Environmental signals, such heat shock and acidosis, induce a structural and functional remodeling of the nucleolus. This process, which depends on the expression of intergenic long noncoding RNA, reversibly converts the nucleolus from a transcriptionally active ribosome factory into a transcriptionally inert prison for proteins.
Abstract
The nucleolus is a plurifunctional organelle in which structure and function are intimately linked. Its structural plasticity has long been appreciated, particularly in response to transcriptional inhibition and other cellular stresses, although the mechanism and physiological relevance of these phenomena are unclear. Using MCF-7 and other mammalian cell lines, we describe a structural and functional adaptation of the nucleolus, triggered by heat shock or physiological acidosis, that depends on the expression of ribosomal intergenic spacer long noncoding RNA (IGS lncRNA). At the heart of this process is the de novo formation of a large subnucleolar structure, termed the detention center (DC). The DC is a spatially and dynamically distinct region, characterized by an 8-anilino-1-naphthalenesulfonate–positive hydrophobic signature. Its formation is accompanied by redistribution of nucleolar factors and arrest in ribosomal biogenesis. Silencing of regulatory IGS lncRNA prevents the creation of this structure and allows the nucleolus to retain its tripartite organization and transcriptional activity. Signal termination causes a decrease in IGS transcript levels and a return to the active nucleolar conformation. We propose that the induction of IGS lncRNA by environmental signals operates as a molecular switch that regulates the structure and function of the nucleolus.
INTRODUCTION
Cellular organelles are composed of distinct subdomains that assemble into functional architectures. The individual proteins that take part in this organization are highly mobile molecules that continuously associate and disassociate in an effort to maintain dynamic frameworks of functional interactions (Vikstrom et al., 1992; Phair and Misteli, 2000; Misteli, 2001a, b; Shav-Tal et al., 2004). A primary example of molecular dynamics occurs within the nucleolus. This subnuclear organelle is composed of three distinct compartments that assemble around ∼400 tandem repeats of ribosomal DNA (rDNA), thereby consolidating the different factors required for ribosomal biogenesis. RNA polymerase (pol) I transcription is believed to take place at the interface between the fibrillar centers (FCs) and dense fibrillar components (DFCs), with further posttranscriptional maturation of newly synthesized rRNA occurring within the DFC. Final processing of rRNA and assembly with ribosomal proteins takes place in the granular component (GC). Through its tremendous output of rRNA, the nucleolus plays a central role in providing the cell with the protein synthesis capabilities necessary to sustain growth and proliferation (Melese and Xue, 1995; Derenzini et al., 1998; Scheer and Hock, 1999; Lempiainen and Shore, 2009).
The remarkable plasticity of the nucleolus is demonstrated during each mammalian cell cycle, with the structure disassembling at the onset of mitosis and reassembling at mitotic exit. It is also evident under conditions of cellular stress. Actinomycin D (ActD)–mediated inhibition of transcription leads to irreversible redistribution of both nucleolar and nucleoplasmic proteins into cap structures at the periphery of the nucleolus (Journey and Goldstein, 1961; Reynolds et al., 1964; Shav-Tal et al., 2005) and a correspondingly dramatic alteration in the nucleolar proteome (Andersen et al., 2005). Changes in the nucleolar proteome are also observed in response to DNA damage and viral infection (Hiscox, 2007; Moore et al., 2011). Reversible disorganization of nucleolar structure can be induced by treatment with the casein kinase 2 (CK2) inhibitor 5,6-dichloro-β-d-ribofuranosylbenzimadole (Scheer et al., 1984; Panse et al., 1999). Under these conditions, RNA pol I remains active, while nucleoli dissociate into substructures and disperse throughout the nucleoplasm as a necklace of transcribing “beads.” Nucleolar reformation upon removal of the drug is CK2 driven and ATP/GTP dependent (Louvet et al., 2006). Although the functional significance of some of these responses is unclear, they demonstrate that the structure of the nucleolus is highly responsive to a variety of stimuli (Olson and Dundr, 2005). Furthermore, the nucleolus harbors many proteins that are unrelated to ribosomal biogenesis, supporting the notion that it is a plurifunctional organelle (Pederson, 1998; Boisvert et al., 2007) involved in cell cycle regulation, senescence, and stress responses (Guarente, 1997; Visintin and Amon, 2000; Olson, 2004; Boulon et al., 2010). In fact, the nucleolus has been reported to associate with additional structural elements, including perinuclear compartments in cancer cells (Matera et al., 1995; Pollock and Huang, 2009; Pollock et al., 2011) and intranucleolar bodies (Hutten et al., 2011).
A striking example of functional nucleolar plasticity involves the capture of proteins by long noncoding RNA (lncRNA) in response to changing environmental conditions (Audas et al., 2012a; Jacob et al., 2012). Increases in temperature (heat shock) trigger the induction of ribosomal intergenic spacer (IGS) RNAs located 16 kb (IGS16RNA) and 22 kb (IGS22RNA) downstream of the rDNA transcription start site (Audas et al., 2012a). Similarly, a decrease in extracellular pH (acidosis) causes the accumulation of a transcript located 28 kb (IGS28RNA) downstream of the cassette start site (Audas et al., 2012a). These RNA pol I transcripts, which are processed into 300–400 nucleotide products, immobilize select protein in the nucleolus, away from their downstream effectors. A specific example is the nucleolar sequestration of the von Hippel– Lindau (VHL) protein (Mekhail et al., 2004a), which allows hypoxia-inducible factors to evade proteasomal degradation (Maxwell et al., 1999) and transcriptionally (Semenza, 2000) and translationally (Uniacke et al., 2012) activate their target genes, thus mediating cellular adaptation to hypoxia (Semenza, 2001; Mekhail et al., 2004b). Of interest, VHL is only one of many targets of IGS lncRNA–induced nucleolar sequestration. Other factors include the heat shock protein 70 kDa (Hsp70), ring finger protein 8 (RNF8), DNA polymerase catalytic subunit δ (POLD1), and DNA methyl transferase 1 (DNMT1; Audas et al., 2012b). Sequestered proteins are generally characterized by the presence of a nucleolar detention sequence (NoDS), a discrete amino acid sequence composed of an arginine motif (R-R-L/I) and at least two hydrophobic triplets (L-X-L/V) (Mekhail et al., 2007).
The nucleolar detention pathway places the nucleolus at the heart of a systemic remodeling of molecular networks, which appears central to the cell's adaptation to environmental stresses (Audas et al., 2012b). The fundamental question, however, of how the nucleolar architecture adapts to fulfill this additional role is unanswered. Here, we report that IGS lncRNAs functionally reorganize the nucleolus by regulating the formation of the nucleolar detention center (DC), a previously uncharacterized compartment that is spatially, dynamically, and biochemically distinct from the classic nucleolar domains. Formation of the DC reversibly alters the distribution of nucleolar factors and induces arrest in the synthesis of rRNA. These data highlight a role for lncRNA in the reorganization of the nucleolar space.
RESULTS
The nucleolus responds structurally to environmental stimuli
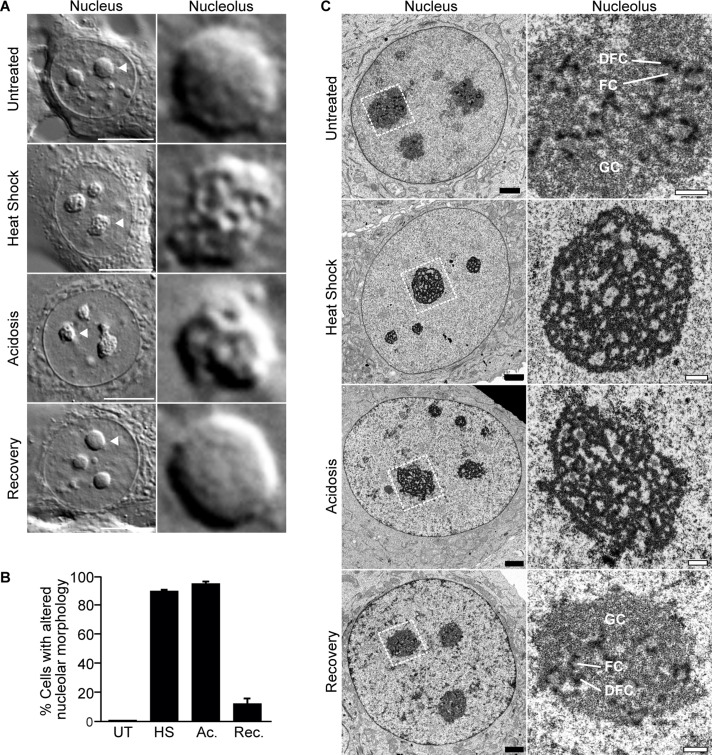
Heat shock and acidosis induce the capture and immobilization of an array of proteins in the nucleolus (Audas et al., 2012a). To investigate how the nucleolar architecture adapts to accommodate this large influx of proteins, we looked for gross morphological changes by differential interference contrast (DIC) microscopy. Of interest, we observed that ∼90% of the nucleoli of heat-shocked or acidotic MCF-7 cells possessed a more irregular shape, perforated with voids, compared with untreated cells (Figure 1, A and B, and Supplemental Figure S1A). Similar observations were made in U-87 MG, PC-3, and NIH/3T3 cells (Supplemental Figure S1, C–E). On return to normal conditions, the nucleoli of these treated cells quickly reverted to their native conformation (Figure 1, A and B, and Supplemental Figure S1A). Cell viability was not affected by these treatments (Supplemental Figure S1B).
FIGURE 1:
The nucleolus responds structurally to environmental stimuli. (A) DIC images of live untreated, heat-shocked, acidotic, or recovered (heat shock followed with 6-h recovery) MCF-7 cells. Scale bars, 10 μm. Right, indicated nucleoli are enlarged. (B) Quantification of untreated, heat-shocked, acidotic, or recovered MCF-7 cells showing altered nucleolar morphology. Columns, mean (n = 3); error bars, SEM. (C) Transmission electron micrograph of untreated, heat-shocked (30 min), acidotic (60 min), or recovered MCF-7 cells. Subnucleolar compartments (GC, DFC, FC) are indicated. Black scale bars, 2 μm; white scale bars, 0.5 μm.
In light of these results, we decided to use transmission electron microscopy to further monitor changes of the nucleolus at the ultrastructural level. Untreated cells possessed the characteristic tripartite nucleolar organization, with FCs and DFCs distributed throughout the GC (Figure 1C). These distinctive features were lost in response to heat shock and acidosis. Unlike the classic organization described earlier, treated cells displayed nucleoli with a noticeably transformed morphology, composed primarily of electron-dense anastomosed sheets in a reticular structure (Figure 1C). On return to normal conditions, the nucleolus regained its original tripartite organization (Figure 1C). These results suggest that the nucleolus undergoes a significant, yet reversible remodeling in response to changing environmental conditions.
The nucleolus is reorganized around the detention center
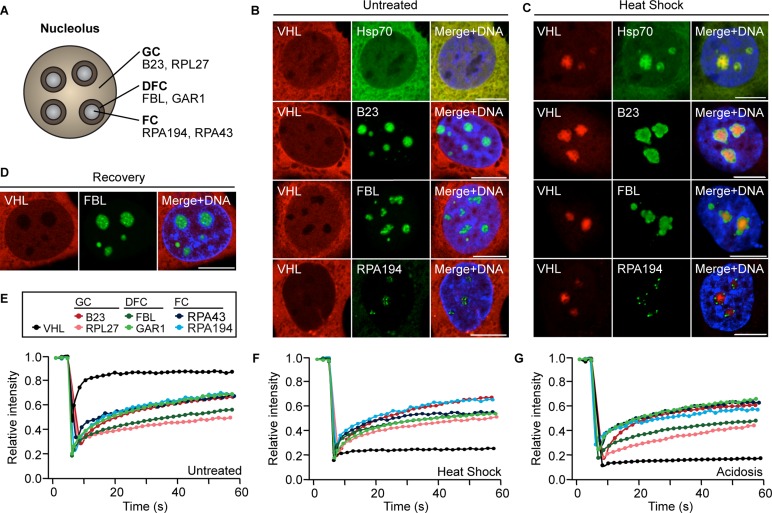
The transformation of the nucleolus described earlier correlates with an influx of proteins from other regions of the cell. To understand the consequences of this process for the functional organization of the nucleolus, we compared the localization of VHL, a well-characterized marker of nucleolar sequestration (Mekhail et al., 2004a, 2005, 2007; Audas et al., 2012a), to several nucleolar proteins. B23 and 60S ribosomal protein L27 (RPL27) were used as GC markers, fibrillarin (FBL) and GAR1 as markers of the DFC, and RNA polymerase I polypeptide A subunits 43 and 194 (RPA43 and RPA194) as markers of the FC (Figure 2A). Under standard growth conditions, VHL exhibited its normal nucleocytoplasmic distribution, whereas B23, RPL27, FBL, GAR1, RPA43, and RPA194 localized primarily to their respective nucleolar compartments (Figure 2B and Supplemental Figure S2A). In response to heat shock, VHL relocalized to the core of the nucleolus, forming a large and irregular structure (Figure 2C and Supplemental Figure S2B). A similar intranucleolar localization was observed for sequestered proteins Hsp70 (Figure 2C) and RNF8 (Supplemental Figure S2B). Surprisingly, the structure containing VHL did not colocalize with any of the known nucleolar compartments, as the resident proteins appeared to relocalize to the nucleolar periphery (Figure 2C and Supplemental Figure S2B). This segregation was also visualized in three dimensions (Supplemental Figure S2C) and with the endogenous proteins Hsp70 and FBL in both MCF-7 (Supplemental Figure S2D) and U-87MG cells (Supplemental Figure S2E). On stimulus termination, VHL was released from the nucleolus and FBL regained its normal distribution (Figure 2D).
FIGURE 2:
The nucleolus is reorganized around the detention center. (A) Diagram of the nucleolus, indicating marker proteins for the three subnucleolar compartments: the fibrillar centers (FC), the dense fibrillar centers (DFC), and the granular component (GC). (B, C) Localization of transfected Cherry-VHL with Hsp70-GFP, EGFP-B23, EGFP-FBL, or EGFP-RPA194 in live untreated (B) or heat-shocked (C) MCF-7 cells. (D) Localization of transfected Cherry-VHL and EGFP-FBL in live recovered cells (heat shock followed by 6-h recovery). (E–G) FRAP analysis of the dynamic profiles of VHL, B23, RPL27, FBL, GAR1, RPA43, and RPA194 in untreated (E), heat-shocked (F), and acidotic (G) cells. (B–D) Cells were stained with Hoechst 33342 to visualize DNA (blue). Scale bars, 10 μm.
Next we compared the dynamic properties of the nucleolar components, as most nuclear proteins are known to be highly mobile (Phair and Misteli, 2000; Chen and Huang, 2001; Misteli, 2001a, b). Fluorescence recovery after photobleaching (FRAP) experiments revealed that the markers of known nucleolar compartments remained mobile under heat shock and acidosis, whereas VHL underwent immobilization by the nucleolus (Figure 2, E–G). These data suggest that protein sequestration is associated with a reorganization of the nucleolus and that the structure in which proteins are immobilized is spatially and dynamically distinct. We refer to this region as the detention center.
The detention center is characterized by a hydrophobic signature
The immobility of proteins within the DC suggests that the structural organization of this compartment relies on a distinctive set of biochemical properties. Of interest, VHL and Hsp70 extracted from heat-shocked and acidotic cells migrated as monomers by SDS–PAGE, even in the absence of reducing agents (Supplemental Figure S3A), suggesting that covalent bonding of these molecules is not necessary for DC integrity. Therefore we hypothesized that high-affinity hydrophobic interactions could be involved in maintaining the DC architecture in a static state, a possibility consistent with the requirement for hydrophobic triplets in the nucleolar detention sequence of sequestered proteins (Mekhail et al., 2007). Using the membrane-permeable fluorescent dye 8-anilinonaphthalene-1-sulfonate (ANS), which highlights misfolded and hydrophobic protein deposits in cells and tissues (Hadley et al., 2011), we stained untreated and treated cells. Untreated cells incubated with ANS exhibited high levels of fluorescence in the cytoplasm and an absence of signal in the nucleus and nucleolus (Figure 3A). This pattern was consistent with previous reports, as the endoplasmic reticulum, Golgi, and lysosomes are rich in misfolded proteins and ANS-binding sites (Hadley et al., 2011). In contrast, large ANS-positive structures formed within the nucleoli of MCF-7 (Figure 3, A and B) and U-87 MG cells (Supplemental Figure S3B) during heat shock and acidosis treatments. On return to standard conditions, nucleolar ANS signal was lost (Figure 3, A and B, Supplemental Figure S3B). To determine the nature of these ANS-positive structures, we compared their localization with that of different nucleolar markers. We found that immobilized VHL and Hsp70 colocalized with ANS in the nucleoli of heat-shocked cells (Figure 3C). FC, DFC, and GC markers such as RPA194, RPA43, FBL, GAR1, Nopp140, and B23 were excluded from ANS-stained areas (Figure 3C and Supplemental Figure S3C). These data indicate that the DC is characterized by a hydrophobic signature that is distinct from other nucleolar compartments, further highlighting its unique biochemical properties. This signature allows ANS to be used as an effective marker of nucleolar reorganization and protein sequestration.
FIGURE 3:
The detention center is characterized by a hydrophobic signature. (A) Live untreated, heat-shocked, acidotic, and recovered (heat shock followed with 6-h recovery) MCF-7 cells stained with ANS. (B) Quantification of untreated, heat-shocked, acidotic, and recovered (1, 6 h) cells showing ANS-positive nucleoli. Columns, mean (n = 3); error bars, SEM. (C) Top, localization of mCherry-VHL, Hsp70-GFP, EGFP-FBL, EGFP-GAR1, EGFP-RPA194, and mCherry-RPA43 in live heat-shocked cells stained with ANS. Bottom, localization of mCherry-VHL and EGFP-FBL in live untreated cells. Scale bars, 10 μm.
The remodeled nucleolus is transcriptionally inactive
Remodeling of the nucleolus involves the immobilization of a wide variety of cellular factors, including E3 ubiquitin ligases, chaperones, and cell cycle regulators (Audas et al., 2012b). Surprisingly, we found that several components of the ribosomal biogenesis machinery were also targets of this pathway. RPA16 and RPA40, two essential subunits of RNA polymerase I and III, were captured in the DC in both heat shock and acidosis, as indicated by colocalization with ANS staining (Figure 4, A and B) and sequestered VHL (Supplemental Figure S4, A and B). On return to normal condition, they regained their punctuate distribution in the FCs (Figure 4, A and B). Moreover, the dynamic profile of these proteins changed from a state of high mobility in the FC to a state of static detention in the DC under both heat shock and acidosis (Figure 4, C and D). Like VHL, these proteins regained mobility upon signal termination (Figure 4, C and D). Similarly, several GC proteins were immobilized in the DC, including PES1, NOP52, RRP1B, NOM1, NOL1, and SENP3 (Supplemental Figure S4, C–E and G–I, and Table 1). The ability of certain nucleolar proteins, such as FBL, B23, and NOPP140, to retain mobility and evade capture by the DC further highlights the specificity of this intranucleolar reorganization process (Supplemental Figure S4, F and J, and Table 1).
FIGURE 4:
Remodeled nucleoli are transcriptionally inactive. (A, B) Localization of EGFP-RPA16 (A) and EGFP-RPA40 (B) in live untreated, heat-shocked, acidotic, and recovered (heat shock followed by 6-h recovery) MCF-7 cells stained with ANS. (C, D) Nucleolar FRAP analysis of the dynamic profile of RPA16 (C) and RPA40 (D) in untreated, heat-shocked, acidotic, and recovered cells. (E) Relative levels of 45S pre-rRNA in untreated, heat-shocked, acidotic, and recovered cells measured by real-time RT-PCR. Actinomycin D (2 h, 50 nM) was used as a control for inhibition of RNA polymerase I–dependent transcription. (F) Left, quantification of untreated, heat-shocked, and recovered cells showing nucleolar incorporation of 5-FU. Right, de novo rRNA synthesis visualized by incorporation of 5-FU in heat-shocked and recovered cells. Scale bar, 10 μm (A, B, F). Columns in E and F, mean (n = 3); error bars, SEM.
TABLE 1:
Spatio-dynamic profile of nucleolar proteins.
| Intranucleolar localization | ||||
|---|---|---|---|---|
| Symbol | Full name | Untreated | Heat shock | Acidosis |
| RPA194 | Polymerase (RNA) I polypeptide A, 194 kDa | FC (mobile) | FC (mobile) | FC (mobile) |
| RPA40 | Polymerase (RNA) I polypeptide C, 40 kDa | FC (mobile) | DC (immobile) | DC (immobile) |
| RPA16 | Polymerase (RNA) I polypeptide D, 16 kDa | FC (mobile) | DC (immobile) | DC (immobile) |
| RPA43 | Polymerase (RNA) I polypeptide subunit 43 kDa | FC (mobile) | FC (mobile) | FC (mobile) |
| FBL | Fibrillarin | DFC (mobile) | DFC (mobile) | DFC (mobile) |
| GAR1 | GAR1 ribonucleoprotein | DFC (mobile) | DFC (mobile) | DFC (mobile) |
| Nopp140 | Nucleolar and coiled body phorosphoprotein 1 | DFC (mobile) | DFC (mobile) | DFC (mobile) |
| B23 | Nucleophosmin B23 | GC (mobile) | GC (mobile) | GC (mobile) |
| NOL1 | NOP2 nucleolar protein homologue | GC (mobile) | DC (immobile) | DC (immobile) |
| NOM1 | Nucleolar protein with MIF4G domain 1 | GC (mobile) | DC (immobile) | DC (immobile) |
| NOP52 | rRNA processing protein 1 homologue A | GC (mobile) | DC (immobile) | DC (immobile) |
| PES1 | Pescadillo | GC (mobile) | DC (immobile) | DC (immobile) |
| RPL27 | 60S ribosomal protein L27 | GC (mobile) | GC (mobile) | GC (mobile) |
| RRP1B | rRNA processing protein 1 homologue B | GC (mobile) | DC (immobile) | DC (immobile) |
| SENP3 | SUMO1/sentrin/SMT3 specific peptidase 3 | GC (mobile) | DC (immobile) | DC (immobile) |
Ribosomal biogenesis relies on a complex and compartmentalized assortment of dynamic factors in which structure and function are intimately linked (Hernandez-Verdun, 2006). Given the drastic remodeling of the nucleolar architecture and the immobilization of several key nucleolar proteins, we asked whether rRNA synthesis could be sustained under conditions of nucleolar sequestration. We used real-time reverse transcription (RT) PCR (qPCR) analysis to measure the levels of 45S pre-rRNA transcripts in untreated and treated cells. Results revealed ∼85% decrease in both heat-shocked and acidotic cells relative to untreated cells (Figure 4E). The 45S pre-rRNA levels returned to their original values upon extracellular signal termination (Figure 4E). In addition to qPCR, we labeled de novo RNA transcription sites with 5-fluorouridine (5-FU). We observed an absence of nucleolar incorporation in ∼95% of heat-shocked cells (Figure 4F), with a return to normal values after signal termination (Figure 4F). Similar results were observed in U-87 MG cells (Supplemental Figure S4K). Incorporation of 5-FU is impractical under acidotic conditions due to the pH of the media. These results suggest that remodeling of the nucleolus is associated with a severe, yet reversible inhibition of ribosomal biogenesis.
Environment-induced remodeling of the nucleolus is not a byproduct of transcriptional inhibition
Transcriptional inhibition induces several alterations to the nucleolar architecture, including the formation of nucleolar caps (Reynolds et al., 1964; Shav-Tal et al., 2005) and the flux of a large number of different factors (Andersen et al., 2005). Given the correlation between structure and transcriptional activity under heat shock and acidosis, we wanted to know whether transcriptional inhibition was sufficient to induce some of the traits described thus far. We found that ActD treatment (5 μg/ml) failed to induce DC formation, as evidenced by the lack ANS-positive nucleoli and the diffuse nucleocytoplasmic distribution of VHL (Figure 5A). Furthermore, using polypyrimidine tract–binding, protein-associated splicing factor (PSF), a common marker of nucleolar caps, we observed that only ActD-mediated transcriptional arrest triggered the formation of these cap structures (Figure 5A). This suggests that this form of reorganization is distinct from that observed in response to environmental stresses, such as heat shock. In addition, whereas RPA16 and RPA40 were immobilized in the DC under heat shock conditions (Figure 5A), both proteins migrated to the nucleolar periphery and retained their mobility during transcriptional arrest (Figure 5, A and B). Lower levels of ActD that specifically inhibit rRNA synthesis (0.04 μg/ml) also failed to induce DC formation (Supplemental Figure S5). These results demonstrate that the remodeling observed in response to environmental cues such as heat shock is not simply a byproduct of transcriptional inhibition, as ActD appears to induce a different reorganizational program. Our data further suggest that the impairment in rRNA synthesis (Figure 4, E and F) is likely a consequence, not a cause, of the nucleolar detention pathway.
FIGURE 5:
The remodeled nucleolus is not a byproduct of transcriptional inhibition. (A) Localization of mCherry-VHL, ANS signal, EGFP-PSF, EGFP-RPA16, and EGFP-RPA194 in live untreated, heat-shocked, and ActD-treated (5 μg/ml) MCF-7 cells. VHL, PSF, RPA16, and RPA194: cells were stained with Hoechst 33342 to visualize DNA (blue). Bottom, DIC images of untreated, heat-shocked, and ActD-treated cells. Scale bars, 10 μm. (B) Nucleolar FRAP analysis of the dynamic profile of RPA16 and RPA40 in ActD-treated cells.
IGS lncRNA is required to remodel the nucleolus
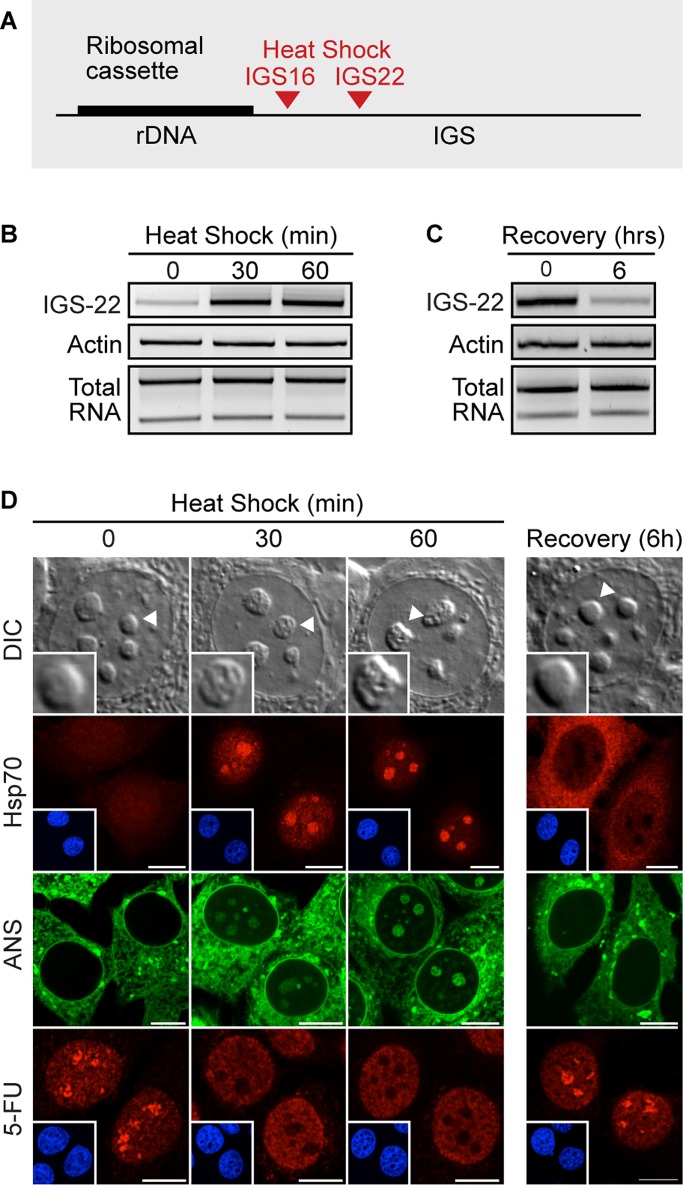
Sequestered proteins are immobilized on the rDNA cassette by several stimulus-specific IGS RNAs, including IGS16RNA and IGS22RNA during heat shock (Audas et al., 2012a; Figure 6A). These transcripts rapidly accumulate in response to environmental cues (Audas et al., 2012a; Figure 6B) and return to basal levels upon signal termination (Figure 6C). Induction of IGS lncRNA readily correlates with remodeling of the nucleolus, rapid sequestration of endogenous Hsp70, formation of ANS-positive DCs, and interruption of nucleolar transcription (Figure 6D). On signal termination, IGS lncRNA expression returns to basal levels (Figure 6C) and the nucleoli regain their original morphology, as indicated by Hsp70 release, loss in nuclear ANS signal, and resumption in nucleolar 5-FU incorporation (Figure 6D). As expected, RNA FISH experiments confirmed the presence of IGS lncRNA transcripts within the DC, as evidenced by colocalization with sequestered VHL (Supplemental Figure S5; Audas et al., 2012a). These IGS lncRNA–containing regions also notably excluded FBL (Supplemental Figure S6). Together these results demonstrate that the DC correlates spatially and temporally with the presence of IGS lncRNA.
FIGURE 6:
Formation of the detention center correlates with the induction of IGS lncRNA. (A) Schematic diagram of the rDNA cassette with the sites of protein immobilization during heat shock treatment (IGS16, IGS22). (B) Semiquantitative RT-PCR analysis of the IGS22RNA levels in MCF-7 cells heat shock treated for the indicated times. (C) Semiquantitative RT-PCR analysis of the IGS22RNA levels in heat-shocked and recovered (6 h) cells. (D) DIC images of live untreated, heat-shocked (30, 60 min), and recovered (6 h) cells. Localization of endogenous Hsp70, ANS signal, and 5-FU incorporation in untreated, heat-shocked (30, 60 min), and recovered (6 h) cells. Scale bars, 10 μm.
Given the presence of IGS lncRNA in the DC, we asked whether these transcripts were required to mediate the nucleolar reorganization process described here. Despite the intricacy of the IGS as a transcriptional unit, we found that cells stably expressing short hairpin RNA (shRNA) targeting IGS22RNA (Figure 7A) effectively delayed the sequestration of proteins during heat shock treatment (Audas et al., 2012a), with near-complete inhibition at the 30-min time point. Stable knockdown of IGS22RNA did not affect the proper localization of the nucleolar factors B23, FBL, RPA16, and RPA194 (Figure 7B), the tripartite ultrastructure of the nucleolus (Supplemental Figure S7), or the expression level of rRNA under standard conditions (Figure 7C), but it impaired the sequestration of Hsp70 during heat shock treatment (Audas et al., 2012a; Figure 7D). We asked whether the formation of the DC would also be compromised by inhibiting IGS22RNA expression. ShIGS22 cells failed to produce any of the previously characterized hallmarks of DC formation. During heat shock treatment these cells did not acquire ANS-positive subnuclear structures (Figure 7D), and most of the nucleoli retained their original appearance by DIC imaging (Figure 7, E and F). These observations were supported by transmission electron micrographs that show that the majority of shIGS22 cells retained their native nucleolar morphology, with clearly visible FCs, DFCs, and GC (Figure 7G).
FIGURE 7:
IGS lncRNA is required to remodel the nucleolus. (A) Parental and MCF-7 cells expressing short hairpin RNA targeted to a scrambled sequence (shControl) or IGS22RNA (shIGS-22) were exposed to heat shock to induce transcription of the IGS22 locus before semiquantitative RT-PCR. (B) Localization of EGFP-B23, EGFP-FBL, EGFP-RPA16, and EGFP-RPA194 in live untreated shIGS-22 cells. Inset, Hoechst 33342–stained DNA. (C) Relative levels of 45S pre-rRNA in untreated parental and shIGS-22 cells measured by real-time RT-PCR. (D) shControl and shIGS-22 cells were heat shocked for 30 min and either immunostained for Hsp70 or stained with ANS (live). Inset, Hoechst 33342–stained DNA. (E) Quantification of untreated and heat-shocked (30 min) shControl and shIGS-22 cells showing altered nucleolar morphology by DIC. (F) DIC images of live heat-shocked (30 min) shControl and shIGS-22 cells. Right, indicated nucleoli are enlarged. (G) Transmission electron micrograph of an shIGS22 cell after a 30-min heat shock treatment. Subnucleolar compartments (GC, DFC, FC) are indicated. Black scale bar, 2 μm; white scale bar, 0.5 μm. (H) Relative levels of 45S pre-rRNA in untreated and heat-shocked (30 min) shControl and shIGS-22 cells measured by real-time RT-PCR. Actinomycin D (2 h, 50 nM) was used as a control for inhibition of RNA polymerase I–dependent transcription. (I) Quantification of untreated and heat-shocked (30 min) shControl and shIGS-22 cells showing nucleolar incorporation of 5-FU. (J) De novo rRNA synthesis visualized by incorporation of 5-FU in untreated and heat-shocked (30 min) shControl and shIGS-22 cells. (B, D, J) Cells were stained with Hoechst 33342 to visualize DNA (blue). Scale bars, 10 μm. (C, E, H, I) Columns, mean (n = 3); error bars, SEM.
Finally, we tested whether an absence of reorganization would allow the nucleolus to retain its transcriptional activity during extracellular stimulation. Using qPCR analysis of the 45S pre-rRNA transcript, we found a 65% decrease in the levels of pre-rRNA in control cells within 30 min of heat shock treatment but no changes in shIGS22 cells (Figure 7H). Consistently, 5-FU incorporation revealed that shIGS22 cells were unable to fully repress nucleolar transcription during heat shock treatment, as 88% of shIGS22-expressing cells retained nucleolar incorporation upon treatment versus 20% of the control cells (Figure 7, I and J). These data demonstrate the requirement for IGS RNA in both structural and functional remodeling of the nucleolus.
DISCUSSION
We propose that the nucleolus readily alternates between two distinct and environment-dependent morphologies: a tripartite, transcriptionally active conformation under normal conditions, and a remodeled, transcriptionally inert conformation under specific environmental settings (Figure 8). The latter is characterized by the presence of the detention center, a region that is distinct from the well-documented and characterized nucleolar compartments (FC, DFC, and GC). These two architectures differ functionally as well as structurally. The former provides the cell with the output of rRNA required to sustain protein synthesis under growth conditions. The latter operates as a molecular prison, detaining cellular proteins away from their associated pathways. Through the inactivation of molecular networks and the interruption of rRNA synthesis, the remodeled nucleolus likely contributes to cell viability under conditions of stress (Olson, 2004; Mayer and Grummt, 2005; Boulon et al., 2010; Audas et al., 2012b). Therefore our results provide a physiological rationale for the inherent plasticity of the nucleolar architecture.
FIGURE 8:
Model: functional remodeling of the nucleolus. The nucleolus readily alternates between two distinct and environment-specific morphologies: a tripartite, transcriptionally active conformation and a remodeled, transcriptionally inert architecture. This transition is regulated by IGS lncRNAs.
An interesting aspect of this transformation is the reliance on a central transcriptional event that produces lncRNA from discrete regions of the IGS. Under standard conditions, IGS transcripts are kept at basal levels, allowing the nucleolus to assume its classic organization and function. In contrast, the cell harnesses the inducible nature of IGS RNA upon signal activation to orchestrate a rapid and drastic remodeling of the nucleolus. This response relies on the simultaneous induction of multiple IGS transcripts, and although silencing IGS22RNA largely inhibits nucleolar remodeling, we expect IGS16RNA to account for part of the phenotype in heat shock. Previous studies showed that IGS RNAs directly interact with NoDS-containing proteins (Audas et al., 2012a), suggesting that they may induce a nucleation event, further stabilized by high-affinity hydrophobic interactions. We suggest that IGS transcripts operate as a macromolecular switch that is of central importance to nucleolar function and organization, although further investigation will be required to fully understand the mechanisms at work in this process. Of interest, parallel studies have shown that another IGS RNA, derived from the promoter region of rDNA, is involved in the recruitment of the chromatin-remodeling complex NoRC and in the control of rRNA synthesis (Mayer et al., 2006, 2008). Homologies may exist between this pathway and the process described here.
The DC described here is emerging as a discrete nuclear component, with a distinct protein composition, spatial location, dynamic profile, hydrophobic signature, and biological function. The intranucleolar relocalization of many nucleolar proteins into the DC represents an unexpected mechanism of ribosomal biogenesis regulation. Synthesis of rRNA in the nucleolus is predominantly controlled through transcription factors and chromatin-remodeling complexes. Additional regulatory mechanisms have been reported at the initiation, elongation, and termination steps (Leary and Huang, 2001; Santoro and Grummt, 2001; Lempiainen and Shore, 2009). The pathway described here differs not only in its mode of action, but also in its generality. The capture of proteins in the DC is a systemic phenomenon that affects a breadth of unrelated factors, including DNA-processing enzymes, ubiquitin ligases, cell cycle regulators, and chaperones (Audas et al., 2012b). The requirement for RPA16 and RPA40 in RNA pol III suggests that IGS RNAs may also control pol III–mediated transcription. The tools introduced here, such as the ANS marker, could be used to rapidly assess whether a protein is specifically inactivated within the DC or otherwise located within the nucleolus. More generally, nucleolar morphology may be indicative of a systemic posttranslational remodeling of molecular networks through the nucleolar detention pathway (Audas et al., 2012b).
Ongoing ribosomal biogenesis is essential to sustaining the growth and proliferation of tumors (Lempiainen and Shore, 2009). In fact, processes that promote the up-regulation of rRNA synthesis are described as oncogenic, whereas those that repress it are considered tumor suppressing (Montanaro et al., 2012). For this reason, enlarged nucleolar morphology is indicative of malignancy (Derenzini et al., 1998, 2000; Maggi and Weber, 2005), and the RNA pol I machinery is emerging as a therapeutic target for the treatment of cancer (Drygin et al., 2010). Of interest, the nucleolar stresses described in this study are relevant to a variety of pathological conditions, including cancer. Extracellular acidosis, the outcome of anaerobic metabolism, is encountered in the core of tumors (Tannock and Rotin, 1989; Engin et al., 1995), during development, and in ischemic tissues, where it has protective effects (Giffard et al., 1990; Currin et al., 1991). Similarly, hyperthermia has been reported in some metabolically hyperactive tumors (Jayasundar and Singh, 2002) and, perhaps not coincidently, is also correlated with neuroprotection of ischemic brain tissues (Laptook et al., 1994; Thoresen et al., 1995). In light of these reports, it is tempting to speculate that the tumor microenvironment may inhibit cellular proliferation by inducing the nucleolar detention pathway. Future research may link IGS lncRNA–dependent remodeling of the nucleolus to various physiological and pathological conditions.
MATERIALS AND METHODS
Cell culture, transfection, treatment, and imaging
MCF-7, U-87 MG, PC-3, and NIH/3T3 cells were grown in monolayer culture in modified DMEM (high glucose) supplemented with 5% (vol/vol) fetal bovine serum and 1% (vol/vol) penicillin–streptomycin. All cultures were maintained at 37°C and 5% CO2 in a humidified atmosphere, and passaged every 2–3 d. Cells were plated 24 h before transfection and allowed to grow to 50–60% confluency before transfection with Effectene (Qiagen, Valencia, CA) as per manufacturer's instructions. shControl and shIGS22 cells were previous described (Audas et al., 2012a). Heat shock was performed by transferring cells to 42°C for 3 h, unless specified otherwise. Hypoxic acidosis was obtained by placing cells in acidic (pH 6.0) DMEM at 1% O2 for 4 h, unless specified otherwise. For transcriptional inhibition, 5 μg/ml actinomycin D was added to cells for 2 h, unless stated otherwise.
Plasmids
Plasmids expressing VHL-GFP, RNF8-GFP, and EGFP-B23 were previously described (Audas et al., 2012a). mCherry-VHL was cloned from plasmid cDNA into an mCherry-c1 empty vector. EGFP-NOM1 (Kathleen Conklin, University of Minnesota, Minneapolis, MN), EGFP-FBL, EGFP-NOPP140, EGFP-PSF, EGFP-RPA40, mCherry-RPA43, EGFP-RPA194, EGFP-NOL1, EYFP-PES1, EYFP-RPL27, and EGFP-RRP1B (Angus Lamond, University of Dundee, Dundee, United Kingdom) were kindly provided. The following plasmids were obtained from Addgene (Cambridge, MA): EGFP-RPA16 (Tom Misteli, National Institutes of Health, Bethesda, MD; Dundr et al., 2002; Addgene plasmid 17657) and EGFP-SENP3 (Mary Dasso, National Institutes of Health, Bethesda, MD; Yun et al., 2008; Addgene plasmid 34554).
Microscopy and image processing
Images were collected by confocal microscopy (Zeiss 200 LSM510 META; Carl Zeiss, Jena, Germany) using a 63× Plan-Apochromat/1.4 numerical aperture (NA) objective. The system was controlled with Zeiss Zen software. DIC images were collected on a Zeiss AxioObserver D1 microscope using a 63× Plan-Apochromat/1.4 NA objective. All postacquisition image processing was performed using Zeiss Zen software. No gamma or other nonlinear adjustments were made to any image.
Fluorescence recovery after photobleaching
Live cells expressing constructs of interest were grown on 35-mm glass-bottom culture dishes and visualized by confocal microscopy (Zeiss 200 LSM510 META) in a 37°C and 5% CO2 environmental chamber (Zeiss Incubator XL S1) using a 63× Plan-Apochromat/1.4 NA objective. The system was controlled by Zeiss Zen software for bleaching and image acquisition. Bleached areas (nucleolus) were subjected to 40 iterations at 100% argon laser strength at 488 nm, and imaging used 5% laser strength. Three prebleach measurements were taken before the photobleaching and 60- to 120-s recovery of the region of interest, as indicated. For nucleolar proteins, a full nucleolus was photobleached. For nucleocytoplasmic proteins, a region of the nucleoplasm was bleached instead. Region intensity measurements were performed by Zen software and normalized as previously described (Misteli, 2001b). All FRAP experiment data were the average of at least 10 cells.
ANS fluorescence imaging
ANS was purchased from Sigma-Aldrich (St. Louis, MO; A1028) and dissolved in PBS, pH 7.4, to a 10 mM stock solution. Culture medium was supplemented with ANS at a concentration of 250 μM for at least 1 h. Media were changed before imaging. ANS was excited at 405 nm and detected using a band pass filter.
Immunofluorescence
Cells were seeded onto 20-mm glass coverslips and fixed with CSK (4% paraformaldehyde) for 15 min. After permeabilization in 0.5% Triton X-100 for 10 min, cells were incubated with primary antibody (1:200), washed three times in PBS, and incubated for 1 h in Alexa Fluor (Invitrogen, Carlsbad, CA) secondary antibody (1:400).
Fluorescence in situ hybridization
Cells were fixed in methanol for 5 min. Samples were blocked in digoxigenin (DIG) hybridization (Roche, Indianapolis, IN) solution and hybridized with 5′ and 3′ DIG-labeled oligonucleotide IGS22RNA antisense (5′-DIG-TACTGCATTGTCGCTGAACGTTCTCCCAAAAGGCCAGAAACCCCCTGACTCAGGTCAAGG-DIG), IGS28RNA antisense (5′-DIG-CCGGCCTTAACAGTTTATGTTGAAGTCGAGGAGACTTATCGGGGAAATAGGAGAAGTACG-DIG), or IGS sense (5′-DIG-CGTACTTCTCCTATTTCCCCGATAAGTCTCCTCGACTTCAACATAAACTGTTAAGGCCGG-DIG) probes in a 50% formamide/DIG hybridization (Roche) solution at 37°C. After hybridization, cells were washed in 0.2% SSC, and probes were detected with an anti-DIG (Roche) antibody.
5-Fluorouridine labeling and detection of nascent rRNA
Live cells were incubated in 1 mM 5-FU (F5130; Sigma-Aldrich) for 15 min before fixation in CSK (4% paraformaldehyde). Cells were then permeabilized in 0.5% Triton X-100 and immunostained using an anti-bromodeoxyuridine mouse antibody (B2531; Sigma-Aldrich) at 1:500, followed by incubation in Alexa Fluor (Invitrogen, Carlsbad, CA) secondary antibody (1:500).
Western blotting
Western blot was performed following standard protocol. Monoclonal antibodies were used to detect FLAG (Sigma-Aldrich), Hsp70 (Santa Cruz Biotechnology, Santa Cruz, CA), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; GeneTex, Irvine, CA). Bands were detected by enhanced chemiluminescence (Pierce, Rockford, IL).
Transmission electron microscopy
Cells were treated and fixed with 2% glutaraldehyde in PBS, pH 7.4. Samples were subsequently washed and postfixed with 1% osmium tetroxide before dehydration in a series of ascending alcohols. Specimens were then embedded in Spurr's epoxy resin onto BEEM (West Chester, PA) capsule molds. Specimen blocks were ultrathin sectioned on a Leica EM UC6 ultramicrotome (Leica, Wetzlar, Germany), and the resulting sections were stained with uranyl acetate and lead citrate. Samples were imaged with a JEOL 1230 transmission electron microscope (JEOL, Peabody, MA) equipped with AMT software (Advanced Microscopy Techniques, Woburn, MA).
RNA isolation, reverse transcription PCR, and real-time PCR
Total RNA was collected using TriPure reagent (Roche) according to manufacturer's specifications. RT-PCR was performed using the two-step High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA), followed by standard PCR conditions. Real-time PCRs were performed using iQ SYBR Green SuperMix (Bio-Rad Laboratories, Carlsbad, CA) and read with Stratagene MX3005P (Stratagene, Santa Clara, CA). Transcripts levels were normalized to actin mRNA. Relative fold change in expression is calculated using the ΔΔCt method. The 45S pre-rRNA and actin mRNA primers were used as previously described (Feng et al., 2010).
Statistical analysis
The p values associated with all comparisons were based on paired two-tailed Student's t tests. Results are mean (n = 3) ± SEM.
Supplementary Material
Acknowledgments
We thank Alan Prescott (University of Dundee, Dundee, United Kingdom) for support with electron microscopy and Danielle Healy for technical support. This work was supported by grants from the Canadian Institute for Health Research to S.L. J.U. is a Research Fellow of the Terry Fox Foundation (Canadian Cancer Society Award 700014).
Abbreviations used:
- Ac
acidosis
- ANS
8-anilo-1-naphthalanesulfonic acid
- DC
detention center
- DFC
dense fibrillar component
- DIC
differencial intereference contrast
- HS
heat shock
- IGS
intergenic spacer
- lncRNA
long noncoding RNA
- Rec
recovered
- UT
untreated
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-04-0223) on July 31, 2013.
The authors declare that they have no conflict of interest and no competing financial interests.
REFERENCES
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell. 2012a;45:147–157. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Audas TE, Jacob MD, Lee S. The nucleolar detention pathway: a cellular strategy for regulating molecular networks. Cell Cycle. 2012b;11:2059–2062. doi: 10.4161/cc.20140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol. 2001;153:169–176. doi: 10.1083/jcb.153.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currin RT, Gores GJ, Thurman RG, Lemasters JJ. Protection by acidotic pH against anoxic cell killing in perfused rat liver: evidence for a pH paradox. FASEB J. 1991;5:207–210. doi: 10.1096/fasebj.5.2.2004664. [DOI] [PubMed] [Google Scholar]
- Derenzini M, Trere D, Pession A, Govoni M, Sirri V, Chieco P. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J Pathol. 2000;191:181–186. doi: 10.1002/(SICI)1096-9896(200006)191:2<181::AID-PATH607>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs RL. Nucleolar function and size in cancer cells. Am J Pathol. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- Engin K, Leeper DB, Cater JR, Thistlethwaite AJ, Tupchong L, McFarlane JD. Extracellular pH distribution in human tumours. Int J Hyperthermia. 1995;11:211–216. doi: 10.3109/02656739509022457. [DOI] [PubMed] [Google Scholar]
- Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol. 2010;17:445–450. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- Giffard RG, Monyer H, Choi DW. Selective vulnerability of cultured cortical glia to injury by extracellular acidosis. Brain Res. 1990;530:138–141. doi: 10.1016/0006-8993(90)90670-7. [DOI] [PubMed] [Google Scholar]
- Guarente L. Link between aging and the nucleolus. Genes Dev. 1997;11:2449–2455. doi: 10.1101/gad.11.19.2449. [DOI] [PubMed] [Google Scholar]
- Hadley KC, Borrelli MJ, Lepock JR, McLaurin J, Croul SE, Guha A, Chakrabartty A. Multiphoton ANS fluorescence microscopy as an in vivo sensor for protein misfolding stress. Cell Stress Chaperones. 2011;16:549–561. doi: 10.1007/s12192-011-0266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D. Nucleolus: from structure to dynamics. Histochem Cell Biol. 2006;125:127–137. doi: 10.1007/s00418-005-0046-4. [DOI] [PubMed] [Google Scholar]
- Hiscox JA. RNA viruses: hijacking the dynamic nucleolus. Nat Rev Microbiol. 2007;5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S, Prescott A, James J, Riesenberg S, Boulon S, Lam YW, Lamond AI. An intranucleolar body associated with rDNA. Chromosoma. 2011;120:481–499. doi: 10.1007/s00412-011-0327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob MD, Audas TE, Mullineux ST, Lee S. Where no RNA polymerase has gone before: novel functional transcripts derived from the ribosomal intergenic spacer. Nucleus. 2012;3:314–319. doi: 10.4161/nucl.20585. [DOI] [PubMed] [Google Scholar]
- Jayasundar R, Singh VP. In vivo temperature measurements in brain tumors using proton MR spectroscopy. Neurol India. 2002;50:436–439. [PubMed] [Google Scholar]
- Journey LJ, Goldstein MN. Electron microscope studies on HeLa cell lines sensitive and resistant to actinomycin D. Cancer Res. 1961;21:929–932. [PubMed] [Google Scholar]
- Laptook AR, Corbett RJ, Sterett R, Burns DK, Tollefsbol G, Garcia D. Modest hypothermia provides partial neuroprotection for ischemic neonatal brain. Pediatric Res. 1994;35:436–442. [PubMed] [Google Scholar]
- Leary DJ, Huang S. Regulation of ribosome biogenesis within the nucleolus. FEBS Lett. 2001;509:145–150. doi: 10.1016/s0014-5793(01)03143-x. [DOI] [PubMed] [Google Scholar]
- Lempiainen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Louvet E, Junera HR, Berthuy I, Hernandez-Verdun D. Compartmentation of the nucleolar processing proteins in the granular component is a CK2-driven process. Mol Biol Cell. 2006;17:2537–2546. doi: 10.1091/mbc.E05-10-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi LB, Jr, Weber JD. Nucleolar adaptation in human cancer. Cancer Invest. 2005;23:599–608. doi: 10.1080/07357900500283085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Frey MR, Margelot K, Wolin SL. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Mayer C, Grummt I. Cellular stress and nucleolar function. Cell Cycle. 2005;4:1036–1038. doi: 10.4161/cc.4.8.1925. [DOI] [PubMed] [Google Scholar]
- Mayer C, Neubert M, Grummt I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 2008;9:774–780. doi: 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Mekhail K, Gunaratnam L, Bonicalzi ME, Lee S. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat Cell Biol. 2004a;6:642–647. doi: 10.1038/ncb1144. [DOI] [PubMed] [Google Scholar]
- Mekhail K, Khacho M, Carrigan A, Hache RR, Gunaratnam L, Lee S. Regulation of ubiquitin ligase dynamics by the nucleolus. J Cell Biol. 2005;170:733–744. doi: 10.1083/jcb.200506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Khacho M, Gunaratnam L, Lee S. Oxygen sensing by H+: implications for HIF and hypoxic cell memory. Cell Cycle. 2004b;3:1027–1029. [PubMed] [Google Scholar]
- Mekhail K, Rivero-Lopez L, Al-Masri A, Brandon C, Khacho M, Lee S. Identification of a common subnuclear localization signal. Mol Biol Cell. 2007;18:3966–3977. doi: 10.1091/mbc.E07-03-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melese T, Xue Z. The nucleolus: an organelle formed by the act of building a ribosome. Curr Opin Cell Biol. 1995;7:319–324. doi: 10.1016/0955-0674(95)80085-9. [DOI] [PubMed] [Google Scholar]
- Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001a;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Protein dynamics: implications for nuclear architecture and gene expression. Science. 2001b;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- Montanaro L, Trere D, Derenzini M. Changes in ribosome biogenesis may induce cancer by down-regulating the cell tumor suppressor potential. Biochim Biophys Acta. 2012;1825:101–110. doi: 10.1016/j.bbcan.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Moore HM, Bai B, Boisvert FM, Latonen L, Rantanen V, Simpson JC, Pepperkok R, Lamond AI, Laiho M. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol Cell Proteomics. 2011;10:M111.009241. doi: 10.1074/mcp.M111.009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MO. Sensing cellular stress: another new function for the nucleolus. Sci STKE. 2004;2004:pe10. doi: 10.1126/stke.2242004pe10. [DOI] [PubMed] [Google Scholar]
- Olson MO, Dundr M. The moving parts of the nucleolus. Histochem Cell Biol. 2005;123:203–216. doi: 10.1007/s00418-005-0754-9. [DOI] [PubMed] [Google Scholar]
- Panse SL, Masson C, Heliot L, Chassery JM, Junera HR, Hernandez-Verdun D. 3-D organization of ribosomal transcription units after DRB inhibition of RNA polymerase II transcription. J Cell Sci. 1999;112:2145–2154. doi: 10.1242/jcs.112.13.2145. [DOI] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Pollock C, Daily K, Nguyen VT, Wang C, Lewandowska MA, Bensaude O, Huang S. Characterization of MRP RNA-protein interactions within the perinucleolar compartment. Mol Biol Cell. 2011;22:858–867. doi: 10.1091/mbc.E10-09-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock C, Huang S. The perinucleolar compartment. J Cell Biochem. 2009;107:189–193. doi: 10.1002/jcb.22107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds RC, Montgomery PO, Hughes B. Nucleolar “caps” produced by actinomycin D. Cancer Res. 1964;24:1269–1277. [PubMed] [Google Scholar]
- Santoro R, Grummt I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol Cell. 2001;8:719–725. doi: 10.1016/s1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- Scheer U, Hugle B, Hazan R, Rose KM. Drug-induced dispersal of transcribed rRNA genes and transcriptional products: immunolocalization and silver staining of different nucleolar components in rat cells treated with 5,6-dichloro-beta-D-ribofuranosylbenzimidazole. J Cell Biol. 1984;99:672–679. doi: 10.1083/jcb.99.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. HIF-1 and human disease: one highly involved factor. Genes Dev. 2000;14:1983–1991. [PubMed] [Google Scholar]
- Semenza GL. HIF-1 and mechanisms of hypoxia sensing. Curr Opin Cell Biol. 2001;13:167–171. doi: 10.1016/s0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock IF, Rotin D. Acid pH in tumors and its potential for therapeutic exploitation. Cancer Res. 1989;49:4373–4384. [PubMed] [Google Scholar]
- Thoresen M, et al. Mild hypothermia after severe transient hypoxia-ischemia ameliorates delayed cerebral energy failure in the newborn piglet. Pediatric Res. 1995;37:667–670. doi: 10.1203/00006450-199505000-00019. [DOI] [PubMed] [Google Scholar]
- Uniacke J, Holterman CE, Lachance G, Franovic A, Jacob MD, Fabian MR, Payette J, Holcik M, Pause A, Lee S. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012;486:126–129. doi: 10.1038/nature11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikstrom KL, Lim SS, Goldman RD, Borisy GG. Steady state dynamics of intermediate filament networks. J Cell Biol. 1992;118:121–129. doi: 10.1083/jcb.118.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Amon A. The nucleolus: the magician's hat for cell cycle tricks. Curr Opin Cell Biol. 2000;12:372–377. doi: 10.1016/s0955-0674(00)00102-2. [DOI] [PubMed] [Google Scholar]
- Yun C, Wang Y, Mukhopadhyay D, Backlund P, Kolli N, Yergey A, Wilkinson KD, Dasso M. Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J Cell Biol. 2008;183:589–595. doi: 10.1083/jcb.200807185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.