Abstract
Recently, we achieved the first in vitro selection of 2′-O,4′-C-methylene bridged/locked nucleic acid (2′,4′-BNA/LNA) aptamers. High-affinity thrombin-binding aptamers (TBAs) were obtained from DNA-based libraries containing 2′-O,4′-C-methylene-bridged/linked bicyclic ribonucleotides (B/L nucleotides) in the 5′-primer region, using the method of capillary electrophoresis systematic evolution of ligands by exponential enrichment (CE-SELEX). Furthermore, a similar selection protocol could provide TBAs that contain B/L nucleotides in both primer and random regions. We review technical challenges involved in the generation of various BNA libraries using analogs of B/L nucleoside-5′-triphosphate and polymerase variants and also discuss applications of these libraries to the selection of BNA (LNA) aptamers, as well as future prospects for their therapeutic and diagnostic uses.
Keywords: 2′,4′-BNA/LNA; KODDNA polymerase; Bridged (locked) nucleic acid aptamer; Capillary electrophoresis-systematic evolution of ligands by exponential enrichment (CE-SELEX); Xeno-nucleic acid (XNA)
Introduction
B/L nucleotides were independently developed by two research groups: Obika and Imanishi et al. at Osaka University and Wengel et al. at the University of Southern Denmark, and these were reported almost simultaneously in the late 1990s.1-3 The nucleotide analogs are widely recognized as LNA, a term coined by the latter group, but are identical to the 2′,4′-BNA named by the former group. 2′,4′-BNA/LNA exhibits high nuclease resistance and low cytotoxicity in addition to excellent hybridization properties so that it forms a very stable complex with complementary oligonucleotides and has been a promising candidate as a material for nucleic acid drugs, such as antisenses, antigenes, siRNAs, and miRNAs.4 Such molecules targeting DNA and RNA can be created by rational molecular design based on known base-pair modes. In contrast, nucleic acid aptamers cannot be rationally designed, although they can target non-nucleic acids such as metabolites, proteins, cells, and viruses.5-8 Therefore, selection methods such as SELEX, using a random library, are thus far the only way to acquire them.
Although various improved SELEX methods have been developed to date,9 they all include a step to amplify active species that can bind to the target using polymerase. This enzymatic step proceeds smoothly when a natural DNA or RNA library is used, but it could become a bottleneck when a modified nucleic acid library is used. This is because the low yield and low accuracy involved in the enzymatic synthesis of modified nucleic acids generally make enrichment of active species very difficult during selection rounds. Therefore, great efforts have been made to create BNA aptamers. Earlier, use of post-SELEX modification methods were mainly reported in studies on BNA aptamer development.10-25 However, in recent years, in vitro selection of BNA aptamers is preferred owing to the discovery of polymerases available for BNA-containing oligonucleotide syntheses and genetic engineering of these polymerases. Last year, Pinheiro et al. developed Tgo DNA polymerase variants,26 which enable transcription and reverse transcription of six different XNAs (xeno-nucleic acids): HNA (1,5-anhydrohexitol nucleic acid), CeNA (cyclohexenyl nucleic acid), ANA (arabinonucleic acid), FANA (2′-fluoroarabinonucleic acid), TNA (α-L-threofuranosyl nucleic acid), and 2′,4′-BNA/LNA (Fig. 1). Furthermore, they successfully obtained HNA aptamers by a conventional SELEX method. However, 2′,4′-BNA/LNA aptamers were not reported, implying that it would be inefficient to recover 2′,4′-BNA/LNA aptamers from a 2′,4′-BNA/LNA library only by improvement of polymerases. Thus, no reports exist on generation of LNA aptamers by in vitro selection, as Wengel et al. mentioned in their review article published in December, 2012.27
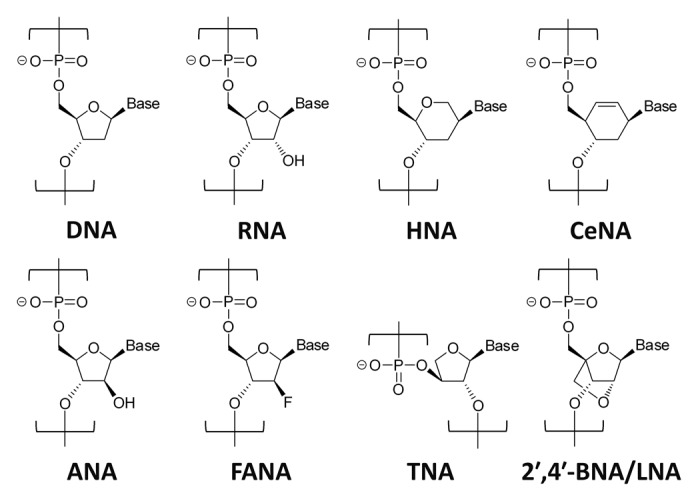
Figure 1. Chemical structure of DNA, RNA, and XNAs. The six XNAs can be transcribed and reverse-transcribed using polymerase variants.
BNA development for therapeutic use
Nucleic acids are readily degraded by various nucleases in vivo; therefore, high biostability is required for nucleic acid drugs. To evade enzymatic degradation, majority of nucleic acids currently in clinical trials are S-oligos,4 which substitute phosphorothioates for phosphates in the backbone structure. However, recently 2′,4′-BNA/LNA has come to the fore as a promising candidate for therapeutic use owing to its high affinity and specificity to DNA/RNA targets as well as its high nuclease resistance. For example, a 12-mer modified oligodeoxynucleotide (ODN), in which six residues are replaced with 2′,4′-BNA/LNA, binds to target RNA much more tightly than the corresponding natural ODN, in addition to its markedly improved biostability.2 Since the invention of 2′,4′-BNA/LNA, various B/L nucleoside analogs have been developed (Fig. 2), and their performances have been analyzed as new drug candidates. Among those, 2′,4′-BNACOC, 2′,4′-BNANC, and 2′,4′-BNANC(Me) were found to exhibit further improved nuclease resistance while still retaining binding affinities to single-stranded RNA and/or double-stranded DNA as a target.28-31
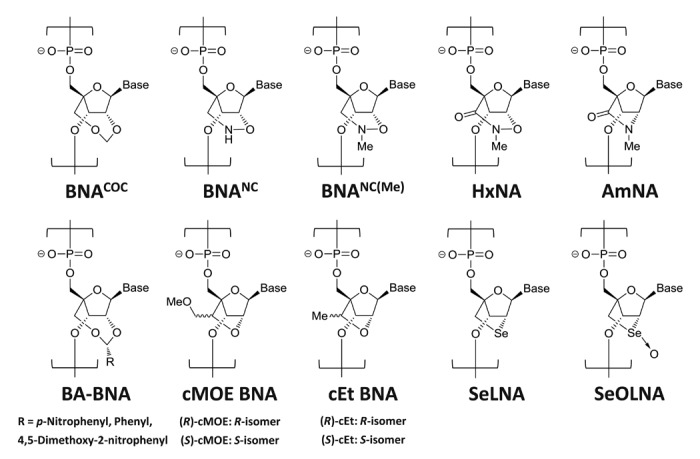
Figure 2. Examples of 2′,4′-BNA/LNA analogs that have been developed to date.
The first model of BNAs, i.e., 2′,4′-BNA/LNA has already reached clinical trials. Santaris Pharma A/S in Demark has developed SPC 3649 (miravirsen) and SPC 4955 that are being tested in phase II and I clinical studies, respectively.32,33 SPC 3649 is a candidate therapeutic drug for hepatitis C; it is a microRNA and targets miR-122, which is closely associated with hepatitis C virus (HCV) replication. It was first observed to lower blood low density lipoprotein cholesterol (LDL-C) levels. However, it was recently observed to greatly reduce HCV levels in the liver and blood using drug dosage tests in chimpanzees with chronic hepatitis C infection. The preclinical test results of 2′,4′-BNA/LNA were quite favorable; its beneficial effects can be maintained for several months in the case with the longest duration, without any remarkable side effects. Miravirsen is of considerable interest not only because the current hepatitis C drugs, such as interferon α and Ribavirin, are inadequate in terms of efficacy but also because it has a different mechanism of action.
Meanwhile, SPC 4955 is a potential antisense nucleic acid drug for hypercholesterolemia therapy. It targets the mRNA of apolipoprotein B (ApoB), a major component of LDL-C, and lowers blood LDL-C levels because of the inhibition of ApoB synthesis in liver. Interestingly, in vivo beneficial effects of SPC 4955 were observed to be superior to those of conventional antisense nucleic acids, which are several to 10 residues longer in length (18–22-mers) because of the excellent hybridization properties of 2′,4′-BNA/LNA.34
Advanced types of BNAs are also developed and being studied for medicinal and/or some other applications. For example, benzylidene acetal type BNAs (BA-BNAs) with 2′,4′-bridged structures that cleaved upon exposure to appropriate external stimuli, i.e., light, acid and reductant, were designed and synthesized.35,36 Cleavage of the bridge between 2′-oxygene and 4′-carbon removed conformational restrictions on the sugar moiety and changed the hybridizing ability and enzymatic stability of the ODNs containing BA-BNAs. It was well known that replacement of 2′-oxygen by the other heteroatoms showed little influence on the binding properties of 2′,4′-BNA/LNA.37-40 Selenomethylene locked nucleic acid (SeLNA), in which 2′-oxygen is replaced by selenium, also showed excellent hybridization affinity, while its oxidized form, selenoxide-bridged nucleic acid (SeOLNA), revealed lower binding affinity compared with SeLNA. Thus, SeLNA enables reversible hybridization to complementary ssDNA or ssRNA strands in response to redox changes.41
Hydroxamate-bridged nucleic acid (HxNA) possesses a six-membered perhydro-1,2-oxazin-3-one ring, which is also the 2′,4′-BNA/LNA analog. The HxNA-modified ODNs showed selectively high affinity toward ssRNA along with superior nuclease resistance. It is of interest that exposure of HxNA-modified ODNs to 3′-exonuclease resulted in gradual opening of the bridge, which stopped further digestion of the ODNs.42 Amido-bridged nucleic acid (AmNA) has a carbonyl functionality at the 2′,4′-bridged structure just like HxNA. Potent and RNA selective hybridizing ability and excellent biostabilities of AmNA-modified ODNs were observed. In addition, the in vitro antisense potencies of AmNA-modified ODNs was tested to reveal improved potencies of AmNAs relative to 2′,4′-BNA/LNA counterparts.43 2′,4′-Constrained 2′-O-methoxyethyl (cMOE) and 2′-O-ethyl (cEt) nucleic acid derivatives were developed by ISIS Pharmaceuticals. These derivatives have a methoxymethyl or methyl substituent at the bridged structure of 2′,4′-BNA/LNA and showed higher nuclease resistance without any decrease in binding affinity.44,45
Thus, these examples demonstrate that BNA-containing oligonucleotides can adequately satisfy the criteria required for use as medicines and strongly encourage us to develop therapeutic BNA aptamers.
2′,4′-BNA/LNA aptamers created by post-SELEX modification
The performance of SELEX-selected aptamers can be improved by post-SELEX modification. A typical, successful example is the first aptamer drug i.e., pegaptanib, which is used for age-related macular degeneration (AMD) therapy.46 The precursory form of pegaptanib was recovered from a modified RNA library containing 2′-fluoropyrimidine nucleotides (U, C) by SELEX, and then all natural purine nucleotides (A, G) except for only two residues in the strand were successfully replaced with the corresponding 2′-methoxy purine nucleotides by post-SELEX modification. However, to date, the only known method for determining which nucleotides are suitable for replacement is by trial and error, i.e., by preparing variants and analyzing their respective binding activities.
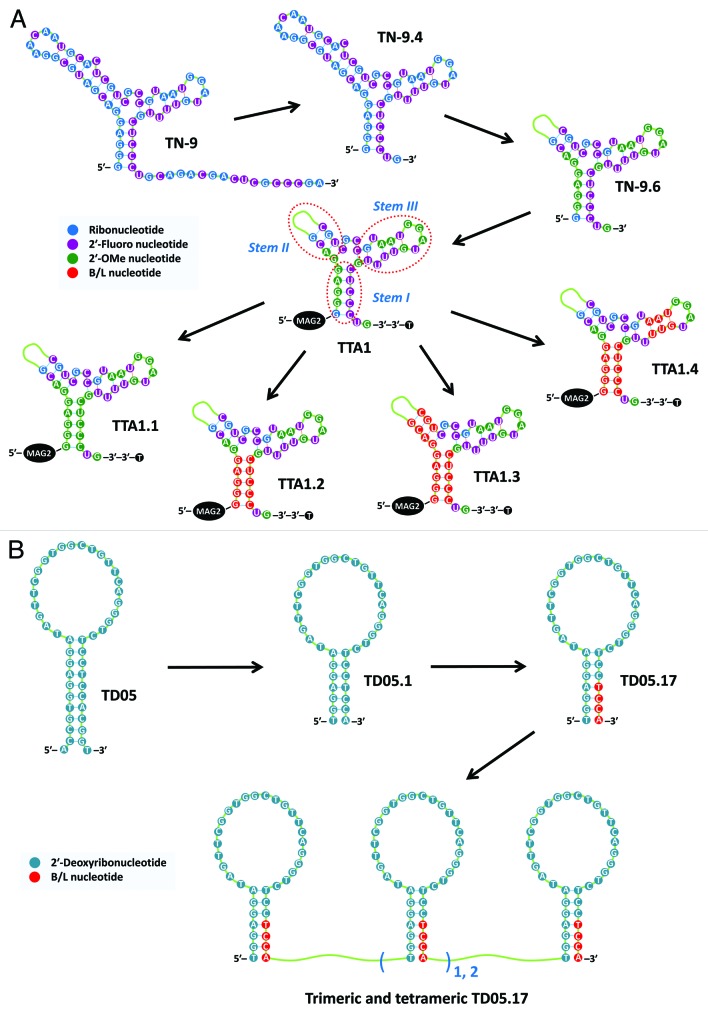
Successful examples of 2′,4′-BNA/LNA aptamers by post-SELEX modification have been reported.10-25 Schmidt et al. developed an LNA aptamer, TTA1, specific for human tenascin-C (TN-C).11 The original form TN-9 was selected from a 2′-fluoropyrimidine RNA library (Fig. 3A). The TN-9 71-mer was truncated by removing 16 nucleotides from the 3′-end, which were selected on the basis of the predicted secondary structure.47 The resulting 55-mer TN-9.4 could form a three-way junction structure. A further deletion of TN-9.4 replaced 17 nucleotides from the 5′-end at positions 10–26 with a single spacer (CH2CH2O)6 to produce the 39-mer TN-9.6. To enhance aptamer biostability, 14 of the 19 purine nucleotides were successfully replaced with 2′-deoxy-2′-OMe nucleotides through structure–activity relationship (SAR) studies. However, it was found that four guanosines at positions 9, 11, 14, and 17 should be retained as the 2′-ribo form to maintain high affinity with the TN-C target. The complete structure of TTA1 comprised a thymidine cap at the 3′-end to prevent digestion by exonucleases; a mercapto-acetyl-glycinyl-glycine (MAG2) chelate conjugated through a hexyl-amino linker at the 5′-end for application in tumor radioimaging; and the abovementioned 39-mer oligonucleotide. Full replacement with the corresponding B/L nucleotides in the stem I moiety (positions 1–5 and 33–37) produced TTA1.2, which was found to exhibit superior stability in human plasma and target binding affinity (t1/2 = 53 h, EC50 = 2.0 nM) compared with TTA1 (t1/2 = 42 h, EC50 = 5.8 nM). In contrast, replacement with 2′-OMe nucleotides at the same positions, which yielded TTA1.1, resulted in a more than 2-fold decrease in binding affinity, although stability was substantially improved (t1/2 = 49 h, EC50 = 13.7 nM). In addition to the stem I modification, replacement with B/L nucleotides in stems II or III produced a loss of binding activity, although extended plasma lifetimes were observed (t1/2 = 69 h and 72 h) for the respective TTA 1.3 and TTA 1.4 aptamers.
Figure 3. Examples of 2′,4′-BNA/LNA aptamers created by post-SELEX modification. RNA-based 2′,4′-BNA/LNA aptamer specific for TN-C (A) and DNA-based 2′,4′-BNA/LNA aptamer specific for B cells (B).
B/L nucleotides are partially compatible with those in DNA aptamers (Fig. 3B). Mallikaratchy et al. created a B/L-nucleotide-containing multivalent DNA aptamer specific to B-cells by engineering a 45-mer DNA aptamer, TD05, which specifically binds to human membrane immunoglobulin M (mIgM) as a B-cell receptor.23 First, TD05, which was predicted to form a hairpin (stem-loop) structure, was truncated to produce a 37-mer, TD05.1, by removing several residues from the 3′- and 5′-stem regions. The four pyrimidine nucleotides at the 3′-end of TD05.1 were successfully replaced with the corresponding B/L nucleotides to produce TD05.17. It exhibited enhanced B-cell binding affinity, which was approximately 8.3- and 1.2-fold greater than that of TD05 and TD05.1, respectively. In contrast, replacement with B/L nucleotides in other regions of TD05.1 produced aptamers with reduced activities. Trimeric and tetrameric TD05.17 aptamers with appropriate lengths of a polyethylene glycol linker exhibited increased conformational stability and nuclease resistance and high avidity for membrane-associated mIgM but not for soluble IgM.
Thus, post-SELEX modification has been successful, but it still requires considerable time and effort, because binding affinities could be markedly decreased or eliminated depending on the position of the replacement. Hence, intense interest has focused on the development of enzymatic BNA syntheses to apply BNA to SELEX methods.
Enzymatic syntheses of BNAs
Enzymatic BNA syntheses have been reported in the literature since the middle of the last decade.48-65 Veedu et al. first tested B/L nucleotide incorporation using Phusion High Fidelity DNA polymerase, 9°Nm DNA polymerase, and Pfu DNA polymerase, and observed that the first and second of these three polymerases could accept B/L nucleoside 5′-triphosphates as substrates and catalyze primer extension reactions to yield DNA-based 2′,4′-BNA/LNA strands.49,50 Subsequently, several other polymerases, such as Taq DNA polymerase, Large (Klenow) fragments, T4 DNA polymerase, Pfx DNA polymerase, Speed STAR HS DNA polymerase, T7 RNA polymerase, E. coli RNA polymerase, AMV reverse transcriptase, and mutant T7 R&DNA polymerase have been similarly assessed.51 From these results, they concluded that Phusion High Fidelity DNA polymerase was the only enzyme that could efficiently incorporate B/L nucleotides. In 2008, we first demonstrated that KOD Dash DNA polymerase, derived from hyperthermophilic archaeon pyrococcus kodakaraensis (KOD), was superior to Phusion High Fidelity DNA polymerase52 because of its reduced 3′,5′ exonuclease activity and applicable not only for the synthesis of 2′,4′-BNA/LNA but also for other types of BNA, i.e., 2′,4′-BNACOC and 2′,4′-BNANC. Previously, we demonstrated the usefulness of KOD DNA polymerases in enzymatic syntheses of base-modified DNAs and investigated their application in SELEX methods since 2001.66-78 Therefore, we readily noticed that KOD-related DNA polymerases are also applicable DNA synthesis involving modified sugars. This knowledge is now being widely accepted among researchers engaged in relevant studies.54,58-60,62,65,79-83 To date, enzymatic syntheses of various modified DNAs using KOD-related DNA polymerases have been reported. A DNA polymerase named KOD XL is commonly used (which is essentially the same as KOD Dash); KOD XL comprises a mixture of approximately 2:98 wild-type KOD DNA polymerase and KOD(exo-) DNA polymerase. In the exo-knockout form, 3′,5′ exonuclease activity is heavily reduced or absent.
Polymerase chain reaction (PCR) and reverse-transcription involving B/L nucleotide-containing oligonucleotide have been examined. Under certain reaction conditions, PCR-amplified DNA-based 2′,4′-BNA/LNA strands, DNA-based 2′,4′-BNA/LNA strands from RNA templates, and DNA strands from RNA-based 2′,4′-BNA/LNA templates have been successfully obtained.53,61 However, these interesting and potentially useful findings have yet to be exploited for in vitro selection of BNA aptamers; this may be because direct PCR amplification of BNA is not absolutely essential for BNA aptamer selection, and RNA-based BNA oligomers are limited because of biostability as well as by a limited variety of available RNA polymerases.
In applying BNA libraries to SELEX methods, yield and fidelity in the polymerase reaction step of BNA syntheses are of high importance. A clever approach is to select desired polymerases from random libraries.84,85 This approach has been successfully implemented and was published in Science in 2012, and a variant of Tgo DNA polymerase derived from Thermococcus gorgonarius, PolC7, was created.26 Another way is to design polymerase variants and assess their catalytic abilities one by one56; we discovered several KOD variants suitable for BNA syntheses in our collaboration with Toyobo Co., Ltd. These polymerases are promising candidates for applications in BNA aptamer selection and will be used as parent enzymes for further improvements.
2′,4′-BNA/LNA aptamers created by SELEX selection
During SELEX rounds, low yield in enzymatic syntheses could cause biased cutoff of so-called difficult sequences containing B/L nucleotide-dense and/or GC-rich alignments even if they are highly active for target binding, while low fidelity of polymerase could interfere with reproduction of selected sequences. Performances of polymerases should be further improved. However, we thought that it might be possible to implement 2′,4′-BNA/LNA aptamer selection using currently available engineered polymerases, if appropriate selection methods were chosen and were able to be sophisticated. We assumed that methodologies that require only a single selection round for enrichment such as Mono-LEX86 and non-SELEX selection87 should be promising because of the minimized number of times of enzymatic oligonucleotide synthesis in the entire process until aptamer acquisition.
To this end, we have first challenged to obtain 2′,4′-BNA/LNA aptamers specific for human α-thrombin using non-SELEX selection, but unfortunately could not attain sufficient enrichment of these aptamers (unpublished work). Then, we attempted the CE-SELEX method88-91 as an original model for non-SELEX selection. Unlike many other alternative SELEX techniques based on the principal of affinity chromatography involving molecular interactions at the solid–liquid interface (Fig. 4A), CE-SELEX can separate active species form non-active species in the liquid phase by non-equilibrium capillary electrophoresis of equilibrium mixtures (NECEEM)92 (Fig. 4B). Furthermore, in NECEEM, electro-osmotic flow (EOF), which occurs from the anode to the cathode, can carry the target–aptamer complexes prior to unbound free oligonucleotides. Hence, CE-SELEX may be one of the best methods to enable efficient aptamer enrichment because it maximally excludes contamination of non-active species; in the typical SELEX methods, species non-specifically bound to the solid support cannot be completely washed out while maintaining the desirable specific binding, resulting in inevitable contamination of non-active species in the elution of active species.
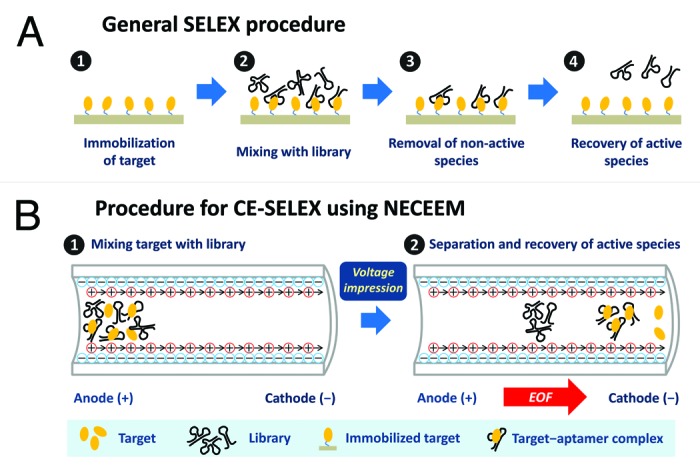
Figure 4. Procedure for standard SELEX based on the principal of affinity chromatography (A), and that for CE-SELEX using NECEEM employed for 2′,4′-BNA/LNA aptamer selection (B).
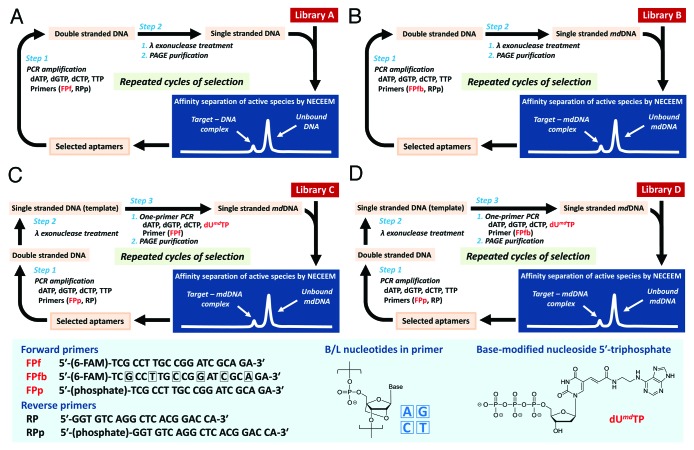
Accordingly, we conducted in vitro selection of aptamers specific for human α-thrombin from four different libraries A–D93 prior to library E selection (Figs. 5 and 6).94 Here, A is a common DNA library, whereas B, D, and E are DNA-based libraries containing B/L nucleotides at intervals of two residues in the forward primer region. Libraries C and D contain the C5-modified thymidine, i.e., (E)-5-(2-{N-[2-(N6-adeninyl)ethyl]}carbamylvinyl)-2′-deoxyuridine (t) instead of the natural thymidines (T’s) in the random region and the reverse primer-binding region at the 3′ end. Library E contains B/L thymidines (T’s) instead of T’s in the non-primer region, except for an 8-mer tail attached at the 3′ end, in addition to that in the primer region.
Figure 5. Schematic illustrations of SELEX experiments using (A) library A, (B) library B, (C) library C, and (D) library D, respectively. The primer P1fb contains six B/L nucleotides (A, G, C, and T), and dUmdTP is a 5′-triphosphate analog of the C5-modified thymidine bearing N6-ethyladenine (t).
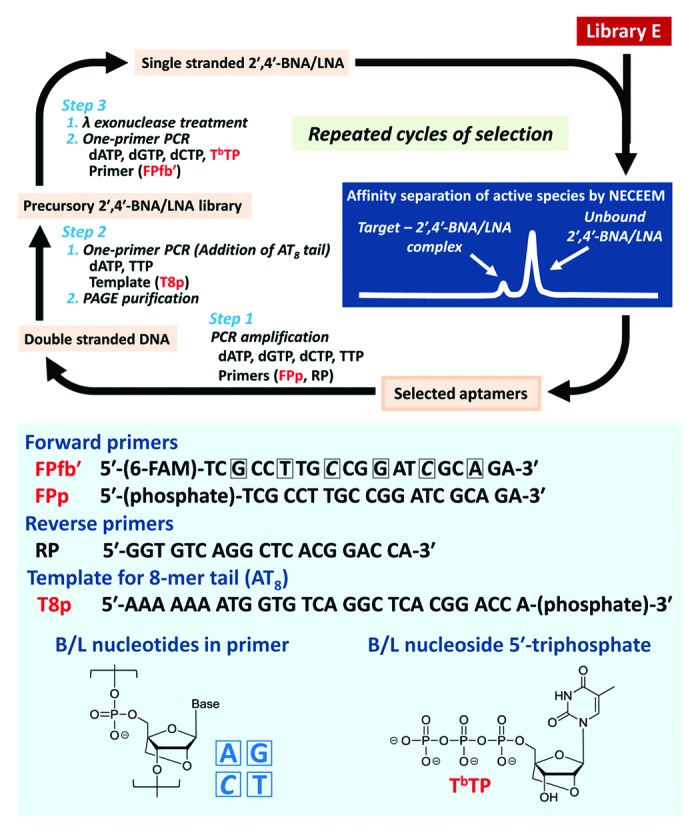
Figure 6. The schematic illustration of library E selection. The primer P1fb′ contains six B/L nucleotides (A, G, 5-methyl C, and T). The 8-mer tail (AT8) was attached for separation of single-stranded BNA/LNA library.
Four cycles of separation and amplification were required for sufficient enrichment of active species in libraries A−C, while six cycles were required for enrichment in library D (Fig. 7). In library A–D selections, peak areas of the thrombin–aptamer complex were significantly increased as selection rounds progressed. Selection outcomes from libraries A and B indicated that B/L nucleotides in the primer region have marginal effects on the binding activity or the biostability of the selected aptamers. However, insertions of t’s into the non-primer region were found to enable acquisition of aptamers that can display binding activities dependent on the presence of B/L nucleotides in the primer region, as observed in selection outcomes from library D. Furthermore, significant enhancement of biostability was observed in aptamers from library D but not in those from the other libraries. Our results indicated that B/L nucleotides introduced in advance into the primer region could influence selection outcomes and contribute to aptamer performances by combination with base-modified nucleotides. This knowledge will provide novel strategies for design and selection of chimeric BNA aptamers with desired functions.
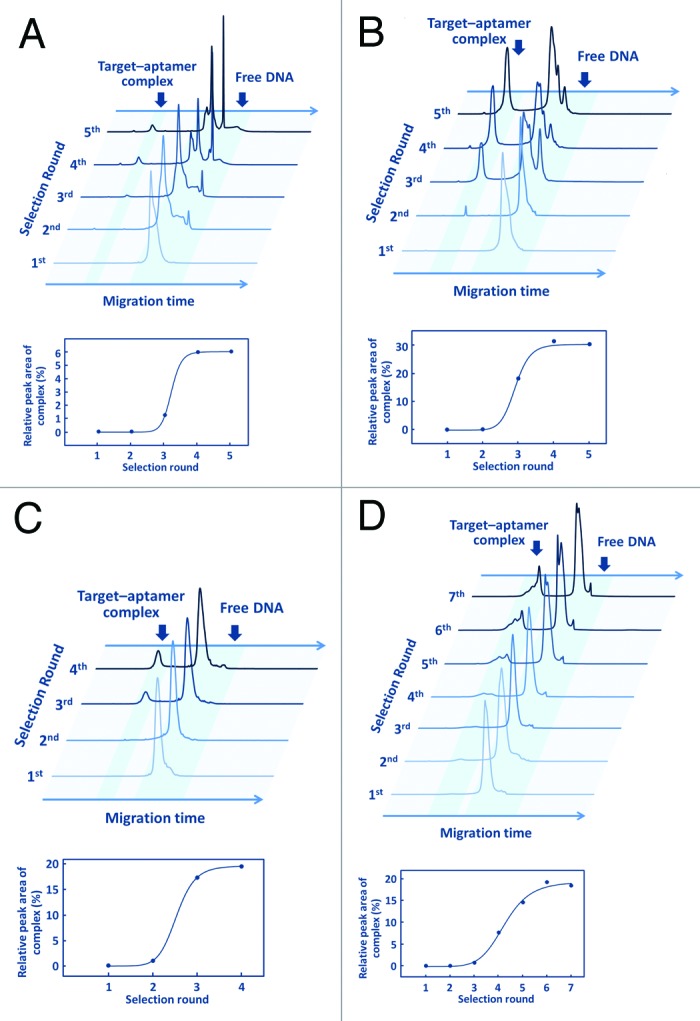
Figure 7. Process of active species enrichment in selection rounds. Capillary electrograms for (A) library A, (B) library B, (C) library C, and (D) library D of each round with human thrombin (upper graphics). All electrograms recorded fluorescent intensity of 5′-labeled 6-FAM vs. migration time. Saturation curves of library enrichment for TBA acquisition (lower graphics).
Unlike library A–D selections, the selection of library E containing B/L nucleotides in the non-primer region was not simple. In 2′,4′-BNA/LNA syntheses, KOD Dash DNA polymerase was used for library A–D selections, while a 19:1 enzyme mix of KOD Dash and KOD mutant DNA polymerases (KOD2)56 was used for library E selection. Unlike in library A–D selections, obvious complex peaks could not be observed in library E selection, and electrograms seemed to remain unchanged during the course of selection (Fig. 8A). However, a substantially broadened peak was observed prior to free 2′,4′-BNA/LNA migration (Fig. 8B). The increase in the peak area at the broadening part was surely saturated after around 10 rounds of selection to yield the enriched pool of active species. Among 40 sequences identified, the binding affinity (Kd) of the best aptamer was 18 nM, while some sequences did not show any binding activities (Table 1). In addition, Kd values of A–D aptamers were in the range of sub-nanomolar to several ten nanomolar levels, implying that conditions of library E selection may not yet be completely sophisticated in spite of our efforts. However, our first demonstration of the BNA (LNA) aptamer selection should have made a great impact on the recent progress in aptamer development.
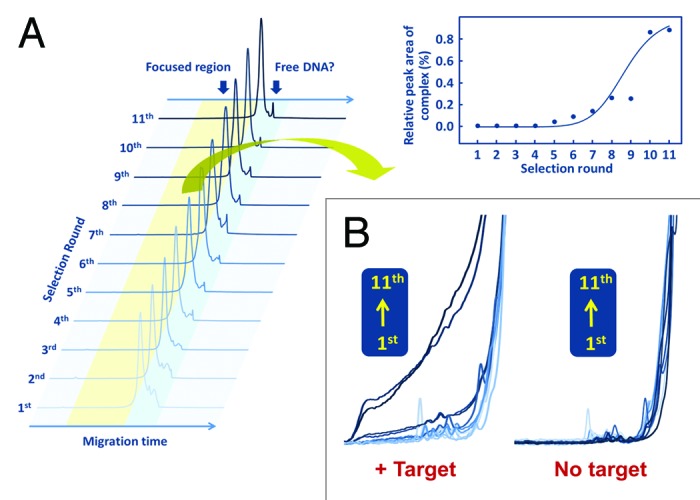
Figure 8. Process of active species enrichment in selection rounds. (A) Capillary electrograms for library E of each round with human thrombin (left graphic). All electrograms recorded fluorescent intensity of 5′-labeled 6-FAM vs. migration time. Saturation curve of library enrichment for TBA acquisition (right graphic). (B) Enlarged and overlapped view of each round of capillary electrograms with or without human thrombin.
Table 1. Sequences and affinities of representative TBAs recovered from libraries A−E.
| Aptamer * |
Sequence† |
Kd (nM)‡ |
|---|---|---|
| Library A: TCGCCTTGCCGGATCGCAGA(random region)TGGTCCGTGAGCCTGACACC | ||
| A#1,2,3,4,6 |
GAATCAGGTTCACGTTGGTTCGGTTGGTAT |
3.1 |
| A#5,14 |
TTGTCAGGTGGCTTCGTGGTTCGGTTGGTG |
1.9 |
| A#8,9,11,12,13,16 | TCGTGGCAGGATCCGTTGGTTTTGGTTGGG | 7.9 |
| Library B: TCGCCTTGCC GGATCGCAGA (random region) TGGTCCGTGA GCCTGACACC | ||
|---|---|---|
| B#1,8 |
TCGTGGCAGGATCCGTTGGTTTTGGTTGGG |
3.7 |
| B#2,15 |
GGGCACTTGGCTGGTTGGTGGGTTTTGCCC |
5.0 |
| B#3,5,6,10,12,14 |
GAATCAGGTTCACGTTGGTTCGGTTGGTAT |
3.3 |
| B#4 |
GTGTGGTGGGTTGGCTCTGGGTGACCTCTG |
1.9 |
| B#11 | AGTGGCCGGCTCCGCGTGGGGGGTTGGGTT | 2.1 |
| Library C: TCGCCTTGCC GGATCGCAGA(random region)tGGtCCGtGA GCCtGACACC | ||
|---|---|---|
| C#1 |
AAGCAGCtCGCGtACGAAtttttGttGGtA |
0.57 |
| C#2,11 |
GtGGGGAtCtCtGtttAtCCACttCAGtGC |
2.6 |
| C#4 |
GGGAGGCACGCGAtGCGAGtttACCCtACG |
2.9 |
| C#5,14,20 |
ttCGGGGGCGCtCCCCtCAtGtttACCCtAG |
10 |
| C#7 |
GGGAGGGCAGCGGCAGAttGGCGGttAGGC |
34 |
| C#8 |
CttGttAtACGGtCtCttGGCCCGGttGGC |
4.3 |
| C#9 |
tGGCCCCCCGtttGCGGttttttCGtAGGC |
7.5 |
| C#10 | ttCtAGGGAAAGGCtCGCtttttGttGGtA | 1.7 |
| Library D: TCGCCTTGCC GGATCGCAGA (random region) tGGtCCGtGA GCCtGACACC | ||
|---|---|---|
| D#1,10 |
ttGGtCAAAGGtCCGACGAGttttGCtAGC |
2.3 |
| D#2,3 |
GCCGtGAGGACGCGCttGttGGGGGGttGt |
4.0 |
| D#6,9 |
ttGGtCGAAGGttCCGGGGtCCttAGtGGt |
0.26 |
| D#7 |
tGCGGAGtGGCCAAtCACtttttGttGGCA |
0.79 |
| D#11 |
ACGGGCGtGGCttCAtAAtttttGttGGtA |
0.50 |
| D#12 |
ttAGtCGAAGGttCCtGGCCACttAtAtCG |
0.093 |
| D#13 | CGGCCGAGGtGGCCAACGtttttGttGGtA | 0.91 |
| Library E: TCGCCTTGCC GGATCGCAGA (random region) TGGTCCGTGA GCCTGACACC ATTTTTTT | ||
|---|---|---|
| E#1,8,11,12,13,30,35,37 |
GGAGCAAGAT GTCACGGTCC GCATGCAGAG |
NB |
| E#3,18 |
CTCTTGACAG GACTCATGCA CGGGGGGGCT |
27 |
| E#6,20,25,33 |
AGTGGCTGGC TCCGCGTGGG GGGTTGGGTT |
NB |
| E#14,38 |
GAGGCAATGT GTGGGTACCG CATTACCCAGT |
NB |
| E#17 |
CTCTTAACAG GACAAGTGAA ACAAACCCGC |
23 |
| E#26 |
CACACAGGTTTGCGGCCGCTTCTGGGCAGC |
NB |
| E#23,28,32 |
CTCTTGACAG GACACATGCA CGCGGGGGCT |
20 |
| E#40 | CTCTTAACAG GACTCATGCA CGGGGGGGCT | 18 |
The name of each aptamer indicates the type of library used and the clone number. †Sequences are aligned in the 5′ to 3′ direction. The B/L nucleotides with adenine, guanine, cytosine, 5-methylcytosine, and thymine are enclosed in squares (A, G, C, C, and T). The C5-modified thymidine is shown in bold letters (t). ‡Kd values were determined by non-equilibrium capillary electrophoresis of equilibrium mixtures (NECEEM). NB, no binding.
Conclusions and future prospects
Using CE-SELEX, we succeeded in in vitro selection of aptamers that contain B/L nucleotides in the primer region (library B and D selections) and in both the random and constant regions (library E selection), respectively. In general, the presence of B/L nucleotides in the primer region only provides marginal effects but can influence selection outcomes if base-modified nucleotides were employed in the non-primer regions. Accordingly, various chimeric BNA aptamers will be developed by combinations with nucleotide analogs in a broad chemical repertoire.
The attempts to reduce the number of selection rounds basically aim for saving labor and time, i.e., production costs. However, as far as BNA aptamer selection is concerned, it would be critical to complement imperfection of polymerase performances.95 In this respect, it may be worthwhile to attempt some single round selection methodologies again. The result would depend on whether active species can be sharply and efficiently separated from non-active ones. Meanwhile, polymerase variants that enable BNA replication, transcription, and reverse transcription with much higher yield and fidelity should be developed to facilitate BNA aptamer development. We believe that progress in both separation technology and polymerase engineering will enable direct selection of BNA aptamers such as those containing B/L nucleotides with high density and those containing improved analogs of 2′,4′-BNA/LNA, which are extremely stable under physiological conditions encountered during medical applications. Finally, as the advanced model of our present system, selection from enzymatically synthesized biopolymer libraries involving various types of modifications in base, sugar, and phosphate moieties will be possible.
Acknowledgments
This work was supported by a Grant for Industrial Technology Research from the New Energy and Industrial Technology Development Organization (NEDO) of Japan and by a Grant in-Aid for Scientific Research (C), No. 25350962, from the Japan Society for the Promotion of Science (JSPS).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/artificialdna/article/25786
References
- 1.Obika S, Nanbu D, Hari Y, Morio K, In Y, Ishida T, et al. Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3′-endo sugar puckering. Tetrahedron Lett. 1997;38:8735–8. doi: 10.1016/S0040-4039(97)10322-7. [DOI] [Google Scholar]
- 2.Obika S, Nanbu D, Hari Y, Andoh J, Morio K, Doi T, et al. Stability and structural features of the duplexes containing nucleoside analogs with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39:5401–4. doi: 10.1016/S0040-4039(98)01084-3. [DOI] [Google Scholar]
- 3.Singh SK, Koshkin AA, Wengel J, Nielsen P. LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem Commun (Camb) 1998;4:455–6. doi: 10.1039/a708608c. [DOI] [Google Scholar]
- 4.Yamamoto T, Nakatani M, Narukawa K, Obika S. Antisense drug discovery and development. Future Med Chem. 2011;3:339–65. doi: 10.4155/fmc.11.2. [DOI] [PubMed] [Google Scholar]
- 5.Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu Rev Biochem. 1995;64:763–97. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 6.Osborne SE, Ellington AD. Nucleic Acid Selection and the Challenge of Combinatorial Chemistry. Chem Rev. 1997;97:349–70. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 7.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–47. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 8.Famulok M, Mayer G, Blind M. Nucleic acid aptamers-from selection in vitro to applications in vivo. Acc Chem Res. 2000;33:591–9. doi: 10.1021/ar960167q. [DOI] [PubMed] [Google Scholar]
- 9.McKeague M, Derosa MC. Challenges and opportunities for small molecule aptamer development. J Nucleic Acids. 2012; 2012748913; PMID23150810. 10.1155/2012/748913 [DOI] [PMC free article] [PubMed]
- 10.Darfeuille F, Hansen JB, Orum H, Di Primo C, Toulmé JJ. LNA/DNA chimeric oligomers mimic RNA aptamers targeted to the TAR RNA element of HIV-1. Nucleic Acids Res. 2004;32:3101–7. doi: 10.1093/nar/gkh636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt KS, Borkowski S, Kurreck J, Stephens AW, Bald R, Hecht M, et al. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004;32:5757–65. doi: 10.1093/nar/gkh862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darfeuille F, Reigadas S, Hansen JB, Orum H, Di Primo C, Toulmé JJ. Aptamers targeted to an RNA hairpin show improved specificity compared to that of complementary oligonucleotides. Biochemistry. 2006;45:12076–82. doi: 10.1021/bi0606344. [DOI] [PubMed] [Google Scholar]
- 13.Förster C, Brauer AB, Brode S, Schmidt KS, Perbandt M, Meyer A, et al. Comparative crystallization and preliminary X-ray diffraction studies of locked nucleic acid and RNA stems of a tenascin C-binding aptamer. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:665–8. doi: 10.1107/S1744309106020343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Primo C, Rudloff I, Reigadas S, Arzumanov AA, Gait MJ, Toulmé JJ. Systematic screening of LNA/2′-O-methyl chimeric derivatives of a TAR RNA aptamer. FEBS Lett. 2007;581:771–4. doi: 10.1016/j.febslet.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 15.Virno A, Randazzo A, Giancola C, Bucci M, Cirino G, Mayol L. A novel thrombin binding aptamer containing a G-LNA residue. Bioorg Med Chem. 2007;15:5710–8. doi: 10.1016/j.bmc.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Lebars I, Richard T, Di Primo C, Toulmé JJ. NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA-modified aptamer. Nucleic Acids Res. 2007;35:6103–14. doi: 10.1093/nar/gkm655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonifacio L, Church FC, Jarstfer MB. Effect of locked-nucleic acid on a biologically active g-quadruplex. A structure-activity relationship of the thrombin aptamer. Int J Mol Sci. 2008;9:422–33. doi: 10.3390/ijms9030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebars I, Richard T, Di Primo C, Toulmé JJ. LNA derivatives of a kissing aptamer targeted to the trans-activating responsive RNA element of HIV-1. Blood Cells Mol Dis. 2007;38:204–9. doi: 10.1016/j.bcmd.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez FJ, Kalra N, Wengel J, Vester B. Aptamers as a model for functional evaluation of LNA and 2′-amino LNA. Bioorg Med Chem Lett. 2009;19:6585–7. doi: 10.1016/j.bmcl.2009.10.039. [DOI] [PubMed] [Google Scholar]
- 20.Barciszewski J, Medgaard M, Koch T, Kurreck J, Erdmann VA. Locked nucleic acid aptamers. Methods Mol Biol. 2009;535:165–86. doi: 10.1007/978-1-59745-557-2_10. [DOI] [PubMed] [Google Scholar]
- 21.Förster C, Oberthuer D, Gao J, Eichert A, Quast FG, Betzel C, et al. Crystallization and preliminary X-ray diffraction data of an LNA 7-mer duplex derived from a ricin aptamer. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65:881–5. doi: 10.1107/S1744309109029145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanwar JR, Roy K, Kanwar RK. Chimeric aptamers in cancer cell-targeted drug delivery. Crit Rev Biochem Mol Biol. 2011;46:459–77. doi: 10.3109/10409238.2011.614592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mallikaratchy PR, Ruggiero A, Gardner JR, Kuryavyi V, Maguire WF, Heaney ML, et al. A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic Acids Res. 2011;39:2458–69. doi: 10.1093/nar/gkq996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen EB, Nielsen JT, Nielsen C, Filichev VV. Enhanced anti-HIV-1 activity of G-quadruplexes comprising locked nucleic acids and intercalating nucleic acids. Nucleic Acids Res. 2011;39:2470–81. doi: 10.1093/nar/gkq1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Förster C, Zydek M, Rothkegel M, Wu Z, Gallin C, Geßner R, et al. Properties of an LNA-modified ricin RNA aptamer. Biochem Biophys Res Commun. 2012;419:60–5. doi: 10.1016/j.bbrc.2012.01.127. [DOI] [PubMed] [Google Scholar]
- 26.Pinheiro VB, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, et al. Synthetic genetic polymers capable of heredity and evolution. Science. 2012;336:341–4. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlsen KK, Wengel J. Locked nucleic acid and aptamers. Nucleic Acid Ther. 2012;22:366–70. doi: 10.1089/nat.2012.0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hari Y, Obika S, Ohnishi R, Eguchi K, Osaki T, Ohishi H, et al. Synthesis and properties of 2′-O,4′-C-methyleneoxymethylene bridged nucleic acid. Bioorg Med Chem. 2006;14:1029–38. doi: 10.1016/j.bmc.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 29.Rahman SM, Seki S, Obika S, Haitani S, Miyashita K, Imanishi T. Highly stable pyrimidine-motif triplex formation at physiological pH values by a bridged nucleic acid analogue. Angew Chem Int Ed Engl. 2007;46:4306–9. doi: 10.1002/anie.200604857. [DOI] [PubMed] [Google Scholar]
- 30.Rahman SM, Seki S, Obika S, Yoshikawa H, Miyashita K, Imanishi T. Design, synthesis, and properties of 2′,4′-BNA(NC): a bridged nucleic acid analogue. J Am Chem Soc. 2008;130:4886–96. doi: 10.1021/ja710342q. [DOI] [PubMed] [Google Scholar]
- 31.Prakash TP, Siwkowski A, Allerson CR, Migawa MT, Lee S, Gaus HJ, et al. Antisense oligonucleotides containing conformationally constrained 2′,4′-(N-methoxy)aminomethylene and 2′,4′-aminooxymethylene and 2′-O,4′-C-aminomethylene bridged nucleoside analogues show improved potency in animal models. J Med Chem. 2010;53:1636–50. doi: 10.1021/jm9013295. [DOI] [PubMed] [Google Scholar]
- 32.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 33.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Straarup EM, Fisker N, Hedtjärn M, Lindholm MW, Rosenbohm C, Aarup V, et al. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 2010;38:7100–11. doi: 10.1093/nar/gkq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morihiro K, Kodama T, Nishida M, Imanishi T, Obika S. Synthesis of light-responsive bridged nucleic acid and changes in affinity with complementary ssRNA. Chembiochem. 2009;10:1784–8. doi: 10.1002/cbic.200900241. [DOI] [PubMed] [Google Scholar]
- 36.Morihiro K, Kodama T, Obika S. Benzylidene acetal type bridged nucleic acids: changes in properties upon cleavage of the bridge triggered by external stimuli. Chemistry. 2011;17:7918–26. doi: 10.1002/chem.201100541. [DOI] [PubMed] [Google Scholar]
- 37.Kumar R, Singh SK, Koshkin AA, Rajwanshi VK, Meldgaard M, Wengel J. The first analogues of LNA (locked nucleic acids): phosphorothioate-LNA and 2′-thio-LNA. Bioorg Med Chem Lett. 1998;8:2219–22. doi: 10.1016/S0960-894X(98)00366-7. [DOI] [PubMed] [Google Scholar]
- 38.Singh SK, Kumar R, Wengel J. Synthesis of Novel Bicyclo[2.2.1] Ribonucleosides: 2′-Amino- and 2′-Thio-LNA Monomeric Nucleosides. J Org Chem. 1998;63:6078–9. doi: 10.1021/jo9806658. [DOI] [PubMed] [Google Scholar]
- 39.Singh SK, Kumer R, Wengel J. Synthesis of 2'-Amino-LNA: A Novel Conformationally Restricted High-Affinity Oligonucleotide Analogue with a Handle. J Org Chem. 1998;63:10035–9. doi: 10.1021/jo9814445. [DOI] [Google Scholar]
- 40.Xu J, Liu Y, Dupouy C, Chattopadhyaya J. Synthesis of conformationally locked Carba-LNAs through intramolecular free-radical addition to C=N. Electrostatic and steric implication of the carba-LNA substituents in the modified oligos for nuclease and thermodynamic stabilities. J Org Chem. 2009;74:6534–54. doi: 10.1021/jo901009w. [DOI] [PubMed] [Google Scholar]
- 41.Morihiro K, Kodama T, Kentefu, Moai Y, Veedu RN, Obika S. Selenomethylene locked nucleic acid enables reversible hybridization in response to redox changes. Angew Chem Int Ed Engl. 2013;52:5074–8. doi: 10.1002/anie.201300555. [DOI] [PubMed] [Google Scholar]
- 42.Shrestha AR, Hari Y, Yahara A, Osawa T, Obika S. Synthesis and properties of a bridged nucleic acid with a perhydro-1,2-oxazin-3-one ring. J Org Chem. 2011;76:9891–9. doi: 10.1021/jo201597e. [DOI] [PubMed] [Google Scholar]
- 43.Yahara A, Shrestha AR, Yamamoto T, Hari Y, Osawa T, Yamaguchi M, et al. Amido-bridged nucleic acids (AmNAs): synthesis, duplex stability, nuclease resistance, and in vitro antisense potency. Chembiochem. 2012;13:2513–6. doi: 10.1002/cbic.201200506. [DOI] [PubMed] [Google Scholar]
- 44.Seth PP, Vasquez G, Allerson CA, Berdeja A, Gaus H, Kinberger GA, et al. Synthesis and biophysical evaluation of 2′,4′-constrained 2’O-methoxyethyl and 2′,4′-constrained 2’O-ethyl nucleic acid analogues. J Org Chem. 2010;75:1569–81. doi: 10.1021/jo902560f. [DOI] [PubMed] [Google Scholar]
- 45.Pallan PS, Allerson CR, Berdeja A, Seth PP, Swayze EE, Prakash TP, et al. Structure and nuclease resistance of 2′,4′-constrained 2′-O-methoxyethyl (cMOE) and 2′-O-ethyl (cEt) modified DNAs. Chem Commun (Camb) 2012;48:8195–7. doi: 10.1039/c2cc32286b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng EW, Shima DT, Calias P, Cunningham ET, Jr., Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5:123–32. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 47.Hicke BJ, Marion C, Chang YF, Gould T, Lynott CK, Parma D, et al. Tenascin-C aptamers are generated using tumor cells and purified protein. J Biol Chem. 2001;276:48644–54. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- 48.Nagashima J, Minezaki S, Obika S, Imanishi T, Kuwahara M, Sawai H. Polymerisation of a DNA strand using oligo-DNA template with modified bases, sugars and phosphates. Nucleic Acids Symp Ser (Oxf) 2007;51:55–6. doi: 10.1093/nass/nrm028. [DOI] [PubMed] [Google Scholar]
- 49.Veedu RN, Vester B, Wengel J. In vitro incorporation of LNA nucleotides. Nucleosides Nucleotides Nucleic Acids. 2007;26:1207–10. doi: 10.1080/15257770701527844. [DOI] [PubMed] [Google Scholar]
- 50.Veedu RN, Vester B, Wengel J. Novel applications of locked nucleic acids. Nucleic Acids Symp Ser (Oxf) 2007;51:29–30. doi: 10.1093/nass/nrm015. [DOI] [PubMed] [Google Scholar]
- 51.Veedu RN, Vester B, Wengel J. Enzymatic incorporation of LNA nucleotides into DNA strands. Chembiochem. 2007;8:490–2. doi: 10.1002/cbic.200600501. [DOI] [PubMed] [Google Scholar]
- 52.Kuwahara M, Obika S, Nagashima J, Ohta Y, Suto Y, Ozaki H, et al. Systematic analysis of enzymatic DNA polymerization using oligo-DNA templates and triphosphate analogs involving 2′,4′-bridged nucleosides. Nucleic Acids Res. 2008;36:4257–65. doi: 10.1093/nar/gkn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veedu RN, Vester B, Wengel J. Polymerase chain reaction and transcription using locked nucleic acid nucleotide triphosphates. J Am Chem Soc. 2008;130:8124–5. doi: 10.1021/ja801389n. [DOI] [PubMed] [Google Scholar]
- 54.Veedu RN, Vester B, Wengel J. Efficient enzymatic synthesis of LNA-modified DNA duplexes using KOD DNA polymerase. Org Biomol Chem. 2009;7:1404–9. doi: 10.1039/b819946a. [DOI] [PubMed] [Google Scholar]
- 55.Veedu RN, Wengel J. Locked nucleic acid nucleoside triphosphates and polymerases: on the way towards evolution of LNA aptamers. Mol Biosyst. 2009;5:787–92. doi: 10.1039/b905513b. [DOI] [PubMed] [Google Scholar]
- 56.Kuwahara M, Takano Y, Kasahara Y, Nara H, Ozaki H, Sawai H, et al. Study on suitability of KOD DNA polymerase for enzymatic production of artificial nucleic acids using base/sugar modified nucleoside triphosphates. Molecules. 2010;15:8229–40. doi: 10.3390/molecules15118229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veedu RN, Wengel J. Locked nucleic acids: promising nucleic acid analogs for therapeutic applications. Chem Biodivers. 2010;7:536–42. doi: 10.1002/cbdv.200900343. [DOI] [PubMed] [Google Scholar]
- 58.Veedu RN, Burri HV, Kumar P, Sharma PK, Hrdlicka PJ, Vester B, et al. Polymerase-directed synthesis of C5-ethynyl locked nucleic acids. Bioorg Med Chem Lett. 2010;20:6565–8. doi: 10.1016/j.bmcl.2010.09.044. [DOI] [PubMed] [Google Scholar]
- 59.Wheeler M, Chardon A, Goubet A, Morihiro K, Tsan SY, Edwards SL, et al. Synthesis of selenomethylene-locked nucleic acid (SeLNA)-modified oligonucleotides by polymerases. Chem Commun (Camb) 2012;48:11020–2. doi: 10.1039/c2cc36464f. [DOI] [PubMed] [Google Scholar]
- 60.Højland T, Veedu RN, Vester B, Wengel J. Enzymatic synthesis of DNA strands containing α-L-LNA (α-L-configured locked nucleic acid) thymine nucleotides. Artif DNA PNA XNA. 2012;3:14–21. doi: 10.4161/adna.19272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crouzier L, Dubois C, Edwards SL, Lauridsen LH, Wengel J, Veedu RN. Efficient reverse transcription using locked nucleic acid nucleotides towards the evolution of nuclease resistant RNA aptamers. PLoS One. 2012;7:e35990. doi: 10.1371/journal.pone.0035990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johannsen MW, Veedu RN, Madsen AS, Wengel J. Enzymatic polymerisation involving 2′-amino-LNA nucleotides. Bioorg Med Chem Lett. 2012;22:3522–6. doi: 10.1016/j.bmcl.2012.03.073. [DOI] [PubMed] [Google Scholar]
- 63.Doessing H, Hansen LH, Veedu RN, Wengel J, Vester B. Amplification and re-generation of LNA-modified libraries. Molecules. 2012;17:13087–97. doi: 10.3390/molecules171113087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lauridsen LH, Rothnagel JA, Veedu RN. Enzymatic recognition of 2′-modified ribonucleoside 5′-triphosphates: towards the evolution of versatile aptamers. Chembiochem. 2012;13:19–25. doi: 10.1002/cbic.201100648. [DOI] [PubMed] [Google Scholar]
- 65.Goubet A, Chardon A, Kumar P, Sharma PK, Veedu RN. Synthesis of DNA oligonucleotides containing C5-ethynylbenzenesulfonamide-modified nucleotides (EBNA) by polymerases towards the construction of base functionalized nucleic acids. Bioorg Med Chem Lett. 2013;23:761–3. doi: 10.1016/j.bmcl.2012.11.096. [DOI] [PubMed] [Google Scholar]
- 66.Sawai H, Ozaki AN, Satoh F, Ohbayashi T, Masud MM, Ozaki H. Expansion of structural and functional diversities of DNA using new 5-substituted deoxyuridine derivatives by PCR with superthermophilic KOD Dash DNA polymerase. Chem Commun (Camb) 2001;24:2604–5. doi: 10.1039/b107838k. [DOI] [Google Scholar]
- 67.Obayashi T, Masud MM, Ozaki AN, Ozaki H, Kuwahara M, Sawai H. Enzymatic synthesis of labeled DNA by PCR using new fluorescent thymidine nucleotide analogue and superthermophilic KOD dash DNA polymerase. Bioorg Med Chem Lett. 2002;12:1167–70. doi: 10.1016/S0960-894X(02)00111-7. [DOI] [PubMed] [Google Scholar]
- 68.Mehedi Masud M, Ozaki-Nakamura A, Kuwahara M, Ozaki H, Sawai H. Modified DNA bearing 5(methoxycarbonylmethyl)-2′-deoxyuridine: preparation by PCR with thermophilic DNA polymerase and postsynthetic derivatization. Chembiochem. 2003;4:584–8. doi: 10.1002/cbic.200200539. [DOI] [PubMed] [Google Scholar]
- 69.Kuwahara M, Takahata Y, Shoji A, Ozaki AN, Ozaki H, Sawai H. Substrate properties of C5-substituted pyrimidine 2′-deoxynucleoside 5′-triphosphates for thermostable DNA polymerases during PCR. Bioorg Med Chem Lett. 2003;13:3735–8. doi: 10.1016/j.bmcl.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 70.Masud MM, Kuwahara M, Ozaki H, Sawai H. Sialyllactose-binding modified DNA aptamer bearing additional functionality by SELEX. Bioorg Med Chem. 2004;12:1111–20. doi: 10.1016/j.bmc.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 71.Ohbayashi T, Kuwahara M, Hasegawa M, Kasamatsu T, Tamura T, Sawai H. Expansion of repertoire of modified DNAs prepared by PCR using KOD Dash DNA polymerase. Org Biomol Chem. 2005;3:2463–8. doi: 10.1039/b504330a. [DOI] [PubMed] [Google Scholar]
- 72.Ohmichi T, Kuwahara M, Sasaki N, Hasegawa M, Nishikata T, Sawai H, et al. Nucleic acid with guanidinium modification exhibits efficient cellular uptake. Angew Chem Int Ed Engl. 2005;44:6682–5. doi: 10.1002/anie.200500904. [DOI] [PubMed] [Google Scholar]
- 73.Kuwahara M, Hanawa K, Ohsawa K, Kitagata R, Ozaki H, Sawai H. Direct PCR amplification of various modified DNAs having amino acids: convenient preparation of DNA libraries with high-potential activities for in vitro selection. Bioorg Med Chem. 2006;14:2518–26. doi: 10.1016/j.bmc.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 74.Kuwahara M, Nagashima J, Hasegawa M, Tamura T, Kitagata R, Hanawa K, et al. Systematic characterization of 2′-deoxynucleoside- 5′-triphosphate analogs as substrates for DNA polymerases by polymerase chain reaction and kinetic studies on enzymatic production of modified DNA. Nucleic Acids Res. 2006;34:5383–94. doi: 10.1093/nar/gkl637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shoji A, Hasegawa T, Kuwahara M, Ozaki H, Sawai H. Chemico-enzymatic synthesis of a new fluorescent-labeled DNA by PCR with a thymidine nucleotide analogue bearing an acridone derivative. Bioorg Med Chem Lett. 2007;17:776–9. doi: 10.1016/j.bmcl.2006.10.072. [DOI] [PubMed] [Google Scholar]
- 76.Shoji A, Kuwahara M, Ozaki H, Sawai H. Modified DNA aptamer that binds the (R)-isomer of a thalidomide derivative with high enantioselectivity. J Am Chem Soc. 2007;129:1456–64. doi: 10.1021/ja067098n. [DOI] [PubMed] [Google Scholar]
- 77.Sawai H, Nagashima J, Kuwahara M, Kitagata R, Tamura T, Matsui I. Differences in substrate specificity of C(5)-substituted or C(5)-unsubstituted pyrimidine nucleotides by DNA polymerases from thermophilic bacteria, archaea, and phages. Chem Biodivers. 2007;4:1979–95. doi: 10.1002/cbdv.200790165. [DOI] [PubMed] [Google Scholar]
- 78.Ohsawa K, Kasamatsu T, Nagashima J, Hanawa K, Kuwahara M, Ozaki H, et al. Arginine-modified DNA aptamers that show enantioselective recognition of the dicarboxylic acid moiety of glutamic acid. Anal Sci. 2008;24:167–72. doi: 10.2116/analsci.24.167. [DOI] [PubMed] [Google Scholar]
- 79.Kuwahara M, Takeshima H, Nagashima J, Minezaki S, Ozaki H, Sawai H. Transcription and reverse transcription of artificial nucleic acids involving backbone modification by template-directed DNA polymerase reactions. Bioorg Med Chem. 2009;17:3782–8. doi: 10.1016/j.bmc.2009.04.045. [DOI] [PubMed] [Google Scholar]
- 80.Vaught JD, Bock C, Carter J, Fitzwater T, Otis M, Schneider D, et al. Expanding the chemistry of DNA for in vitro selection. J Am Chem Soc. 2010;132:4141–51. doi: 10.1021/ja908035g. [DOI] [PubMed] [Google Scholar]
- 81.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kasahara Y, Kuwahara M. Artificial specific binders directly recovered from chemically modified nucleic Acid libraries. J Nucleic Acids. 2012;2012156482; PMID23094139. 10.1155/2012/156482 [DOI] [PMC free article] [PubMed]
- 83.Dubois C, Campbell MA, Edwards SL, Wengel J, Veedu RN. Stepping towards highly flexible aptamers: enzymatic recognition studies of unlocked nucleic acid nucleotides. Chem Commun (Camb) 2012;48:5503–5. doi: 10.1039/c2cc31316b. [DOI] [PubMed] [Google Scholar]
- 84.Ghadessy FJ, Ong JL, Holliger P. Directed evolution of polymerase function by compartmentalized self-replication. Proc Natl Acad Sci U S A. 2001;98:4552–7. doi: 10.1073/pnas.071052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuwahara M. CSJ Curr. Rev. 2011;6:78–85. [Google Scholar]
- 86.Nitsche A, Kurth A, Dunkhorst A, Pänke O, Sielaff H, Junge W, et al. One-step selection of Vaccinia virus-binding DNA aptamers by MonoLEX. BMC Biotechnol. 2007;7:48. doi: 10.1186/1472-6750-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Berezovski M, Musheev M, Drabovich A, Krylov SN. Non-SELEX selection of aptamers. J Am Chem Soc. 2006;128:1410–1. doi: 10.1021/ja056943j. [DOI] [PubMed] [Google Scholar]
- 88.Mendonsa SD, Bowser MT. In vitro evolution of functional DNA using capillary electrophoresis. J Am Chem Soc. 2004;126:20–1. doi: 10.1021/ja037832s. [DOI] [PubMed] [Google Scholar]
- 89.Mendonsa SD, Bowser MT. In vitro selection of aptamers with affinity for neuropeptide Y using capillary electrophoresis. J Am Chem Soc. 2005;127:9382–3. doi: 10.1021/ja052406n. [DOI] [PubMed] [Google Scholar]
- 90.Mosing RK, Mendonsa SD, Bowser MT. Capillary electrophoresis-SELEX selection of aptamers with affinity for HIV-1 reverse transcriptase. Anal Chem. 2005;77:6107–12. doi: 10.1021/ac050836q. [DOI] [PubMed] [Google Scholar]
- 91.Drabovich AP, Berezovski M, Okhonin V, Krylov SN. Selection of smart aptamers by methods of kinetic capillary electrophoresis. Anal Chem. 2006;78:3171–8. doi: 10.1021/ac060144h. [DOI] [PubMed] [Google Scholar]
- 92.Geiger M, Hogerton AL, Bowser MT. Capillary electrophoresis. Anal Chem. 2012;84:577–96. doi: 10.1021/ac203205a. [DOI] [PubMed] [Google Scholar]
- 93.Kasahara Y, Irisawa Y, Fujita H, Yahara A, Ozaki H, Obika S, et al. Capillary electrophoresis-systematic evolution of ligands by exponential enrichment selection of base- and sugar-modified DNA aptamers: target binding dominated by 2′-O,4′-C-methylene-bridged/locked nucleic acid primer. Anal Chem. 2013;85:4961–7. doi: 10.1021/ac400058z. [DOI] [PubMed] [Google Scholar]
- 94.Kasahara Y, Irisawa Y, Ozaki H, Obika S, Kuwahara M. 2′,4′-BNA/LNA aptamers: CE-SELEX using a DNA-based library of full-length 2′-O,4′-C-methylene-bridged/linked bicyclic ribonucleotides. Bioorg Med Chem Lett. 2013;23:1288–92. doi: 10.1016/j.bmcl.2012.12.093. [DOI] [PubMed] [Google Scholar]
- 95.Kuwahara M, Sugimoto N. Molecular evolution of functional nucleic acids with chemical modifications. Molecules. 2010;15:5423–44. doi: 10.3390/molecules15085423. [DOI] [PMC free article] [PubMed] [Google Scholar]