER–Golgi intermediate compartment (ERGIC) or omegasomes in mammalian cells, which function as structural platforms and/or a specialized membrane source for phagophore nucleation and expansion (Figure 8A). Consistent with a role of an ERGIC-like compartment, we observed physical interactions of the autophagy machinery with Emp46 and Emp47, the yeast orthologues of the mammalian ERGIC marker ERGIC-53 (Satoh et al., 2006). Atg9-marked membranes may also recruit COPII vesicles directly to pre-phagophore membranes or the edges of expanding phagophores to drive maturation and growth, respectively (Figure 8B). Consistent with this possibility, peripheral Atg9 membranes are also found at the edges of nascent phagophores and contain factors such as the Rab GTPase Ypt1 and the autophagy-specific Ypt1 effector complex TRAPPIII subunit, Trs85, which are required for the targeting of secretory vesicles (Kakuta et al., 2012; Suzuki et al., 2013).
FIGURE 8:
Model for the functional specialization of the ER for autophagosomal biogenesis. 1) Initiation: on autophagy induction, the Atg1 kinase complex localizes to PAS. 2) Phagophore nucleation: coinciding with recruitment of Atg14, ERES generate COPII-derived membrane compartments that function as pre-phagophore membranes and/or as structural platforms for Atg9-driven phagophore assembly (A). Alternatively, fusion of ERES-dependent COPII vesicles with pre-phagophores derived from Atg9 vesicles drives phagophore maturation (B). 3, 4) Phagophore expansion is initiated by Atg2-18 complex, Atg5-12/16 complex, and Atg8 recruitment and is driven by the fusion of COPII vesicle and potentially other membrane sources at the edge of cup-shaped phagophores. Expansion of the phagophore separates the autophagy machinery into at least two distinct groups: the Atg2-18 complex is associated with ERES at the edge; other components remain at punctate structures in proximity to the vacuole. Phagophores are tethered simultaneously to ERES and vacuoles by separate tethering structures. 5, 6) On completion, autophagy components dissociate, and mature autophagosomes are released from ERES tethers and fuse with vacuolar membranes in a temporally coordinated manner.
Our data indicate that both ERES and vacuoles remain spatially linked with expanding phagophores and autophagosomes, suggesting the existence of two molecular tethers that stably connect ER/ERES and vacuoles to autophagosomal structures, respectively (Figure 8). In yeast, these tethers likely function to temporally coordinate autophagosomal maturation with autophagosome–vacuole fusion. As a consequence, at steady state, autophagosomes not tethered to ER/ERES are detected only under conditions in which autophagosome–vacuolar fusion is blocked. In contrast, in mammalian cells, the formation of autophagosomes is not strictly spatially linked to lysosomes, and active transport of autophagosomes is required to the perinuclear region of cells, where lysosomes are enriched (Kimura et al., 2008; Korolchuk et al., 2011). Indeed, nonspecific forms of autophagy depend on an intact cytoskeleton in mammalian cells but not in yeast cells (Reggiori et al., 2005). Thus our data suggest that, as a general feature, the release of autophagosomes from ERES is a permissive event for their subsequent fusion with the vacuole/lysosome. Although the molecular features of the autophagosome-ER/ERES and -vacuole tethers are not known, their existence raises the question of whether ERES are modified in their localization, structure, composition, and/or protein cargoes to specifically function in autophagosome biogenesis. Regardless of potential adaptations, the generally distributive and dynamic nature of ERES is ideally suited to localize autophagosome formation to diverse sites and cargoes throughout the cell in response to various signals.
MATERIALS AND METHODS
Cloning and plasmids
pFA6a-link-3xyEGFP-CaURA3 was generated by replacing a HindIII-link-yEGFP-SacI fragment in pFA6a-link-yEGFP-CaURA3 (Sheff and Thorn, 2004) with HindIII-link-yEGFP-PstI, PstI-link1-yEGFP-BamHI, and BamHI-link1-yEGFP-SacI fragments (link: 5′-GGT GAC GGT GCT GGT TTA ATT AAC-3′; link1: 5′-GGT GCT GGT GCT-3′). pRS306-2xyEGFP-ATG8 was constructed by inserting an EcoRI-prATG8-XmaI fragment (−223 to −1 base pair 5′-region of ATG8), an XmaI-link-yEGFP-link1-yEGFP-BamHI fragment, and a BamHI-ATG8-NotI fragment (ORF + 176–base pair 3′-region) into pRS306 using EcoRI/NotI sites. pRS304-yEmCherry-ATG8 was generated by inserting an EcoRI-prATG8-XmaI fragment (−223 to −1 base pair 5′-region of ATG8), an XmaI-yEmCherry-BamHI fragment (Lackner et al., 2013), and a BamHI-ATG8-NotI fragment (ORF + 176–base pair 3′-region) into pRS304 using EcoRI/NotI sites. pRS305-2xyEGFP-ATG8-NATMX6-ER-dsRed was constructed by inserting an XhoI-prTPI-Kar2(1-135)-dsRed-HDEL-terminatorCYC1-HindIII fragment (ER-dsRed) in pRS305 using SalI/HindIII sites. A prATG8-2xyEGFP-ATG8-NATMX6 fusion fragment generated by PCR using pRS306-2xyEGFP-ATG8 was integrated into pRS305-ER-dsRed via SpeI/HindIII sites. pRS305-prPGK1-3xtagBFP-PHO8 (vac-BFP) was generated by cloning a SalI-pPGK1-PstI fragment (−982 to −1 base pair 5′-region of PGK1), a PstI-tagBFP-link2-tagBFP-link3-tagBFP-XmaI fragment in which tagBFPs contained no stop codons (link2: 5′-GGT GGT GCT TCT TCT GCT CAA GGT GGT-3′; link3: 5′-TCT GGT CAT GCT CAA TCT GGT TCT GCT-3′), and a XmaI-PHO8-BamHI fragment (ORF + 338–base pair 3′-region) into pRS305 using SalI/BamHI sites.
pFA6a-link-yEGFP-CaURA3, YIplac204/TKC-dsRed-HDEL (ER-dsRed), and pYX142-mt-dsRed have been described previously (Sheff and Thorn, 2004; Onischenko et al., 2009; Friedman et al., 2011).
mCerulean-Sec61β was generated by first PCR amplifying mCerulean and cloning it into the NheI/XhoI sites of pAcGFP1-C1 (replacing GFP) and then cutting a fragment containing Sec61β from GFP-Sec61β (Shibata et al., 2008) and cloning into the BglII/PstI sites of that vector. mCitrine-LC3 was generated by first PCR amplifying mCitrine and cloning it into the NheI/XhoI sites of pAcGFP1-C1 (replacing GFP) and then PCR amplifying a fragment containing mouse Map1LC3b from EGFP-LC3 (Martens et al., 2005) and cloning into the XhoI/BamHI sites of that vector. mCherry-Sec16 was generated by cloning a fragment containing the short isoform of human Sec16 from GFP-Sec16S (Bhattacharyya and Glick, 2007); plasmid 15575, purchased from AddGene, Cambridge, MA) into the BglII/SalI sites of mCherry-Sec61β (Zurek et al., 2011), replacing Sec61β.
Strains and media
All strains used in this study are derivatives of W303, with the exceptions of rsy263 and rsy252, and are listed in Table 1. Gene deletions were obtained by replacing the complete ORF of genes by the indicated cassettes using PCR-based targeted homologous recombination (Δatg7::KanMX6; Δatg19::HIS3MX3; Δvam7::NATMX6). Functional C-terminally tagged strains were constructed by PCR-based targeted homologous recombination using pFA6a-link-yEGFP-CaURA3 or pFA6a-link-3xyEGFP-CaURA. Haploid double-tagged strains were constructed by crossing, followed by sporulation or PCR-based targeted homologous recombination. The temperature-sensitive allele sec12-4 (Kaiser and Schekman, 1990) was transferred into haploid W303 strains by PCR-based amplification and 3′-fusion of a NATMX6 cassette generating sec12-4-NATMX6, replacement of a complete Δsec12::CaURA3 deletion in a heterozygous diploid W303 strain, and subsequent sporulation. The strain Δatg19::prATG8-2xGFP-ATG8-NATMX6 was constructed by PCR-based targeted homologous recombination of a prATG8-2xGFP-ATG8-NATMX6 fragment amplified from pRS306- prATG8-2xGFP-ATG8 fused at its 3′-end to a NATMX6 cassette with flanking 40 base pairs of 5′- and 3′-region of ATG19.
Strains were grown in synthetic complete glucose medium (0.7% [wt/vol] yeast nitrogen base, 2% [wt/vol] glucose) at 30°C as standard temperature. In case of the temperature-sensitive strains expressing sec12-4, strains were grown at permissive temperature of 23°C and shifted to nonpermissive temperature of 37°C. Autophagy was induced by the addition of rapamycin (400 ng/ml final concentration).
Mammalian cell growth, transfection, and rapamycin treatment
Cos-7 cells (American Type Culture Collection, Manassas, VA) were grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (PAA Laboratories, Pasching, Austria) and penicillin/streptomycin (Invitrogen). Cells were seeded in six-well plates at a density of 2 × 105 cells/well ∼16 h before transfection. Cells were transfected for ∼5 h in OPTI-MEM (Invitrogen) containing 100 ng of mCitrine-LC3, 1 μg of mCherry-Sec16, and 1 μg of mCerulean-Sec61β, as well as 5 μl of Lipofectamine 2000 (Invitrogen). Cells were then reseeded in glass-bottom microscope dishes (MatTek, Ashland, MA) at a density of 2 × 105 cells/dish in normal growth media and allowed to recover ∼16 h before treatment. Cells were treated with rapamycin (218 ng/ml) and imaged between 4 and 6 h posttreatment.
Proteomic analysis
Strains (Δatg19, Δatg19 expressing C-terminally 3xGFP-tagged ATG1, ATG2, ATG6, ATG9, ATG11, ATG13, ATG14, ATG17, ATG18, ATG23, ATG27, ATG29, ATG31, VPS34, VPS15, Δatg19 expressing 2xGFP-ATG8, or Δatg19 expressing C-terminally GFP-tagged SEC13, SEC23, or SEC24) were grown at 30°C in synthetic complete glucose medium to OD600 of 1.0. Rapamycin was added to a final concentration of 400 ng/ml, and cells were incubated at 30°C for 1 h. Cells corresponding to 2000 OD600 units were pelleted, washed in distilled H2O, and resuspended in IPLB (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid–KOH, pH 7.4, 150 mM KOAc, 2 mM Mg(Ac)2, 1 mM ethylene glycol tetraacetic acid, and 0.6 M sorbitol) containing 1× Protease Inhibitor Mixture I (PIC; Calbiochem, La Jolla, CA), 1 mM NaF, and 20 mM β-phosphoglycerol. Yeast cell-buffer pellets created in liquid N2 were stored at −80°C. Frozen cells were lysed using a Freezer/Mill (SPEX Metuchen, NJ; settings, T1 = 1 min; T2 = 2 min; T3 = 2 min; cycles, 2; rate, 7). Resulting cell lysates were thawed at 23°C, 1× PIC was added, and lysates were cleared with a low-speed spin, 500 × g for 10 min at 4°C. For chemical cross-linking, 1 mM dithiobis(succinimidyl propionate) was added to supernatants, and samples were incubated for 30 min on ice. To quench cross-linking reactions, 100 mM Tris (pH 7.5) was added to samples, followed by 15 min of incubation on ice. To solubilize membranes, digitonin (final concentration of 1% [wt/vol]) was added, and samples were incubated for 30 min on ice. Lysates were cleared with a 12,000 × g spin for 10 min at 4°C, and supernatants were incubated for 30 min on ice with 50 μl of μMACS anti-GFP MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany). Beads were isolated using Miltenyi μ columns and a μMACS separator (Miltenyi Biotec), washed three times with 800 μl of IPLB plus 0.1% (wt/vol) digitonin plus PIC, and washed twice with 500 μl of IPLB. For on-bead digestion, beads were incubated in 25 μl of elution buffer I (2 M urea, 50 mM Tris, pH 7.5, 1 mM dithiothreitol, and 5 μg/ml trypsin) for 30 min at 23°C. Samples were eluted twice with 50 μl of elution buffer II (2 M urea, 50 mM Tris, pH 7.5, and 5 mM chloroacetamide). After overnight incubation at 23°C, 1 μl of trifluoroacetic acid was added to collected elutions to stop the reactions. Samples were analyzed using a high-performance linear trap quadrupole Orbitrap Velos Pro mass spectrometer (Thermo Scientific, Waltham, MA). Resulting MS/MS spectra were subsequently matched against yeast protein sequences using the SEQUEST database search engine, and match quality was evaluated using spectral (i.e., peptide) counts and probability scores generated by the STATQUEST algorithm (Kislinger et al., 2003; Babu et al., 2009).
Functional sorting and network analysis
In total, 768 unique proteins were identified by the proteomics approach that passed the cutoff defined by probability scores of ≥70% and not being detected in control samples derived from the untagged Δatg19 strain processed in parallel. To identify functional groups, the list of 768 proteins was sorted according to functional annotations using the DAVID bioinformatic resource with medium classification stringency, similarity term overlap of 3, similarity threshold of 3, initial and final group membership of 3, multiple linkage threshold of 0.5, and enrichment threshold of 1.0 (Huang da et al., 2009a, b), giving rise to the functional group “vesicle-mediated transport” containing 134 proteins in total. A physical interaction network was generated based on the sum of total peptides identified in up to three purifications for each bait–prey interaction within the functional group “vesicle-mediated transport” (Supplemental Table S1), using Cytoscape, version 2.3.8. For simplification in Figure 2A, prey interactions of bait proteins of the same complex are shown as single edges between the complex and prey proteins according to the sum of total peptides.
Cytological analysis
Yeast cells.
Yeast cells were grown to log phase in synthetic complete glucose medium. After addition of rapamycin and further incubation as indicated, cells were concentrated by centrifugation. Cell monolayers were generated by mounting cells on a 3.5% (wt/vol) agarose pad. Whole-cell z-series with a step size of 0.2 μm were captured using a DeltaVision-Real Time microscope (Applied Precision, Issaquah, WA) fitted with a 60×, 1.4 numerical aperture (NA) objective and CoolSnap HQ camera (Photometrics, Tucson, AZ). Capture and postcapture image processing was done using softWoRx software (Applied Precision) and Photoshop (Adobe, San Jose, CA). Deconvolved images are shown. Colocalization analyses were performed manually using a complete z-series. Nokia three-dimensional secondary ion mass analysis was performed using an Eclipse Ti microscope (Nikon, Melville, NY) fitted with an Apo TIRF 100×/1.48 NA oil objective and 405-, 488-, 561-, and 640-nm lasers. Capture and postcapture image processing was done using Nokia NIS-Elements AR imaging software v4.11 (Nikon).
Mammalian cells.
Single planes of transiently transfected Cos-7 cells were imaged using the spinning-disk module of a Marianas SDC Real Time 3D Confocal-TIRF microscope (Intelligent Imaging Innovations, Denver, CO) fitted with a 100×, 1.46 NA objective and an electron-multiplying charge-coupled device camera (Photometrics). Image capture was performed using SlideBook5 software (Intelligent Imaging Innovations), and ImageJ (National Institutes of Health, Bethesda, MD), and Photoshop was used to for image processing. LC3 and Sec16 puncta colocalization analysis was manually performed in regions of the cell periphery where tubular ER could be resolved and Sec16 localization to the ER could be verified.
Whole-cell extraction and Western blot analysis.
At indicated time points, cells corresponding to 0.25 OD600 unit were collected and lysed by alkaline whole-cell extraction (0.255 M NaOH, 1% β-mercaptoethanol). Protein extracts were analyzed by SDS–PAGE and immunoblotting (α-GFP, monoclonal; UC Davis/NIH NeuroMab Facility, University of California, Davis, CA [P42212; clone 86/8]; α-Atg13, polyclonal; D. J. Klionsky, University of Michigan) and visualized with appropriate secondary antibodies conjugated to IRDye (800CW; LI-COR Biosciences, Lincoln, NE) using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Supplementary Material
Acknowledgments
We thank Brian C. Cook and Chris Baeh for excellent technical assistance and Daniel Klionsky and Randy Schekman for providing the Atg13 antibody and the sec12-4 strain, respectively. All imaging was conducted in the University of California, Davis, Molecular and Cellular Biology Light Microscopy Imaging Facility, with technical advice from Michael Paddy. J.N. is supported by National Institutes of Health Grants R01GM062942 and R01GM097432, and J.N. and M.B. are supported by Canadian Institutes of Health Research Grant MOP-125952. J.R.F. is supported by the Jane Coffin Childs Memorial Fund for Medical Research.
Abbreviations used:
- AMP
adenosine monophosphate
- COPII
coat protein complex II
- GTP
guanosine triphosphate
- TRAPP
transport protein particle
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-07-0381) on July 31, 2013.
*These authors contributed equally to this work.
The authors have no financial or nonfinancial competing interest.
REFERENCES
- Abeliovich H, Dunn WA, Jr, Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M, Krogan NJ, Awrey de Emili A, Greenblatt JF. Systematic characterization of the protein interaction network and protein complexes in Saccharomyces cerevisiae using tandem affinity purification and mass spectrometry. Methods Mol Biol. 2009;548:187–207. doi: 10.1007/978-1-59745-540-4_11. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Schekman R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya D, Glick BS. Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization. Mol Biol Cell. 2007;18:839–849. doi: 10.1091/mbc.E06-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon GA, Watanabe R, Taylor M, Schwabe TM, Riezman H. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic. 2009;10:186–200. doi: 10.1111/j.1600-0854.2008.00857.x. [DOI] [PubMed] [Google Scholar]
- Cebollero E, Reggiori F. Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2009;1793:1413–1421. doi: 10.1016/j.bbamcr.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:1845–1846. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- Dancourt J, Barlowe C. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, Dibenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno K, Ishikawa T, Akasaki K, Lee S, Nishimura Y, Tsuji H, Himeno M, Kato K. Immunocytochemical study of the surrounding envelope of autophagic vacuoles in cultured rat hepatocytes. Exp Cell Res. 1990;189:261–268. doi: 10.1016/0014-4827(90)90245-6. [DOI] [PubMed] [Google Scholar]
- Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2257–2269. doi: 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Hamasaki M, Noda T, Ohsumi Y. The early secretory pathway contributes to autophagy in yeast. Cell Struct Funct. 2003;28:49–54. doi: 10.1247/csf.28.49. [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen D, Schekman R. COPII-mediated vesicle formation at a glance. J Cell Sci. 2011;124:1–4. doi: 10.1242/jcs.069773. [DOI] [PubMed] [Google Scholar]
- Kaiser CA, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Kakuta S, Yamamoto H, Negishi L, Kondo-Kakuta C, Hayashi N, Ohsumi Y. Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J Biol Chem. 2012;287:44261–44269. doi: 10.1074/jbc.M112.411454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. TOR-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. TOR directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct. 2008;33:109–122. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislinger T, Rahman K, Radulovic D, Cox B, Rossant J, Emili A. PRISM, a generic large scale proteomic investigation strategy for mammals. Mol Cell Proteomics. 2003;2:96–106. doi: 10.1074/mcp.M200074-MCP200. [DOI] [PubMed] [Google Scholar]
- Knodler LA, Celli J. Eating the strangers within: host control of intracellular bacteria via xenophagy. Cell Microbiol. 2011;13:1319–1327. doi: 10.1111/j.1462-5822.2011.01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13:453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen EL, Thumm M. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J Cell Biol. 2010;190:965–973. doi: 10.1083/jcb.201002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL, Ping H, Graef M, Murley A, Nunnari J. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc Natl Acad Sci USA. 2013;110:E458-E467. doi: 10.1073/pnas.1215232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi SK, Bhattacharyya D, Strack RL, Austin JR, 2nd, Glick BS. The yeast GRASP Grh1 colocalizes with COPII and is dispensable for organizing the secretory pathway. Traffic. 2010;11:1168–1179. doi: 10.1111/j.1600-0854.2010.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci USA. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S, Parvanova I, Zerrahn J, Griffiths G, Schell G, Reichmann G, Howard JC. Disruption of Toxoplasma gondii parasitophorous vacuoles by the mouse p47-resistance GTPases. PLoS Pathog. 2005;1:e24. doi: 10.1371/journal.ppat.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes–Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21:3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Kurokawa K, Matsuura-Tokita K, Saito C, Hirata R, Nakano A. High-curvature domains of the ER are important for the organization of ER exit sites in Saccharomyces cerevisiae. J Cell Sci. 2012;125:3412–33420. doi: 10.1242/jcs.100065. [DOI] [PubMed] [Google Scholar]
- Onischenko E, Stanton LH, Madrid AS, Kieselbach T, Weis K. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J Cell Biol. 2009;185:475–4791. doi: 10.1083/jcb.200810030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010a;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010b;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Monastyrska I, Shintani T, Klionsky DJ. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Wang CW, Nair U, Shintani T, Abeliovich H, Klionsky DJ. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Sato TK, Darsow T, Emr SD. Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol Cell Biol. 1998;18:5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Sato K, Kanoh A, Yamashita K, Yamada Y, Igarashi N, Kato R, Nakano A, Wakatsuki S. Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p. J Biol Chem. 2006;281:10410–10419. doi: 10.1074/jbc.M512258200. [DOI] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells. 2009;14:525–538. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem. 2008;283:18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindiapina P, Barlowe C. Requirements for transitional endoplasmic reticulum site structure and function in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:1530–1545. doi: 10.1091/mbc.E09-07-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci USA. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Van der Vaart A, Griffith J, Reggiori F. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2270–2284. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek N, Sparks L, Voeltz G. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 2011;12:28–41. doi: 10.1111/j.1600-0854.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.