The low-complexity Drosophila genome includes previously uncharacterized SOCS36E, an ancestral SOCS4/5 homologue. It is shown that SOCS36E suppresses JAK/STAT signaling through two separate mechanisms: via receptor stability, mediated by the conserved SOCS-box domain, and via suppression of receptor phosphorylation that requires the N-terminal domain.
Abstract
Conserved from humans to Drosophila, the Janus kinase/signal transducer and activators of transcription (JAK/STAT) signaling cascade is essential for multiple developmental and homeostatic processes, with regulatory molecules controlling pathway activity also highly conserved. We characterize the Drosophila JAK/STAT pathway regulator SOCS36E and show that it functions via two independent mechanisms. First, we show that Drosophila Elongin B/C and Cullin-5 act via the SOCS-box of SOCS36E to reduce pathway activity specifically in response to ligand stimulation—a process that involves endocytic trafficking and lysosomal degradation of the Domeless (Dome) receptor. Second, SOCS36E also suppresses both stimulated and basal pathway activity via an Elongin/Cullin-independent mechanism that is mediated by the N-terminus of SOCS36E, which is required for the physical interaction of SOCS36E with Dome. Although some human SOCS proteins contain N-terminal kinase-inhibitory domains, we do not identify such a region in SOCS36E and propose a model wherein the N-terminal of SOCS36E blocks access to tyrosine residues in Dome. Our biochemical analysis of a SOCS-family regulator from a lower organism highlights the fundamental conserved roles played by regulatory mechanisms in signal transduction.
INTRODUCTION
The JAK/STAT signaling pathway plays a central role in many developmental processes and is a key regulator of homeostasis and immune responses (reviewed in Rawlings et al., 2004b; Arbouzova and Zeidler, 2006; Tamiya et al., 2011). The central component of the JAK/STAT pathway is the Janus kinase (JAK) family of proteins, which are associated with single-pass, transmembrane receptors at the plasma membrane (Haan et al., 2006). On ligand binding to their extracellular domains, receptors undergo conformational changes, resulting in activation of associated cytosolic JAKs and subsequent phosphorylation of the effectors of the pathway, the signal transducer and activators of transcription (STATs). Aberrant function of any element of the cascade can compromise the entire pathway and has been associated with both solid tumors and hematological malignancies (James et al., 2005; Levine and Wernig, 2006; Valentino and Pierre, 2006; Staerk et al., 2007; Jatiani et al., 2010; Tamiya et al., 2011). As a consequence, numerous regulators of pathway activity have emerged. Of these, the protein inhibitors of activated STATs, protein tyrosine phosphatases, and suppressors of cytokine signaling (SOCS) families represent the major classes of proteins involved in negative regulation and termination of JAK/STAT pathway signaling (Arbouzova and Zeidler, 2006; Stec and Zeidler, 2011; Tamiya et al., 2011; Dittrich et al., 2012; Alicea-Velázquez et al., 2013).
The mammalian SOCS family of proteins consists of eight members, SOCS1–7 and CIS, which act in a classical negative-feedback loop (Croker et al., 2008; Piessevaux et al., 2008; Yoshimura, 2009; Delgado-Ortega et al., 2013). At the molecular level, all SOCS proteins are characterized by a C-terminally located SOCS-box motif, via which Elongin B and Elongin C interact and subsequently recruit Cullin-5 and Rbx 1 (Babon et al., 2009). This complex, termed the Elongin-Cullin-SOCS (ECS) complex, acts as an E3 ubiquitin ligase mediating the transfer of a ubiquitin moiety from a donor E2 protein onto the substrate (Linossi and Nicholson, 2012). Thanks to a centrally located Src homology 2 (SH2) domain that mediates interaction with phosphorylated tyrosine (pTyr) residues, SOCS molecules fulfill the role of substrate recognition in the ECS complex. By contrast, the N-termini of SOCS proteins do not contain any recognizable domains and share low levels of conservation between family members. Exceptions to this are SOCS1 and SOCS3, which have a characteristic kinase-inhibitory region located immediately upstream of the SH2 domain (Nicholson et al., 1999; Sasaki et al., 1999; Piganis et al., 2011; Doti et al., 2012). Recently crystallographic studies have shown that the SOCS3 kinase-inhibitory region prevents substrates such as the associated cytokine receptor from binding to the activation loop of JAK2 (Kershaw et al., 2013).
The fruit fly Drosophila melanogaster represents a low-complexity model for a wide range of developmental, cellular, and molecular mechanisms, including the JAK/STAT pathway (Arbouzova and Zeidler, 2006). The Drosophila JAK/STAT pathway consists of a single receptor termed Domeless (Dome; Brown et al., 2001), a single JAK called Hopscotch (Hop; Binari and Perrimon, 1994), and a single STAT termed STAT92E (Hou et al., 1996; Yan et al., 1996). Furthermore, the Drosophila genome also encodes multiple negative regulators of pathway signaling, including three putative SOCS proteins, termed SOCS16D, SOCS36E, and SOCS44A (Callus and Mathey-Prevot, 2002; Rawlings et al., 2004a; reviewed in Stec and Zeidler, 2011). Of these, SOCS36E is the best-characterized family member, with closest homology to mammalian SOCS5. Transcription of socs36E mRNA is JAK/STAT pathway regulated (Karsten et al., 2002), and the resulting protein has been shown to negatively regulate both JAK/STAT and epidermal growth factor receptor (EGFR) signaling in vivo (Silver et al., 2005; Almudi et al., 2009; Herranz et al., 2012).
However, no molecular characterization of SOCS36E has been undertaken to date. In this paper, we describe our analysis of Drosophila SOCS36E and its two separable functions as a negative regulator of both basal and activated JAK/STAT pathway signaling.
RESULTS
Dome undergoes lysosomal degradation
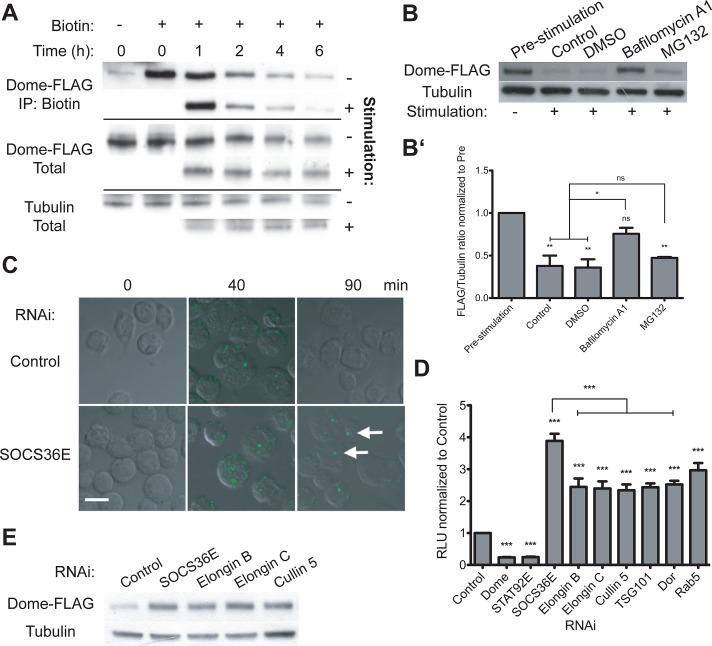
Ligand-mediated endocytosis of cytokine receptors results in either their proteosomal or lysosomal degradation or their recycling to the plasma membrane following ligand dissociation (Grant and Donaldson, 2009; Raiborg and Stenmark, 2009; Platta and Stenmark, 2011). Consistent with this, previous reports have indicated that binding of ligands to Dome on the plasma membrane is rapidly followed by endocytosis of the complex (Devergne et al., 2007; Vidal et al., 2010). Although both reports showed localization of the ligand–receptor complex to the late endosome, cargo localized in late endosomes can still be recycled to the trans-Golgi network via Rab7b and Rab9 (reviewed in Pfeffer, 2009). As a result, the ultimate fate of internalized Dome remains unclear. To study this aspect of Dome endocytosis, we treated cells expressing Dome-FLAG with biotin to label plasma membrane–localized Dome before stimulation with either Upd2–green fluorescent protein (Upd2-GFP) or mock-conditioned media while the addition of cyclohexamide prevented de novo protein synthesis throughout the experiment. We found that total Dome levels decrease at a similar rate with and without stimulation by Upd2-GFP–conditioned media, with an average decrease of 38% at 2 h (Figure 1A, middle two panels, n = 3). Considering the biotinylated fraction of Dome that was present at the plasma membrane during ligand treatment, a difference in degradation rate following stimulation with mock or ligand-conditioned media was evident at 2-h (18 vs. 30% decrease, respectively) and 6-h (58 vs. 65% decrease) time points (Figure 1A, top two panels). Overall, the time frame for endocytosis observed is in line with previous reports on ligand degradation (Devergne et al., 2007) and demonstrates that the degradation of Dome present at the plasma membrane is enhanced following ligand-mediated pathway stimulation.
FIGURE 1:
Dome stability and the role of Elongin B/C, Cullin-5, and SOCS36E. (A) Levels of total and biotinylated Dome-FLAG present in cyclohexamide-treated cells following stimulation with either mock or Upd2-GFP–conditioned medium at the indicated time points. (B and B′) Levels of Dome-FLAG following a 2-h stimulation, as indicated. By comparison with untreated and DMSO carrier controls, pre-incubation with 0.2 μM bafilomycin A1 prevented ligand-induced degradation of Dome, while treatment with 10 μM MG132 had no significant effect. (B′) The average (and SEM; **, p < 0.001; *, p < 0.05) of three experiments, with (B) showing a representative example. (C) Drosophila Kc167 cells treated with the indicated dsRNAs prior to stimulation with Upd2-GFP (green). After allowing for ligand internalization, low-pH washes removed remaining plasma-membrane-bound ligand prior to incubation for the indicated times. By contrast with controls, cells lacking SOCS36E still include ligand structures (white arrows), even 90 min after stimulation. (D) JAK/STAT pathway activity as shown by the 6×2xDrafLuc reporter is modulated by dsRNAs targeting the indicated genes. Knockdown of Elongin B/C, Cullin-5, and proteins required for multiple stages of endocytic trafficking all results in broadly equivalent increases in pathway activity. Dome and STAT92E knockdowns act as controls. Significance is shown for selected combinations with ***, p < 0.0001; **, p < 0.001; *, p < 0.05. (E) By comparison with control cells, knockdown of SOCS36E, Elongin B/C, and Cullin-5 stabilize Dome under steady-state conditions.
The turnover and destruction of cell surface receptors can occur via both lysosomal and proteosomal degradation (Thrower et al., 2000; Peng et al., 2003; Pickart and Fushman, 2004; Lauwers et al., 2009). We therefore investigated the fate of Dome following ligand-mediated pathway stimulation by using a pharmacological approach. By comparison with prestimulation levels, ligand stimulation of both otherwise untreated and dimethyl sulfoxide (DMSO) carrier–treated cells reduces Dome levels (Figure 1B). Similarly, addition of the proteosomal inhibitor MG132 does not have any significant effect on the receptor levels. By contrast, treatment with the lysosomal inhibitor bafilomycin A1 prevented ligand-induced degradation, resulting in increased Dome protein levels relative to controls (Figure 1B, quantified in B′). Given the presence of cyclohexamide in these experiments, we surmise that changes in protein levels equate to changes in Dome protein stability and suggest that degradation of the Dome receptor occurs in the lysosome, a finding consistent with a previous report that showed Upd2-GFP also trafficking to the lysosomal compartment (Vidal et al., 2010).
SOCS36E regulates Dome stability
Mammalian SOCS4 and SOCS5 have been shown to affect the stability of cell surface receptors (Kario et al., 2005; Bullock et al., 2007). We therefore set out to test whether the homologous Drosophila SOCS36E may play a similar role in the regulation of Dome. Using an assay previously developed to study ligand–GFP:receptor complex endocytosis (Vidal et al., 2010), we investigated whether RNA interference (RNAi)-mediated knockdown of SOCS36E (the effectiveness of which is shown in Supplemental Figure S1) has an effect on the rate at which endocytosed ligand is cleared from cells. Strikingly, while cells treated with double-stranded RNA (dsRNA) targeting control and socs36E mRNA contain similar levels of internalized ligand 40 min after stimulation (Figure 1C, 40 min), knockdown of SOCS36E resulted in delayed clearing of ligand at later time points (Figure 1C, 90 min). This effect is similar to that observed upon knockdown of proteins involved in endocytic processing, such as Rab5, TSG101, or Dor (Vidal et al., 2010), and suggests that SOCS36E is required for the trafficking of Upd2-GFP:Dome complexes to the lysosome.
On the basis of reports that SOCS proteins can form multi-protein E3 ubiquitin ligase complexes (Babon et al., 2009; Linossi and Nicholson, 2012), we next set out to identify potential components of the Drosophila ECS complex. While not previously characterized biochemically, Drosophila Elongin B, Elongin C, and Cullin-5 (Aso and Conrad, 1997; Kugler et al., 2010) were selected based on their similarity to human homologues and were tested for their expression in Kc167 cells and the efficiency of RNAi-based knockdown (Figure S1). Using a luciferase-based transcriptional assay to report JAK/STAT pathway activity (Müller et al., 2005), we confirmed that RNAi-mediated knockdown of endogenously expressed Dome, STAT92E, and SOCS36E modify pathway activity as expected (Figure 1D). In addition, we found that knockdown of each of the potential ECS components also increases reporter activity compared with controls (Figure 1D), a finding that indicates that Elongin B/C and Cullin-5 act as negative regulators of the JAK/STAT pathway. Although care must be taken making quantitative comparisons, increases in pathway activity following knockdown of Elongin B/C and Cullin-5 were broadly similar to knockdown of TSG101, Dor, and Rab5 (Figure 1D), components of the trafficking machinery previously shown to be involved in the endocytosis of JAK/STAT pathway components (Vidal et al., 2010). Given the conservation of pathway regulatory mechanisms between flies and vertebrates, our results suggest that the Drosophila homologues of Elongin B/C and Cullin-5 are likely to represent bona fide ECS components.
Given the role of Drosophila ECS components as negative regulators of JAK/STAT signaling, we hypothesized that the ECS complex may be involved in the regulation of Dome stability. Consistent with this, RNAi-mediated knockdown of ECS components significantly increases levels of the receptor under steady-state conditions (Figure 1E), suggesting that the ECS complex may affect JAK/STAT pathway activity via regulation of Dome stability.
SOCS36E can negatively regulate pathway signaling independently of the ECS
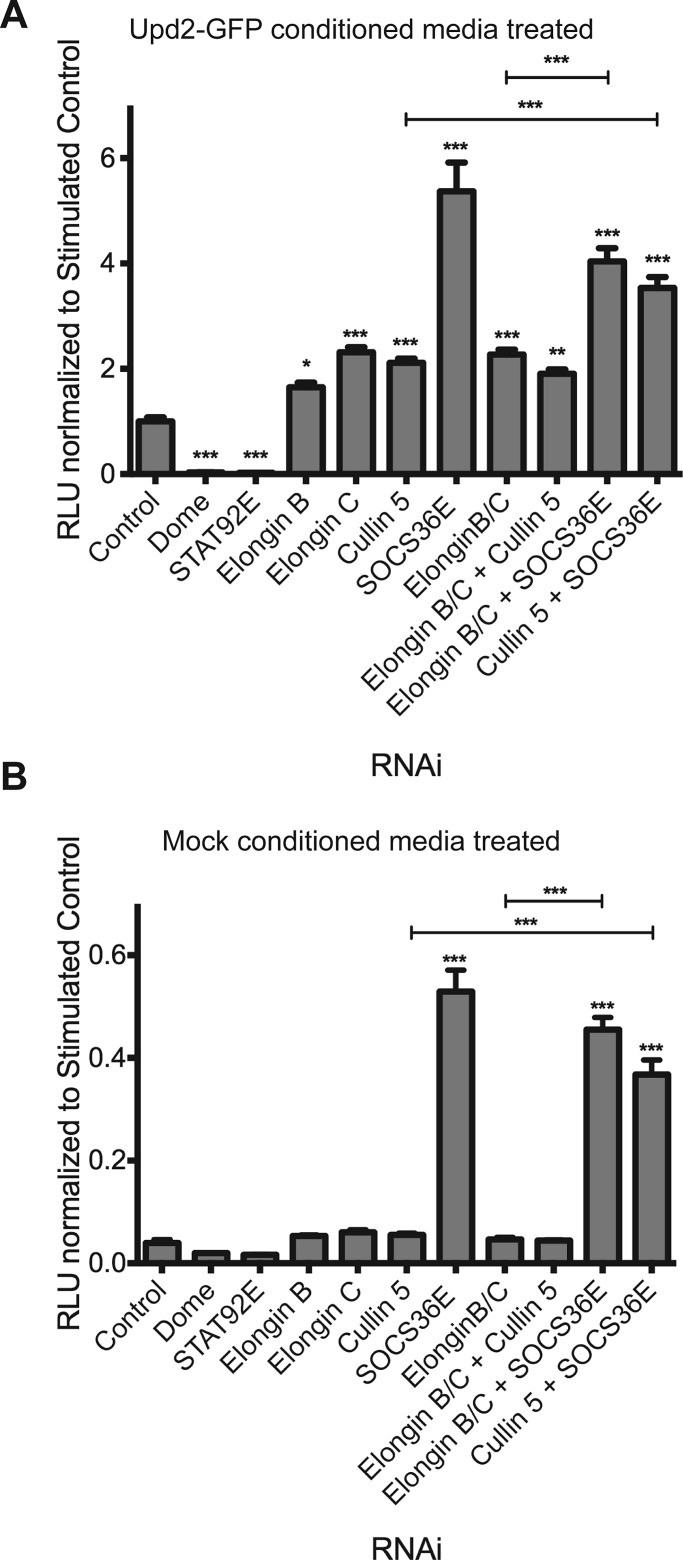
Increases in JAK/STAT pathway reporter activity following RNAi treatment consistently indicate that knockdown of SOCS36E itself results in a more potent increase in signaling than that elicited by knockdown of other ECS complex components (Figure 1D). One possible interpretation is that SOCS36E acts via a combination of two negative regulatory activities, with the second mechanism acting independently of other ECS components. To address this possibility, we tested the effect of combinatorial knockdown of SOCS36E and other ECS components in an experimental design that maintained constant levels of dsRNA targeting each component (see Materials and Methods). Consistent with previous results, cells treated with dsRNA targeting Elongin B, Elongin C, and Cullin-5 in isolation show increased levels of pathway reporter activity that are lower than that produced by SOCS36E knockdown (Figure 2A). Furthermore, simultaneous knockdown of Elongin B and C or Elongin B, C, and Cullin-5 did not result in additive effects following ligand stimulation of the pathway (Figure 2A). This result is consistent with a model in which all components of the ECS are required to assemble an active complex (Babon et al., 2009). However, knockdown of SOCS36E on its own or in combination with Elongin B/C or Cullin-5 resulted in additional, statistically significant increases in pathway activity compared with individual, Elongin B/C, or Elongin B/C and Cullin-5 combinatorial knockdowns (Figure 2A), indicating that knockdown of SOS36E exerts an additional, additive negative effect, even in the absence of other ECS components.
FIGURE 2:
SOCS36E can suppress the JAK/STAT pathway in a manner independent of Elongin B/C and Cullin-5. (A and B) JAK/STAT pathway activity as indicated by the 6×2xDrafLuc reporter in cells treated with dsRNAs targeting the indicated genes in different combinations. Individual genes were targeted by the same quantity of dsRNA, with the total amount of dsRNA kept constant by the addition of control dsRNA. Cells were treated with Upd2-GFP–conditioned (A) or mock-conditioned (B) media, and pathway activity was measured using the 6×2xDrafLuc reporter. Significance is shown for selected combinations with ***, p < 0.0001, **, p < 0.001, *, p < 0.05.
Further insight into ECS-independent activities of SOCS36E are also provided by the effect of pathway regulators on low-level “basal” pathway activity constitutively present in cells that have not been stimulated with exogenous ligand (Figure 2B). Under these conditions, no increase in pathway activity was observed upon individual or combinatorial knockdown of Elongin B/C and Cullin-5 (Figure 2B). By contrast, knockdown of SOCS36E by itself or together with the ECS components resulted in a highly significant increase in reporter activity. This suggests that, under steady-state conditions, SOCS36E acts to suppress the basal pathway activity in a manner independent of Elongin B/C and Cullin-5.
Taken together, these data suggest that SOCS36E is able to act via two separate mechanisms to suppress the Drosophila JAK/STAT pathway with the ECS cofactors being specifically used to provide negative regulatory modulation of JAK/STAT pathway signaling following ligand-mediated activation.
Both the N- and C-termini of SOCS36E have roles in regulation of the JAK/STAT pathway
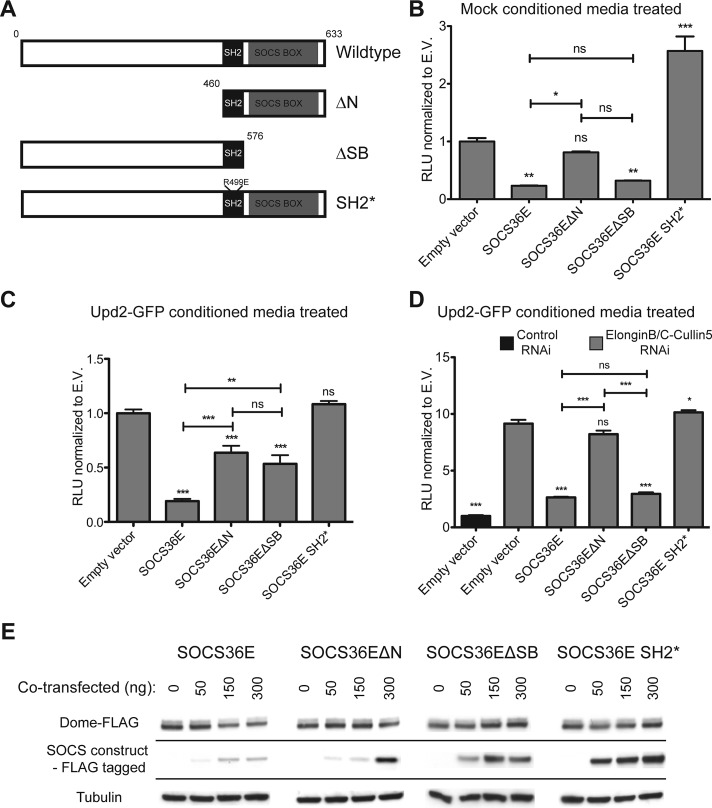
Given the evidence suggesting that SOCS36E can regulate the JAK/STAT pathway signaling via an ECS-independent mechanism, we used a structure–function approach to better characterize this second mechanism. In addition to full-length wild-type SOCS36E, we generated an N-terminal truncation (termed ΔN) that removes the first 460 amino acids up to the SH2 domain and a C-terminal truncation (termed ΔSB) that removes the SOCS-box region (Figure 3A). In addition, we also generated a full-length molecule with an R499E mutation in the conserved SH2 domain. This mutation, termed SOCS36E SH2*, has previously been shown to negate effects of SOCS36E overexpression in vivo (Callus and Mathey-Prevot, 2002; Almudi et al., 2009) and has been molecularly shown to prevent binding of the SH2* domain to pTyr residues in other contexts (Marengere and Pawson, 1992; Kamura et al., 1998). The molecular weight and protein expression levels of these constructs were confirmed by Western blotting (Figure S2), with all subsequent experiments normalized to express equivalent levels of protein.
FIGURE 3:
Both N- and C-termini of SOCS36E are able to suppress JAK/STAT pathway activity. (A) Schematic representation of SOCS36E constructs. (B–D) JAK/STAT pathway activity as indicated by the 6×2xDrafLuc reporter in cells expressing the SOCS36E constructs shown in (A) and either stimulated with mock-conditioned media (B) or Upd2-GFP–conditioned media (C and D). In each case, values are normalized to empty vector (E.V.) controls, with error bars representing the SE of four experimental replicates. In (D), samples indicated in gray were treated with dsRNA targeting Elongin B/C and Cullin-5 mRNA, while column 1 (black) was treated with the same quantity of control dsRNA. Significance is shown for selected combinations with ***, p < 0.0001; **, p < 0.001; *, p < 0.05. (E) The steady-state levels of Dome-FLAG expressed in cells transfected with the indicated levels of SOCS36E constructs shown in (A). Levels of protein expressed by the SOCS36E constructs and levels of tubulin used as a loading control are also shown.
We first examined the effect of overexpressing SOCS36E variants on the basal JAK/STAT pathway activity that is not modulated by Elongin B/C or Cullin-5. We found that expression of both full-length SOCS36E and SOCS36EΔSB (lacking its SOCS-box region) decreased basal pathway activity (Figure 3B). By contrast, expression of SOCS36EΔN did not significantly affect pathway activity (Figure 3B), suggesting that the N-terminal region of SOCS36E is necessary for the suppression of basal JAK/STAT activity. Finally, expression of SOCS36E SH2* resulted in increased pathway activity. Although not investigated further in this study, it is possible that this is due to sequestration of cofactors that would otherwise have interacted with endogenous SOCS36E.
We next examined the consequences of SOCS36E expression on stimulated pathway activity (Figure 3C). As expected, expression of full-length SOCS36E strongly reduced pathway activity (Figure 3C). In addition, expression of both the ΔN and ΔSB SOCS36E truncations also decreased the STAT92E activity compared with the controls (Figure 3C). However, while each truncation was expressed at similar levels (Figure S2), the ability of the ΔN and ΔSB truncations to suppress pathway signaling was quantitatively less than that exerted by the full-length protein (Figure 3C). Finally, the inability of SOCS36E SH2* overexpression to change pathway activity (Figure 3C) suggests that a functional SH2 domain able to bind pTyr is required to regulate pathway activity. Taken together, these results indicate that both the N- and the C-termini of SOCS36E independently, and probably additively, negatively regulate JAK/STAT pathway activity following ligand-mediated stimulation in an SH2 domain–dependent manner.
We also tested the activity of the SOCS36E truncations for their effect on pathway activity in an ECS-independent manner by expressing each construct in cells treated with dsRNA targeting Elongin B/C and Cullin-5 prior to stimulation with ligand-conditioned media (Figure 3D). As previously shown, knockdown of Elongin B/C and Cullin-5 significantly increased pathway activity (Figure 3D), an increase that is countered by the expression of both full-length SOCS36E and SOCS36EΔSB. By contrast, expression of SOCS36EΔN, which was previously able to suppress pathway activity in the presence of endogenous ECS components (Figure 3C), was not able to do so under ECS-depleted conditions (Figure 3D). These results indicate that the negative regulation of JAK/STAT pathway activity mediated by the ECS complex functions via the SOCS-box of SOCS36E. Furthermore, our data also suggest that the N-terminal of SOCS36E functions as a negative regulator of the pathway via a mechanism that is independent of ECS complex assembly.
FIGURE 4:
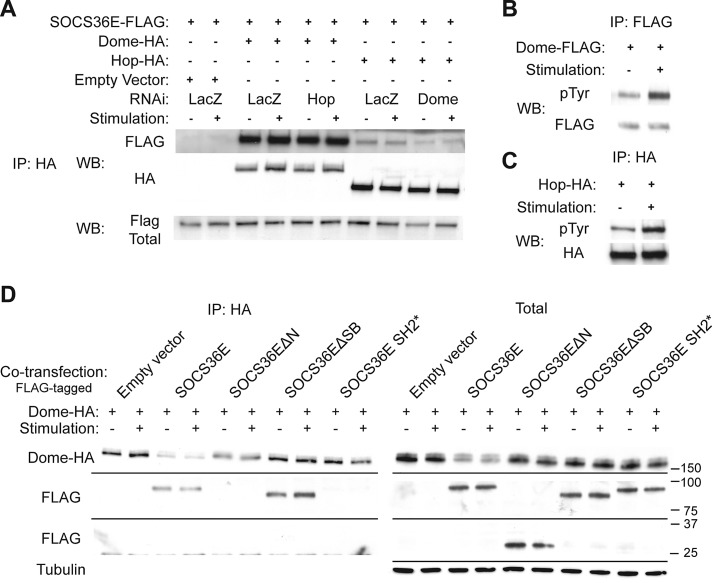
The N-terminal and SH2 domain of SOCS36E are required for binding to Dome. (A) Coimmunoprecipitation of SOCS36E-FLAG following pull down of Dome and Hop is shown in the presence and absence of exogenous ligand. Cells were also treated with the indicated dsRNAs targeting LacZ, Hop, or Dome. (B and C) Levels of tyrosine phosphorylation detected following immunoprecipitation of Dome (B) and Hop (C) are shown under stimulated and unstimulated conditions. (D) Coimmunoprecipitation of FLAG-tagged SOCS36E constructs following Dome-HA pull down. While each of the indicated SOCS36E constructs (which migrate at different molecular weights) is expressed (right), only full-length and SOCS36EΔSB coimmunoprecipitate with the receptor (left).
In an independent approach to characterize the separate mechanisms of pathway suppression mediated by the N- and C-termini of SOCS36E, we next investigated the effects of cotransfecting increasing concentrations of SOCS36E truncation constructs on the steady-state concentration of Dome. We found that even relatively low levels of full-length wild-type SOCS36E are able to reduce the receptor levels (by 46% in Figure 3E, left panel). Similarly, overexpression of SOCS36EΔN also reduced receptor concentration, although higher expression levels were required to see this effect (39% reduction in Figure 3E, center left). By contrast, the SOCS36EΔSB and SH2* forms had no detectable effect on steady-state Dome stability, even when expressed at high levels (Figure 3E). This indicates that both the SOCS-box and a functional SH2 domain are required to reduce the steady-state levels of Dome and suggests that the decrease in pathway activity mediated by the N-terminal region of SOCS36E is not the result of a change in receptor stability.
SOCS36E interaction with Dome requires N-terminal and SH2 domains
Previous reports have indicated that mammalian SOCS molecules display differing affinities for JAKs, their associated receptors, and the JAK:receptor complex as a whole (Piganis et al., 2011; Kershaw et al., 2013; reviewed in Yoshimura, 2009; Lachance et al., 2012). To better understand how Drosophila SOCS36E functions, we set out to identify its binding partners, using coimmunoprecipitation techniques with a focus on potential binding to Dome and Hop. We found that SOCS36E can be efficiently immunoprecipitated with Dome, an interaction that appeared to be much stronger than the Hop:SOCS36E interactions tested in parallel under identical conditions (Figure 4A). Furthermore, the coprecipitation of SOCS36E with Dome was not affected by RNAi targeting Hop, implying that the interaction with the receptor is direct. By contrast, the weaker interaction of SOCS36E with Hop was decreased following Dome knockdown, suggesting that SOCS36E coprecipitation is dependent on Dome, a phenomenon also previously described for mammalian SOCS proteins (Starr and Hilton, 1998; Yoshimura, 2009).
The activation of the JAK/STAT signaling pathway activity depends on tyrosine phosphorylation of the receptor, JAK, and STAT pathway components by the JAK kinase. In particular, phosphorylation of the receptor is a prerequisite for protein:protein interactions with STAT and SOCS molecules, which bind via their conserved SH2 domains (reviewed in Kaneko et al., 2012; Liu et al., 2012; Tinti et al., 2013). As interactions between Dome or Hop and SOCS36E appeared to be independent of ligand-mediated pathway stimulation (Figure 4A), we investigated how pTyr modification of Dome and Hop changed upon treatment with Upd2-GFP–conditioned media (Figure 4, B and C). In both cases, we detected constitutive tyrosine phosphorylation of Dome and Hop under steady-state conditions, with a twofold increase following ligand stimulation. This finding may explain both the basal pathway activity shown in Figure 2B and the ability of SOCS36E to bind to Dome and Hop under unstimulated conditions (Figure 4A).
Finally, we set out to identify the domains of SOCS36E necessary for its interaction with Dome, using the SOCS36E truncations. We found that both full-length SOCS36E and SOCS36EΔSB are coimmunoprecipitated with Dome, even under unstimulated conditions (Figure 4D). In addition, SOCS36E SH2* did not interact with Dome, confirming that Dome:SOCS36E interaction requires a functional SH2 domain. More strikingly, we also found that SOCS36EΔN is not coprecipitated with Dome, suggesting that the N-terminal region of SOCS36E is required for interaction (Figure 4D). This result is also consistent with the higher levels of SOCS36EΔN required to destabilize the receptor previously observed in Figure 3E. Taken together, these results demonstrate that SOCS36E:Dome interactions require both the SH2 domain and the N-terminal region of SOSC36E.
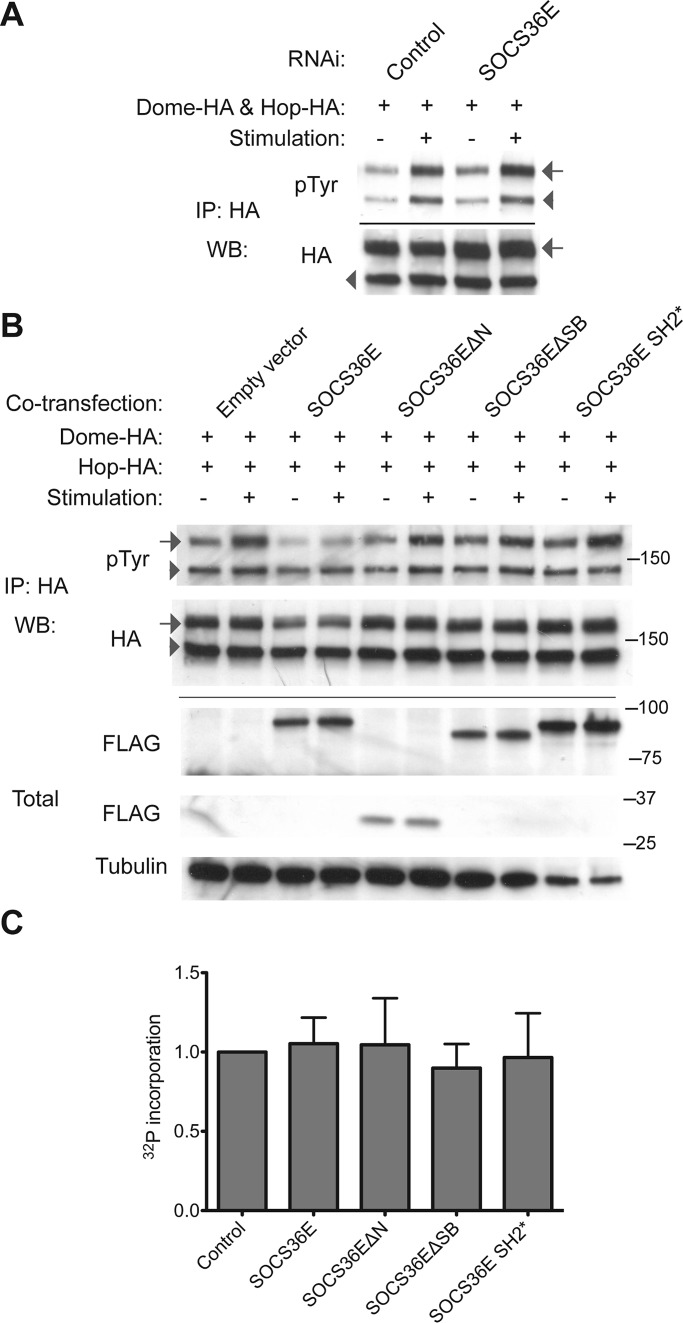
Dome and Hop phosphorylation is affected by SOCS36E and its truncations
Although intriguing, the requirement of the SOCS36E N-terminal region for Dome binding does not directly explain why overexpressed SOCS36EΔSB is able to suppress pathway activity (Figure 3). Two mechanisms that could account for this activity are hypothesized. Either the binding of SOCS36E to Dome acts to block Hop kinase from accessing key tyrosine residues, or the N-terminal of SOCS36E is able to regulate kinase activity itself. As cytokine receptors are substrates of their associated kinases, and given that JAKs are themselves regulated by autophosphorylation (Saharinen et al., 2000; Matsuda et al., 2004; Funakoshi-Tago et al., 2006), we analyzed the pTyr levels of both Dome and Hop following knockdown of SOCS36E (Figure 5A). As expected, stimulation with ligand-conditioned media resulted in increased tyrosine phosphorylation of Hop and Dome in control cells, while knockdown of SOCS36E was sufficient to elevate pTyr levels of Dome both in the presence and absence of pathway stimulation (by 25 and 50% respectively; Figure 5A). By contrast, levels of Hop phosphorylation are not noticeably affected following loss of endogenous SOCS36E (Figure 5A). In the converse experiment, we analyzed Dome and Hop pTyr levels following coexpression of the SOCS36E truncation constructs (Figure 5B). Expression of full-length SOCS36E almost completely prevented Dome phosphorylation in response to pathway stimulation and also mildly reduced phosphorylation of Hop. By contrast, both ΔN and ΔSB truncations, as well as the SH2* construct, had no effect on phosphorylation of either Dome or Hop. These results suggest a role for SOCS36E in regulation of phosphorylation levels of receptor complex components that requires both N- and C-termini and an intact SH2 domain.
FIGURE 5:
SOCS36E can suppress phosphorylation of Dome but does not affect Hop kinase activity. (A) Total protein levels and the levels of tyrosine phosphorylation detectable for Dome (arrows) and Hop (arrowheads) are shown following ligand stimulation and treatment with the indicated dsRNAs. (B) Total protein levels and the levels of tyrosine phosphorylation of Dome (arrows) and Hop (arrowheads) following ligand stimulation and coexpression of the indicated SOCS36E constructs illustrated in Figure 3A. (C) In vitro measurement of 32P incorporation (derived from [32P]γ-ATP) into Hop (via autophosphorylation) in the presence of normalized concentrations of control protein and the indicated SOCS36E truncations (see Materials and Methods for details). Incorporation is normalized to control, and error bars represent the SE of three independent experiments. Differences are not statistically significant.
Finally, given the possibility that SOCS36E might directly affect the catalytic activity of Hop, we established an in vitro kinase activity assay assessing the ability of Hop to autophosphorylate in a context that is likely to lack the normal Hop:Dome complex present in vivo (Figure 5C). Although kinase activity could be detected under these conditions, addition of de novo synthesized full-length SOCS36E or its truncations did not have any effect on the levels of 32P incorporation into Hop. Indeed, it is possible that SOCS36E and Hop do not physically interact in this assay, given the weak interactions detected in Figure 4A. Although based principally on negative results, these findings tentatively suggest that the N-terminal domain of SOCS36E does not directly block kinase activity. This, in turn, suggests that the alternative substrate-blocking hypothesis, dependent on an intact N-terminal and SH2 domain, may explain the second ECS-independent activity of SOSC36E.
DISCUSSION
This study presents detailed molecular characterization of SOCS36E, a negative regulator of Drosophila JAK/STAT pathway signaling. We show that Elongin B/C, Cullin-5, and SOCS36E negatively regulate the stability of Dome and act as negative regulators of the ligand-activated JAK/STAT pathway. This activity may be mediated by a sorting mechanism that directs receptor complexes to the lysosome and requires both the SOCS-box and an intact SH2 domain. We also show that SOCS36E is able to negatively regulate both basal and ligand-induced activity of the JAK/STAT pathway in a manner independent of SOCS-box, Elongin B/C, and Cullin-5 via its N-terminal region. Although the exact molecular mechanism by which the N-terminus of SOCS36E operates remains unresolved, it is likely that this activity is linked to the interactions of the N-terminal region of SOCS36E with Dome, which may act to prevent the phosphorylation of key tyrosine residues by Hop. Taking the results together, we show that SOCS36E negatively regulates both basal and activated activity of the Drosophila JAK/STAT pathway via two independent and separable mechanisms.
Components of the ECS complex affect JAK/STAT activity and Dome stability
Mammalian SOCS proteins have been shown to affect internalization and endocytosis of cytokine receptors by more than one mechanism. For example, the granulocyte colony-stimulating factor receptor is targeted for degradation via ubiquitination by an Elongin–Cullin–SOCS3 complex (Hörtner et al., 2002; Zhuang et al., 2005; Wölfler et al., 2009). Our results indicate that, in Drosophila, Elongin B/C, Cullin-5, and SOCS36E are also involved in the regulation of Dome stability (Figure 1E) and pathway activity (Figure 1D). In light of our previous work identifying endocytosis as a negative regulator of the Drosophila JAK/STAT pathway (Vidal et al., 2010), we propose that the ECS complex represents an integral component of SOCS-mediated pathway regulation that functions via interactions with the endocytic machinery. By analogy to mammalian systems, it seems possible that the Drosophila ECS complex might mediate interactions with the endocytic machinery via its hypothesized E3 ubiquitin ligase activity and the subsequent ubiquitination of Dome.
N-terminal of SOCS36E has a role in suppression of JAK/STAT pathway activity
A number of studies have focused on mammalian SOCS molecules, characterizing their molecular structure and function and their involvement in disease and cancer (reviewed in Croker et al., 2008; Yoshimura, 2009; Delgado-Ortega et al., 2013). However, only a handful of reports have identified a role for the relatively divergent N-terminal regions of SOCS-family members. The N-terminal of SOCS5 has been shown to interact with IL-4 receptor in a phosphotyrosine-independent manner (Seki et al., 2002), and SOCS5 has been reported to associate with EGFR in an N-terminal–dependent manner (Kario et al., 2005; Nicholson et al., 2005). A recent report from Feng and colleagues has also identified a conserved motif in the N-terminus of SOCS4 and SOCS5 that has a potential role in protein interaction (Feng et al., 2011). Considering that SOCS36E is most closely related to mammalian SOCS4 and 5, our results provide further support for a conserved functional role of the long N-terminus present in these SOCS molecules. We have shown that both the N-terminal of SOCS36E and its SH2 domain are required for efficient binding of SOCS36E to Dome (Figure 4D), although it is not known whether the SOCS36E N-terminal itself is sufficient for this interaction or whether it stabilizes the interaction in a manner similar to the N-terminal of SOCS3. In the case of SOCS3, an N-terminally extended SH2 domain (N-ESS) has been described that is involved in orientating interactions with phosphorylated tyrosine residues (Sasaki et al., 1999; Yasukawa et al., 1999; Babon et al., 2006). Truncation of the N-terminal of SOCS36E could have affected any N-ESS region that might be present and could thus destabilize the association of SOCS36E with Dome, explaining why higher levels of SOCS36EΔN overexpression were required to decrease receptor levels in Figure 3E.
We have also shown that, in addition to playing a role in protein:protein interactions, the N-terminal of SOCS36E is able to suppress JAK/STAT pathway activity irrespective of ligand stimulation. This function is independent of Elongin B/C, Cullin-5, or the SOCS-box domain (Figure 3) and is consistent with previous genetic analysis showing that the N-terminal of SOCS36E is able to inhibit both the EGFR and JAK/STAT pathways in vivo (Callus and Mathey-Prevot, 2002; Silver et al., 2005; Almudi et al., 2009). We also show that endogenous SOCS36E regulates the phosphorylation level of receptor complex components (Figure 5A), a finding that correlates with its role in suppressing basal pathway activity under steady-state conditions (Figure 2), and we show that full-length SOCS36E also reduces Dome phosphorylation following ligand binding (Figure 5, A and B). While not a line of enquiry followed in this study, the ability of SOCS36E to bind to receptors and modulate their tyrosine phosphorylation may also represent the mechanism via which SOCS36E is able to modulate EGFR activity in vivo (Callus and Mathey-Prevot, 2002; Almudi et al., 2009; Herranz et al., 2012).
Finally, our finding that overexpression of SOCS36E or its deletions were unable to significantly affect the phosphorylation of Hop (Figure 5B) and does not affect Hop kinase activity in vitro (Figure 5C) suggests SOCS36E functions as a steric inhibitor of Hop that prevents interactions with its substrate Dome. This proposed function for the N-terminal of SOCS36E also suggests that the unstructured N-terminal regions of other mammalian SOCS molecules, including the most closely homologous SOCS4 and SOCS5, might also act via similar mechanisms, possibly in a receptor-specific manner to suppress pathway activity under steady-state conditions. However, we have not excluded the possibility that the N-terminal of SOCS36E mediates interaction with yet unknown cofactors that might directly or indirectly affect receptor phosphorylation levels. Irrespective of which hypothesis is true, further research into mammalian SOCS-family members and their homologues could provide insight into mechanisms of receptor:JAK complex inhibition, potentially laying the groundwork for novel drug development approaches in the future.
MATERIALS AND METHODS
Cell culture
S2R+ and KC167 Drosophila cell lines were cultured, and conditioned media was generated as described by Vidal et al. (2010). Cell transfections were performed using Effectene (Qiagen, Valencia, CA) according to the manufacturer's instructions. Pathway activity was measured using 6×2xDrafLuc reporter assay, as described by Müller et al. (2005) and Vidal et al. (2010), and is presented as the Firefly (reporter) to Renilla (constitutively expressed cell density control) luciferase ratio.
dsRNA and quantitative PCR
Synthesis of dsRNA was performed as described by Vidal et al. (2010), using the same primers for dor, TSG101, and Rab5. Primer pairs used to generate dsRNA fragments targeting dome, hop, stat92E, SOCS36E, Elongin B, Elongin C, and Cullin-5 mRNA are shown in Supplemental Table S1. Knockdowns were performed over 4 d, using 1.5 μg of total dsRNA per well in a 96-well plate, 6 μg per well in a 12-well plate, or 12 μg per well in a six-well plate. Effectiveness of selected dsRNA treatments was tested by qPCR (Figure S1).
RNA for qPCR was isolated as described by Chomczynski and Mackey (1995). Reverse transcription was performed using the High Capacity RNA-to-cDNA Kit (Applied Biosystems, Bedford, MA), and cDNA quantification was performed using SYBR Green JumpStart Taq Ready Mix for Quantitative PCR (Sigma-Aldrich, St. Louis, MO) and a C1000 Touch Thermal Cycler (Bio-Rad, Hercules, CA).
Generation of SOCS36E truncations and other constructs
Wild-type full-length SOCS36E coding region was obtained from SD04308 cDNA Drosophila Genomics Resource Center (DRGC, Bloomington, IN). Truncation constructs were generated by PCR using primers ATGCACTGCCTGGTTCCCGATCT and TACATTGCCGTAGTACGGCATCG to generate ΔN and ATGGGTCATCACCTTAGCAAGTTCTCAGCA and GGAGAAGGTCTGCCTTCTGTGCAG to generate ΔSB constructs. SH2* mutant was obtained by directed mutagenesis of arginine at position 499 to glutamine acid (R499E), using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). All constructs were cloned into the Gateway System entry vector using pENTR Directional TOPO Cloning Kit (Invitrogen, Carlsbad, CA), and subsequently into destination vectors pAWF or pAWH using Gateway LR Clonase II Enzyme Mix (Invitrogen). Gateway destination vectors were obtained from the DRGC. Hop and Dome constructs were prepared in similar manner, using the Gateway System.
Immunoprecipitation and Western blots
Cells batch transfected in six-well plates with appropriate constructs were split 2 h prior to assay into 12- or 24-well plates. When necessary, mock or Upd2-GFP–conditioned media (prepared as described in Vidal et al., 2010) was added to the media overlying cells for 10 min; this was followed by lysis for 30 min at 4°C on a horizontal shaker. In experiments focusing on protein phosphorylation, kinase immunoprecipitation lysis buffer (kinase IPLB; 50 mM Tris-HCl, pH 7.4, 1 mM ethylene glycol tetraacetic acid [EGTA], 1 mM EDTA, 5 mM β-glycerophosphate, 2.5 mM Na-pyrophosphate, 1 mM Na-orthovanadate, 0.5% Triton X-100 supplemented freshly with a Complete Mini EDTA-Free protease inhibitor cocktail tablet [Roche, Indianapolis, IN]) was used; in all other cases, standard lysis buffer (50 mM Tris-HCl, pH 7.4, 1 mM EGTA, 1 mM EDTA, 0.5% Triton X-100 freshly supplemented in Complete Mini EDTA-Free tablet protease inhibitor cocktail tablet [Roche]) was used. Lysates were cleared by centrifugation at 7000 × g for 5 min at 4°C and incubated with 1:200 anti-hemagglutinin (anti-HA) High Affinity (clone 3F10; Roche) or 1:200 Monoclonal M2 anti-FLAG (Sigma-Aldrich) antibody for 4 h at 4°C with gentle agitation; this was followed by incubation with 1:5 Dynabeads (Novex, Invitrogen) overnight at 4°C with gentle agitation. Proteins were eluted into 3× Laemmli buffer by boiling and were resolved by SDS–PAGE. For Western blot analysis, Mini-PROTEAN TGX 4–15% gradient gels (Bio-Rad) were used for sample separation. Nitrocellulose membranes (GE Healthcare, Waukesha, WI) were blocked with 5% horse serum (Sigma-Aldrich) in 0.5% TBS-Tween-20 for 30 min at room temperature and incubated with primary antibodies in blocking solution. The following antibodies were used: 1:2500 Monoclonal M2 anti-FLAG (Sigma-Aldrich), 1:2500 anti-HA High Affinity (Roche), 1:400 anti-pTyrosine PY20 (Calbiochem, San Diego, CA), 1:5000 Monoclonal anti–α-tubulin (Sigma-Aldrich); all secondary antibodies (Dako, Carpinteria, CA) were applied for 2 h at room temperature at 1:5000 in blocking solution. Where indicated, blots were scanned and quantified using Quantity One software (Bio-Rad).
Biotinylation and pharmacological agents
Cells transfected with Dome-FLAG were biotinylated using 0.25 mg/ml EZ-Link Sulfo-NHS-Biotin (Thermo Fisher Scientific, Lafayette, CO) in cold phosphate-buffered saline, according to the manufacturer's protocol. Biotin pull down was performed using Streptavidin Magnetic Beads (New England BioLabs, Ipswich, MA). Where indicated, cells were incubated with 10 μg/ml cyclohexamide (Sigma-Aldrich), 10 μM MG132 (Tocris Bioscience, Bristol, UK), 0.1 μM bafilomycin A1 (Sigma-Aldrich), and appropriate carrier (DMSO) control for 30 min prior to the experiment and for the duration of the experiment.
Statistics and reproducibility
All experiments were performed independently at least three times. All Western blots shown are representative blots. All graphs were produced from pooling results from three or more independent experiments. Quantifiable data were analyzed for statistical significance using two-way analysis of variance with Bonferroni posttest (*, p < 0.05; **, p < 0.01; ***, p < 0.001. All error bars represent the SEM.
Kinase activity assays
Hop-HA bound to beads was obtained by immunoprecipitation performed in batches into tubes containing Dynabeads (Invitrogen) used in the assay. Beads were washed three times with kinase IPLB and three times with kinase activity assay reaction buffer (KAAB: 20 mM HEPES, pH 7.4, 10 mM NaCl, 20 mM MgCl2, 100 mM NaF, 0.2 mM NaOVa, 2 mM dithiothreitol, 10 mM MnCl2) and resuspended in 25 μl of 2× KAAB. Twenty microliters of water or synthesized protein was added to the reaction, and samples were incubated at 30°C with agitation for 10 min. Enzymatic reaction was initialized by addition of 5 μl of 1 mM ATP spiked with [32P]γ-ATP (Perkin Elmer-Cetus, Waltham, MA) to approximately 1 × 106 cpm/μl. Reactions were incubated at 30°C for 20 min with occasional agitation and were terminated by the addition of 200 μl of 20 mM EDTA. Three washes with kinase IPLB preceded elution into 3× Laemmli buffer by boiling. Eluted Hop was resolved by SDS–PAGE, transferred to nitrocellulose, quantified using Molecular Imager FX (Bio-Rad), and analyzed with Quantity One software (Bio-Rad). Protein levels were determined by blotting of the nitrocellulose for HA. SOCS36E and its truncation constructs were cloned into pRSET A (Invitrogen) and synthesized in vitro using PURExpress (New England BioLabs), according to the manufacturer's instructions. Constructs were labeled with [35S]methionine (Perkin Elmer-Cetus).
Supplementary Material
Acknowledgments
The authors thank Elizabeth Smythe, Robert Piggott, Kirsty Johnstone, Maria Fragiadaki, and Steve Winder for antibodies, reagents, technical assistance, and helpful comments. M.P.Z. is a Cancer Research-UK Senior Cancer Research Fellow, and W.S. holds a Cancer Research-UK Ph.D. fellowship.
Abbreviations used:
- DMSO
dimethyl sulfoxide
- Dome
Domeless
- DRGC
Drosophila Genomics Resource Center
- dsRNA
double-stranded RNA
- ECS
Elongin-Cullin-SOCS
- EGFR
epidermal growth factor receptor
- EGTA
ethylene glycol tetraacetic acid
- HA
hemagglutinin
- Hop
Hopscotch
- JAK
Janus kinase
- KAAB
kinase activity assay reaction buffer
- kinase IPLB
kinase immunoprecipitation lysis buffer
- RNAi
RNA interference
- SH2
Src homology 2
- SOCS
suppressors of cytokine signaling
- STAT
signal transducer and activators of transcription
- Upd2-GFP
Upd2–green fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0275) on July 24, 2013.
The authors declare that they have no conflict of interest.
*Present address: Laboratory of Obesity and Metabolic Diseases, National Heart, Lung and Blood Institute, 31 Center Drive MSC 2486, Bethesda, MD 20892.
W.S. designed and undertook experiments, interpreted results, and wrote the paper. O.V. designed and undertook experiments. M.P.Z. designed experiments, interpreted results, and wrote the paper.
REFERENCES
- Alicea-Velázquez NL, Jakoncic J, Boggon TJ. Structure-guided studies of the SHP-1/JAK1 interaction provide new insights into phosphatase catalytic domain substrate recognition. J Struct Biol. 2013;181:243–251. doi: 10.1016/j.jsb.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almudi I, Stocker H, Hafen E, Corominas M, Serras F. SOCS36E specifically interferes with Sevenless signaling during Drosophila eye development. Dev Biol. 2009;326:212–223. doi: 10.1016/j.ydbio.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- Aso T, Conrad MN. Molecular cloning of DNAs encoding the regulatory subunits of elongin from Saccharomyces cerevisiae and Drosophila melanogaster. Biochem Biophys Res Commun. 1997;241:334–340. doi: 10.1006/bbrc.1997.7819. [DOI] [PubMed] [Google Scholar]
- Babon JJ, et al. The structure of SOCS3 reveals the basis of the extended SH2 domain function and identifies an unstructured insertion that regulates stability. Mol Cell. 2006;22:205–216. doi: 10.1016/j.molcel.2006.03.024. [DOI] [PubMed] [Google Scholar]
- Babon JJ, Sabo JK, Zhang JG, Nicola NA, Norton RS. The SOCS box encodes a hierarchy of affinities for Cullin5: implications for ubiquitin ligase formation and cytokine signalling suppression. J Mol Biol. 2009;387:162–174. doi: 10.1016/j.jmb.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 1994;8:300–312. doi: 10.1101/gad.8.3.300. [DOI] [PubMed] [Google Scholar]
- Brown S, Hu N, Hombría JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. 2001;11:1700–1705. doi: 10.1016/s0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- Bullock AN, Rodriguez MC, Debreczeni JE, Songyang Z, Knapp S. Structure of the SOCS4-ElonginB/C complex reveals a distinct SOCS box interface and the molecular basis for SOCS-dependent EGFR degradation. Structure. 2007;15:1493–1504. doi: 10.1016/j.str.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callus BA, Mathey-Prevot B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21:4812–4821. doi: 10.1038/sj.onc.1205618. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Mackey K. Short technical reports. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques. 1995;19:942–945. [PubMed] [Google Scholar]
- Croker BA, Kiu H, Nicholson SE. SOCS regulation of the JAK/STAT signalling pathway. Semin Cell Dev Biol. 2008;19:414–422. doi: 10.1016/j.semcdb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Ortega M, Marc D, Dupont J, Trapp S, Berri M, Meurens F. SOCS proteins in infectious diseases of mammals. Vet Immunol Immunopathol. 2013;151:1–19. doi: 10.1016/j.vetimm.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devergne O, Ghiglione C, Noselli S. The endocytic control of JAK/STAT signalling in Drosophila. J Cell Sci. 2007;120:3457–3464. doi: 10.1242/jcs.005926. [DOI] [PubMed] [Google Scholar]
- Dittrich A, Quaiser T, Khouri C, Görtz D, Mönnigmann M, Schaper F. Model-driven experimental analysis of the function of SHP-2 in IL-6-induced Jak/STAT signaling. Mol Biosyst. 2012;8:2119–2134. doi: 10.1039/c2mb05488d. [DOI] [PubMed] [Google Scholar]
- Doti N, Scognamiglio PL, Madonna S, Scarponi C, Ruvo M, Perretta G, Albanesi C, Marasco D. New mimetic peptides of the kinase-inhibitory region (KIR) of SOCS1 through focused peptide libraries. Biochem J. 2012;443:231–240. doi: 10.1042/BJ20111647. [DOI] [PubMed] [Google Scholar]
- Feng Z, Chandrashekaran I, Low A, Speed T, Nicholson SE, Norton RS. The N-terminal domains of SOCS proteins: a conserved region in the disordered N-termini of SOCS4 and 5. Proteins. 2011;80:946–957. doi: 10.1002/prot.23252. [DOI] [PubMed] [Google Scholar]
- Funakoshi-Tago M, Pelletier S, Matsuda T, Parganas E, Ihle JN. Receptor specific downregulation of cytokine signaling by autophosphorylation in the FERM domain of Jak2. EMBO J. 2006;25:4763–4772. doi: 10.1038/sj.emboj.7601365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B, Donaldson J. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan C, Kreis S, Margue C, Behrmann I. Jaks and cytokine receptors—an intimate relationship. Biochem Pharmacol. 2006;72:1538–1546. doi: 10.1016/j.bcp.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Herranz H, Hong X, Hung NT, Voorhoeve PM, Cohen SM. Oncogenic cooperation between SOCS family proteins and EGFR identified using a Drosophila epithelial transformation model. Genes Dev. 2012;26:1602–1611. doi: 10.1101/gad.192021.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtner M, Nielsch U, Mayr LM, Johnston JA, Heinrich PC, Haan S. Suppressor of cytokine signaling-3 is recruited to the activated granulocyte-colony stimulating factor receptor and modulates its signal transduction. J Immunol. 2002;169:1219–1227. doi: 10.4049/jimmunol.169.3.1219. [DOI] [PubMed] [Google Scholar]
- Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- James C, Ugo V, Casadevall N, Constantinescu SN, Vainchenker W. A JAK2 mutation in myeloproliferative disorders: pathogenesis and therapeutic and scientific prospects. Trends Mol Med. 2005;11:546–554. doi: 10.1016/j.molmed.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Jatiani SS, Baker SJ, Silverman LR, Reddy EP. JAK/STAT pathways in cytokine signaling and myeloproliferative disorders: approaches for targeted therapies. Genes Cancer. 2010;1:979–993. doi: 10.1177/1947601910397187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Sato S, Haque D, Liu L, Kaelin WG, Conaway RC, Conaway JW. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Joshi R, Feller SM, Li SS. Phosphotyrosine recognition domains: the typical, the atypical and the versatile. Cell Commun Signal. 2012;10:32. doi: 10.1186/1478-811X-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kario E, Marmor MD, Adamsky K, Citri A, Amit I, Amariglio N, Rechavi G, Yarden Y. Suppressors of cytokine signaling 4 and 5 regulate epidermal growth factor receptor signaling. J Biol Chem. 2005;280:7038–7048. doi: 10.1074/jbc.M408575200. [DOI] [PubMed] [Google Scholar]
- Karsten P, Häder S, Zeidler MP. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech Dev. 2002;117:343–346. doi: 10.1016/s0925-4773(02)00216-2. [DOI] [PubMed] [Google Scholar]
- Kershaw NJ, Murphy JM, Liau NP, Varghese LN, Laktyushin A, Whitlock EL, Lucet IS, Nicola NA, Babon JJ. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nat Struct Mol Biol. 2013;20:469–476. doi: 10.1038/nsmb.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler JM, Lem C, Lasko P. Reduced cul-5 activity causes aberrant follicular morphogenesis and germ cell loss in Drosophila oogenesis. PLoS One. 2010;5:e9048. doi: 10.1371/journal.pone.0009048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance C, Goupil S, Leclerc P. StatticV, a STAT3 inhibitor, affects human spermatozoa through regulation of mitochondrial activity. J Cell Physiol. 2012;228:704–713. doi: 10.1002/jcp.24215. [DOI] [PubMed] [Google Scholar]
- Lauwers E, Jacob C, André B. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J Cell Biol. 2009;185:493–502. doi: 10.1083/jcb.200810114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL, Wernig G. Role of JAK-STAT signaling in the pathogenesis of myeloproliferative disorders. Hematology Am Soc Hematol Educ Program. 2006;2006:233–239, 510. doi: 10.1182/asheducation-2006.1.233. [DOI] [PubMed] [Google Scholar]
- Linossi EM, Nicholson SE. The SOCS box—adapting proteins for ubiquitination and proteasomal degradation. IUBMB Life. 2012;64:316–323. doi: 10.1002/iub.1011. [DOI] [PubMed] [Google Scholar]
- Liu BA, Engelmann BW, Nash PD. The language of SH2 domain interactions defines phosphotyrosine-mediated signal transduction. FEBS Lett. 2012;586:2597–2605. doi: 10.1016/j.febslet.2012.04.054. [DOI] [PubMed] [Google Scholar]
- Marengere LE, Pawson T. Identification of residues in GTPase-activating protein Src homology 2 domains that control binding to tyrosine phosphorylated growth factor receptors and p62. J Biol Chem. 1992;267:22779–22786. [PubMed] [Google Scholar]
- Matsuda T, Feng J, Witthuhn BA, Sekine Y, Ihle JN. Determination of the transphosphorylation sites of Jak2 kinase. Biochem Biophys Res Commun. 2004;325:586–594. doi: 10.1016/j.bbrc.2004.10.071. [DOI] [PubMed] [Google Scholar]
- Müller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- Nicholson SE, et al. Suppressor of cytokine signaling (SOCS)-5 is a potential negative regulator of epidermal growth factor signaling. Proc Natl Acad Sci USA. 2005;102:2328–2333. doi: 10.1073/pnas.0409675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson SE, Willson TA, Farley A, Starr R, Zhang JG, Baca M, Alexander WS, Metcalf D, Hilton DJ, Nicola NA. Mutational analyses of the SOCS proteins suggest a dual domain requirement but distinct mechanisms for inhibition of LIF and IL-6 signal transduction. EMBO J. 1999;18:375–385. doi: 10.1093/emboj/18.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Multiple routes of protein transport from endosomes to the trans Golgi network. FEBS Lett. 2009;583:3811–3816. doi: 10.1016/j.febslet.2009.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Piessevaux J, Lavens D, Peelman F, Tavernier J. The many faces of the SOCS box. Cytokine Growth Factor Rev. 2008;19:371–381. doi: 10.1016/j.cytogfr.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Piganis RA, De Weerd NA, Gould JA, Schindler CW, Mansell A, Nicholson SE, Hertzog PJ. Suppressor of cytokine signaling (SOCS) 1 inhibits type I interferon (IFN) signaling via the interferon α receptor (IFNAR1)-associated tyrosine kinase Tyk2. J Biol Chem. 2011;286:33811–33818. doi: 10.1074/jbc.M111.270207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta HW, Stenmark H. Endocytosis and signaling. Curr Opin Cell Biol. 2011;23:393–403. doi: 10.1016/j.ceb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Rawlings JS, Rennebeck G, Harrison SM, Xi R, Harrison DA. Two Drosophila suppressors of cytokine signaling (SOCS) differentially regulate JAK and EGFR pathway activities. BMC Cell Biol. 2004a;5:38. doi: 10.1186/1471-2121-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004b;117:1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol. 2000;20:3387–3395. doi: 10.1128/mcb.20.10.3387-3395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Yasukawa H, Suzuki A, Kamizono S, Syoda T, Kinjyo I, Sasaki M, Johnston JA, Yoshimura A. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes Cells. 1999;4:339–351. doi: 10.1046/j.1365-2443.1999.00263.x. [DOI] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Matsumoto A, Seki N, Tsukada J, Ransom J, Naka T, Kishimoto T, Yoshimura A, Kubo M. Expression of the suppressor of cytokine signaling-5 (SOCS5) negatively regulates IL-4-dependent STAT6 activation and Th2 differentiation. Proc Natl Acad Sci USA. 2002;99:13003–13008. doi: 10.1073/pnas.202477099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Geisbrecht ER, Montell DJ. Requirement for JAK/STAT signaling throughout border cell migration in Drosophila. Development. 2005;132:3483–3492. doi: 10.1242/dev.01910. [DOI] [PubMed] [Google Scholar]
- Staerk J, Kallin A, Royer Y, Diaconu CC, Dusa A, Demoulin JB, Vainchenker W, Constantinescu SN. JAK2, the JAK2 V617F mutant and cytokine receptors. Pathol Biol. 2007;55:88–91. doi: 10.1016/j.patbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Starr R, Hilton DJ. SOCS: suppressors of cytokine signalling. Int J Biochem Cell Biol. 1998;30:1081–1085. doi: 10.1016/s1357-2725(98)00067-3. [DOI] [PubMed] [Google Scholar]
- Stec WJ, Zeidler MP. Drosophila SOCS proteins. J Signal Transduct. 2011:894510. doi: 10.1155/2011/894510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamiya T, Kashiwagi I, Takahashi R, Yasukawa H, Yoshimura A. Suppressors of cytokine signaling (SOCS) proteins and JAK/STAT pathways: regulation of T-cell inflammation by SOCS1 and SOCS3. Arterioscler Thromb Vasc Biol. 2011;31:980–985. doi: 10.1161/ATVBAHA.110.207464. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinti M, et al. The SH2 domain interaction landscape. Cell Rep. 2013;3:1293–1305. doi: 10.1016/j.celrep.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentino L, Pierre J. JAK/STAT signal transduction: regulators and implication in hematological malignancies. Biochem Pharmacol. 2006;71:713–721. doi: 10.1016/j.bcp.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Vidal OM, Stec W, Bausek N, Smythe E, Zeidler MP. Negative regulation of Drosophila JAK-STAT signalling by endocytic trafficking. J Cell Sci. 2010;123:3457–3466. doi: 10.1242/jcs.066902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfler A, Irandoust M, Meenhuis A, Gits J, Roovers O, Touw IP. Site-specific ubiquitination determines lysosomal sorting and signal attenuation of the granulocyte colony-stimulating factor receptor. Traffic. 2009;10:1168–1179. doi: 10.1111/j.1600-0854.2009.00928.x. [DOI] [PubMed] [Google Scholar]
- Yan R, Small S, Desplan C, Dearolf CR, Darnell JE. Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- Yasukawa H, et al. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. EMBO J. 1999;18:1309–1320. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A. Regulation of cytokine signaling by the SOCS and Spred family proteins. Keio J Med. 2009;58:73–83. doi: 10.2302/kjm.58.73. [DOI] [PubMed] [Google Scholar]
- Zhuang D, Qiu Y, Haque SJ, Dong F. Tyrosine 729 of the G-CSF receptor controls the duration of receptor signaling: involvement of SOCS3 and SOCS1. J Leukoc Biol. 2005;78:1008–1015. doi: 10.1189/jlb.0105032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.