The modulation of inositol pentakisphosphate (IP5) and hexakisphosphate (IP6) intracellular levels controls the differentiation and survival of PC12 cells and primary neurons. These mechanisms are controlled by the levels of the protein kinase IP5-2K responsible for the conversion of IP5 into IP6.
Abstract
The binding of neurotrophins to tropomyosin receptor kinase receptors initiates several signaling pathways, including the activation of phospholipase C-γ, which promotes the release of diacylglycerol and inositol 1,4,5-trisphosphate (IP3). In addition to recycling back to inositol, IP3 serves as a precursor for the synthesis of higher phosphorylated inositols, such as inositol 1,3,4,5,6-pentakisphosphate (IP5) and inositol hexakisphosphate (IP6). Previous studies on the effect of neurotrophins on inositol signaling were limited to the analysis of IP3 and its dephosphorylation products. Here we demonstrate that nerve growth factor (NGF) regulates the levels of IP5 and IP6 during PC12 differentiation. Furthermore, both NGF and brain-derived neurotrophic factor alter IP5 and IP6 intracellular ratio in differentiated PC12 cells and primary neurons. Neurotrophins specifically regulate the expression of IP5-2 kinase (IP5-2K), which phosphorylates IP5 into IP6. IP5-2K is rapidly induced after NGF treatment, but its transcriptional levels sharply decrease in fully differentiated PC12 cells. Reduction of IP5-2K protein levels by small interfering RNA has an effect on the early stages of PC12 cell differentiation, whereas fully differentiated cells are not affected. Conversely, perturbation of IP5-2K levels by overexpression suggests that both differentiated PC12 cells and sympathetic neurons require low levels of the enzyme for survival. Therefore maintaining appropriate intracellular levels of inositol polyphosphates is necessary for neuronal survival and differentiation.
INTRODUCTION
Neurotrophins comprise a family of peptide growth factors that regulate many aspects of neuronal development and function, including neuronal precursor proliferation and survival, axon and dendrite growth, membrane trafficking, and synapse formation, to cite a few (reviewed in Reichardt, 2006). Neurotrophins interact with two distinct classes of receptors, the p75 neurotrophin receptor (p75NTR) and the tropomyosin receptor kinase (Trk) family of tyrosine kinase receptors. Whereas p75NTR has been shown to bind each of the neurotrophins with similar affinity (Rodriguez-Tebar et al., 1990; Frade and Barde, 1998), the three members of the Trk subfamily (TrkA, TrkB, TrkC) specifically interact with different neurotrophins. Nerve growth factor (NGF) activates TrkA, brain-derived neurotrophic factor (BDNF) and NT-4 activate TrkB, and NT-3 preferentially binds to TrkC (and to the other Trk receptors, albeit with lower affinity). On interaction with neurotrophins, Trk receptors undergo autophosphorylation, which triggers activation of several signaling pathways, including Ras-MAPK, PI3-kinase, phospholipase C-γ1 (PLC-γ1), and their downstream effectors (Chao, 2003). Of interest, NGF-TrkA receptor complexes are internalized as signaling endosomes and transported along the axons to cell bodies, where they activate transcription (Bhattacharyya et al., 1997; Riccio et al., 1997; Zweifel et al., 2005).
Activation of phospholipases, including PLC-γ1, induces the synthesis of the second messenger inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG), with subsequent IP3-dependent mobilization of Ca2+ from internal stores and activation of DAG-regulated protein kinases (Berridge, 2005). IP3 serves as a precursor for the synthesis of the higher phosphorylated forms of inositol. It is a substrate of the inositol polyphosphate multikinase (IPMK), which generates inositol 1,3,4,5,6-pentakisphosphate (IP5) from IP3. In a subsequent metabolic step, the inositol 1,3,4,5,6-pentakisphosphate 2-kinase (IP5-2K, also known as IPPK or IPK1 in yeast) generates the fully phosphorylated inositol hexakisphosphate (IP6) from IP5 (York et al., 1999; Irvine and Schell, 2001). Although IP5 and IP6 are the most abundant forms of inositol phosphates in eukaryotic cells and have been associated with numerous cellular functions (Irvine and Schell, 2001; Shears, 2001; Monserrate and York, 2010), changes in their cellular levels have rarely being associated with a physiological event (Guse et al., 1993; Mountford et al., 1994; Gao and Wang, 2007). This has led to the suggestion that both IP5 and IP6 are metabolically lethargic molecules (Menniti et al., 1993). More recent studies, however, suggest that IP5 and IP6 are present in cells in multiple pools, such that there are both stable reservoirs and signal-induced, rapidly changing pools (Otto et al., 2007). In this way, IP6 may serve as both a second messenger and a precursor to other signaling molecules such as the inositol pyrophosphates (Burton et al., 2009; Saiardi, 2012).
Despite both the importance of IP5 and IP6 in mediating cellular functions and their synthesis after PLC activation, a link between neurotrophin signaling and these molecules has not been demonstrated. In fact, two reports in which NGF was shown to increase the levels of lower phosphorylated forms of inositol date back more than 20 years (Contreras and Guroff, 1987; Altin and Bradshaw, 1990), and even recent studies of the involvement of inositol phosphates in growth cone formation do not investigate beyond the second messenger IP3 (Akiyama et al., 2009). Here we demonstrate that a balance between the intracellular levels of IP5 and IP6 is necessary for neurotrophin-dependent neuronal survival and differentiation. Of importance, we show that activation of the gene that regulates the levels of IP5 and IP6 is essential for cell survival and neurite outgrowth in PC12 cells and primary cortical and sympathetic neurons.
RESULTS
Analysis of IP5 and IP6 levels during neurotrophin-dependent differentiation
We initially investigated whether neurotrophins regulate inositol phosphates levels using PC12 cells, a model in which NGF signaling has been extensively characterized.
The analysis of IP5 and IP6 cellular levels requires the experiment to be performed after the myo-[3H]inositol labeling procedure reaches a state of metabolic equilibrium in which all inositol phosphate pools, especially the highly phosphorylated IP5 and IP6 species, reach steady-state labeling levels. Thus, undifferentiated PC12 cells were labeled with myo-[3H]inositol and the ratio between the amount of radiolabeled IP6 and the total lipid fraction (TotLipid; Supplemental Figure S1A), as well as the absolute values of IP5 and IP6 (Supplemental Figure S1C), were determined at 24-h intervals. The ratio IP6/TotLipid reaches equilibrium after 2 d of labeling (Supplemental Figure S1, A and B); however, the absolute value of radioactivity incorporated in IP6 needed 72 h to reach stable levels (Supplemental Figure S1C). Accordingly, all further experiments were performed in cells labeled for a minimum of 5 d to be certain that inositol phosphates had reached metabolic equilibrium.
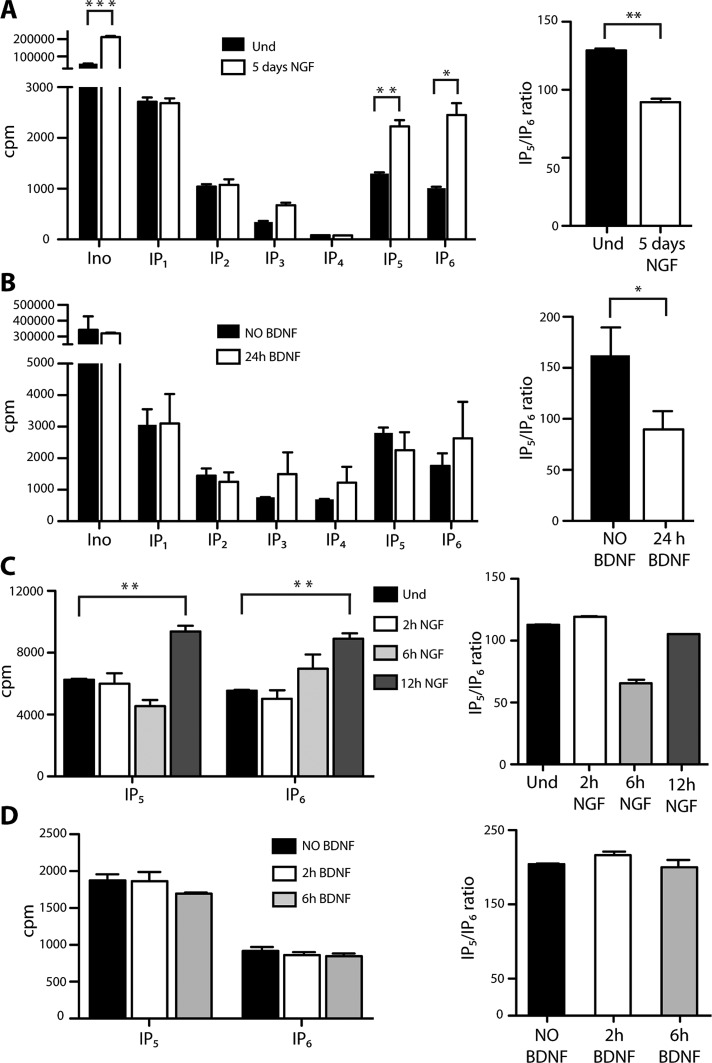
After labeling for 5 d, PC12 cells were differentiated using NGF (100 ng/ml) with constant presence of myo-[3H]inositol. After 5 d of differentiation, inositol phosphates were acid extracted and resolved using strong anion exchange, high-performance liquid chromatography (SAX-HPLC; see Materials and Methods for protocol details). As expected, exposure of PC12 cells to NGF for 5 d increased significantly the levels of myo-inositol, probably due to its function as an osmolite during cell differentiation (Figure 1A), a process that leads to an increase in the overall cell volume. Although levels of inositol monophosphate (IP1) to IP4 were not changed, we observed a robust increase of IP5 and IP6 in differentiated PC12 cells (Figure 1A, left). The use of radiolabeled IP5 standard established that these cells possess the isomer I(1,3,4,5,6)P5 (Supplemental Figure S1C). Similar results were obtained when the data were represented as the percentage of each inositol phosphate to the total lipid fraction (Supplemental Figure S1D). Moreover, the IP5/IP6 ratio was 30% lower due to a greater increase of IP6 compared with IP5 (Figure 1A, right, and Supplemental Figure S1C). Similar changes in IP5/IP6 ratio were observed when rat primary cortical neurons were exposed to the neurotrophin BDNF for 24 h, thereby indicating a common mechanism that controls neurotrophin-dependent levels of IP5 and IP6 (Figure 1B, right). This change was mainly due to a decrease in the levels of IP5 and an increase in the levels of IP6, but no significant increases in the absolute levels of IP5 and IP6 were seen when cortical neurons were treated with BDNF (Figure 1B, left), likely because the absolute increase of IP5 and IP6 is associated with neurite growth during differentiation. Instead, cortical neurons are already fully differentiated before treatment with BDNF, whose function is to induce only a modest increase of dendritic growth (McAllister et al., 1995).
FIGURE 1:
Neurotrophins increase the levels of IP5 and IP6 in PC12 cells and primary cortical neurons. (A) Left, comparison of HPLC analysis of inositol phosphates between undifferentiated (Und) and 5-d NGF-differentiated PC12 cells. Right, ratio between IP5 and IP6 levels in PC12 cells after 5 d of NGF differentiation. (B) Left, comparison of HPLC analysis of inositol phosphates between untreated (NO BDNF) and 24-h BDNF-treated primary cortical neurons. Right, ratio between IP5 and IP6 levels in primary cortical neurons after 24 h of BDNF treatment. (C) Left, time-course analysis of IP5 and IP6 levels in PC12 cells NGF differentiated for 2, 6, and 12 h. Right, time-course analysis of IP5 and IP6 ratio in PC12 cells NGF differentiated for 2, 6, and 12 h. (D) Left, time course analysis of IP5 and IP6 levels in primary cortical neurons BDNF treated for 2 and 6 h. Right, time-course analysis of IP5 and IP6 ratio in primary cortical neurons BDNF treated for 2 and 6 h. *p < 0.05, **p < 0.01, ***p < 0.001; error bars represent ± SD, n = 3.
We next determined the time course of the changes in IP5 and IP6 intracellular levels in response to NGF. Naive PC12 cells were exposed to NGF for 2, 6, or 12 h, and inositol phosphates were analyzed with SAX-HPLC chromatography. As shown in Figure 1C, left, 12 h after addition of NGF, levels of both IP5 and IP6 were increased compared with PC12 cells maintained in control conditions, and the IP5/IP6 ratio also changed (Figure 1C, right). No differences in IP5 or IP6 levels were observed in already differentiated cortical neurons stimulated with BDNF for 2 and 6 h (Figure 1D).
Taken together, these findings demonstrate that NGF changes the cellular levels of IP5 and IP6 both at early stages of differentiation and in fully differentiated PC12 cells. In addition, the ratio between IP5 and IP6 is modified upon addition of neurotrophic factors in both PC12 cells and primary cortical neurons.
Neurotrophins regulate expression of the gene responsible for regulation of IP5 and IP6 levels
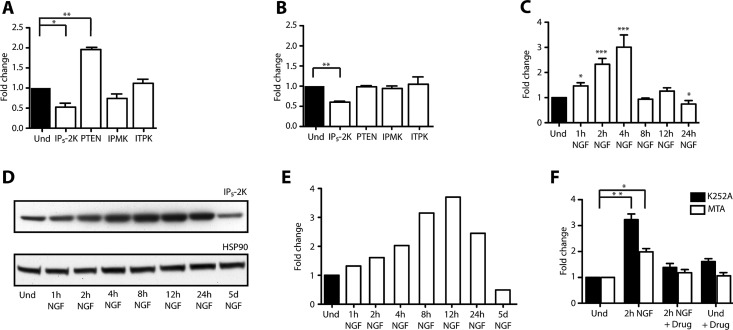
To identify the kinases responsible for both the increase in IP5 and IP6 levels and the change in their ratio, we treated PC12 cells and cortical neurons with NGF and BDNF for 5 d and 24 h, respectively. The mRNA was extracted and reverse transcribed to cDNA, and quantitative real-time (RT) PCR was performed. The mRNA levels of the genes IPMK, ITPK, and IP5-2K, responsible for the synthesis of inositol polyphosphates, were determined. As a control, the levels of the inositol phosphatase and tensin homologue (PTEN) were also measured. As shown in Figure 2, A and B, only one gene, IP5-2K, the sole kinase to use I(1,3,4,5,6)P5 as a substrate for the synthesis of IP6 (Verbsky et al., 2002), was differentially expressed after exposure to neurotrophins. This difference was calculated as a twofold down-regulation in both PC12 cells treated with NGF for 5 d (Figure 2A) and primary cortical neurons treated with BDNF for 24 h (Figure 2B). The expression of the PTEN was also increased in PC12 cells treated with NGF, as previously described (Lachyankar et al., 2000), validating our experimental protocols. To determine the kinetics of NGF-dependent expression or stability of IP5-2K, we treated PC12 cells with NGF for various times. We observed that mRNA levels of IP5-2K increased after 1 h of treatment with NGF and continued to rise for a further 3 h up to threefold compared with undifferentiated cells (Figure 2C). Prolonged exposure to NGF resulted in a decrease of IP5-2K mRNA to basal levels (Figure 2A). We next studied whether the changes in the mRNA levels were mirrored by protein levels. A similar time course was performed, and endogenous protein levels were analyzed by Western blot. Analysis of IP5-2K protein levels revealed a gradual increase of IP5-2K expression for ∼24 h of NGF treatment and a sharp decrease from 24 h onward, thereby reflecting the transcriptional changes (Figure 2, D and E).
FIGURE 2:
Neurotrophins modulate the levels of IP5-2K in PC12 cells and primary cortical neurons. (A) Quantitative RT-PCR analysis of inositol phosphate kinases in PC12 cells differentiated for 5 d with NGF. Bars are a representation of fold change levels in comparison to the undifferentiated sample (Und) normalized to GAPDH. (B) As in A, but in primary cortical neurons treated with BDNF for 24 h. (C) Quantitative RT-PCR analysis of IP5-2K in PC12 cells differentiated with NGF for 1, 2, 4, 8, 12, and 24 h. Bars are a representation of fold change levels in comparison to the undifferentiated sample (Und) normalized to GAPDH. (D, E) Western blot analysis (D) and quantification (E) of IP5-2K protein levels in PC12 cells differentiated with NGF for 1, 2, 4, 8, 12, and 24 h and 5 d. Protein levels were compared with the undifferentiated sample (Und). (F) Quantitative RT-PCR analysis of IP5-2K in PC12 cells differentiated with NGF for 2 h. Undifferentiated PC12 cells were treated with either K252A (200 nM) or MTA (1 mM) for 3 h before differentiation. PC12 cells were also treated with the specific drug and not differentiated in order to monitor the toxicity levels. mRNA levels were compared with the undifferentiated sample (Und) normalized to GAPDH. *p < 0.05, **p < 0.01, ***p < 0.001; error bars represent ± SD, n = 3.
To confirm that NGF directly regulates IP5-2K expression, naive PC12 cells were treated with the TrkA inhibitor K252A and the differentiation blocker 5′-S-methyl thioadenosine (MTA) for 3 h before adding NGF for a further 2 h, and IP5-2K mRNA levels were determined by quantitative RT-PCR. MTA was previously described as an inhibitor of phospholipid methylation necessary for the binding of ligands to cell surface receptors (Seeley et al., 1984). Based on its inhibitory properties, MTA is widely used to block NGF-mediated neurite outgrowth (Borasio, 1990; Maher, 1993). In PC12 cells, when NGF signaling was inhibited, we fail to observe any increase in IP5-2K mRNA levels (Figure 2F), confirming that TrkA-dependent NGF signaling regulates IP5-2K expression.
Inhibition of IP5-2K affects early stages of neuronal differentiation
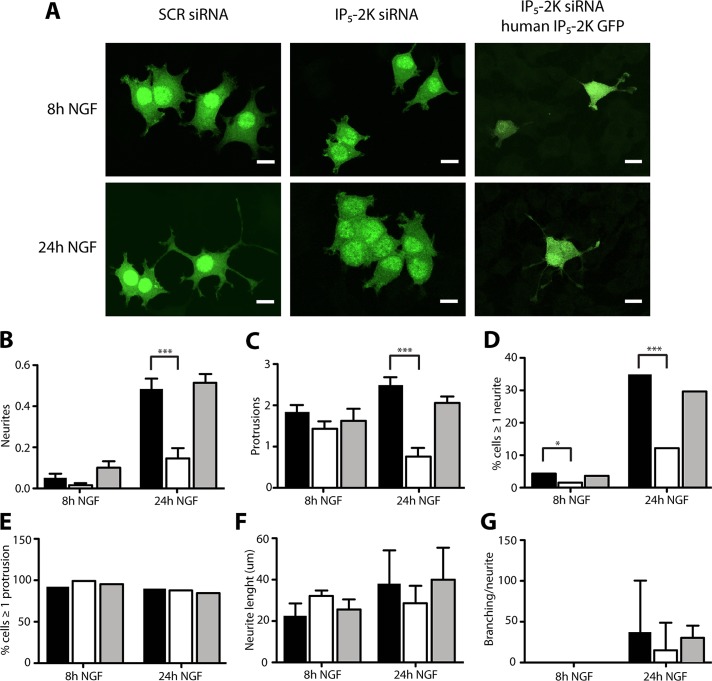
The critical role of NGF in fine-tuning IP5 and IP6 cellular level suggests that alteration of their metabolism will affect normal neuronal development. Therefore, to investigate the role of IP6 in mediating NGF-dependent functions, we reduced IP5-2K levels using a specific rat small interfering RNA (siRNA). In PC12 cells, ∼60% reduction of the endogenous level of IP5-2K was obtained (Supplemental Figure S2A). IP5-2K siRNA also efficiently silenced a rat IP5-2K green fluorescent protein (GFP)–tagged construct expressed in human HEK293T cells (Supplemental Figure S2B). Of interest, in contrast to the nuclear localization of the yeast orthologue Ipk1, IP5-2K GFP was localized in the cytoplasm, as previously described (Fujii and York, 2005). Because IP5-2K expression was highest between 2 and 24 h of NGF exposure, naive PC12 cells were transfected with IP5-2K siRNA for 48 h and treated with NGF for either 8 or 24 h, and several morphological features were analyzed (Figure 3, B–G). Confocal microscopy analysis showed that PC12 cells with reduced levels of IP5-2K and exposed to NGF for 24 h have fewer neurites than PC12 cells with normal levels of IP5-2K (Figure 3A). Extensive quantitative analysis demonstrated that the number of neurites per cell in PC12 cells transfected with IP5-2K siRNA was reduced by 2.5-fold after 24 h of exposure to NGF (Figure 3B). A lower number of protrusions/cell (Figure 3C) and a decrease in the fraction of cells with at least one neurite (Figure 3D) were also observed. In contrast, the percentage of cells with at least one protrusion, the total length of neurites/cell, and neurite branching all remained unchanged (Figure 3, E–G). In all cases, cotransfection of rat IP5-2K siRNA and a human IP5-2K GFP construct rescued the phenotypes seen during IP5-2K down-regulation (Figure 3, B–G, gray columns).
FIGURE 3:
IP5-2K knockdown affects PC12 cells differentiation. (A) Microscopy analysis of PC12 cells transfected with scramble (SCR) siRNA and GFP (left), IP5-2K siRNA and GFP (middle), or IP5-2K GFP and human IP5-2K GFP (right) differentiated with NGF for 8 (top) and 24 h (bottom). Scale bar, 10 μm. (B–G) Comparison between 8- and 24-h NGF-differentiated PC12 cells transfected with either SCR siRNA and GFP (black), IP5-2K siRNA and GFP (white), or IP5-2K GFP and human IP5-2K GFP (gray) for several phenotypic traits (n = 100). Number of neurites (B), number of protrusions (C), percentage of cells with minimum one neurite (D), percentage of cells with minimum one protrusion (E), length of neurites (F), and percentage of branches/neurite (G) were analyzed. All data were normalized to the undifferentiated sample. *p < 0.05, ***p < 0.001; error bars represent ± SD, n = 3.
We next examined whether inhibition of IP5-2K affected cell survival of both differentiated PC12 cells and superior cervical ganglion (SCG) neurons maintained in NGF for 5 d before siRNA IP5-2K electroporation. Of interest, IP5-2K did not affect the morphology or the viability of either cell type (Supplemental Figure S3, A and B). These findings indicate that although increased levels of IP5-2K are necessary for mediating the initial phases of NGF-mediated differentiation, a decrease in its level in both fully differentiated PC12 cells and sympathetic neurons does not affect cell survival.
Expression of IP5-2K affects differentiated PC12 cell and sympathetic neuron survival
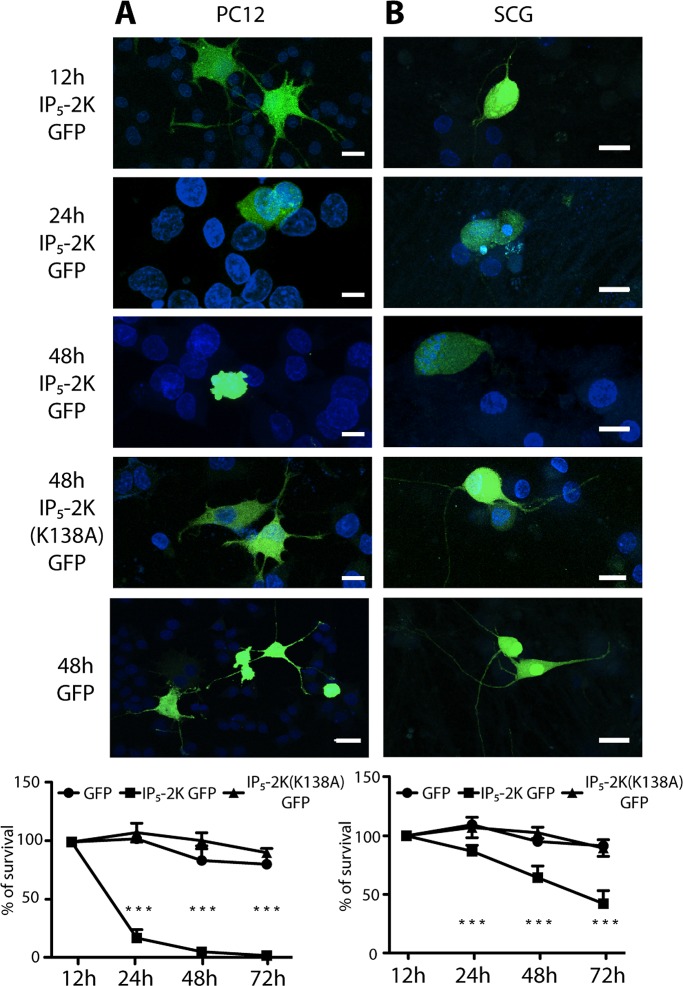
Because in differentiated PC12 cells and in sympathetic neurons exposure to neurotrophins lowers IP5-2K levels (Figure 2, A and B), up-regulation of this enzyme, which alters the balance between IP5 and IP6, might affect neuronal physiology. To test this hypothesis, we expressed GFP-tagged IP5-2K in both differentiated PC12 cells and sympathetic neurons. Confocal microscopy analysis revealed a gradual decrease in the number of cells expressing the GFP-tagged construct. In both PC12 cells (Figure 4A) and SCG neurons (Figure 4B), disappearance of neurites in PC12 cells and axons in sympathetic neurons preceded nuclear fragmentation as early as 24 h after transfection, reaching a nearly total loss of transfected cells after 48 h. To prove that overexpression of IP5-2K triggered a change in the intracellular levels of IP5 and IP6, we transfected myo-[3H]inositol–labeled HEK293T cells with IP5-2K GFP for 48 h and analyzed the levels of IP5 and IP6 and their ratio by HPLC. IP5-2K indeed caused a dramatic decrease in the level of IP5 (Supplemental Figure S4A) and the IP5/IP6 ratio (Supplemental Figure S4B) as previously reported (Fujii and York, 2005; Verbsky et al., 2005b).
FIGURE 4:
IP5-2K overexpression affects the survival of differentiated PC12 cells and sympathetic neurons. (A) Microscopy analysis of 5-d-differentiated PC12 cells transfected with IP5-2K GFP for 12, 24, or 48 h or IP5-2K(K138A) GFP or GFP for 48 h. Scale bar, 10 μm. Bottom, cell survival analysis of cells transfected for 12, 24, 48, and 72 h (n = 100). (B) As in A, but in 5-d-differentiated superior cervical ganglion neurons. ***p < 0.001; error bars represent ± SD, n = 3.
To confirm the importance of IP5-2K catalytic activity during neurotrophin-mediated differentiation, we transfected primary SCG neurons and fully differentiated PC12 cells with the kinase-dead mutant IP5-2K(K138A) GFP construct for a minimum of 48 h. Neurons expressing this construct did not show any sign of axonal degeneration and nuclear fragmentation (Figure 4, A and B), as for the neurons transfected with an empty eGFP plasmid (Figure 4, A and B).
To rule out a generic nonspecific toxic effect of IP5-2K expression, we transfected the vector IP5-2K GFP into HEK293T cells for a minimum of 48 h. No significant degeneration or nuclear fragmentation was observed (Supplemental Figure S5A).
Transfection of differentiated PC12 with a plasmid expressing IP6K1, the kinase responsible for the synthesis of the pyrophosphate IP7 from IP6, did not reveal any significant change in the morphology and viability of the cells (Supplemental Figure S5B), proving that a correct balance between the levels of IP5 and IP6 and not the downstream products is necessary for the survival of PC12 cells and primary neurons.
We next examined the effect of IP5-2K expression in naive PC12 cells treated with NGF for 6, 12, or 24 h. Microscopy analysis of the cell phenotype and survival rate did not show any difference between control cells and cells overexpressing IP5-2K (Supplemental Figure S6). Neurite degeneration and nuclear fragmentation, however, appeared in PC12 cells expressing IP5-2K that were differentiated for 24 h, similar to the results seen in 5-d-differentiated PC12 cells (Figure 4A). Taken together, these data suggest that tight control of the levels of IP5-2K is essential during neuronal differentiation and perturbation of such levels affects cellular differentiation and viability.
Expression of IP5-2K triggers activation of caspase 3 and consequent apoptosis in PC12 cells
To establish whether the axonal degeneration and nuclear fragmentation seen during expression of the IP5-2K GFP construct in PC12 cells and primary neurons was a result of cell death, we paraformaldehyde fixed IP5-2K GFP-transfected PC12 cells at different time points and costained them for the cell apoptosis indicator caspase 3. As shown in Figure 5A, PC12 cells expressing IP5-2K and with signs of nuclear fragmentation showed increased cleaved caspase 3 signal, of intensity similar to that of PC12 cells, in which apoptosis was induced using the cytotoxic quinoline alkaloid camptothecin (4 μM; Figure 5A, bottom).
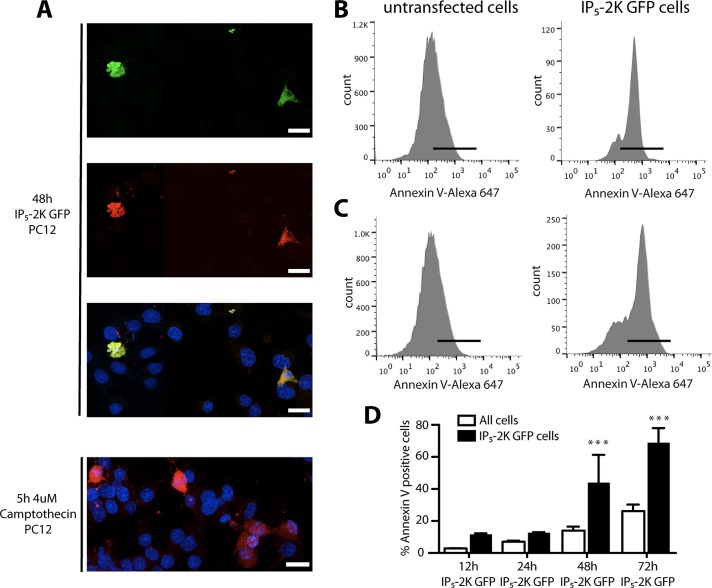
FIGURE 5:
IP5-2K overexpression triggers caspase 3 activation and consequent apoptosis. (A) Microscopy analysis of 5-d-differentiated PC12 cells transfected with IP5-2K GFP and analyzed for caspase 3 activation using an anti–cleaved caspase 3 antibody (red). Bottom, 5–d-differentiated PC12 cells treated with the apoptosis inducer camptothecin for 5 h and stained using an anti–cleaved caspase 3 antibody (red). (B) Fluorescence-activated cell sorting (FACS) analysis of annexin V–positive cells in 48-h IP5-2K GFP–transfected cells. Comparison between IP5-2K GFP–gated cells (right) and untransfected cells (left). Black bar represents annexin V–positive cells. (C) As in B, but in 72-h-transfected cells. (A) Quantification of FACS analysis of annexin V–positive cells in 12-, 24-, 48-, and 72-h IP5-2K GFP–transfected cells. White, untransfected cells; black, IP5-2K GFP–transfected cells. ***p < 0.001; error bars represent ± SD, n = 3.
To support this finding, we analyzed differentiated PC12 cells expressing IP5-2K GFP at different time points for annexin V staining using flow cytometry. After transfection, the intensity of annexin V was compared between the cells expressing IP5-2K GFP and the untransfected cells. Figure 5B shows that ∼60% of the cells expressing IP5-2K GFP for 48 h were positive for annexin V, whereas only 15% of the untransfected cells were positive. Similarly, 72 h after transfection, 70% of the cells expressing IP5-2K GFP were annexin V positive versus 25% of the untransfected cells (Figure 5C). Quantification for all the time points revealed a gradual increase in the percentage of IP5-2K GFP-expressing cells positive for annexin V (Figure 5D), confirming an increase of apoptotic events when differentiated PC12 cells were expressing IP5-2K at high levels.
DISCUSSION
The change in IP5 and IP6 levels in PC12 cells after differentiation with NGF represents one of very few instances known—the others being during cell cycle progression (Guse et al., 1993), hematopoietic cell differentiation (Mountford et al., 1994), and activation of Frizzled-1 by Wnt3a (Gao and Wang, 2007)—in which a physiological stimulus regulates intracellular IP5 and IP6 concentrations in mammalian cells.
In neurons, besides changes in the absolute levels, NGF also modulates the intracellular ratio of IP5 and IP6. Whereas addition of NGF to PC12 cells triggers rapid morphological changes and neurite extension, differentiation of cortical neurons does not require exogenous neurotrophins, and addition of BDNF has been shown to induce only a modest increase of dendritic growth (McAllister et al., 1995), mainly promoting synaptic functions (Binder and Scharfman, 2004). This may explain why in cortical neurons we failed to observe absolute changes of IP5 and IP6, although we consistently observed changes in their relative cellular levels. The observation that both NGF, through TrKA, and BDNF, through TrKB, mediate changes in the ratio between IP5 and IP6 suggests a fundamental role for such regulatory mechanisms. The ideal system in which to study the effect of neurotrophin-dependent differentiation on inositol signaling would be the primary SCG neurons. Unfortunately we found it extremely difficult to obtain a sufficient amount of labeled inositol phosphates from SCG primary cultures for detection using our HPLC system.
Evidence for involvement of inositol polyphosphates in neurotrophin signaling was provided in the late 1980s when two independent groups showed that exposure to NGF for a time as short as 15 s increased IP1, IP2, and IP3 levels (Contreras and Guroff, 1987; Altin and Bradshaw, 1990). The techniques used at that time, however, were not suitable for the detection of higher phosphorylated isoforms. More recently, a study in which NGF-driven growth cone formation was investigated focused primarily on the effect of the second messengers Ca2+ and IP3 but not higher phosphorylated isoforms of inositol (Akiyama et al., 2009). Our data demonstrate that in PC12 cells, IP5 and IP6 levels increase after exposure to NGF. It would be interesting to determine whether this leads to changes in inositol pyrophosphate metabolism. Unfortunately, the sensitivity of the HPLC technology used does not allow the detection of inositol pyrophosphates.
Analysis of the enzymes responsible for IP5 and IP6 metabolism revealed that the kinase IP5-2K is transcriptionally regulated by neurotrophins. This enzyme was found up-regulated at early stages of differentiation, consistent with the observed increases in the cellular levels of IP5 and IP6. At later stages of differentiation, however, IP5-2K seemed to reach an expression level lower than that of the undifferentiated counterpart, which clearly reflected the changes in the ratio between IP5 and IP6 noted during our analysis. Our data also demonstrate that such changes depend on TrK-mediated signaling.
The HPLC elution profile revealed the presence of only the I(1,3,4,5,6)P5 isomer of IP5 in PC12 cells. This molecule is the sole substrate of IP5-2K, which phosphorylates position 2 of the inositol ring into the fully phosphorylated IP6.
Microscopy analysis of a GFP-tagged IP5-2K construct transfected into both PC12 cells and sympathetic neurons showed ubiquitous cytoplasmic localization with only a low level of IP5-2K GFP in the nucleus, as previously demonstrated in Rat-1 cells (Fujii and York, 2005). This localization is in contrast with what was described in the yeast Saccharomyces cerevisiae, in which tagged versions of the IP5-2K orthologue, Ipk1, were found predominantly in the nucleus (York et al., 1999).
These data suggest that IP5 and IP6 might regulate the first stages of differentiation. Down-regulation of IP5-2K levels during the early stages of PC12 differentiation induced a decrease of both the number of neurites and the number of cells with at least one neurite. At later stages of differentiation, IP5-2K mRNA levels decreased significantly, although IP5 and IP6 levels remained high. The low levels of IP5-2K found in fully differentiated PC12 cells and the fact that its expression in both differentiated PC12 cells and sympathetic neurons triggers caspase 3 activation with consequent apoptosis suggest that maintaining the appropriate balance of IP5 and IP6 is necessary to promote neuronal cell survival. We cannot exclude the possibility, however, that it is the absolute change in IP5 or IP6, and not their relative ratio, that is responsible for the phenotypes described. Conversely, in nonneuronal cell lines, such as HEK293T, overexpression of IP5-2K does not induce apoptotic processes, as previously demonstrated (Verbsky and Majerus, 2005).
Of interest, deletion of IP5-2K in mouse resulted in early embryonic lethality (embryonic day 8.5), with the major anatomical anomaly described as a defect in migration of neural crest cells and neuronal tube formation (Verbsky et al., 2005a).
Several cellular pathways have been linked to IP6 signaling. Studies from Wente and colleagues demonstrated a role of IP6 in mediating the transport of mRNA from the nucleus to the cytoplasm (Folkmann et al., 2011). During neuronal differentiation, several morphological changes occur, including the extension of axons over long distances driven by their growth cone. During this stage, several mRNA transcripts are locally transported from the nucleus along the axons and translated at this specific subcellular region (Lin and Holt, 2007; Andreassi et al., 2010). The fact that high levels of IP6 are required during the early stages of differentiation suggests that mRNA transport and axonal development may be related. Of importance, the finding that changes of IP5 and IP6 levels occur rapidly partly contradicts the hypothesis that these molecules are in a metabolically lethargic state (Menniti et al., 1993). The development of novel assays that will allow an accurate measurement of both the abundance and localization of IP5 and IP6 in differentiating cells will help to clarify the role of these molecules in neurons. It will be especially important to understand whether a general or a local increase of IP5 and IP6 levels is required for neuronal differentiation.
MATERIALS AND METHODS
Reagents and antibodies
The following primary antibodies were used: goat polyclonal anti–IP5-2K (1:1000 for Western blot; Sigma-Aldrich, St. Louis, MO), rabbit monoclonal anti–cleaved caspase 3 (1:500 for immunofluorescence; Cell Signaling Technology, Beverly, MA). Annexin V, Alexa Fluor 647 conjugated, was used for flow cytometry analysis according to the manufacturer's protocol (Life Technologies, Carlsbad, CA). 5′-S-methyl adenosine, K252A TrkA inhibitor, and (S)-(+)-camptothecin were purchased from Sigma-Aldrich. Rat IP5-2K siRNA was purchased from Dharmacon RNAi Technologies (Lafayette, CO; L-090228-01).
Cell culture
PC12 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich), 5% horse donor serum, HS (Sigma-Aldrich), and 2 mM l-glutamine (Sigma-Aldrich) in a humidified incubator at 37°C and 10% CO2. To induce differentiation, cells were incubated with serum-deprived medium containing 2 mM l-glutamine and 100 ng/ml NGF (Sigma-Aldrich).
Western blot analysis
Protein extracts were obtained using RIPA extraction buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% sodium deoxycholate, 1 mM EDTA, 0.1% SDS, 1% NP-40, protease inhibitor cocktail). Protein extracts were heated to 100°C for 5 min in the presence of 5% 2-mercaptoethanol. Proteins were separated on a 4–12% gradient polyacrylamide gel (Invitrogen/Life Technologies) and then electrotransferred to polyvinylidene fluoride membranes (Bio-Rad, Hercules, CA). The membranes were incubated with 5% nonfat milk in phosphate-buffered saline (PBS) and 0.3% Tween-20 for 1 h. The antigens were incubated with the primary antibody overnight at 4°C and then for 1 h with the specific secondary antibody. Antigen detection was obtained by incubation with Luminata Crescendo Western HRP substrate (Millipore, Billerica, CA) and scanned using a film developer.
Primary culture of cortical and sympathetic neurons
The Institutional Animal Care and Use Committees at University College London approved all animal studies. SCG neurons were dissected from postnatal day 1 Sprague–Dawley rats. The ganglia were enzymatically dissociated and plated on glass coverslips. SCG explants and primary sympathetic neurons were maintained in DMEM supplemented with 10% FBS (Sigma-Aldrich) and 2 mM l-glutamine (Sigma-Aldrich) in a humidified incubator at 37°C and 10% CO2. Cytosine arabinoside (10 μM) was added 24 h after plating to block the proliferation of nonneuronal cells.
Dissociated cultures of cortical neurons were prepared as described (Threadgill et al., 1997) and grown in DMEM containing FBS (10%).
Inositol polyphosphate analysis of PC12 cells and primary neurons
PC12 cells or cortical neurons grown on supplemented DMEM medium were labeled for 5 d with 30 μl/ml myo-[3H]inositol, 30 Ci/mmol (American Radiolabeled Chemicals, St. Louis, MO). Half of the medium was replaced with fresh DMEM containing 30 μl/ml myo-[3H]inositol every 3 d. After the various treatments, the radioactive medium was carefully removed, and cells were washed three times with 2 ml of warm DMEM. After addition of 500 μl of extraction buffer (1 M perchloric acid, 1 mM EDTA), cells were incubated on ice for 8 min. The extraction buffer was subsequently collected on an Eppendorf tube. Equal counts of inositol phosphates were subjected to neutralization before SAX-HPLC analysis, which was performed as previously described (Azevedo and Saiardi, 2006). Data are represented as absolute values in cpm or as percentage of total radioactivity measured in the lipid fraction, which was obtained by overnight incubation of the cells with a solution of 1 M NaOH and 0.1% Triton X-100. The [3H]I(1,3,4,5,6) standard preparation was performed as previously described (Maffucci et al., 2005).
RNA isolation, reverse transcription, and quantitative RT-PCR
RNA isolation and reverse transcription were performed using TRIzol and SuperScriptIII (Invitrogen), respectively, according to the manufacturer's instructions.
PCRs (25 μl) contained 12.5 μl of PCR Sybr Green mix (New England BioLabs, Ipswich, MA) and 0.3 μM primers. All reactions were performed in triplicate with an Opticon 2 System (MJ Research, Cambridge, MA), and each experiment included a standard curve, a no–reverse transcription control, and a no-template control. Standard template consisted of known concentration of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA, and each standard curve consisted of five serial dilutions of template. At the end of 40 cycles of amplification, a dissociation curve was performed in which Sybr Green was measured at 1°C intervals between 50 and 100°C to generate a melting curve. Results were normalized to GAPDH and expressed as fold changes over unstimulated samples.
Cloning of rat and human IP5-2K
Rat IP5-2K was amplified using primers GCACTCGAGTCATGGAAGAGGGGAAAATGGACG and GCAGGATCCTTAGACCTTATGGAGAACTAATG from rat brain cDNA. Human IP5-2K was amplified using primers GCACTCGAGTCATGGAAGAGGGGAAGATGGAC and TTAGACCTTGTGGAGAACTAATG from human cDNA. The PCR product was then cut using the restriction enzymes XhoI and BamHI and fused into the plasmid pEGFP-C1 (Clontech, Mountain View, CA) using standard DNA ligation.
GFP-tagged, kinase-dead mutants of rat IP5-2K with point mutations at K138A were obtained by site-direct mutagenesis of the plasmid IP5-2K GFP using primers IP5-2K(K138A)5F, CTGTGTGTGGAGATTGCGCCAAAATGTGGGTT, and IP5-2K(K138A)3R, AACCCACATTTTGGCGCAATCTCCACACACAG (Gonzalez et al., 2010).
PC12 cell and primary neuron transfection and microscopy analysis
PC12 cells were transfected with the indicated constructs or siRNA using Lipofectamine2000 (Invitrogen) according to the manufacturer's protocol.
Sympathetic neurons were electroporated using the Cellaxess CX1 system (Cellectricon, Mölndal, Sweden) as described previously (Andreassi et al., 2010).
Confocal images were acquired with an SP5 confocal system (Leica, Wetzlar, Germany) using LAS AF software and automated tiling over several z-stacks to cover the whole thickness and length of the cells and neurons. Maximal intensity projections were processed with ImageJ software (National Institutes of Health, Bethesda, MD).
Quantitative analysis of neurite outgrowth
PC12 cells were fixed for 20 min using 4% paraformaldehyde in PBS at the desired time point after differentiation and/or transfection. Cells were subsequently mounted on a coverslip using Dako mounting medium (Dako, Carpinteria, CA). 4′,6-Diamidino-2-phenylindole dye was included in the fixative to label cell nuclei. Slides were then analyzed using a confocal microscope for which fluorescence images were produced using a 40× oil objective. The acquired images were analyzed for a number of morphological parameters, including total neurite length per cell, neurites per cell, protrusions per cell, and branch points. A cell was defined as possessing a neurite when this had a length greater than the cell body diameter. When the length of the neurite was shorter than the cell body diameter it was annotated as a protrusion.
Quantification of cell viability
PC12 cell and sympathetic neuron viability was quantified in cells grown on glass-bottom dishes (Zell-kontakt, Nörten-Hardenberg, Germany). Cells transfected with either siRNA or GFP-tagged proteins were labeled with Hoechst 33258 dye for 30 min at 37°C. Subsequently, live images were taken at the desired time point using a confocal microscope with a chamber heated to 37°C at 10% CO2. Fluorescent images were taken for the same area during all time points, and the number of cells considered alive was counted. As alive, we considered any cell possessing intact nuclei and neurites/axons. Data were analyzed and represented as percentage of cell survival normalized to the control sample.
Annexin V staining for flow cytometry
IP5-2K GFP-transfected cells (2 × 106) were resuspended in binding buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 140 mM NaCl, 2.5 mM CaCl2, pH 7.4), labeled with Alexa Fluor 647–conjugated annexin V, and incubated at room temperature for 15 min. Subsequently, the cells were washed and resuspended in binding buffer before flow cytometry analysis using an LSRII cytometer (BD Biosciences, San Diego, CA) equipped with 488- and 633-nm lasers.
Cells expressing IP5-2K GFP were gated for analysis of annexin V activation and compared with the untransfected cells. Subsequent analyses were conducted using FlowJo software (TreeStar, Ashland, OR).
Statistics
Data are expressed as averages ± SD. A minimum of three replicates were analyzed for each experiment. Unless otherwise noted, unpaired two-tailed Student's t tests or two-way analysis of variance were used to test for statistical significance, which was placed at p < 0.05.
Supplementary Material
Acknowledgments
This work was supported by the Medical Research Council (funding to the Cell Biology Unit) and a Human Frontier Science Program Grant (RGP0048/2009-C).
Abbreviations used:
- BDNF
brain-derived neurotrophic factor
- GFP
green fluorescent protein
- IP5
inositol 1,3,4,5,6-pentakisphosphate
- IP5-2K
inositol pentakisphosphate-2 kinase
- IP6
inositol hexakisphosphate
- MTA
5′-S-methyl thioadenosine
- NGF
nerve growth factor
- SCG
superior cervical ganglion
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-04-0198) on July 17, 2013.
REFERENCES
- Akiyama H, Matsu-ura T, Mikoshiba K, Kamiguchi H. Control of neuronal growth cone navigation by asymmetric inositol 1,4,5-trisphosphate signals. Sci Signal. 2009;2:ra34. doi: 10.1126/scisignal.2000196. [DOI] [PubMed] [Google Scholar]
- Altin JG, Bradshaw RA. Production of 1,2-diacylglycerol in PC12 cells by nerve growth factor and basic fibroblast growth factor. J Neurochem. 1990;54:1666–1676. doi: 10.1111/j.1471-4159.1990.tb01220.x. [DOI] [PubMed] [Google Scholar]
- Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, Saiardi A, Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13:291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- Azevedo C, Saiardi A. Extraction and analysis of soluble inositol polyphosphates from yeast. Nat Protoc. 2006;1:2416–2422. doi: 10.1038/nprot.2006.337. [DOI] [PubMed] [Google Scholar]
- Berridge MJ. Unlocking the secrets of cell signaling. Annu Rev Physiol. 2005;67:1–21. doi: 10.1146/annurev.physiol.67.040103.152647. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A, Watson FL, Bradlee TA, Pomeroy SL, Stiles CD, Segal RA. Trk receptors function as rapid retrograde signal carriers in the adult nervous system. J Neurosci. 1997;17:7007–7016. doi: 10.1523/JNEUROSCI.17-18-07007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder DK, Scharfman HE. Brain-derived neurotrophic factor. Growth Factors. 2004;22:123–131. doi: 10.1080/08977190410001723308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borasio GD. Differential effects of the protein kinase inhibitor K-252a on the in vitro survival of chick embryonic neurons. Neurosci Lett. 1990;108:207–212. doi: 10.1016/0304-3940(90)90732-o. [DOI] [PubMed] [Google Scholar]
- Burton A, Hu X, Saiardi A. Are inositol pyrophosphates signalling molecules? J Cell Physiol. 2009;220:8–15. doi: 10.1002/jcp.21763. [DOI] [PubMed] [Google Scholar]
- Chao MV. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- Contreras ML, Guroff G. Calcium-dependent nerve growth factor-stimulated hydrolysis of phosphoinositides in PC12 cells. J Neurochem. 1987;48:1466–1472. doi: 10.1111/j.1471-4159.1987.tb05687.x. [DOI] [PubMed] [Google Scholar]
- Folkmann AW, Noble KN, Cole CN, Wente SR. Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus. 2011;2:540–548. doi: 10.4161/nucl.2.6.17881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frade JM, Barde YA. Nerve growth factor: two receptors, multiple functions. Bioessays. 1998;20:137–145. doi: 10.1002/(SICI)1521-1878(199802)20:2<137::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Fujii M, York JD. A role for rat inositol polyphosphate kinases rIPK2 and rIPK1 in inositol pentakisphosphate and inositol hexakisphosphate production in rat-1 cells. J Biol Chem. 2005;280:1156–1164. doi: 10.1074/jbc.M412006200. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wang HY. Inositol pentakisphosphate mediates Wnt/beta-catenin signaling. J Biol Chem. 2007;282:26490–26502. doi: 10.1074/jbc.M702106200. [DOI] [PubMed] [Google Scholar]
- Gonzalez B, Banos-Sanz JI, Villate M, Brearley CA, Sanz-Aparicio J. Inositol 1,3,4,5,6-pentakisphosphate 2-kinase is a distant IPK member with a singular inositide binding site for axial 2-OH recognition. Proc Natl Acad Sci USA. 2010;107:9608–9613. doi: 10.1073/pnas.0912979107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse AH, Greiner E, Emmrich F, Brand K. Mass changes of inositol 1,3,4,5,6-pentakisphosphate and inositol hexakisphosphate during cell cycle progression in rat thymocytes. J Biol Chem. 1993;268:7129–7133. [PubMed] [Google Scholar]
- Irvine RF, Schell MJ. Back in the water: the return of the inositol phosphates. Nat Rev Mol Cell Biol. 2001;2:327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Lachyankar MB, et al. A role for nuclear PTEN in neuronal differentiation. J Neurosci. 2000;20:1404–1413. doi: 10.1523/JNEUROSCI.20-04-01404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Holt CE. Local translation and directional steering in axons. EMBO J. 2007;26:3729–3736. doi: 10.1038/sj.emboj.7601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffucci T, et al. Inhibition of the phosphatidylinositol 3-kinase/Akt pathway by inositol pentakisphosphate results in antiangiogenic and antitumor effects. Cancer Res. 2005;65:8339–8349. doi: 10.1158/0008-5472.CAN-05-0121. [DOI] [PubMed] [Google Scholar]
- Maher PA. Inhibition of the tyrosine kinase activity of the fibroblast growth factor receptor by the methyltransferase inhibitor 5′-methylthioadenosine. J Biol Chem. 1993;268:4244–4249. [PubMed] [Google Scholar]
- McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- Menniti FS, Miller RN, Putney JW, Jr, Shears SB. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J Biol Chem. 1993;268:3850–3856. [PubMed] [Google Scholar]
- Monserrate JP, York JD. Inositol phosphate synthesis and the nuclear processes they affect. Curr Opin Cell Biol. 2010;22:365–373. doi: 10.1016/j.ceb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Mountford JC, Bunce CM, French PJ, Michell RH, Brown G. Intracellular concentrations of inositol, glycerophosphoinositol and inositol pentakisphosphate increase during haemopoietic cell differentiation. Biochim Biophys Acta. 1994;1222:101–108. doi: 10.1016/0167-4889(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Otto JC, Kelly P, Chiou ST, York JD. Alterations in an inositol phosphate code through synergistic activation of a G protein and inositol phosphate kinases. Proc Natl Acad Sci USA. 2007;104:15653–15658. doi: 10.1073/pnas.0705729104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio A, Pierchala BA, Ciarallo CL, Ginty DD. An NGF-TrkA-mediated retrograde signal to transcription factor CREB in sympathetic neurons. Science. 1997;277:1097–1100. doi: 10.1126/science.277.5329.1097. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Tebar A, Dechant G, Barde YA. Binding of brain-derived neurotrophic factor to the nerve growth factor receptor. Neuron. 1990;4:487–492. doi: 10.1016/0896-6273(90)90107-q. [DOI] [PubMed] [Google Scholar]
- Saiardi A. Cell signalling by inositol pyrophosphates. Subcell Biochem. 2012;59:413–443. doi: 10.1007/978-94-007-3015-1_14. [DOI] [PubMed] [Google Scholar]
- Seeley PJ, Rukenstein A, Connolly JL, Greene LA. Differential inhibition of nerve growth factor and epidermal growth factor effects on the PC12 pheochromocytoma line. J Cell Biol. 1984;98:417–426. doi: 10.1083/jcb.98.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB. Assessing the omnipotence of inositol hexakisphosphate. Cell Signal. 2001;13:151–158. doi: 10.1016/s0898-6568(01)00129-2. [DOI] [PubMed] [Google Scholar]
- Threadgill R, Bobb K, Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- Verbsky J, Lavine K, Majerus PW. Disruption of the mouse inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene, associated lethality, and tissue distribution of 2-kinase expression. Proc Natl Acad Sci USA. 2005a;102:8448–8453. doi: 10.1073/pnas.0503656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbsky J, Majerus PW. Increased levels of inositol hexakisphosphate (InsP6) protect HEK293 cells from tumor necrosis factor (alpha)- and Fas-induced apoptosis. J Biol Chem. 2005;280:29263–29268. doi: 10.1074/jbc.M503366200. [DOI] [PubMed] [Google Scholar]
- Verbsky JW, Chang SC, Wilson MP, Mochizuki Y, Majerus PW. The pathway for the production of inositol hexakisphosphate in human cells. J Biol Chem. 2005b;280:1911–1920. doi: 10.1074/jbc.M411528200. [DOI] [PubMed] [Google Scholar]
- Verbsky JW, Wilson MP, Kisseleva MV, Majerus PW, Wente SR. The synthesis of inositol hexakisphosphate. Characterization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. J Biol Chem. 2002;277:31857–31862. doi: 10.1074/jbc.M205682200. [DOI] [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- Zweifel LS, Kuruvilla R, Ginty DD. Functions and mechanisms of retrograde neurotrophin signalling. Nat Rev Neurosci. 2005;6:615–625. doi: 10.1038/nrn1727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.