P21-activated kinase 1 (PAK1) activation is achieved through relief of autoinhibition. The authors show that this event is not sufficient to bring about PAK1 activation but is required for CK2-dependent PAK1S223 phosphorylation, which enables PAK1 to be catalytically competent. S223 phosphorylation is crucial for PAK1 GTPase-dependent and GTPase-independent activation.
Abstract
Activation of the p21-activated kinase 1 (PAK1) is achieved through a conformational change that converts an inactive PAK1 dimer to an active monomer. In this paper, we show that this change is necessary but not sufficient to activate PAK1 and that it is, rather, required for CK2-dependent PAK1S223 phosphorylation that converts a monomeric PAK1 into a catalytically active form. This phosphorylation appears to be essential for autophosphorylation at specific residues and overall activity of PAK1. A phosphomimetic mutation (S223E) bypasses the requirement for GTPases in PAK1 activation, whereas the constitutive activity of the PAK1 mutant (PAK1H83,86L), postulated to mimic GTPase-induced structural changes, is abolished by inhibition of S223 phosphorylation. Thus, S223 is likely accessible to CK2 upon conformational changes of PAK1 induced by GTPase-dependent and GTPase-independent stimuli, suggesting that S223 phosphorylation may play a key role in the final step of the PAK1 activation process. The physiological significance of this phosphorylation is reinforced by the observations that CK2 is responsible for epidermal growth factor–induced PAK1 activation and that inhibition of S223 phosphorylation abrogates PAK1-mediated malignant transformation of prostate epithelial cells. Taken together, these findings identify CK2 as an upstream activating kinase of PAK1, providing a novel mechanism for PAK1 activation.
INTRODUCTION
P21-activated kinases (PAKs) are downstream effectors of the small GTPases Cdc42 and Rac, molecular switches that transduce various extracellular signals into intracellular responses (Manser et al., 1994). To date, six PAK family members have been identified and subdivided into two groups on the basis of their structural and biochemical features (Bokoch, 2003; Arias-Romero and Chernoff, 2008): group I (PAK1–3) and group II (PAK4–6). PAK1, the best-characterized member of the PAK family, is regulated by autoinhibition imposed on the C-terminal catalytic domain by the N-terminal autoinhibitory domain (AID; Lei et al., 2000; Parrini et al., 2002). This autoinhibition is relieved upon binding of activated GTPases to the p21-binding domain/Cdc42/Rac-interacting domain (PBD/CRIB domain) that partially overlaps with AID, leading to PAK1 activation (Morreale et al., 2000). However, group II PAKs do not contain the AID, and thus GTPases have little effect on kinase activity (Pandey et al., 2002).
Group I PAKs are also activated by various mechanisms independent of GTPases (Field and Manser, 2012). PAK1 is recruited to the plasma membrane via the SH3-containing proteins Nck and Grb2 (Bokoch et al., 1996), where it is activated by signaling molecules such as PDK1 kinase (King et al., 2000), sphingolipids (Zenke et al., 1999), and PIP2 (Strochlic et al., 2010). PAK2 is activated through proteolytic cleavage by caspases, liberating the N-terminal AID and thereby generating an active C-terminal catalytic domain (Rudel and Bokoch, 1997). PAK3 is unique among PAKs because it exists as four alternatively spliced isoforms activated in different manners (Kreis et al., 2008). Thus, PAKs are likely regulated by different mechanisms that operate independently.
PAK1 plays fundamental roles in various cellular processes, including actin cytoskeleton dynamics (Edwards et al., 1999), apoptosis (Vadlamudi et al., 2004), histone modification (Li et al., 2002), and cell cycle progression (Zhao et al., 2005). PAK1 does so by acting as a convergence point of many signaling pathways that are frequently dysregulated in human cancer and appears to be widely overexpressed and hyperactivated in various cancer cells (Kumar et al., 2006). Consistent with these observations, overexpression of an activated mutant form of PAK1 leads to the formation of mammary gland tumors in a transgenic mouse model (Wang et al., 2006a). However, in some cell types, including prostate cells, there is no significant difference in PAK1 mRNA expression between normal and malignant tissues (GeneCards, www.genecards.org), suggesting the existence of a distinct mechanism for the conversion of PAK1 from an inactive to an active form through posttranslational mechanisms in those cells.
CK2 is a ubiquitously expressed protein kinase and regulates a wide variety of biological processes, including cell growth and survival, gene expression, and actin cytoskeleton organization (Meggio and Pinna, 2003). CK2 is overexpressed in cancer cells and promotes tumorigenesis by regulating the stability and/or activity of tumor suppressors and by perpetuating multiple cell survival signaling pathways. CK2 is also membrane targeted and acts directly on actin or actin-associated proteins such as WASP (Cory et al., 2003) and CRN2 (Xavier et al., 2012), regulating actin polymerization. This study reveals that CK2 kinase plays a key role in regulating PAK1 activity. The prevailing view is that PAK1 is activated through conversion of PAK1 from an inactive dimer to an active monomer. In this paper, we provide evidence that this conversion is necessary but not sufficient to bring about full activation of PAK1 and that it is, rather, required for CK2-dependent PAK1 phosphorylation, enabling the kinase domain of PAK1 to be catalytically competent.
RESULTS
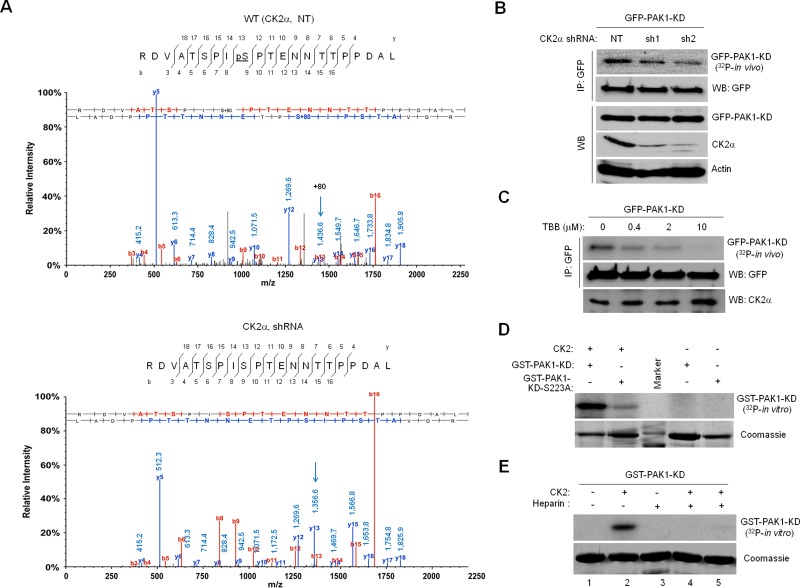
CK2 phosphorylates PAK1 at S223
The observations that CK2 and PAK1 both are often activated by the same external stimuli, including growth factors, and that PAK1 contains several putative CK2 phosphorylation sites (Meggio and Pinna, 2003) support a hypothesis that CK2 may directly phosphorylate and activate PAK1. For testing this hypothesis, green fluorescent protein (GFP)-PAK1 expressed in 293T cells with or without CK2α knockdown was immunoprecipitated with anti-GFP antibody and processed for liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis (Figure 1 and Supplemental Figure S1A). Data analysis with MASCOT and Scaffold software identified a peptide that matches 215RDVATSPISPTENNTTPPDAL235, indicating that S223 (underlined) is modified by phosphorylation (Figure 1A, top, arrow). The observation that this phosphorylation is not detected in the peptide derived from PAK1 expressed in CK2α knockdown cells (Figure 1A, bottom) indicated that S223 phosphorylation is CK2 dependent. Consistent with this notion, S223 lies within a canonical CK2 phosphorylation site, S/T-X-X-E/D/Ps (Meggio and Pinna, 2003; Diella et al., 2008) and appears to be conserved in mammalian PAK1 proteins (Figure S1B). Interestingly, however, S223 is not found in other members of the PAK family but is unique to PAK1 (Figure S1C).
FIGURE 1:
CK2 phosphorylates PAK1 on S223. (A) Mass spectrometry identification of CK2-catalyzed PAK1 phosphorylation at S223. The 293T cells treated with a nontargeting shRNA (NT, top) or a CK2α-specific shRNA (shRNA, bottom) were infected with lentivirus expressing GFP-PAK1. GFP-PAK1 was immunoprecipitated with anti-GFP antibody, resolved by SDS–PAGE, and digested with trypsin or chymotrypsin/elastase. The resulting eluted peptides were analyzed directly by LC-MS/MS. The m/z difference between y-13s (arrows) of the peptides derived from 293T cells with (bottom) or without (top) CK2α knockdown indicates a phosphoserine at position 223. (B) Effect of CK2α knockdown on PAK1 phosphorylation in vivo. Western blot analysis of expression of GFP-PAK1-KD, CK2α, and actin in 293T cells with (sh1 and sh2) or without (NT) CK2α knockdown (WB). The 293T cells expressing GFP-PAK1-KD were metabolically labeled with [32P]orthophosphate. GFP-PAK1-KD was immunoprecipitated with anti-GFP antibody and analyzed by autoradiography. (C) Effect of the CK2 inhibitor TBB on PAK1 phosphorylation in vivo. The 293T cells expressing GFP-PAK1-KD were treated with different concentrations of TBB, as indicated above each lane, and metabolically labeled with [32P]orthophosphate. (D) In vitro phosphorylation of PAK1 by CK2. Bacterially expressed GST-PAK1-KD or GST-PAK1-KD-S223A was incubated with purified CK2 holoenzyme (20 ng) and [32P]γ-ATP and analyzed by autoradiography. (E) Effect of the CK2 inhibitor heparin on PAK1 phosphorylation in vitro. Bacterially expressed GST-PAK1-KD was incubated with purified CK2 holoenzyme (20 ng) and [32P]γ-ATP in the absence and presence of different amounts of heparin (lane 3: 25 μg; lane 4: 5 μg; lane 5: 1 μg) for 30 min and analyzed by autoradiography.
To obtain further evidence of CK2-catalyzed PAK1S223 phosphorylation, we infected 293T cells with lentivirus expressing GFP-PAK1 and metabolically labeled them with [32P]orthophosphate. Because wild-type PAK1 possesses autophosphorylation activity (Manser et al., 1994), the kinase-dead (KD) PAK1 (PAK1K299R) was generated with a point mutation (GST-PAK1-KD) and used for phosphorylation assays. We found that CK2α knockdown does not affect PAK1 expression but significantly decreases PAK1 phosphorylation (Figure 1B) and that phosphorylation of GFP-PAK1-KD is inhibited by treatment of the CK2 inhibitor TBB (Figure 1C). Purified CK2α was shown to phosphorylate bacterially produced wild-type PAK1 protein (GST-PAK1-KD) but not mutant PAK1 protein (GST-PAK1S223A-KD) in vitro (Figure 1D); however, this phosphorylation was also significantly inhibited by treatment of the CK2 inhibitor heparin in a dose-dependent manner (Figure 1E).
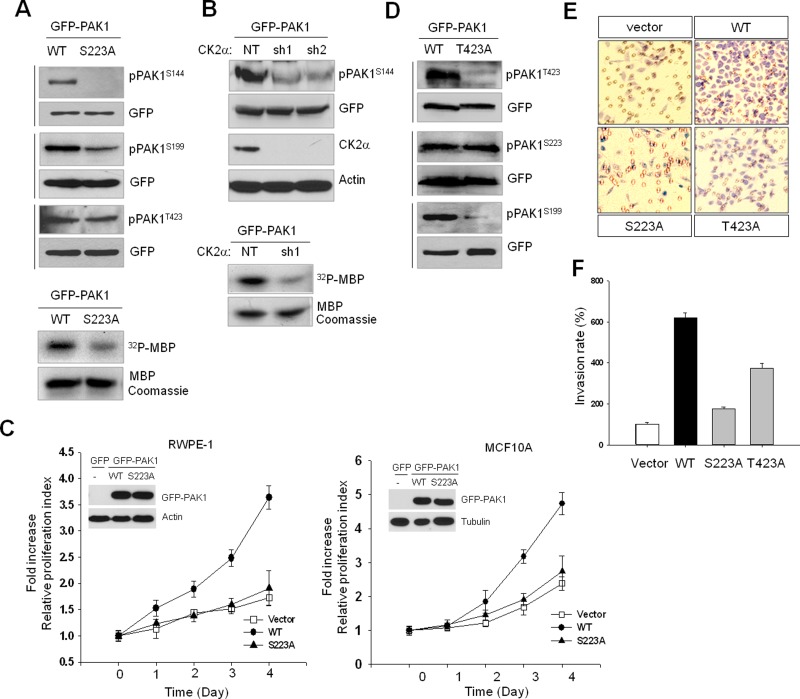
PAK1 activity is critically regulated by CK2-dependent S223 phosphorylation
Given that increased PAK1 activity is associated with autophosphorylation of specific sites, including S144 and S199 (Chong et al., 2001), we examined whether S223 phosphorylation affects the kinase activity of PAK1. Our results showed that S223A mutation or CK2α knockdown inhibits phosphorylation of those residues (Figure 2, A and B, top panels) and reduces PAK1 kinase activity, as determined by in vitro phosphorylation assay using myelin basic protein (MBP) as a substrate (Figure 2, A and B, bottom panels). We further examined the significance of PAK1S223 phosphorylation in cell growth and found that proliferation of human prostate RWPE-1 (Figure 2C, left) and human breast MCF10A (Figure 2C, right) cells was enhanced by expression of wild-type PAK1 but not of mutant PAK1 (PAK1S223A). Similar observations were made in mouse NIH 3T3 and Rat1 cells (Figure S2), suggesting that S223 phosphorylation may be a general requirement for PAK1 activation. Phosphorylation of T423 in the activation loop of PAK1 is important for PAK1 activation, but this phosphorylation does not readily occur; rather, it is catalyzed by PDK1 at the plasma membrane in a GTPase-independent manner (King et al., 2000). S223A mutation did not affect T423 phosphorylation (Figure 2A); T423A mutation also did not regulate S223 phosphorylation (Figure 2D), suggesting that S223 and T423 are phosphorylated by different signaling pathways. However, in a comparison of the effects of S223A and T423A mutations on PAK1-induced cell invasion, we found that S223A mutation is more severe than T423A mutation (Figure 2, E and F).
FIGURE 2:
CK2-catalyzed PAK1S223 phosphorylation is required for autophosphorylation and activity of PAK1. The 293T cells were transiently transfected with wild-type or mutant PAK1 protein, and cell extracts were subjected to Western blot analysis with phospho-specific PAK1 antibodies. The same blots were stripped and reprobed with anti-GFP antibody to control for equal protein loading. (A) Western blot analysis of phosphorylation of S144, S199, and T423 of GFP-PAK1 (WT) or GFP-PAK1S223A. The kinase activity of PAK1 (GFP-PAK1 [WT]) and GFP-PAK1S223A) was assessed by in vitro phosphorylation assay using MBP as a substrate (bottom). (B) Western blot analysis of phosphorylation of GFP-PAK1 expressed in 293T cells with (sh1 and sh2) or without (NT) CK2α knockdown with anti-pPAK1S144 antibody (pPAK1S144). Top, Western blot analysis of expression of CK2α (CK2α). Actin was used as a loading control (Actin). Bottom, in vitro phosphorylation of MBP by GFP-PAK1 immunoprecipitated with anti-GFP antibody from extracts of 293T cells with (sh1) or without (NT) CK2α knockdown. (C) MTT proliferation assays of RWPE-1 (left) and MCF 10A (right) cells. Cell viability over time was expressed as fold increase over untreated control at day 1. Error bars represent mean ± SD (n = 5). Insets: Western blot analysis of expression of GFP-PAK1 (WT) or GFP-PAK1S223A. (D) Western blot analysis of phosphorylation of T423, S223, and S199 of GFP-PAK1 (WT) or GFP-PAK1T423A. (E) PAK1-induced cell invasion. The benign prostate RWPE-1 cells infected with lentivirus expressing the vector plasmid, GFP-PAK1 (WT), GFP-PAK1S223A (S223A), or GFP- PAK1T423A (T423A) were used for invasion assays. (F) The invasion rate was determined by counting the cells that migrated through BME-coated inserts in the Transwell Boyden chamber and expressed as the percentage relative to control (vector). Each bar represents the mean ± SD of five fields counted.
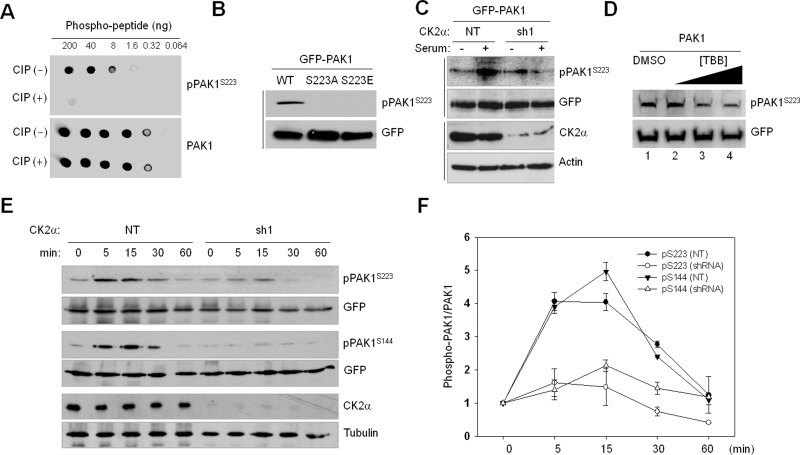
To directly assess the phosphorylation state of PAK1S223, we generated a phospho-PAK1 (S223) antibody (pPAK1S223) and determined the specificity of the antibody by dot blot analysis (Figure 3A) and by Western blot analysis of S223 phosphorylation of PAK1 (WT), PAK1S223A, and PAK1S223E (Figure 3B). Serum-stimulated phosphorylation of S223 was shown to be inhibited by CK2α knockdown (Figure 3C) or TBB treatment (Figure 3D), confirming that S223 of PAK1 is a CK2 target in vivo. We further examined the time course of PAK1 phosphorylation by CK2 and found that phosphorylation of S223 and S144 is enhanced by treatment of serum but inhibited by CK2α knockdown (Figure 3E) in a similar manner (Figure 3F).
FIGURE 3:
Verification of CK2-dependent PAK1S223 phosphorylation by Western blot analysis using anti-pPAK1S223 antibody. (A) Dot blot analysis of a newly generated phospho-PAK1 (S223) antibody (pPAK1S223). The phospho-PAK1 peptide (214–227; TRDVATSPI-pSer-PTEN) was untreated or treated with calf intestinal phosphatase and incubated with anti-pPAKS223 and anti-PAK1 antibodies. Signals were detected by chemiluminescence detection reagents. (B) Western blot analysis of phosphorylation of S223 of PAK1 (WT), PAK1S223A, and PAK1S223E. (C) GFP-PAK1–expressing 293T cells were treated with (sh1) or without (NT) CK2α knockdown. The cells were incubated for 1 d in the absence of serum and then cultured in the presence (+) or absence (−) of serum for 15 min. Cell lysates were subjected to Western blot analysis with anti-pPAK1S223. (D) Western blot analysis of phosphorylation of PAK1S223 in 293T cells treated with TBB (lane 2: 1 μM; lane 3: 5 μM; lane 4: 20 μM) with anti-pPAK1S223 or anti-GFP antibody. (E) 293T cells with (sh1) or without (NT) CK2α knockdown were infected with lentivirus expressing GFP-PAK1. Cells were incubated for 1 d in the absence of serum and then cultured in the presence of serum for different periods of time, as indicated above each lane, and cell extracts were subjected to Western blot analysis with anti-pPAK1S223 or anti-pPAK1S144 antibody. The same membrane was then stripped and reprobed with anti-PAK1 antibody. (F) Densitometry analysis of results shown in (E). The signals of pPAK1S223 in the immunoblot were quantified by scanning densitometry and normalized to the signals of total GFP-PAK1. Each point represents the mean value of three independent assays, with error bars indicating the SD.
CK2-catalyzed PAK1S223 phosphorylation may constitute a novel mechanism for regulation of PAK1 activity
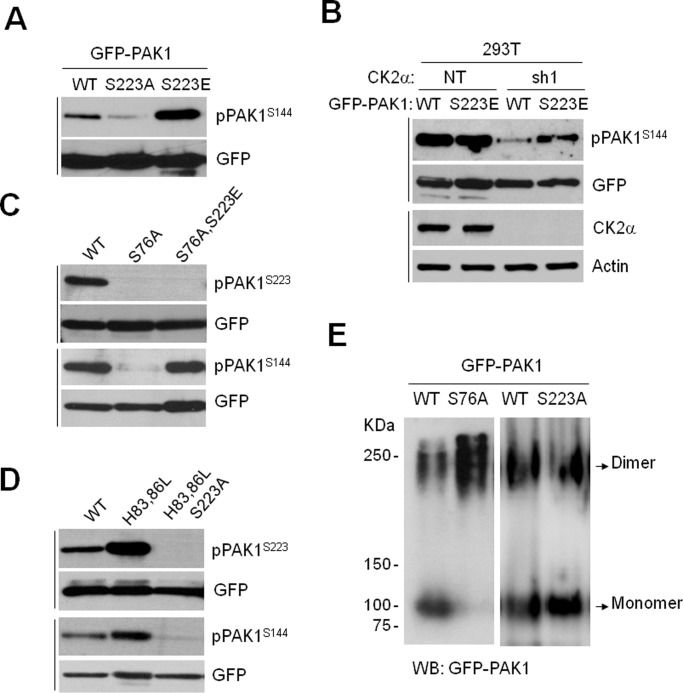
To understand the role of PAK1S223 phosphorylation in the regulation of PAK1 activity, we first assessed the autophosphorylation activity of the PAK1 mutant with glutamic acid substituted at S223 (PAK1S223E). The results showed that S144 phosphorylation of PAK1S223E is strongly detected (Figure 4A) and, more importantly, that this phosphorylation occurs independent of CK2 (Figure 4B). Thus, the S223E mutation likely mimics phosphorylation of S223, suggesting that this mutation may convert PAK1 into a constitutively active form. The binding of PAK1 to Cdc42 and Rac to the CRIB domain results in release of the autoinhibition and conversion of the dimeric PAK1 to a monomeric form (Lei et al., 2000). Therefore, a mutation of S76 (S76A) located in the CRIB domain renders PAK1 inactive by impairing its binding to GTPases (Zhao et al., 1998). We found that the S76A mutation impairs phosphorylation of S223 and S144, but that this inhibition is abolished by the S76A/S223E double mutation (Figure 4C, pPAK1S144). Notably, the dimer-to-monomer conversion of PAK1 was inhibited by the S76A mutation, whereas it was not affected by the S223A mutation (Figure 4E). Therefore, it is likely that S223 becomes available for phosphorylation upon the binding of GTPases to the CRIB domain, leading to release of the autoinhibitory conformation of PAK1. In addition, we confirmed that the S223A mutation does not impair the binding of PAK1 to Rac1 (Figure S3A) and that CK2 does not regulate Rac1 activity (Figure S3B).
FIGURE 4:
CK2-dependent PAK1S223 phosphorylation is required for PAK1 activation by both GTPase-dependent and GTPase-independent mechanisms. (A) Western blot analysis of phosphorylation of S144 of PAK1 (WT), PAK1S223A, and PAK1S223E. The same blots were stripped and reprobed with anti-GFP antibody to control for equal protein loading. (B) GFP-PAK1 (WT) or GFP-PAK1S223E was expressed in 293T cells with (sh1) and without (NT) CK2α knockdown, and cell extracts were subjected to Western blot analysis with anti-pPAK1S144 or anti-GFP antibody. (C) Western blot analysis of phosphorylation of S144 and S223 of GFP-PAK1 (WT), GFP-PAK1S76A, and GFP-PAK1S76A/S223E. (D) Western blot analysis of phosphorylation of S144 and S223 of GFP-PAK1 (WT), GFP-PAK1H83,86L, or GFP-PAK1H83,86L/S223A. (E) The 293T cells were transiently transfected with GFP-PAK1 (WT), GFP-PAK1S76A, or with GFP-PAK1S223A, and cell extracts were resolved in 8% native PAGE gels. The immunoblots were probed with anti-GFP antibody.
We also examined whether S223 phosphorylation is required for PAK1 activation in a GTPase-independent manner. The H83,86L double mutation impairs the binding of PAK1 to GTPases but does not inhibit its activity, illustrating that this mutation mimics activation by GTPases and that PAK1 can also be activated in a GTPase-independent manner (Sells et al., 1997). We found that the effect of the double mutation (H83,86L) on S144 phosphorylation is abolished by the additional mutation of S223A (Figure 4D, H83,86L/S223A).
CK2 is responsible for epidermal growth factor (EGF)-induced PAK1 activation
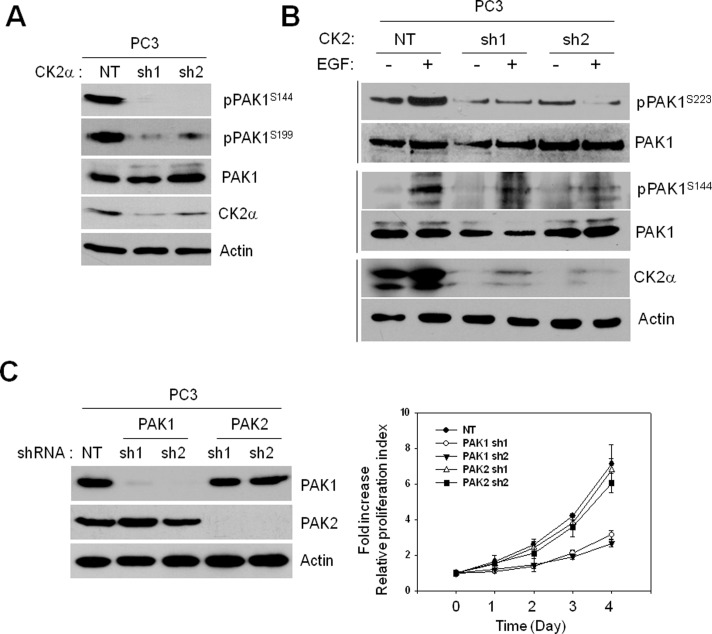
To understand the regulation of PAK1 activity by S223 phosphorylation in cancer cell growth, we first assessed autophosphorylation of endogenous PAK1 in the prostate cancer cell line PC3. CK2α knockdown was shown to decrease phosphorylation of S144 and S199, while it did not affect PAK1 expression (Figure 5A). Recent studies show that CK2 is a component of the kinase suppressor of Ras (KSR1) scaffolding complex associated with the plasma membrane that facilitates the ERK cascade (Ritt et al., 2007) and that the activity of a subcellular portion of CK2 is enhanced by epidermal growth factor receptor (EGFR)-ERK signaling (Ji et al., 2009). Thus, we examined the possible connection between PAK1 and the EGFR-ERK-CK2 pathway by determining the deletion effect of CK2α on the PAK1 autophosphorylation in EGF-stimulated PC3 cells. The results showed that CK2α knockdown inhibits EGF-induced PAK1 phosphorylation at both S223 and S144 (Figure 5B).
FIGURE 5:
CK2-dependent PAK1 phosphorylation is critical for growth of the prostate cancer PC3 cells. (A) Cell extracts of PC3 cells with (sh1) or without (NT) CK2α knockdown were subjected to Western blot analysis with anti-pPAK1S144, anti-PAK1S199, anti-PAK1, anti-CK2α, or anti-actin antibody. (B) PC3 cells with (sh1) or without (NT) CK2α knockdown were incubated for 1 d in the absence of serum and then cultured in the presence (+) or absence (−) of EGF (100 ng/ml) for 1 h, and total cell extracts were subjected to Western blot analysis with anti-pPAK1S223, anti-PAK1S144, anti-CK2α, or anti-actin antibody. (C) Western blot analysis of expression of PAK1 and PAK2 in PC3 cells with (sh1 and sh2) or without (NT) PAK1 or PAK2 knockdown (left) and MTT proliferation assay of the PC3 cells (right).
PAK1 is the major isoform of PAKs expressed in most cancers (Ye and Field, 2012). The fact that S223 of PAK1 is not conserved in other PAK family members (Figure S1C) led to an idea that CK2-mediated PAK1 phosphorylation enables PAK1 to be the principal PAK isoform in cancer cells. To test the relative significance of PAKs in tumor cell growth, we investigated the effect of PAK1 and PAK2 knockdowns on the growth of the prostate cancer cell line PC3. The results indicated that knockdown of PAK1, but not of PAK2, is sufficient to inhibit the growth of PC3 cells (Figure 5C) and are consistent with previous reports that PAK1 promotes prostate tumor growth (Goc et al., 2013).
Inhibition of S223 phosphorylation abrogates PAK1-mediated malignant transformation
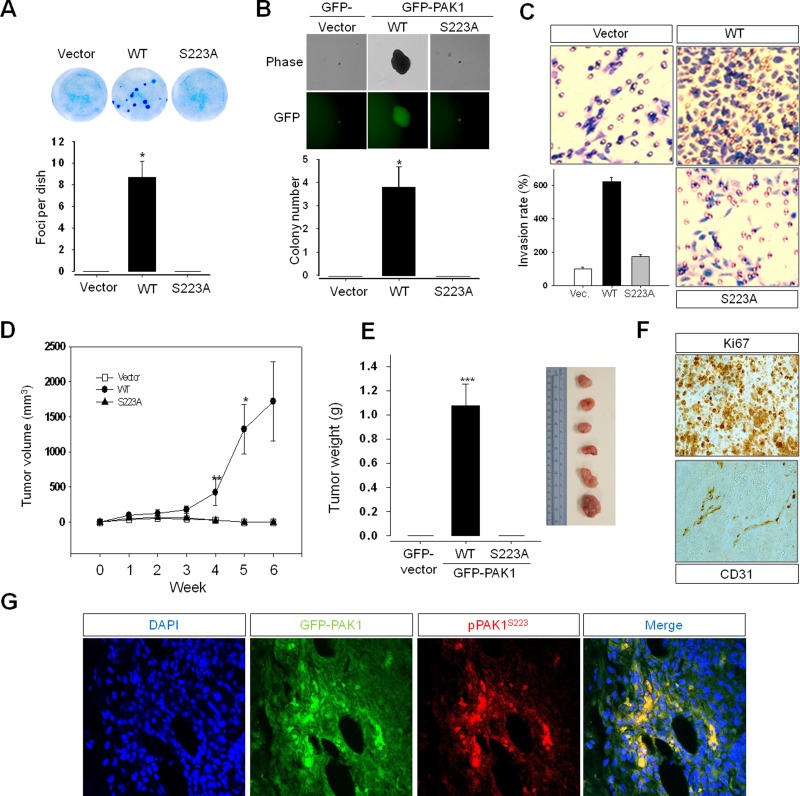
Overexpression of an activated mutant form of PAK1 leads to the stimulation of anchorage-independent growth in breast cancer cell lines (Vadlamudi et al., 2000) and is sufficient for the formation of mammary gland tumors in a transgenic mouse model (Wang et al., 2006b). To further understand the significance of S223 phosphorylation in PAK1-mediated cellular transformation, we compared the ability of wild-type (GFP-PAK1) and mutant (GFP-PAK1-S223A) PAK1 proteins to transform the human prostate epithetical cell line RWPE-1 to a tumorigenic phenotype in culture (Figure 2C, left, inset). Cell proliferation was enhanced by expression of wild-type PAK1 but not of mutant PAK1 (PAK1S223A), as shown in 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT; Figure 2C, left) and foci-forming (Figure 6A) assays, respectively. It was also demonstrated that, in contrast to expression of PAK1, expression of PAK1S223A is neither able to induce anchorage-independent growth of (Figure 6B) nor to confer an invasive phenotype to (Figure 6C) RWPE-1 cells. For investigation of the transforming activity of PAK1 in vivo, RWPE-1 cells expressing PAK1 or PAK1S223A were subcutaneously injected into the flanks of nude mice (Figure S4), and the mice were killed 6 wk afterward. Progressive tumor growth was observed in mice injected with RWPE-1 cells expressing PAK1 but not PAK1S223A (Figure 6, D and E). Immunohistochemical analysis of the tumor tissues dissected from the mice demonstrated the expression of GFP-PAK1, the proliferation marker Ki67 and the angiogenic marker CD31 (Figure 6F), and the phosphorylation of PAK1S223 (Figure 6G). These results establish for the first time that PAK1 overexpression is sufficient for the malignant transformation of epithelial prostate cells and support the view that S223 phosphorylation is critical for the transforming activity of PAK1.
FIGURE 6:
S223A mutation abrogates PAK1-mediated malignant transformation of prostate epithelial cells in vitro and in vivo. The benign prostate RWPE-1 cells infected with lentivirus expressing the vector plasmid, GFP-PAK1, or GFP-PAK1S223A (S223A) were used for experiments shown in (A) through (E). (A) Focus-formation assays. Quantitative analysis of foci numbers (mean ± SD) in week 3 (n = 4). (B) Anchorage-independent growth assay. Cells (RWPE-1) expressing GFP-vector, GFP-PAK1 (WT), or GFP-PAK1S223A (S223A) were visualized by phase-contrast (Phase) and fluorescence (GFP) microscopy. Quantitative analysis of colony numbers (mean ± SD) in week 3 is shown (n = 4). (C) Invasion assay. The invasion rate was determined by counting the cells that migrated through BME-coated inserts in the Transwell Boyden chamber and expressed as the percentage relative to control (vector). Each bar represents the mean ± SD of five fields counted. (D) Tumor growth determined by measuring the volumes of each tumor every 7 d after injection, as described in Materials and Methods. (E) The average tumor weight (left) and the representative tumors (right) of the GFP-PAK1 (WT) group mice killed 6 wk after injection. (F and G) Tumor tissues dissected from the mice (E) were stained with anti-Ki67, anti-CD31, or anti-pPAK1S223 antibody. Nuclei were stained with DAPI (blue).
DISCUSSION
The functional significance of PAK1 in cell migration and survival has long been appreciated (Kumar et al., 2006), but the underlying mechanism of PAK1 activation is not clearly understood. This is mainly due to the fact that PAK1 can be activated in response to a variety of external signals by GTPase-dependent and GTPase-independent mechanisms. The critical question in these mechanisms is how these signals are integrated to bring about efficient activation of PAK1. In this study, we provide evidence that CK2 has an important role in integrating these signals and functions as an upstream activating kinase of PAK1: 1) CK2 phosphorylates PAK1 at S223 in vivo and in vitro (Figure 1); 2) S223 phosphorylation plays a key role in the final step of the PAK1 activation process (Figures 2–4); 3) this phosphorylation is unique to PAK1 among PAK family members, but sufficient for oncogenic conversion of PAK1 (Figures 5 and 6). PAK1, as a key regulator of cell adhesion and motility, is targeted to the plasma membrane upon stimulation (Parrini et al., 2005). Once recruited, PAK1 associates with Cdc42 and Rac1 and is activated at the membrane. CK2 is ubiquitously expressed but redistributed to various cellular compartments upon stimulation (Meggio and Pinna, 2003); its recruitment to the plasma membrane is mediated by the CK2α interacting partner CKIP-1 (Canton et al., 2006). Thus, CK2-dependent PAK1 activation may provide a framework for spatiotemporal regulation of PAK1, as proposed previously (Parrini et al., 2005). Sphingosine positively regulates CK2 activity (McDonald et al., 1991) and activates PAK1 in a GTPase-independent manner (Zenke et al., 1999); moreover, sphingosine 1-phosphate, a derivative of sphingosine, activates ERK1/2 (Maupas-Schwalm et al., 2004; Catarzi et al., 2011), which in turn phosphorylates and activates CK2 (Ji et al., 2009). These observations may support our findings that CK2 is responsible for EGF-induced PAK1 activation and the view that CK2 plays a key role in integrating both GTPase-dependent and GTPase-independent signals to regulate PAK1 activity.
The process of PAK1 activation entails two major events: 1) conversion of a PAK1 dimer to a monomer and relief of autoinhibition; and 2) activation of the catalytic domain and autophosphorylation at specific residues for full activation of PAK1. A phosphomimetic mutation (S223E) bypasses the requirement for GTPases in PAK1 activation, suggesting that CK2 phosphorylation of PAK1S223 may be a key regulatory step for PAK1 activation. In addition, membrane-targeted PAK1 can be activated in the absence of stimulation of Rho family GTPases (Lu et al., 1997), supporting the view that aberrant activation of this phosphorylation may bypass the requirement for GTPases. The PAK1 mutant (PAK1H83,86L) postulated to mimic GTPase-induced structural changes is constitutively active (Sells et al., 1997); however, the kinase activity of the activated PAK1 mutant is abolished by inhibiting S223 phosphorylation of the protein (Figure 4). Thus, S223 likely becomes available for phosphorylation by CK2 after conformational changes induced by GTPase-dependent and GTPase-independent stimuli. This notion may be supported by the observation that PAK1 contains potential phosphorylation sites that become accessible upon activation by Cdc42 or Rac (Hoffman et al., 2000). Based on our observations, we surmise that S223 phosphorylation may be important for locking PAK1 in a catalytically competent conformation and keeping it in an activated state by serving as a priming event for subsequent phosphorylation. We anticipate that a structural analysis would yield a better understanding of the exact role of S223 phosphorylation in PAK1 activation.
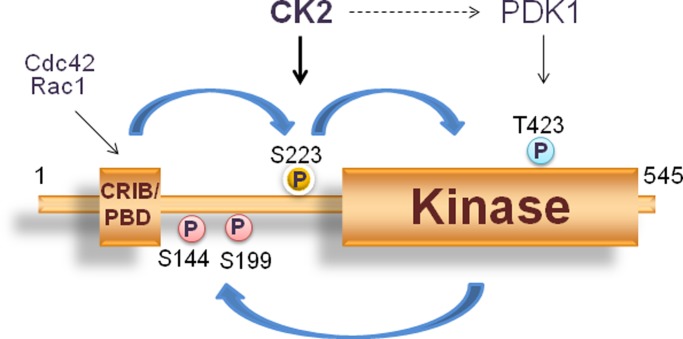
PAK1 phosphorylation at T423 in the activation loop in the kinase domain, which is mediated by PDK1 (Zenke et al., 1999; King et al., 2000), plays a role in stabilizing the active site (Lei et al., 2005). The tumor suppressor phosphatase and tensin homologue deleted on chromosome 10 (PTEN) dephosphorylates phosphatidylinositol-3,4,5-phosphate, down-regulating the PI3K/Akt signaling; CK2 phosphorylates PTEN and reduces PTEN activity (Vazquez et al., 2000; Torres and Pulido, 2001; Al-Khouri et al., 2005), leading to activation of PDK1 (Ruzzene and Pinna, 2010). Hence, it may be possible that, in addition to phosphorylation at S223, CK2 also regulates PAK1 activity by promoting phosphorylation of T423 via the PI3K/Akt signaling (Figure 7).
FIGURE 7:
A proposed model for the role of PAK1S223 phosphorylation in PAK1 activation. PAK1 activation involves conformational changes that lead to the conversion of an inactive dimeric PAK1 to an active monomeric form. This conversion, however, is not sufficient to bring about PAK1 activation; it is required for PAK1 phosphorylation at S223 by CK2. This phosphorylation is thought to lock the PAK1 kinase domain in a catalytically competent form. S223 phosphorylation does not directly affect phosphorylation of T423; however, the possibility is not excluded that CK2 might activate PDK1 by activating the PI3K/Akt signaling via PTEN and positively regulate T423 phosphorylation.
Activation of oncogenic kinases by elevated expression is a common feature of human primary tumors. However, there is no significant difference in PAK1 expression between normal and cancerous prostate cells. A similar pattern of PAK1 expression is also observed in various cell types, including pancreas, kidney, lung, thymus, bone marrow, and brain (GeneCards). These observations suggest that hyperactivation of PAK1 in these cancer cells is not due to overexpression of PAK1, but rather is due to the conversion of PAK1 to a hyperactive form, and thus strongly support our finding that this conversion is mediated by CK2-dependent PAK1 phosphorylation. It is interesting to note that PDK1 phosphorylation of PAK1 at T423 is involved in actin polymerization in normal cells (King et al., 2000), but does not significantly affect tumor cell growth (Lu et al., 2010). In addition, overexpression of the artificially activated PAK1 mutant, PAK1T423E, leads to stimulation of anchorage-independent growth in breast cancer cell lines (Vadlamudi et al., 2000) and breast carcinomas in transgenic mice (Wang et al., 2006a); however, it does not suffice to draw clinically relevant conclusions (Kichina et al., 2010). These observations might highlight the significance of CK2 phosphorylation of S223 in regulating PAK1 activity. PAK1 overexpression appears to be sufficient to induce malignant transformation of prostate epithelial cells, but blocking PAK1S223 phosphorylation leads to growth inhibition of the tumor cells (Figure 6). These findings reinforce the view that PAK1 activation in prostate cancer cells may rely on CK2 activity. CK2 level is increased in most tumor tissues and acts as an anti-apoptotic and prosurvival agent that confers a number of selective advantages to tumor cells. In this sense, our findings establish that PAK1 phosphorylation by CK2 may play a fundamental role in the activation and perpetuation of PAK1 oncogenic signaling, as expression of PAK1 targets is robust in cancer cells.
MATERIALS AND METHODS
Cell culture
Benign prostate epithelial cell line RWPE-1 cells were grown in keratinocyte serum-free medium (K-SFM) containing bovine pituitary extract and EGF, as described previously (Shin and Kim, 2012). The human embryonic kidney cell line 293T and the human prostate cancer cell line PC3 were cultured in RPMI 1640 or DMEM containing 10% fetal bovine serum (FBS) and penicillin/streptomycin at 37°C in a humidified atmosphere of 5% CO2.
Cell invasion
Cell invasion assay was performed using the cell invasion kit (Transwell Boyden's chamber with Transwell Permeable Support Inserts Coated with Cultrex basement membrane extract [BME]; Corning Costar, Cambridge, MA) according to the manufacturer's instruction (Shin and Kim, 2012).
Soft agar assay
For soft agar assays, anchorage-independent growth was determined in six-well plates. A lower-layer plug of 0.6% agar solution in K-SFM containing 2 μg/ml of puromycin was prepared. This was overlaid with 3 ml of a 0.3% agar solution, also in RWPE-1, with 3 × 104 lentivirally infected cells. Cells were then placed in a 37°C and 5% CO2 incubator. Colonies were allowed to grow over the course of 2 wk. After MTT treatment for 4 h, colonies were counted under a microscope (Nikon Eclipse Ti; Melville, NY). All assays were performed in triplicate.
Generation of a phospho-PAK1 (S223) antibody
Mouse studies were performed in accordance with the policies of the George Washington University Institutional Animal Care and Use Committee (IACUC protocol A257). Peptides based on the human PAK1 sequence (214–227; TRDVATSPI-pSer-PTEN) were synthesized by solid state synthesis, conjugated to keyhole limpet hemocyanin (KLH; Biomatik), and used for immunization. All mice were obtained from the Jackson Laboratory (Bar Harbor, ME). After three intraperitoneal injections, antiserum was obtained from the cardiac blood of the mice and tested with indirect enzyme-linked immunosorbent assay against free peptide, peptide-KLH, and KLH.
Lentiviral infection
Lentiviral expression vectors for wild-type PAK1-GFP and mutant PAK1S223A-GFP were constructed by subcloning corresponding cDNAs into pLV-puro lentiviral vector, as described previously (Shin and Kim, 2012). Lentiviral short hairpin RNAs (shRNAs) specific for CK2α (NM_177559) were obtained from Sigma-Aldrich (MISSION shRNA; St. Louis, MO). Biologically active shRNAs were generated from the pLKO.1-puro vector, utilizing the Polymerase III U6-RNA promoter. Transfection was performed as described previously (Shin and Kim, 2012).
Protein phosphorylation and Western blot assays
For in vitro protein phosphorylation assay, GST-PAK1 proteins were harvested from Escherichia coli cell extracts with glutathione Sepharose 4B beads. After being equilibrated with kinase buffer (20 mM HEPES, pH 7.0, 10 mM MgCl2, 1 mM dithiothreitol, 100 μM cold ATP), the beads were mixed with purified CK2 kinase (20 ng) and 0.5 μCi of [32P]γ-ATP and incubated at 30°C for 30 min. For in vivo protein phosphorylation assay, 293T cells were first starved for phosphate in phosphate-free DMEM containing 10% FBS for 2 h. The medium was again replaced with fresh phosphate-free medium and 500 μCi of [32P]orthophosphate per 100-mm dish, and cells were incubated at 37°C for 6 h. GST-PAK1 proteins were resolved by SDS–PAGE and subjected to autoradiography. Western blot analysis was performed, as described previously (Shin and Kim, 2012).
MTT cell proliferation and primary focus formation assays
For MTT cell proliferation assay, cell growth was evaluated by replacing the culture media with 200 μl of 0.5 mg/ml MTT media solution after incubation for 1–4 d. For primary-focus formation assays, RWPE-1 cells were plated at a density of 2 × 105 cells/well on a six-well plate. The next day, cells were transiently transfected with 100 ng of plasmids encoding GFP-PAK1 or GFP-PAK1S223A, and the appearance of PAK1-induced foci of transformed cells was quantitated after 14 d of transfection. Cell invasion was performed as described previously (Shin and Kim, 2012).
Tumor xenograft assay
All mouse studies were performed in accordance with the policies of the George Washington University Institutional Animal Care and Use Committee (IACUC protocol A244). Xenografts were carried out by a flank subcutaneous injection of 5- to 6-wk-old athymic (nu/nu) nude mice (Jackson Laboratory) with 4 × 106 of RWPE-1 cells (200 μl) infected with control, GFP-PAK1, or GFP-PAK1S223A plasmid (n = 8 for each group). For injection, RWPE-1 cells were suspended in a 1:1 solution of growth media Matrigel matrix. Tumor size was measured, and tumor volumes were calculated using the formula: tumor volume = (W2 × L)/2, where W is width and L is length in mm. Xenograft tumor tissues were Formalin-fixed and embedded in paraffin, as described previously (Wang et al., 2006a).
Immunohistochemistry
Xenograft tumor tissues were Formalin-fixed and embedded in paraffin, as described previously (Shin and Kim, 2012). Seven-micron-thick serial sections were cut from paraffin blocks, mounted on acid-cleaned glass slides, and heated at 55°C for 60 min. After deparaffinization and rehydration, tissue section slides were blocked for endogenous peroxidase in 3% H2O2 in methanol for 20 min. Slides were incubated with 10 mM citrate buffer (pH 6.0) at 95–100°C for 20 min and allowed to cool for 30 min. After being washed, slides were incubated with 5% goat serum to reduce nonspecific background staining. Finally, the slides were incubated with anti-Ki67 and CD31 antibodies. Tumor tissue images were taken by a Nikon Eclipse Ti inverted microscope and analyzed using the NIS-Elements BR software. For immunofluorescence staining, the slides were incubated with anti-pPAK(Ser223) antibody for 2 d at 4°C. After being washed, they were incubated with Alexa Fluor 594 goat anti-mouse immunoglobulin G at a 1/2000 dilution for 1 h. After being mounted with mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI [H-1200]; Vector Laboratories, Burlingame, CA), the cells were photographed using a fluorescence microscope (IX71 microscope; Olympus Imaging America).
Statistics
In all experiments, statistical significance was defined by p values: *, p < 0.05; **, p < 0.005; ***, p < 0.001 (as compared with control).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant GM087470 (J.-H.K.) from the National Institute of General Medical Science.
Abbreviations used:
- AID
autoinhibitory domain
- BME
basement membrane extract
- CRIB domain
Cdc42/Rac-interacting domain
- DAPI
4′,6-diamidino-2-phenylindole
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- KD
kinase-dead
- KLH
keyhole limpet hemocyanin
- K-SFM
keratinocyte serum-free medium
- KSR1
kinase suppressor of Ras
- MBP
myelin basic protein
- MTT
3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide
- PAK
P21-activated kinase
- PBD
p21-binding domain
- PTEN
phosphatase and tensin homologue deleted on chromosome 10
- shRNA
short hairpin RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-04-0204) on July 24, 2013.
REFERENCES
- Al-Khouri AM, Ma Y, Togo SH, Williams S, Mustelin T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3β. J Biol Chem. 2005;280:35195–35202. doi: 10.1074/jbc.M503045200. [DOI] [PubMed] [Google Scholar]
- Arias-Romero LE, Chernoff J. A tale of two Paks. Biol Cell. 2008;100:97–108. doi: 10.1042/BC20070109. [DOI] [PubMed] [Google Scholar]
- Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- Bokoch GM, Wang Y, Bohl BP, Sells MA, Quilliam LA, Knaus UG. Interaction of the Nck adapter protein with p21-activated kinase (PAK1) J Biol Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- Canton DA, Olsten ME, Niederstrasser H, Cooper JA, Litchfield DW. The role of CKIP-1 in cell morphology depends on its interaction with actin-capping protein. J Biol Chem. 2006;281:36347–36359. doi: 10.1074/jbc.M607595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarzi S, Romagnoli C, Marcucci G, Favilli F, Iantomasi T, Vincenzini MT. Redox regulation of ERK1/2 activation induced by sphingosine 1-phosphate in fibroblasts: involvement of NADPH oxidase and platelet-derived growth factor receptor. Biochim Biophys Acta. 2011;1810:446–456. doi: 10.1016/j.bbagen.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Chong C, Tan L, Lim L, Manser E. The mechanism of PAK activation. Autophosphorylation events in both regulatory and kinase domains control activity. J Biol Chem. 2001;276:17347–17353. doi: 10.1074/jbc.M009316200. [DOI] [PubMed] [Google Scholar]
- Cory GO, Cramer R, Blanchoin L, Ridley AJ. Phosphorylation of the WASP-VCA domain increases its affinity for the Arp2/3 complex and enhances actin polymerization by WASP. Mol Cell. 2003;11:1229–1239. doi: 10.1016/s1097-2765(03)00172-2. [DOI] [PubMed] [Google Scholar]
- Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho.ELM: a database of phosphorylation sites–update 2008. Nucleic Acids Res. 2008;36:D240–D244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- Field J, Manser E. The PAKs come of age: celebrating 18 years of discovery. Cell Logist. 2012;2:54–58. doi: 10.4161/cl.22084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goc A, Al-Azayzih A, Abdalla M, Al-Husein B, Kavuri S, Lee J, Moses K, Somanath PR. P21 activated kinase-1 (Pak1) promotes prostate tumor growth and microinvasion via inhibition of transforming growth factor β expression and enhanced matrix metalloproteinase 9 secretion. J Biol Chem. 2013;288:3025–3035. doi: 10.1074/jbc.M112.424770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GR, Nassar N, Cerione RA. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- Ji H, et al. EGF-induced ERK activation promotes CK2-mediated disassociation of α-catenin from β-catenin and transactivation of β-catenin. Mol Cell. 2009;36:547–559. doi: 10.1016/j.molcel.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichina JV, Goc A, Al-Husein B, Somanath PR, Kandel ES. PAK1 as a therapeutic target. Expert Opin Ther Targets. 2010;14:703–725. doi: 10.1517/14728222.2010.492779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CC, Gardiner EM, Zenke FT, Bohl BP, Newton AC, Hemmings BA, Bokoch GM. p21-activated kinase (PAK1) is phosphorylated and activated by 3-phosphoinositide-dependent kinase-1 (PDK1) J Biol Chem. 2000;275:41201–41209. doi: 10.1074/jbc.M006553200. [DOI] [PubMed] [Google Scholar]
- Kreis P, Rousseau V, Thevenot E, Combeau G, Barnier JV. The four mammalian splice variants encoded by the p21-activated kinase 3 gene have different biological properties. J Neurochem. 2008;106:1184–1197. doi: 10.1111/j.1471-4159.2008.05474.x. [DOI] [PubMed] [Google Scholar]
- Kumar R, Gururaj AE, Barnes CJ. p21-activated kinases in cancer. Nat Rev Cancer. 2006;6:459–471. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- Lei M, Lu W, Meng W, Parrini MC, Eck MJ, Mayer BJ, Harrison SC. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–397. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
- Lei M, Robinson MA, Harrison SC. The active conformation of the PAK1 kinase domain. Structure. 2005;13:769–778. doi: 10.1016/j.str.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Li F, Adam L, Vadlamudi RK, Zhou H, Sen S, Chernoff J, Mandal M, Kumar R. p21-activated kinase 1 interacts with and phosphorylates histone H3 in breast cancer cells. EMBO Rep. 2002;3:767–773. doi: 10.1093/embo-reports/kvf157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Katz S, Gupta R, Mayer BJ. Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Curr Biol. 1997;7:85–94. doi: 10.1016/s0960-9822(06)00052-2. [DOI] [PubMed] [Google Scholar]
- Lu Z, Cox-Hipkin MA, Windsor WT, Boyapati A. 3-Phosphoinositide-dependent protein kinase-1 regulates proliferation and survival of cancer cells with an activated mitogen-activated protein kinase pathway. Mol Cancer Res. 2010;8:421–432. doi: 10.1158/1541-7786.MCR-09-0179. [DOI] [PubMed] [Google Scholar]
- Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- Maupas-Schwalm F, Auge N, Robinet C, Cambus JP, Parsons SJ, Salvayre R, Negre-Salvayre A. The sphingomyelin/ceramide pathway is involved in ERK1/2 phosphorylation, cell proliferation, and uPAR overexpression induced by tissue-type plasminogen activator. FASEB J. 2004;18:1398–1400. doi: 10.1096/fj.03-1123fje. [DOI] [PubMed] [Google Scholar]
- McDonald OB, Hannun YA, Reynolds CH, Sahyoun N. Activation of casein kinase II by sphingosine. J Biol Chem. 1991;266:21773–21776. [PubMed] [Google Scholar]
- Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- Morreale A, Venkatesan M, Mott HR, Owen D, Nietlispach D, Lowe PN, Laue ED. Structure of Cdc42 bound to the GTPase binding domain of PAK. Nat Struct Biol. 2000;7:384–388. doi: 10.1038/75158. [DOI] [PubMed] [Google Scholar]
- Pandey A, Dan I, Kristiansen TZ, Watanabe NM, Voldby J, Kajikawa E, Khosravi-Far R, Blagoev B, Mann M. Cloning and characterization of PAK5, a novel member of mammalian p21-activated kinase-II subfamily that is predominantly expressed in brain. Oncogene. 2002;21:3939–3948. doi: 10.1038/sj.onc.1205478. [DOI] [PubMed] [Google Scholar]
- Parrini MC, Lei M, Harrison SC, Mayer BJ. Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol Cell. 2002;9:73–83. doi: 10.1016/s1097-2765(01)00428-2. [DOI] [PubMed] [Google Scholar]
- Parrini MC, Matsuda M, de Gunzburg J. Spatiotemporal regulation of the Pak1 kinase. Biochem Soc Trans. 2005;33:646–648. doi: 10.1042/BST0330646. [DOI] [PubMed] [Google Scholar]
- Ritt DA, Zhou M, Conrads TP, Veenstra TD, Copeland TD, Morrison DK. CK2 is a component of the KSR1 scaffold complex that contributes to Raf kinase activation. Curr Biol. 2007;17:179–184. doi: 10.1016/j.cub.2006.11.061. [DOI] [PubMed] [Google Scholar]
- Rudel T, Bokoch GM. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science. 1997;276:1571–1574. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- Ruzzene M, Pinna LA. Addiction to protein kinase CK2: a common denominator of diverse cancer cells. Biochim Biophys Acta. 2010;1804:499–504. doi: 10.1016/j.bbapap.2009.07.018. [DOI] [PubMed] [Google Scholar]
- Sells MA, Knaus UG, Bagrodia S, Ambrose DM, Bokoch GM, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- Shin YJ, Kim JH. The role of EZH2 in the regulation of the activity of matrix metalloproteinases in prostate cancer cells. PLoS One. 2012;7:e30393. doi: 10.1371/journal.pone.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, Viaud J, Rennefahrt UE, Anastassiadis T, Peterson JR. Phosphoinositides are essential coactivators for p21-activated kinase 1. Mol Cell. 2010;40:493–500. doi: 10.1016/j.molcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J, Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J Biol Chem. 2001;276:993–998. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Adam L, Wang RA, Mandal M, Nguyen D, Sahin A, Chernoff J, Hung MC, Kumar R. Regulatable expression of p21-activated kinase-1 promotes anchorage-independent growth and abnormal organization of mitotic spindles in human epithelial breast cancer cells. J Biol Chem. 2000;275:36238–36244. doi: 10.1074/jbc.M002138200. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Li F, Barnes CJ, Bagheri-Yarmand R, Kumar R. p41-Arc subunit of human Arp2/3 complex is a p21-activated kinase-1-interacting substrate. EMBO Rep. 2004;5:154–160. doi: 10.1038/sj.embor.7400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RA, Zhang H, Balasenthil S, Medina D, Kumar R. PAK1 hyperactivation is sufficient for mammary gland tumor formation. Oncogene. 2006a;25:2931–2936. doi: 10.1038/sj.onc.1209309. [DOI] [PubMed] [Google Scholar]
- Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, Goswami S, Bresnick AR, Condeelis JS. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol. 2006b;173:395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier CP, et al. Phosphorylation of CRN2 by CK2 regulates F-actin and Arp2/3 interaction and inhibits cell migration. Sci Rep. 2012;2:241. doi: 10.1038/srep00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye DZ, Field J. PAK signaling in cancer. Cell Logist. 2012;2:105–116. doi: 10.4161/cl.21882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke FT, King CC, Bohl BP, Bokoch GM. Identification of a central phosphorylation site in p21-activated kinase regulating autoinhibition and kinase activity. J Biol Chem. 1999;274:32565–32573. doi: 10.1074/jbc.274.46.32565. [DOI] [PubMed] [Google Scholar]
- Zhao ZS, Lim JP, Ng YW, Lim L, Manser E. The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Mol Cell. 2005;20:237–249. doi: 10.1016/j.molcel.2005.08.035. [DOI] [PubMed] [Google Scholar]
- Zhao ZS, Manser E, Chen XQ, Chong C, Leung T, Lim L. A conserved negative regulatory region in αPAK, inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol Cell Biol. 1998;18:2153–2163. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.