Abstract
Background
Strains of Staphylococcus aureus with an intermediate level of resistance to vancomycin (vancomycin-intermediate S. aureus, or VISA) or which contain subpopulations of mixed susceptibility (heterogeneous VISA, or hVISA) have been reported worldwide. However, the prevalence of VISA and hVISA infections in Northeast China is unknown. From 2007 through 2010, we surveyed the vancomycin susceptibility of methicillin-resistant and methicillin-sensitive S. aureus (MRSA and MSSA, respectively) clinical isolates in Northeast China.
Methods
S. aureus clinical isolates (369 MRSA and 388 MSSA) were screened for hVISA and VISA on brain heart infusion agar containing 3 μg/mL vancomycin, and their identity confirmed using a modified population analysis profile-area under the curve method and broth microdilution. All hVISA and VISA isolates were characterized genotypically and phenotypically.
Results
Ten percent and 0.5 percent of the isolates were hVISA and VISA, respectively. The proportion of hVISA among MSSA isolates for the entire study period was 4.1%, but increased significantly year-by-year, from 1.2% in 2007 to 7.2% in 2010. The predominant sources of hVISA and VISA isolates were sputum (56.3%), pus (18.8%), and blood (8.8%). Molecular typing of hVISA and VISA strains revealed that, taken together, 80% contained the accessory gene regulator (agr) group II, and of these, 85.7% of the MR-hVISA and MR-VISA strains were staphylococcal cassette chromosome mec (SCCmec) type II. The adherence ability of all hVISA and VISA strains was reduced compared with that of vancomycin-susceptible strains, shown by biofilm assay.
Conclusions
The percentage of hVISA strains was high and increased each year. The proportion of hVISA among MSSA specifically also increased significantly each year. In isolates collected from diverse infection sites, hVISA and VISA strains were found predominantly in sputum, pus, and blood, in descending order. Testing for vancomycin susceptibility should include both MRSA and MSSA isolates collected from different clinical sites.
Introduction
Staphylococcus aureus is a ubiquitous bacterium responsible for both community-associated and hospital-acquired infections, which range in severity from non-pathogenic to life threatening [1]. These infections, especially those due to methicillin-resistant S. aureus (MRSA), have been treated for more than a half-century primarily with the glycopeptide antibiotic vancomycin. Unfortunately, increased incidence of nosocomial MRSA infections has led worldwide to the development of S. aureus strains with varying degrees of resistance to vancomycin [2]–[6], first reported in 1997 [7], [8].
Reduced susceptibility to vancomycin in S. aureus is complex and difficult to detect in clinical microbiology laboratories [9], [10]. In January 2006, to improve the correlation between in vitro susceptibility and clinical response, the Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) redefined resistance breakpoints for vancomycin against S. aureus [9]. The current CLSI guidelines suggest that S. aureus strains should be categorized as susceptible, intermediate, or resistant when the vancomycin minimum inhibitory concentration (MIC) is ≤2 μg/mL, 4 to 8 μg/mL, or ≥16 μg/mL, respectively.
Clinical treatment failures with vancomycin have been linked to strains whose susceptibility to vancomycin falls within the intermediate MIC range (vancomycin-intermediate S. aureus, or VISA) and also strains that contain subpopulations of mixed susceptibility (heterogeneous VISA, or hVISA). To characterize VISA strains, determinations of vancomycin broth microdilution MICs performed in accordance with CLSI criteria should be used [9]–[11]. However, the breakpoints that differentiate hVISA strains from vancomycin-susceptible S. aureus (VSSA) have not been established. [10], [11]. While the vancomycin MICs of hVISA fall within the susceptible range when tested by routine methods, these strains stably produce subsets of cells (typically one organism per 105 to 106) in the intermediate range; they are assumed precursors of VISA [8].
A variety of screening assays have been developed to detect hVISA [4], [8], [12], [13], but to date there is no standardized approach that is convenient and reliable. The gold standard for confirmation of hVISA is the population analysis profile-area under the curve (PAP-AUC) method, which is time-consuming, labor-intensive, and costly for routine clinical laboratories [10], [14]. More recently, a novel agar with 3 μg/mL vancomycin was advocated for screening vancomycin non-susceptible S. aureus isolates, with a reported 100% sensitivity and 65% specificity based on evaluation of 100 S. aureus isolates (55 VSSA and 45 VISA) [15]. We also evaluated a screening procedure using brain heart infusion agar containing 3 μg/mL vancomycin (BHIA-3V) for detection of hVISA with PAP-AUC, and found that this method had 100% sensitivity to hVISA (data not shown). In light of the excellent sensitivity of the BHIA-3V screening procedure for detection of hVISA and VISA [15]–[17], in the present study we employed this approach to screen S. aureus isolates with decreased susceptibility to vancomycin.
Since the first descriptions of hVISA (Mu3) and VISA (Mu50) strains by Hiramatsu et al. from Japan in 1997 [7], [8], clinical isolates of S. aureus with decreased susceptibility to vancomycin have been reported in many countries, including the United States [2], [18], Canada [6], China [4], Japan [7], [8], [19], South Korea [20], Australia [21], United Kingdom [3], [14], Italy [5], and France [22], and their occurrence has become a major concern throughout the world. However, although China accounts for one-sixth of the world's population, there are only a few reports from mainland China regarding hVISA or VISA isolates specifically [4], [20], [23]. In one study, 1012 vancomycin-susceptible MRSA isolates obtained from the years 2005 to 2007 in 14 cities in China were investigated, and the estimated prevalence of hVISA was 13% to 16% [4]. With this in mind, we hypothesized that the incidence rate of hVISA and VISA infections might be high in hospitals located in Northeast China, one of the most populated regions in the world. Moreover, although hVISA and VISA have been reported predominately in MRSA strains [2]–[8], they are also present among MSSA, suggesting the need to screen both MRSA and MSSA isolates if a true prevalence is to be determined [24], [25].
Hence, we began the present study, the first such in Northeast China, by investigating the prevalence of hVISA and VISA strains among clinical isolates of S. aureus collected in two large teaching hospitals over a 4-year period. Our study may provide incentive for a larger-scale investigation and prevention program, not only in China but also within all international biomedical and epidemiological communities.
Materials and Methods
Bacterial strains and culture conditions
From January 2007 to January 2011, a total of 757 consecutive S. aureus isolates were collected at the First Affiliated Hospital (2249 beds) and Second Affiliated Hospital (4818 beds) of China Medical University in Shenyang, the capital city of one of the three provinces of Northeast China. These two hospitals are large tertiary hospitals with 237,000 annual admissions (100,000 and 137,000 respectively); 13,280 staff personnel (4000 and 9280); and 57,799 students; and can be considered representative of large metropolitan hospitals in China, serving patients mainly from all over Northeast China.
One S. aureus isolate from each patient was included in this retrospective study. The Medical Ethics Committee of China Medical University, Shenyang, China (Chairperson Prof Qun Zhao) granted ethical approval for this study on 12 February 2011 (Ethical Committee No. 11021207). Each patient who was enrolled in this study signed the informed consent form. Isolates of S. aureus were identified with the positive tube coagulase test and a VITEK 2 automated system (bioMérieux, France). MRSA isolates were confirmed using the cefoxitin disk diffusion method and correlated with the presence of the mecA gene by PCR [26], [27]. All strains were routinely stored at −80°C in brain heart infusion (BHI) broth (Becton Dickinson, Sparks, MD, USA) containing 20% (v/v) glycerol, and 48 h prior to testing were subcultured twice onto Columbia agar containing 5% sheep blood (Oxoid, Basingstoke, UK).
Detection of hVISA and VISA
All S. aureus isolates were screened for hVISA and VISA strains on BHIA-3V plates as previously described [15], [16]. The culture was considered positive if there was growth of one or more colonies after 48 h. The agar plates were prepared in-house daily and the screening tests were performed in duplicate. Isolates which displayed a hVISA/VISA profile on the BHIA-3V screening plates were further confirmed by the PAP-AUC approach using the technique of Wootton et al. [14]. An isolate was considered hVISA if the ratio of the AUC of the test strain to that of Mu3 was ≥0.9; an isolate with a ratio of <0.9 was defined as VSSA. The VSSA strain ATCC 29213 was used as a negative control. The hVISA strain Mu3 (ATCC 700698) and VISA strain Mu50 (ATCC 700699) were used as positive controls. The results from each experiment were recorded only when positive and negative controls were confirmed.
Antimicrobial susceptibility testing and reagents
The routine disk diffusion method with antibiotic disks (Oxoid) was used to determine the susceptibility of all screen-positive isolates to oxacillin, clindamycin, gentamicin, chloramphenicol, trimethoprim-sulfamethoxazole (TMP-SMX), erythromycin, ciprofloxacin, levofloxacin, tetracycline, and rifampin in accordance with the CLSI guidelines [28]. In addition, MICs of vancomycin, teicoplanin, linezolid, daptomycin, ceftobiprole, and tigecycline were determined by broth microdilution using cation-supplemented Mueller-Hinton broth (Oxoid), as recommended by the CLSI guidelines [28].
The results were independently interpreted by two investigators. Isolates for which the vancomycin MIC was 4 to 8 μg/mL were considered VISA, in accordance with the criteria of the CLSI [11]. Vancomycin, teicoplanin, and daptomycin were obtained from Sigma Chemical, St. Louis, MO, USA; linezolid and tigecycline were obtained from Pfizer, NewYork, NY, USA.
DNA extraction and molecular typing
DNA was prepared by the small-scale phenol extraction method, as described previously [29]. The DNA was used as the template in all PCRs described below. All hVISA and VISA isolates identified were analyzed by staphylococcal protein A (spa) typing as previously described [30]. One strain representative of each major spa type was further characterized by multilocus sequence typing (MLST) [31], [32]. All hVISA and VISA isolates were analyzed by accessory gene regulator (agr) grouping using multiplex PCR in accordance with the published procedure [33]. All methicillin-resistant hVISA (MR-hVISA) and methicillin-resistant VISA (MR-VISA) isolates were typed by staphylococcal cassette chromosome mec (SCCmec) using multiplex PCR as previously described [34]. All hVISA and VISA isolates were detected for the Panton-Valentine leukocidin (PVL) toxin gene in accordance with the protocols of prior reports [35], [36].
Delta-hemolysin expression, biofilm assay, and autolysis assay
All screen-positive isolates by BHIA-3V were subjected to delta-hemolysin expression, biofilm assay, and autolysis assay. The function of the agr gene cluster was determined by delta-hemolysin production as described by Sakoulas et al. [37]. The ability of S. aureus to adhere to a polystyrene microtiter plate was evaluated as described previously [21], [37]. The absorbance was measured at 570 nm, and each assay was performed five times. Autolysis of S. aureus in Triton X-100 was performed as described previously [21]. The OD600 was measured each hour for 6 h, and each assay was performed five times.
Statistical analyses
All statistical analyses were performed using SPSS for Windows, version 12.0 (SPSS, Chicago, IL, USA). Categorical variables were compared using the chi-squared (χ2) or Fisher's exact probability tests, including prevalence rates and antimicrobial susceptibility of hVISA/VISA. A p-value of <0.05 was considered statistically significant.
Results
Prevalence rates of hVISA and VISA isolates in clinical specimens
In total, 757 non-duplicate S. aureus isolates were investigated in this study. The proportion of MRSA and MSSA isolates were 48.7% (369/757) and 51.3% (388/757), respectively (Table 1). The number of isolates tested in each year were: 2007, n = 171 (88 MRSA and 83 MSSA); 2008, n = 183 (89 MRSA and 94 MSSA); 2009, n = 197 (97 MRSA and 100 MSSA) and 2010, n = 206 (95 MRSA and 111 MSSA). The isolates were recovered from sputum (n = 249), pus (n = 208), blood (n = 137), secretions (n = 84), drainage (n = 35), wounds (n = 23), or other (n = 21).
Table 1. All hVISA and VISA isolates identified from different clinical specimens a.
| Confirmed by PAP-AUC c | Isolates from different clinical specimens (n = 80) | |||||||||
| Isolates | BHIA-3V+ b | hVISA | VISA | Sputum | Pus | Blood | Secretion | Drainage | Other | |
| MRSA | 369 (48.7) | 120 (32.5) | 60 (16.3) | 3 (0.8) | 43 (53.8) | 6 (7.5) | 6 (7.5) | 4 (5) | 2 (2.5) | 2 (2.5) |
| MSSA | 388 (51.3) | 89 (22.9) | 16 (4.1) | 1 (0.3) | 2 (2.5) | 9 (11.3) | 1 (1.3) | 1 (1.3) | 1 (1.3) | 3 (3.8) |
| Total | 757 | 209 (27.6) | 76 (10.0) | 4 (0.5) | 45 (56.3)d | 15 (18.8) | 7 (8.8) | 5 (6.3) | 3 (3.8) | 5 (6.3) |
All data are presented as number (%).
BHIA-3V is the brain heart infusion agar containing 3 μg/mL vancomycin; 757 isolates were screened for hVISA and VISA on BHIA-3V plates.
PAP-AUC is the population analysis profile-area under the curve; 209 screen-positive isolates were further confirmed by PAP-AUC.
Prevalence of hVISA and VISA isolated from sputum versus other specimens (p<0.001).
Among the 757 isolates, 209 (120 MRSA and 89 MSSA) grew on BHIA-3V screening plates within 48 h (i.e., screen-positive isolates). Seventy-six (60 MRSA and 16 MSSA) of the 209 isolates were confirmed as hVISA via the PAP-AUC approach, and four (3 MRSA and 1 MSSA) were identified as VISA with the broth microdilution in accordance with the CLSI criteria.
Accordingly, the prevalence rates of hVISA and VISA among all S. aureus isolates from different specimens were 10.0% (76/757) and 0.5% (4/757), respectively. In particular, the hVISA prevalence was higher in MRSA strains (60/369, 16.3%) compared with MSSA (16/388, 4.1%; p<0.001). In addition, the predominant sources of hVISA and VISA isolates in decreasing order were sputum (n = 45, 56.3%; p<0.001), pus (n = 15, 18.8%), blood (n = 7, 8.8%), secretions (n = 5, 6.3%), drainage (n = 3, 3.8%), and other (n = 5, 6.3%).
In the year 2007, the percentage of hVISA was 8.2% (13 MR-hVISA, 7.6%; 1 methicillin-sensitive hVISA [MS-hVISA], 0.6%; p = 0.001). In 2008, 2009, and 2010, the percentages of hVISA were 9.3% (15 MR-hVISA, 8.2%; 2 MS-hVISA, 1.1%; p = 0.001), 10.6% (16 MR-hVISA, 8.1%; 5 MS-hVISA, 2.5%; p = 0.009), and 11.7% (16 MR-hVISA, 7.8%; 8 MS-hVISA, 3.9%; p = 0.032), respectively. Of note, the proportion of hVISA among MSSA isolates was 4.1% and increased from 1.2% (1/83) in 2007 to 7.2% (8/111) in 2010. Thus during the four year period examined, hVISA increased gradually year-by-year, and the proportion of MS-hVISA increased rapidly.
Antimicrobial susceptibility
Antimicrobial activity of 15 antimicrobial agents against 209 screen-positive isolates (129 VSSA, 76 hVISA, and 4 VISA) in vitro showed that all these strains were susceptible to linezolid, daptomycin, and tigecycline (Table 2). MR-hVISA and MR-VISA strains were more resistant to several non-β-lactam antibiotics than were MR-VSSA strains, including chloramphenicol, TMP-SMX, and rifampin. Furthermore, MS-hVISA and MS-VISA strains were more likely to be resistant to all the antibiotics tested than were MS-VSSA strains.
Table 2. Antimicrobial activity of 15 antimicrobial agents against 209 screen-positive isolates in vitro.
| VSSA isolates, n (%) | hVISA/VISA isolates, n (%) | |||||||
| MRSA a | MSSA b | Total c | MRSA d | MSSA e | Total f | p-value g | ||
| Vancomycin MIC | 0.5 μg/mL | 10 (17.5) | 16 (22.2) | 26 (20.2) | 0 (0) | 0 (0) | 0 (0) | |
| 1.0 μg/mL | 25 (43.9) | 29 (40.3) | 54 (41.9) | 19 (30.2) | 2 (11.8) | 21 (26.3) | ||
| 2.0 μg/mL | 22 (38.6) | 27 (37.5) | 49 (38.0) | 41 (65.1) | 14 (82.4) | 55 (68.8) | ||
| 4.0 μg/mL | 0 (0) | 0 (0) | 0 (0) | 3 (4.8) | 1 (5.9) | 4 (5) | ||
| Teicoplanin MIC | 1 μg/mL | 17 (29.8) | 33 (45.8) | 50 (38.8) | 2 (3.2) | 2 (11.8) | 4 (5) | |
| 2 μg/mL | 24 (42.1) | 25 (34.7) | 49 (38.0) | 18 (28.6) | 8 (47.1) | 26 (32.5) | ||
| 4 μg/mL | 13 (22.8) | 12 (16.7) | 25 (19.4) | 31 (49.2) | 5 (29.4) | 36 (45.0) | ||
| 8 μg/mL | 3 (5.3) | 2 (2.8) | 5 (3.9) | 11 (17.5) | 2 (11.8) | 13 (16.3) | ||
| 16 μg/mL | 0 (0) | 0 (0) | 0 (0) | 1 (1.6) | 0 (0) | 1 (1.3) | ||
| Susceptibility | Oxacillin | 0 (0) | 72 (100) | 72 (55.8) | 0 (0) | 17 (100) | 17 (21.3) | |
| Clindamycin | 5 (8.8) | 40 (55.6) | 45 (34.9) | 0 (0) | 0 (0) | 0 (0) | <0.001 | |
| Gentamicin | 2 (3.5) | 52 (72.2) | 54 (41.9) | 0 (0) | 0 (0) | 0 (0) | <0.001 | |
| Chloramphenicol | 44 (77.2) | 70 (97.2) | 114 (88.4) | 30 (47.6) | 4 (23.5) | 34 (42.5) | <0.001 | |
| TMP-SMX h | 47 (82.5) | 71 (98.6) | 118 (91.5) | 34 (54.0) | 9 (52.9) | 43 (53.8) | <0.001 | |
| Erythromycin | 1 (1.8) | 6 (8.3) | 7 (5.4) | 0 (0) | 0 (0) | 0 (0) | 0.085 | |
| Ciprofloxacin | 1 (1.8) | 66 (91.7) | 67 (51.9) | 0 (0) | 14 (82.4) | 14 (17.5) | <0.001 | |
| Levofloxacin | 1 (1.8) | 66 (91.7) | 67 (51.9) | 0 (0) | 14 (82.4) | 14 (17.5) | <0.001 | |
| Tetracycline | 2 (3.5) | 59 (81.9) | 61 (47.3) | 0 (0) | 12 (70.6) | 12 (15) | <0.001 | |
| Rifampin | 39 (68.4) | 72 (100) | 111 (86.0) | 25 (39.7) | 14 (82.4) | 39 (48.8) | <0.001 | |
| Linezolid | 57 (100) | 72 (100) | 129 (100) | 63 (100) | 17 (100) | 80 (100) | ||
| Daptomycin | 57 (100) | 72 (100) | 129 (100) | 63 (100) | 17 (100) | 80 (100) | ||
| Tigecycline | 57 (100) | 72 (100) | 129 (100) | 63 (100) | 17 (100) | 80 (100) | ||
n = 57; b n = 72; c n = 129; d n = 63; e n = 17; f n = 80; g No. of total VSSA isolates versus no. of total hVISA/VISA isolates; h TMP-SMX, trimethoprim-sulfamethoxazole.
The vancomycin MIC range for VSSA was 0.5 to 4 μg/mL, for hVISA was 1 to 2 μg/mL, and for VISA was 4 to 4 μg/mL (Table 3). The number of tested isolates with vancomycin MICs of 0.5, 1, 2, and 4 μg/mL were 26 (12.4%), 75 (35.9%), 104 (49.8%), and 4 (1.9%), respectively, as determined by broth microdilution. MIC50/90 of VSSA, hVISA, and VISA were 1/2, 2/2, and 4/4, respectively.
Table 3. MIC50/90 determined by CLSI broth microdilution and the prevalence of agr-dysfunction.
| Screen-positive S. aureus a | VSSA b | hVISA c | VISA d | |
| Vancomycin range (μg/mL) | 0.5–4 | 0.5–4 | 1–2 | 4–4 |
| Vancomycin MIC50/90 | 2/2 | 1/2 | 2/2 | 4/4 |
| Teicoplanin range (μg/mL) | 1–16 | 1–8 | 1–8 | 4–16 |
| Teicoplanin MIC50/90 | 2/4 | 2/4 | 4/8 | 8/16 |
| No. (%) of agr -dysfunction | 99 (47.4) | 29 (22.5) | 66 (86.8) | 4 (100) e |
n = 209; b n = 129; c n = 76; d n = 4; e Prevalence of agr dysfunction in hVISA/VISA isolates versus VSSA isolates (p<0.001).
The teicoplanin MIC range for VSSA was 1 to 8 μg/mL, for hVISA was 1 to 8 μg/mL, and for VISA was 4 to 16 μg/mL. The number of tested isolates with teicoplanin MICs of 1, 2, 4, 8, and 16 μg/mL were 54 (25.8%), 75 (35.9%), 61 (29.2%), 18 (8.6%), and 1 (0.5%), respectively. MIC50/90 of VSSA, hVISA, and VISA were 2/4, 4/8, and 8/16, respectively.
The percentage of hVISA isolates with a vancomycin MIC of 2 μg/mL (55/76, 72.4%) was significantly higher than the percentage of VSSA isolates with the same MIC (49/129, 38.0%; p<0.001).
Spa typing, MLST analysis, SCCmec typing, PVL gene detection, agr grouping and delta-hemolysin expression
Sequence analysis of the PCR products of the spa gene revealed 12 spa types (t002, t030, t037, t045, t105, t437, t034, t163, t189, t386, t548, and t2592) in the 80 hVISA/VISA isolates (Table 4). The most frequently encountered spa types were t002 (59/80, 73.8%), t030 (6/80, 7.5%), t037 (2/80, 2.5%), t045 (2/80, 2.5%), t105 (2/80, 2.5%), t386 (2/80, 2.5%), and t437 (2/80, 2.5%). One isolate each was identified for the five remaining spa types.
Table 4. Molecular characteristics of all hVISA and VISA isolates.
| agr group (n) | SCCmec type (n) | ||||||||||||
| hVISA/VISA (n) | spa type (n) | MLST-CC | I | II | III | IV | I | II | III | IV | V | NT * | PVL+ |
| MR-hVISA (60) | t002 (47) | ST5-CC5 | 47 | 47 | |||||||||
| t030 (4) | ST239-CC8 | 3 | 1 | 4 | |||||||||
| t037 (2) | ST239-CC8 | 2 | 2 | ||||||||||
| t105 (2) | ST5-CC5 | 2 | 1 | 1 | |||||||||
| t045 (2) | ST5-CC5 | 2 | 2 | ||||||||||
| t437 (1) | ST59-CC59 | 1 | 1 | ||||||||||
| t189 (1) | ST188-CC1 | 1 | 1 | ||||||||||
| t548 (1) | ST5-CC5 | 1 | 1 | ||||||||||
| MS-hVISA (16) | t002 (8) | ST5-CC5 | 8 | ||||||||||
| t030 (2) | ST239-CC8 | 2 | |||||||||||
| t386 (2) | ST1-CC1 | 1 | |||||||||||
| t437 (1) | ST59-CC59 | 1 | 2 | ||||||||||
| t034 (1) | ST398-CC398 | 1 | 1 | ||||||||||
| t163 (1) | ST59-CC59 | 1 | |||||||||||
| t2592 (1) | ST88-CC88 | 1 | |||||||||||
| MR-VISA (3) | t002 (3) | ST5-CC5 | 3 | ||||||||||
| MS-VISA (1) | t002 (1) | ST5-CC5 | 1 | 3 | |||||||||
NT, could not be typed.
The most frequent MLST type was ST5 (64/80, 80%), followed by ST239 (8/80, 10%), ST59 (3/80, 3.8%), and ST1 (2/80, 2.5%). ST5, ST239, and ST59 were found in both MRSA and MSSA isolates.
SCCmec types were identified in 63 MR-hVISA and MR-VISA isolates. Among them, the predominant type was SCCmec type II (54/63, 85.7%), followed by SCCmec type III (5/63, 7.9%), SCCmec type V (2/63, 3.2%), and SCCmec type IV (1/63, 1.6%). One MR-hVISA isolate could not be assigned to any SCCmec type using the established protocols.
PVL genes were found in one MR-hVISA strain and 2 MS-hVISA strains. One MR-hVISA strain was PVL-positive, belonging to ST59-MRSA-IV.
Regarding agr groups, agr group II was dominant (64/80, 80%), followed by agr group I (13/80, 16.3%), agr group III (2/80, 2.5%), and agr group IV (1/80, 1.3%). In addition, the expression of delta-hemolysin was evaluated for the 209 screen-positive isolates (129 VSSA, 76 hVISA, and 4 VISA). Overall, the percentages of agr dysfunctional phenotypes in VSSA, hVISA, and VISA were 22.5% (29/129), 86.8% (66/76), and 100% (4/4), respectively (Table 3). The prevalence of the agr dysfunctional phenotype was significantly higher in hVISA/VISA strains compared with that of the VSSA (p<0.001).
Biofilm assay
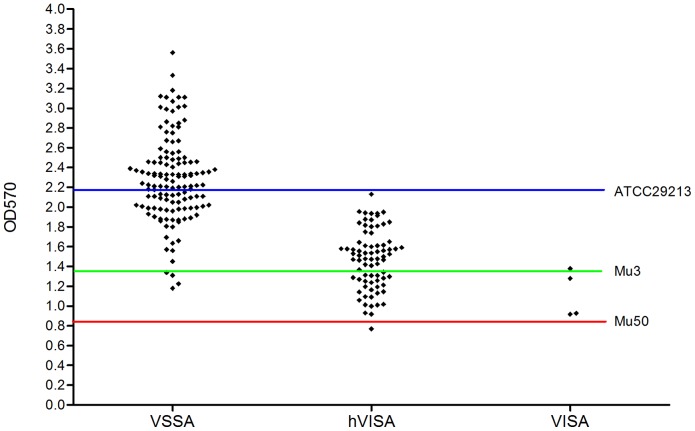
We performed the biofilm assay to compare the ability of the 209 screen-positive isolates (129 VSSA, 76 hVISA, and 4 VISA) to adhere to 96-well polystyrene microtiter plates (Figure 1). The adherence ability of these isolates is reflected by mean optical density values (ODs), which ranged overall from 0.769 to 3.560. The mean ODs for the VSSA, hVISA, and VISA isolates were 2.283 (range 1.180 to 3.560), 1.469 (range 0.769 to 2.129), and 1.125 (range 0.917 to 1.377), respectively. The ODs of most VSSA isolates (77/129, 59.7%) scattered above that of ATCC 29213 (the negative control), and the ODs of the majority of hVISA isolates (48/76, 63.2%) scattered between those of ATCC29213 and Mu3 (a positive control; Figure 1). On the other hand, the ODs of the four VISA isolates scattered between those of Mu3 and Mu50 (a positive control).
Figure 1. Quantification of biofilm formation was determined by crystal violet staining and read as an OD value.
Each rhombus represents the mean OD value of one strain from five independent experiments. The ODs of most VSSA isolates (77/129, 59.7%) scattered above that of ATCC 29213 (the blue line; OD570 = 2.180), and the ODs of the majority of hVISA isolates (48/76, 63.2%) scattered between those of ATCC29213 and Mu3 (the green line; OD570 = 1.350). On the other hand, the ODs of the four VISA isolates scattered between those of Mu3 and Mu50 (the red line; OD570 = 0.848).
Autolysis assay
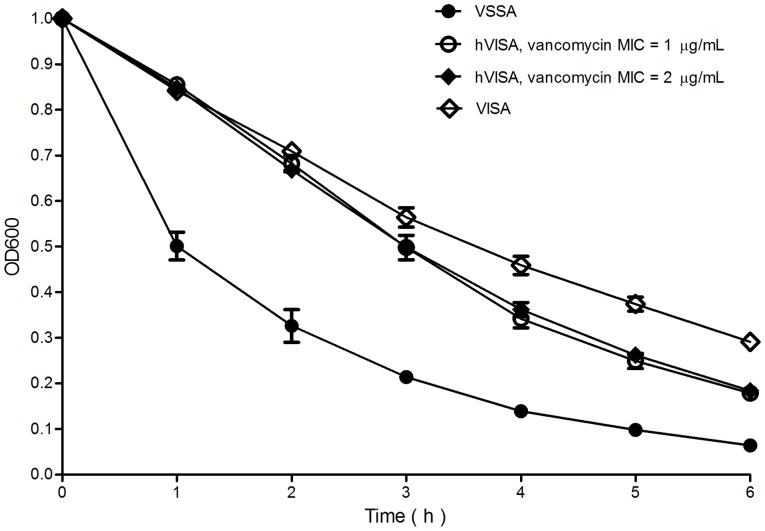
We tested 209 screen-positive isolates (129 VSSA, 76 hVISA, and 4 VISA isolates) for autolytic activity and found that the autolytic activity of hVISA and VISA isolates was significantly less than that of VSSA isolates (Figure 2). No significant difference in autolytic activity was noted between hVISA isolates and hVISA isolates, which have a vancomycin MIC of 1 μg/mL and 2 μg/mL, respectively.
Figure 2. Autolysis assay results for 129 VSSA, 76 hVISA, and 4 VISA isolates.
The results are expressed as the mean OD values of five independent experiments. Black circles indicate the mean OD values of 129 VSSA by hour; white circles and black rhombuses indicate the mean OD values of 21 hVISA with a vancomycin MIC of 1 μg/mL and the mean OD values of 55 hVISA with a vancomycin MIC of 2 μg/mL by hour, respectively; white rhombuses indicate the mean OD values of 4 VISA by hour.
Discussion
In the present study, during a four-year period (2007–2010) we surveyed 757 non-repetitive S. aureus isolates to determine the prevalence of hVISA and VISA. The data showed that in our hospitals in Northeast China, VISA clinical isolates were rare (0.5%), while the overall prevalence of hVISA (in MRSA and MSSA combined) was relatively high (10.0%). Furthermore, hVISA increased gradually during the four-year period examined, from 8.2% in 2007 to 11.7% in 2010. The high percentage and consistent increase of hVISA suggests a high potential for the development of complete drug resistance. We also found that the occurrence of hVISA was much higher (16.3%) in MRSA strains, which is similar to the results obtained by Sun et al. [4].
In an Asian surveillance study conducted in 2004 [20], a total of 1357 MRSA isolates obtained from 1997 to 2000 were investigated, and the hVISA rate was 4.3% (ranging from 0 to 8.2% among 12 countries). Additionally, in a systematic review published in 2011 of 43 epidemiological studies [38], the overall hVISA frequency in MRSA isolates was 1.3%, with a wide range from 0 to 73.7%. The reason for such a discrepancy in prevalence could be attributed to several factors, including differences in test strategies, geographic regions, and study populations.
The majority of reported hVISA and VISA isolates evolved from MRSA strains [2]–[8], yet in the present study we found that the proportion of hVISA among MSSA isolates was 4.1% and increased from 1.2% in 2007 to 7.2% in 2010, a 6-fold increase in four years. Liu and Chambers [24] in 2003 summarized data from 14 previous studies, and found that the prevalence of hVISA in 1868 MSSA isolates was 0.05%. Additionally, a study at one French hospital in 2006 which screened 2300 S. aureus isolates using a three-step approach, revealed that seven (0.3%) of these isolates were MS-hVISA strains [22]. In our study we found a significantly higher percentage of MS-hVISA, which increased rapidly each year. Hence, we believe that it is as important to detect reduced vancomycin susceptibility in MSSA isolates as in MRSA, and there is an alarming need to pay attention to the MS-hVISA population.
Most previous studies have focused on hVISA and VISA strains isolated from blood [2]–[6], whereas we found in the present study that hVISA and VISA strains were identified from diverse infection sites, and the predominant source of hVISA and VISA isolates was sputum (56.3%, p<0.001), followed by pus (18.8%), blood (8.8%), secretions 6.3%), drainage (3.8%), and other (6.3%). Therefore, in an effort to prevent the emergence and spread of vancomycin resistance, we recommend that S. aureus isolates from diverse clinical sites should be included when testing for reduced vancomycin susceptibility.
The antimicrobial susceptibility assay revealed all hVISA and VISA isolates were fully susceptible to the three alternative antistaphylococcal agents (linezolid, daptomycin, and tigecycline) in vitro. In addition, hVISA and VISA isolates differed from VSSA isolates by an overall higher frequency of drug resistance to multiple antibiotics, including clindamycin, chloramphenicol, TMP-SMX, and tetracycline. Interestingly, hVISA/VISA strains were significantly more likely to be resistant than VSSA strains to rifampin (51.3% compared with 14.0%, p<0.001). A previous study found that rifampin resistance developed more frequently in patients with hVISA than in those with VSSA during treatment of bacteremia [39], which suggested that rifampicin resistance was independently associated with the hVISA phenotype. Moreover, it was clearly demonstrated that hVISA strains tend to have higher vancomycin MICs within the susceptible range. A study by Leonard et al. in 2009, which evaluated a new epsilometer test (Etest) method for detection of hVISA using PAP-AUC, found that 49% of the hVISA strains had a vancomycin MIC of 2 μg/mL [40]. In the present study, no hVISA isolate with MIC <1 μg/mL was detected, and 72.4% (55/76) of the hVISA strains had a vancomycin MIC = 2 μg/mL, as determined by broth microdilution. This suggested that we should closely monitor the effect of vancomycin treatment to avoid therapy failures when treating S. aureus infections with a vancomycin MIC ∼2 μg/mL.
The results of molecular typing showed that the hVISA phenotype was prevalent among SCCmec type II and agr group II isolates. Previous studies have indicated that S. aureus isolates exhibiting reduced vancomycin susceptibility were more likely to harbor SCCmec type II [41], [42]. Furthermore, SCCmec type II has been associated with increased mortality in MRSA [42], [43]. On the other hand, agr group II has been frequently associated with reduced vancomycin susceptibility, as well as vancomycin treatment failure [37], [44]. Given this, further understanding of factors that determine virulence will be of importance for preventing the dissemination of S. aureus with reduced vancomycin susceptibility and treating the related infections. We also note that the majority (80%) of hVISA and VISA isolates were from clonal complex 5, in particular ST5 (CC5), which is consistent with prior evidence [7], [8], [45]. Of interest, we found a ST398-MSSA isolate identified as a hVISA by PAP-AUC with vancomycin MIC of 1 μg/mL. Since ST398-MSSA has been suggested as the precursor of livestock-associated ST398-MRSA, this indicates that we should pay more attention to the prevalence of such S. aureus strains [46]. Most notably, two MR-hVISA strains were SCCmec type V, and one PVL-positive MR-hVISA strain was SCCmec type IV, and therefore several hVISA and VISA strains might come from the community [47]; the community MRSA clone USA300 with a VISA phenotype, found in San Francisco and Kansas, has been described [48], [49].
Several changes in phenotypes have been described in studies of clinical or laboratory-induced hVISA and VISA strains [8], [21], [37]. In the present study, hVISA and VISA isolates displayed higher prevalence of agr dysfunction, reduced adherence ability, and reduced autolytic activity compared to VSSA isolates. We note a strong association between reduced vancomycin susceptibility and agr dysfunction, which is similar to previous data [37], [50]. Loss of agr function has been associated with the development of vancomycin resistance [37], [44] and prolonged bacteremia [51], but the mechanism underlying reduced levels of agr expression in hVISA/VISA strains is not completely understood and requires further study. Interestingly, our data from the biofilm assay showed that adherence ability was reduced in all hVISA and VISA strains compared with that of the VSSA strains. This differs from the report of Sakoulas et al. [37] but is similar to the findings of Howden et al. [21]. Although biofilm formation differs between laboratory-derived strains and clinical strains [21], the mechanism of altered biofilm formation in clinical strains remains unclear, and further work is needed to understand this.
The results of this study carry several implications. First, clinicians and microbiologists should be aware that all S. aureus isolates, obtained from diverse clinical sites, have the potential to develop varying degrees of susceptibility to vancomycin, or to produce subpopulations of mixed susceptibility. Therefore, in future studies we recommend that S. aureus isolates from different clinical specimens should be screened for susceptibility to vancomycin. Second, the two-step algorithm we adopted in this study appears to be an improved method for detecting hVISA. By detecting hVISA, clinicians can optimize treatment strategies, reduce associated mortality, shorten the length of stay, and lower patients' hospital charges. Finally, this study should act as a warning of the commonness of hVISA strains. Certainly the finding that hVISA strains are relatively common in Northeast China has alerted us to the need to monitor closely the effect of vancomycin when treating severe S. aureus infections. This study may provide incentive for a larger-scale investigation and prevention program, not only in China but also within all international biomedical and epidemiological communities.
We note several limitations of the present study. The period of collecting isolates was short and the sample size was small. Therefore, our report may only hint at the possible prevalence of hVISA and VISA. Furthermore, a recent study found that the MICs of isolates were inversely associated with time in cold storage [52]. In the present retrospective study, all the isolates were recovered from −80°C storage for MIC determination 1 to 4 years after collection, and we cannot guarantee that the MICs were not affected. We also were unable to obtain medical records regarding demographics, underlying diseases, history of exposure to antimicrobials, outcomes, and so forth. However, the results we have reported herein are of sufficient portent to warrant a prospective study of longer duration, with more isolates and complete patient demographic and clinicopathological data.
In summary, this study conducted in Northeast China from 2007 to 2010 shows that VISA strains were rare, but hVISA strains were unexpectedly more common. In addition, combined use of BHIA-3V and PAP-AUC can improve the accuracy of detection and reduce the number of false-positive results. In an effort to prevent the emergence and spread of vancomycin resistance, S. aureus isolates from diverse clinical sites should be included when testing for reduced vancomycin susceptibility. Further studies concerning optimal laboratory detection methods and the clinical significance of reduced vancomycin susceptibility in S. aureus will be helpful in controlling the development of vancomycin resistance and improving treatment strategies.
Funding Statement
This work was supported by the National Natural Science Foundation (Grant No. 30972520) and the Ministry of Education in China (Grant No. [2009] 1001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lowy FD (2011) How Staphylococcus aureus adapts to its host. N Engl J Med 364: 1987–1990. [DOI] [PubMed] [Google Scholar]
- 2. Richter SS, Satola SW, Crispel EK, Heilmann KP, Dohrn CL, et al. (2011) Detection of Staphylococcus aureus isolates with heterogeneous intermediate-level resistance to vancomycin in the United States. J Clin Microbiol 49: 4203–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kirby A, Graham R, Williams NJ, Wootton M, Broughton CM, et al. (2010) Staphylococcus aureus with reduced glycopeptide susceptibility in Liverpool, UK. J Antimicrob Chemother 65: 721–724. [DOI] [PubMed] [Google Scholar]
- 4. Sun W, Chen H, Liu Y, Zhao C, Nichols WW, et al. (2009) Prevalence and characterization of heterogeneous vancomycin-intermediate Staphylococcus aureus isolates from 14 cities in China. Antimicrob Agents Chemother 53: 3642–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campanile F, Borbone S, Perez M, Bongiorno D, Cafiso V, et al. (2010) Heteroresistance to glycopeptides in Italian meticillin-resistant Staphylococcus aureus (MRSA) isolates. Int J Antimicrob Agents 36: 415–419. [DOI] [PubMed] [Google Scholar]
- 6. Adam HJ, Louie L, Watt C, Gravel D, Bryce E, et al. (2010) Detection and characterization of heterogeneous vancomycin-intermediate Staphylococcus aureus isolates in Canada: results from the Canadian Nosocomial Infection Surveillance Program, 1995–2006. Antimicrob Agents Chemother 54: 945–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, et al. (1997) Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother 40: 135–136. [DOI] [PubMed] [Google Scholar]
- 8. Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, et al. (1997) Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350: 1670–1673. [DOI] [PubMed] [Google Scholar]
- 9. Tenover FC, Moellering RC Jr (2007) The rationale for revising the Clinical and Laboratory Standards Institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin Infect Dis 44: 1208–1215. [DOI] [PubMed] [Google Scholar]
- 10. Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML (2010) Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev 23: 99–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute (CLSI) (2012) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard-ninth edition. CLSI document M07-A9. WaynePA: CLSI.
- 12. Wootton M, MacGowan AP, Walsh TR, Howe RA (2007) A multicenter study evaluating the current strategies for isolating Staphylococcus aureus strains with reduced susceptibility to glycopeptides. J Clin Microbiol 45: 329–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Hal SJ, Wehrhahn MC, Barbagiannakos T, Mercer J, Chen D, et al. (2011) Performance of various testing methodologies for detection of heteroresistant vancomycin-intermediate Staphylococcus aureus in bloodstream isolates. J Clin Microbiol 49: 1489–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wootton M, Howe RA, Hillman R, Walsh TR, Bennett PM, et al. (2001) A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J Antimicrob Chemother 47: 399–403. [DOI] [PubMed] [Google Scholar]
- 15. Burnham CA, Weber CJ, Dunne WM Jr (2010) Novel screening agar for detection of vancomycin-nonsusceptible Staphylococcus aureus. J Clin Microbiol 48: 949–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kosowska-Shick K, Ednie LM, McGhee P, Smith K, Todd CD, et al. (2008) Incidence and characteristics of vancomycin nonsusceptible strains of methicillin-resistant Staphylococcus aureus at Hershey Medical Center. Antimicrob Agents Chemother 52: 4510–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Riederer K, Shemes S, Chase P, Musta A, Mar A, et al. (2011) Detection of intermediately vancomycin-susceptible and heterogeneous Staphylococcus aureus isolates: comparison of Etest and agar screening methods. J Clin Microbiol 49: 2147–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rybak MJ, Leonard SN, Rossi KL, Cheung CM, Sadar HS, et al. (2008) Characterization of vancomycin-heteroresistant Staphylococcus aureus from the metropolitan area of Detroit, Michigan, over a 22-year period (1986 to 2007). J Clin Microbiol 46: 2950–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kawaguchiya M, Urushibara N, Kuwahara O, Ito M, Mise K, et al. (2011) Molecular characteristics of community-acquired methicillin-resistant Staphylococcus aureus in Hokkaido, northern main island of Japan: identification of sequence types 6 and 59 Panton-Valentine leucocidin-positive community-acquired methicillin-resistant Staphylococcus aureus. Microb Drug Resist 17: 241–250. [DOI] [PubMed] [Google Scholar]
- 20. Song JH, Hiramatsu K, Suh JY, Ko KS, Ito T, et al. (2004) Emergence in Asian countries of Staphylococcus aureus with reduced susceptibility to vancomycin. Antimicrob Agents Chemother 48: 4926–4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Howden BP, Johnson PD, Ward PB, Stinear TP, Davies JK (2006) Isolates with low-level vancomycin resistance associated with persistent methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob Agents Chemother 50: 3039–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garnier F, Chainier D, Walsh T, Karlsson A, Bolmstroˇm A, et al. (2006) A 1 year surveillance study of glycopeptide-intermediate Staphylococcus aureus strains in a French hospital. J Antimicrob Chemother 57: 146–149. [DOI] [PubMed] [Google Scholar]
- 23. Li H, Zhao C, Chen H, Zhang F, He W, et al. (2013) Identification of Gene Clusters Associated with Host Adaptation and Antibiotic Resistance in Chinese Staphylococcus aureus Isolates by Microarray-Based Comparative Genomics. PLoS One 8: e53341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu C, Chambers HF (2003) Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob Agents Chemother 47: 3040–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pillai SK, Wennersten C, Venkataraman L, Eliopoulos GM, Moellering RC Jr, et al. (2009) Development of Reduced Vancomycin Susceptibility in Methicillin-Susceptible Staphylococcus aureus. Clin Infect Dis 49: 1169–1174. [DOI] [PubMed] [Google Scholar]
- 26. Ma XX, Sun DD, Hu J, Wang EH, Luo EJ (2011) Epidemiological and molecular characterization of Staphylococcus haemolyticus strains, from a hematology ward, with decreased susceptibility to glycopeptides. Can J Microbiol 57: 476–484. [DOI] [PubMed] [Google Scholar]
- 27. Ma XX, Wang EH, Liu Y, Luo EJ (2011) Antibiotic susceptibility of coagulase-negative staphylococci (CoNS): emergence of teicoplaninnon-susceptible CoNS strains with inducible resistance to vancomycin. J Med Microbiol 60: 1661–1668. [DOI] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute (CLSI) (2012) Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement. CLSI document M100-S22. WaynePA: CLSI.
- 29. Unal S, Hoskins J, Flokowitsch JE, Wu CY, Preston DA, et al. (1992) Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J Clin Microbiol 30: 1685–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aires-de-Sousa M, Boye K, de Lencastre H, Deplano A, Enright MC, et al. (2006) High Interlaboratory Reproducibility of DNA Sequence-Based Typing of Bacteria in a Multicenter Study. J Clin Microbiol 44: 619–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG (2000) Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of staphylococcus aureus. J Clin Microbiol 38: 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG (2004) Eburst: Inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186: 1518–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lina G, Boutite F, Tristan A, Bes M, Etienne J, et al. (2003) Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Appl Environ Microbiol 69: 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, et al. (2007) Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother 51: 264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma XX, Ito T, Kondo Y, Cho M, Yoshizawa Y, et al. (2008) Two Different Panton-Valentine Leukocidin Phage Lineages Predominate in Japan. J Clin Microbiol 46: 3246–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma XX, Sun DD, Wang S, Wang ML, Li M, et al. (2011) Nasal carriage of methicillin-resistant Staphylococcus aureus among preclinical medical students: epidemiologic and molecular characteristics of methicillin-resistant S. aureus clones. Diagn Microbiol Infect Dis 70: 22–30. [DOI] [PubMed] [Google Scholar]
- 37. Sakoulas G, Eliopoulos GM, Moellering RC Jr, Wennersten C, Venkataraman L, et al. (2002) Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob Agents Chemother 46: 1492–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Hal SJ, Paterson DL (2011) Systematic review and meta-analysis of the significance of heterogeneous vancomycin-intermediate Staphylococcus aureus isolates. Antimicrob Agents Chemother 55: 405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maor Y, Hagin M, Belausov N, Keller N, Ben-David D, et al. (2009) Clinical features of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia versus those of methicillin-resistant S. aureus bacteremia. J Infect Dis 199: 619–624. [DOI] [PubMed] [Google Scholar]
- 40. Leonard SN, Rossi KL, Newton KL, Rybak MJ (2009) Evaluation of the Etest GRD for the detection of Staphylococcus aureus with reduced susceptibility to glycopeptides. J Antimicrob Chemother 63: 489–492. [DOI] [PubMed] [Google Scholar]
- 41. Musta AC, Riederer K, Shemes S, Chase P, Jose J, et al. (2009) Vancomycin MIC plus heteroresistance and outcome of methicillin-resistant Staphylococcus aureus bacteremia: trends over 11 years. J Clin Microbiol 47: 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han JH, Edelstein PH, Lautenbach E (2012) Reduced vancomycin susceptibility and staphylococcal cassette chromosome mec (SCCmec) type distribution in methicillin-resistant staphylococcus aureus bacteraemia. J Antimicrob Chemother 67: 2346–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Davis SL, Rybak MJ, Amjad M, Kaatz GW, McKinnon PS (2006) Characteristics of patients with healthcare-associated infection due to SCCmec type methicillin-resistant Staphylococcus aureus. Infect Control Hosp Epidemiol 27: 1025–1031. [DOI] [PubMed] [Google Scholar]
- 44. Sakoulas G, Moellering RC Jr, Eliopoulos GM (2006) Adaptation of methicillin-resistant Staphylococcus aureus in the face of vancomycin therapy. Clin Infect Dis 42: S40–50. [DOI] [PubMed] [Google Scholar]
- 45. Howe RA, Monk A, Wootton M, Walsh TR, Enright MC (2004) Vancomycin susceptibility within methicillin-resistant Staphylococcus aureus lineages. Emerg Infect Dis 10: 855–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rijnders MI, Deurenberg RH, Boumans ML, Hoogkamp-Korstanje JA, Beisser PS, et al. (2009) Population structure of Staphylococcus aureus strains isolated from intensive care unit patients in the Netherlands over an 11-year period (1996 to 2006). J Clin Microbiol 47: 4090–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Popovich KJ, Weinstein RA (2009) The graying of methicillin-resistant Staphylococcus aureus. Infect Control Hosp Epidemiol 30: 9–12. [DOI] [PubMed] [Google Scholar]
- 48. Graber CJ, Wong MK, Carleton HA, Perdreau-Remington F, Haller BL, et al. (2007) Intermediate vancomycin susceptibility in a community-associated MRSA clone. Emerg Infect Dis 13: 491–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hageman JC, Patel J, Franklin P, Miscavish K, McDougal L, et al. (2008) Occurrence of a USA300 vancomycin-intermediate Staphylococcus aureus. Diagn Microbiol Infect Dis 62: 440–442. [DOI] [PubMed] [Google Scholar]
- 50. Harigaya Y, Ngo D, Lesse AJ, Huang V, Tsuji BT (2011) Characterization of heterogeneous vancomycin-intermediate resistance, MIC and accessory gene regulator (agr) dysfunction among clinical bloodstream isolates of staphyloccocus aureus. BMC Infect Dis 11: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fowler VG, Sakoulas G, McIntyre LM, Meka VG, Arbeit RD, et al. (2004) Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J Infect Dis 190: 1140–1149. [DOI] [PubMed] [Google Scholar]
- 52. Ludwig F, Edwards B, Lawes T, Gould IM (2012) Effects of storage on vancomycin and daptomycin MIC in susceptible blood isolates of methicillin-resistant Staphylococcus aureus. J Clin Microbiol 50: 3383–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]