Abstract
In Latin America, Bothrops snakes account for most snake bites in humans, and the recommended treatment is administration of multispecific Bothrops antivenom (SAB – soro antibotrópico). However, Bothrops snakes are very diverse with regard to their venom composition, which raises the issue of which venoms should be used as immunizing antigens for the production of pan-specific Bothrops antivenoms. In this study, we simultaneously compared the composition and reactivity with SAB of venoms collected from six species of snakes, distributed in pairs from three distinct phylogenetic clades: Bothrops, Bothropoides and Rhinocerophis. We also evaluated the neutralization of Bothrops atrox venom, which is the species responsible for most snake bites in the Amazon region, but not included in the immunization antigen mixture used to produce SAB. Using mass spectrometric and chromatographic approaches, we observed a lack of similarity in protein composition between the venoms from closely related snakes and a high similarity between the venoms of phylogenetically more distant snakes, suggesting little connection between taxonomic position and venom composition. P-III snake venom metalloproteinases (SVMPs) are the most antigenic toxins in the venoms of snakes from the Bothrops complex, whereas class P-I SVMPs, snake venom serine proteinases and phospholipases A2 reacted with antibodies in lower levels. Low molecular size toxins, such as disintegrins and bradykinin-potentiating peptides, were poorly antigenic. Toxins from the same protein family showed antigenic cross-reactivity among venoms from different species; SAB was efficient in neutralizing the B. atrox venom major toxins. Thus, we suggest that it is possible to obtain pan-specific effective antivenoms for Bothrops envenomations through immunization with venoms from only a few species of snakes, if these venoms contain protein classes that are representative of all species to which the antivenom is targeted.
Author Summary
Snakebite envenomation is a serious health issue in Latin America, particularly in the Amazon, where antivenom administration may be delayed due to logistic constraints. Bothrops snakes are involved in most of the snakebite-related accidents in Brazil. This work reports a comparative study of the toxin composition and antigenicity of the Bothrops venoms used to prepare the commercial antivenom and its effectiveness against the venom from Bothrops atrox, a prevalent Amazon species that is not included in the pool. Our data show a lack of connection between Bothrops taxonomic identity and venom composition. We also show that different toxins display distinct reactivity with the tested antivenom. However, the antivenom reacted similarly with each class of toxin present in the venoms of the different snakes studied. Important evidence was the neutralization of the major toxic effects of B. atrox venom, not included in the mixture of antigens used to produce the antivenom. Based on the observed antigenicity of the distinct protein classes of toxins, we suggest that it is possible to obtain pan-specific and efficient Bothrops antivenoms via immunization with venoms from a few species of snakes that are representative of the protein composition of a large number of targeted species.
Introduction
Envenomation by snakebites, which is incorporated by the World Health Organization (WHO) in its list of neglected tropical diseases, constitutes an important worldwide public health concern, particularly in the rural areas of tropical countries as Africa, Asia and Latin America, affecting mostly agricultural workers and children [1]. The estimated number of global envenoming events exceed 400,000, with more than 20,000 fatalities [2]. In Brazil, the incidence is above 25,000 accidents/year, and the incidence in the northern region was 52.6 accidents/100,000 inhabitants in 2008 [3]. Most of the Brazilian accidents with species notification are due to vipers of the genera Bothrops (83.8%), Crotalus (8.5%) and Lachesis (3.4%), with only 3.4% of accidents related to the Elapidae snakes of the genus Micrurus [3]. Antivenoms raised in horses are the recommended treatment in Brazil.
Based on early reports [4], it was accepted that the efficacy of a specific antivenom covers bites by those snake groups with venom represented in the pool of antigens used for horse immunization for the production of that specific antivenom. Recently, the knowledge of venom toxins has increased considerably, especially due to the characterization of detailed composition of venom proteomes based on mass spectrometry. In 2007, the concept of ‘venomics’ was introduced by Calvete et al. [5] and the method was important to describe the venom composition from a great number of snake species, as revised recently [6], [7]. Then, it was possible to characterize the families of venom toxins represented in the venoms of different species of snakes [6], [7]. The implications of venomics in the rational necessary for the development of antivenoms was further supported by the ‘antivenomics’ [8], [9], that allowed the identification of venom proteins bearing epitopes recognized by one antivenom and the toxins not covered by the immune response of the hyperimmunized animal. The importance of venomics and antivenomics was readily incorporated in antivenom development, indicating the possibility of a rational design of pan-specific antivenoms combining distinct protein families in immunization pools [10]–[12].
The venom composition of many species of Bothrops complex is already known by venomics [13]–[27] or indirectly by transcriptomics [28]–[32]. From these studies, it has become clear that a limited number of protein families compose the venoms of Bothrops snakes, with snake venom metalloproteinases (SVMPs), snake venom serine proteinases (SVSPs) and phospholipases A2 (PLA2s) being the most abundant and most frequently correlated with the clinical symptoms of envenoming. SVSPs are generally thrombin-like enzymes that are involved in the coagulation disturbances observed in most patients [33]. PLA2s are involved in local effects and the myotoxicity observed in bites with some species [34]. SVMPs are multifunctional enzymes involved in the local and systemic symptoms of bites, such as the induction of local hemorrhage, inflammatory reaction, activation of coagulation factors and inhibition of platelet aggregation [35]. The variability in venom composition is notable and can be correlated with phylogeny [36], [37], age [38], [39], sex [40], geographical distribution [13], [40], [41] and diet [42]–[44] of the snake. However, venom variability is mostly related to the expression level of each group of toxin rather than to the presence or absence of major families of venom proteins. Moreover, within the same protein family, variability in the toxic properties may also occur when distinct functional motifs are introduced in structurally related toxins, increasing the diversity of targets that can be affected by venom toxins [45], [46]. Thus, the relevance of variability in venom composition should also be reflected in the reactivity with antivenom and its efficacy.
This problem particularly affects Bothrops snakes, which are diverse in their morphological and ecological traits and are distributed in different habitats throughout Latin America [47]. Due to the great diversity of Bothrops snakes, the systematics and phylogenetic relationships of this group are not completely resolved, and the distinction of snakes in different genera is often suggested. Based on morphology and mtDNA sequences, a broad classification of the Bothrops complex by Wüster et al. recognized Bothrops and Bothrocophias as independent genera [48]; furthermore, Castoe and co-authors [49] have proposed the classification of Bothrops, Bothrocophias and also Bothriopsis as independent genera. More recently, the Bothrops genus was further divided into three independent genera by Fenwick et al. [50]: Bothropoides, Rhinocerophis and Bothrops, representing the groups of “jararaca/neuwiedi”, “alternatus” and “jararacussu/atrox” snakes, respectively, previously recognized by Wüster et al. [48]. This classification was further questioned by Carrasco and collaborators [47], and the maintenance of Bothrocophias as an independent genus and synonymizing Bothriopsis, Bothropoides and Rhinocerophis within the Bothrops genus was suggested. However, according to the emerging methodology of DNA sequencing for cladistic analyses, it is reasonable to expect that further revisions of Bothrops systematics will be offered in the near future.
Following the classification of Fenwick and coworkers [50], several species of Bothropoides, Rhinocerophis and Bothrops groups are involved in snakebite envenomings, contributing to the high number of reported incidents in Brazil [3]. Antibothropic antivenoms are used in the treatment of these patients and are produced in Brazil by horse immunization with the venoms of five species of these snakes: Bothropoides jararaca, Bothropoides neuwiedi, Rhinocerophis alternatus, Bothrops moojeni and Bothrops jararacussu. In spite of venomics evidences showing the venom composition of several species, there are still concerns about the efficacy of Bothrops antivenoms in the treatment of envenomings inflicted by species whose venom is not used for animal immunization. These objections include mostly the accidents by Bothrops atrox, which is the snake responsible for the majority of snake bites in the Amazon, whose venom is not included in the immunization mixture. Most of these concerns arise because, in previous studies, the venoms were independently analyzed and, also, by the lack of comparative neutralization assays in the few papers showing antivenomics data for Brazilian Bothrops [16], [19], [26]. Thus, the complexity of the Bothrops group and the relevance of these species from a public health viewpoint justify the need for a multifaceted study comparing the venoms of the most relevant species and their reactivity with antivenoms in the light of recent proteomics studies.
In this study, we used a shotgun approach that allowed a simultaneous comparison of the composition of venoms collected from six species of snakes from the Bothrops complex, distributed in pairs from three distinct genera [50]. Fractionated venom components were tested for reactivity with the widely-used antivenom (SAB). The efficacy of the antivenom was then assessed for the neutralization of relevant symptoms of experimental envenomings by (a) B. jararaca, which accounts for 50% of venom composition in the immunization pool and is prevalent in the southeastern Brazil, and (b) B. atrox, which is not present in the immunization pool although representing a common cause of snakebite in the Amazon. The venom analysis showed that phylogenetic classification per se is not directly linked to venom composition. Furthermore, the antivenoms reacted equally with the toxins from the same protein family, regardless of snake phylogeny or the presence of the venom in the immunization pool used for antivenom production, highlighting new priorities when considering the selection of venoms to be used in the production of antivenoms.
Materials and Methods
Venoms
The venoms of Bothropoides jararaca, Bothropoides neuwiedi (B. n. pauloensis, B. n. matogrossensis, B. n. marmoratus, B. n. neuwiedi and B. n. diporus subspecies), Rhinocerophis alternatus, Rhinocerophis cotiara, Bothrops jararacussu and Bothrops atrox were obtained from adult snakes of both sexes kept in captivity at the Laboratório de Herpetologia, Instituto Butantan, Brazil. The venoms from more than 10 specimens of each species were pooled, freeze-dried and stored at −20°C until use. Venoms from snakes kept under captivity represented as close as possible the same pools of venoms used for antivenom production and were used for proteomics and immunoreactivity assays. For experiments involving the neutralization of B. atrox venom toxic activities, we used venoms from wild B. atrox snakes collected at the Amazonian Floresta Nacional (FLONA) do Tapajós, Pará, Brazil, under SISBio license 32098-1, aiming to get venom samples as close as possible to the ones responsible for human accidents. Eight snakes were collected in pitfalls or by active search (five males and three females, with sizes ranging from 82 to 110 cm). The snakes were extracted in the herpetarium of Faculdades Integradas do Tapajós, Santarém, Pará, Brazil, and the venom from each snake was individually lyophilized and stored frozen until use, for which a pool was generated with equal proportions of venom from each snake. The chromatographic profile of the pool of venoms from snakes collected at Floresta Nacional do Tapajós was similar to that described below for the B. atrox venom pooled from snakes kept under captivity (data not shown).
Antivenoms
The antibothropic serum (SAB) was produced at the Instituto Butantan, São Paulo, Brazil in horses immunized with a mixture of the following venoms: B. jararaca (50%), B. neuwiedi (12.5%), R. alternatus (12.5%), B. moojeni (12.5%) and B. jararacussu (12.5%). The final preparation consists of soluble IgG F(ab′)2 fragments: 1 mL neutralizes the lethality of 5 mg standard B. jararaca venom (according to the manufacturer). Anti-jararhagin monoclonal antibodies (MAJar-3) were produced in hybridomas previously selected and maintained in our laboratory, as previously described [51]. The MAJar-3 antibodies are IgG1 isotypes and recognize conformational epitopes located on the jararhagin disintegrin-like domain. MAJar-3 neutralizes jararhagin collagen binding and hemorrhagic activity and cross-reacts with hemorrhagins from venoms of different species of viper snakes [52].
Venomic characterization by shotgun mass spectrometry
Fifty micrograms of each venom were subjected to trypsin digestion, as previously described [53]. The tryptic digests were desalted with in-lab-generated columns packed with Poros R2 resin (Life Technologies, USA). Each of the 12 venom digests generated (6 venoms in duplicate) were analyzed in triplicate by nanoLC-MS/MS. The separation was performed on a 75 µm×30 cm column packed with a 5-µm, 200 A Magic C-18 AQ matrix (Michrom Bioresources, USA). The eluted peptides were directly injected into an LTQ/Orbitrap XL mass spectrometer (Thermo, USA) for analysis. The MS1 spectra were acquired using the orbitrap analyzer (300 to 1,700 m/z) at a 60,000 resolution (for m/z 445.1200). For each spectrum, the 10 most intense ions were subjected to CID fragmentation, followed by MS2 acquisition on a linear trap analyzer. The tandem mass spectra were extracted by RAW Xtractor (version 1.9.9.2) [54]. All of the MS/MS samples were analyzed using ProLuCID (version 1.3.1) [55]. ProLuCID was set up to search a database (forward + reverse decoy) that was built from the protein entries contained in the NCBI non-redundant database from April 29, 2012 that satisfied the following search terms criteria: “serpentes OR snakes OR snake OR venom OR venoms OR bothrops OR bothriopsis OR bothrocophias OR rhinocerophis OR bothropoides”. The database was comprised of 87,384 entries (43,692 “forward” and 43,692 “reverse decoy”). The ProLuCID search was performed with a fragment ion mass tolerance of 600 ppm and a parent ion tolerance of 70 ppm. Cysteine carbamidomethylation was specified as a fixed modification. Scaffold version 4.0.4 (Proteome Software Inc., USA) was used to validate the MS/MS-based peptide and protein identifications. The peptide identifications were accepted if they could be established at greater than 99.0% probability by the Peptide Prophet algorithm [56], with Scaffold delta-mass correction, and the protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. The protein probabilities were assigned by the Protein Prophet algorithm [57]. The acceptable false discovery rates, at the peptide and protein levels, were less than or equal to 1%.
Venom fractionation
The venoms were fractionated by reverse-phase high-performance liquid chromatography (HPLC) according to previously described reports [16], with some modifications. Samples of 5 mg of crude lyophilized venom were dissolved in 250 µL 0.1% trifluoroacetic acid (TFA), and the insoluble material was removed by centrifugation at 18,400×g for 10 min at room temperature. The proteins in the soluble material were applied to a Vydac C-18 column (4.6×250 mm, 10-µm particle size) coupled to an Agilent 1100 HPLC system. The column was eluted at 1 mL/min with a gradient of 0.1% TFA in water (solution A) and 0.1% TFA in acetonitrile (solution B) (5% B for 10 min, followed by 5–15% B over 20 min, 15–45% B over 120 min, 45–70% B over 20 min and 70–100% B over 10 min). The separations were monitored at 214 nm, and the peaks were collected manually and dried in a Speed-Vac (Savant). The fractions were resuspended in PBS, and the protein concentration was estimated by OD at 280 nm in a NanoVue plus spectrophotometer (GE Healthcare).
Venom clustering
The venoms were classified according to their toxin composition by hierarchical clustering of observations constructed using nearest neighbor linkage method (minimum Euclidean distance between items in different clusters), considering initially each observation as an individual cluster. The degrees of similarity between observations were expressed in terms of a cluster tree (dendrogram). We performed also a Principal Component Analysis (PCA) in order to understand the key toxins responsible for the venom clustering. The principal components 1 (PC1) and 2 (PC2), which were responsible for explaining more than 70% of the total variability, were calculated using the covariance matrix. The toxin composition loadings and venom scores were expressed in terms of loading and score plots. These procedures were performed in Minitab 16 software.
The variables used for clustering and PCA were the relative concentrations of each toxin family, accessed by shotgun mass spectrometry. The mean of each protein family spectral counts was normalized by the total venom counting [1,891 (B. atrox); 1,727 (B. jararacussu); 2,719 (B. jararaca); 2,287 (B. neuwiedi); 1,252 (R. alternatus) and 1,767 (R. cotiara)], distributed within the identified protein families: SVMP-I, -II and –III (snake venom metalloproteinase - classes P-I, P-II and P-III); PLA2 (phospholipase A2); SVSP (snake venom serine proteinase); CLEC (C-type lectin); CLECL (C-type lectin-like); LAAO (L-amino acid oxidase); NGF (nerve growth factor); HYALU (hyaluronidase); VEGF (vascular endothelial growth factor); CRISP (cysteine-rich secretory protein); PDIEST (phosphodiesterase 1); ECTONT (ecto-5′-nucleotidase); PLB (phospholipase B); GLUTCYC (glutaminyl cyclase) and ACTIN (actin).
The venoms were also analyzed by the relative mAU of the highest peaks collected in C-18 reverse-phase chromatography in the elution time intervals of 56–57, 57–58, 58–60, 67–71, 108–112, 113–116, 121–123, 124–127, 128–129, 130–132, 134–136, 136–138, 139–140, 140–150, 150–152, 153–155, 157–159, 160–162, 163–164, 164–166, 166–168, 169–170, and 171–172 minutes. The mAU values of the peaks were normalized in % by the mAU of the highest peak eluted in the chromatography, taken as 100%.
ELISA assays
Samples containing 100 µL whole venom (10 µg/mL) or isolated fractions (1 µg/mL), in carbonate buffer (pH 9.6), were used to coat maxisorb microplates (Nunc). To determine the antibody titers, plates coated with whole venom were incubated with serial dilutions of SAB (from 1∶10,000), followed by incubation with anti-horse IgG labeled with peroxidase (1∶2,000). For assessing the antigenicity of the fractions, the plates were incubated with a fixed dilution of SAB (1∶1,000) or MAJar-3 (1∶50), followed by incubation with anti-horse IgG (1∶1,000) or anti-mouse IgG (1∶1,000) labeled with peroxidase. The reactions were developed with ortho-phenylenediamine/H2O2 as the enzyme substrate, and the products were detected at 490 nm. The reactions were performed in duplicates in three independent experiments. The results of antivenom titration are expressed as mean ± sd of the six OD values. The results of fraction antigenicity were calculated as mean of the six OD values after normalization using as 100% the maximal OD value obtained in each of the independent experiments [(Fraction OD/maximal OD of the test)×100].
Western blotting
Samples of crude venom (10 µg) were subjected to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under non-reducing conditions. After SDS-PAGE, the separated proteins were transferred to nitrocellulose membranes, which were then immersed in a blocking solution (5% non-fat milk in Tris-saline). The membranes were incubated with SAB (1∶1,000) as the primary antibody and then with peroxidase-labeled goat anti-horse IgG (1∶1,000). The reactive bands were detected by incubation with 4-chloro-α-naphthol and H2O2. The results shown represent three independent experiments.
Antivenom efficacy
For accessing the neutralization of the lethal and hemorrhagic venom activities, Swiss mice bred and maintained at the Instituto Butantan (Brazil) animal house were used as an animal model.
For the neutralization of hemorrhagic activity, doses of 10 µg B. jararaca or B. atrox venom were incubated with SAB at ratios of 1, 2 or 4 times the SAB volume required to neutralize 10 µg of reference venom, according to the manufacturer. The mixtures were incubated at 37°C for 30 min, and a 50-µL aliquot of each mixture was injected intradermically in the dorsa of a group of 5 mice. The control groups included mice injected with PBS or with venom incubated with PBS. At three hours after the injection, the mice were sacrificed by CO2 inhalation; the skin of the dorsa was removed, and the hemorrhagic spots were measured (longest diameter multiplied by the diameter perpendicular to it). The results represent the values obtained for 5 different mice and are expressed as the % neutralization using as 100% activity the value obtained after an injection with venom incubated with PBS.
For the neutralization of lethal activity, the LD50 values of B. jararaca and B. atrox venoms were estimated according to previous studies [58] to avoid unnecessary animal sacrifice. In all experiments, 3 LD50 doses of B. jararaca (105 µg) or B. atrox (225 µg) venom were incubated with SAB at ratios of 1, 2 and 4 times the potency reference value (1 mL/5 mg venom). The mixtures were incubated at 37°C for 30 min, and 500-µL aliquots were injected intraperitoneally in groups of 5 mice. Control groups included mice injected with PBS or with venom incubated with PBS. Lethality was recorded over a period of 48 hours. The results shown represent the values obtained in 3 independent experiments and are expressed as the % neutralization considering the number of dead/live mice after 48 hours.
The neutralization of the coagulant activity was determined as previously described [59], with some modifications. Samples containing 2 minimum coagulant doses of B. jararaca (71.3 µg/mL) or B. atrox (21.7 µg/mL) venom were incubated with several dilutions of SAB for 30 min at 37°C. Each mixture was added to 100 µL bovine plasma, and the clotting times were recorded using a model ST4 mechanical coagulometer (Diagnostica Stago). Neutralization was expressed as the effective dose (ED), defined as the antivenom/venom ratio at which the clotting time was increased threefold when compared to the clotting time of plasma incubated with venom alone.
Ethics statement
All experiments involving mice were approved by the Ethical Committee for Animal Research of the Instituto Butantan (CEUAIB), São Paulo, Brazil, (application approval number 752/10), who certified its agreement with the Ethical Principles in Animal Research adopted bt the Brazilian College of Animal Experimentation (COBEA).
Results and Discussion
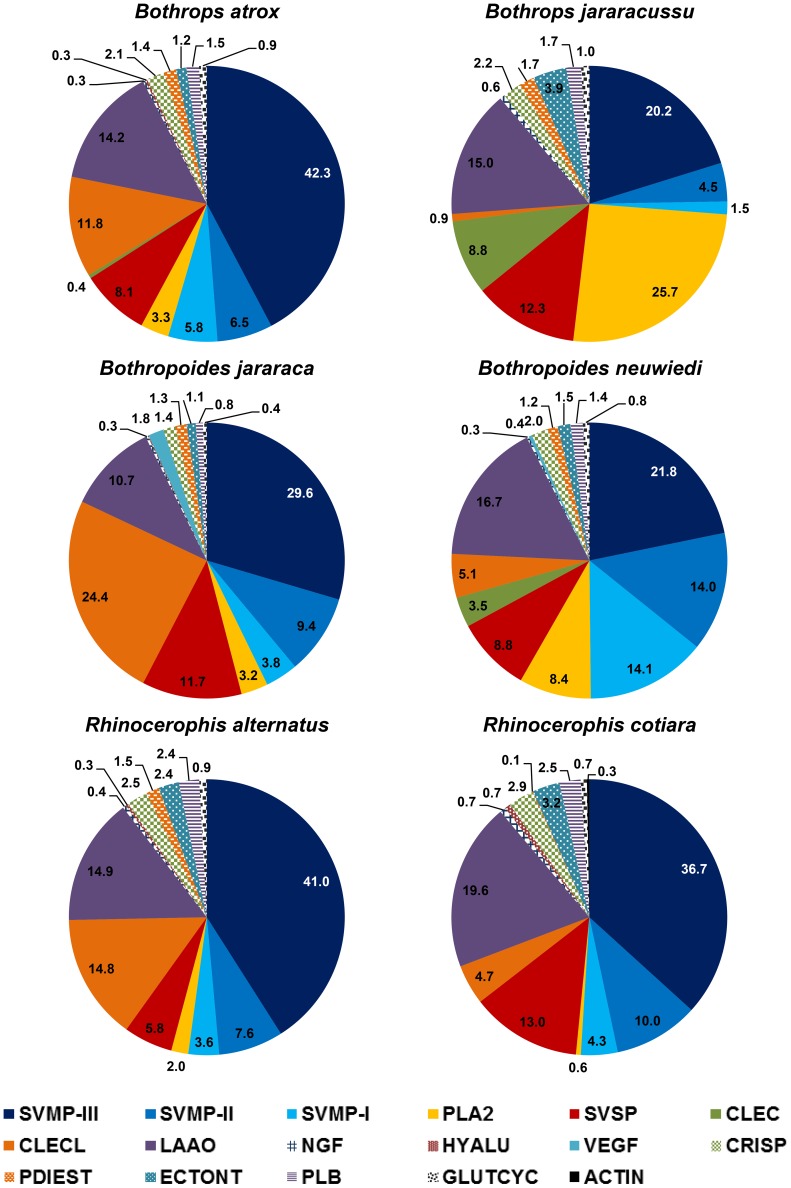
To evaluate the relationship between venom composition and phylogenetic position of the species, we analyzed the proteome of the venoms from the six selected species using shotgun nanoESI-LTQ/Orbitrap. The distribution of the protein families in selected venoms was calculated according to the normalized total spectral counts. As shown in Figure 1, the data analysis revealed 15 different protein groups in different proportions: SVMP-I, -II and –III (snake venom metalloproteinase - classes P-I, P-II and P-III); PLA2 (phospholipase A2); SVSP (snake venom serine proteinase); CLEC (C-type lectin); CLECL (C-type lectin-like); LAAO (L-amino acid oxidase); NGF (nerve growth factor); HYALU (hyaluronidase); VEGF (vascular endothelial growth factor); CRISP (cysteine-rich secretory protein); PDIEST (phosphodiesterase 1); ECTONT (ecto-5′-nucleotidase); PLB (phospholipase B); GLUTCYC (glutaminyl cyclase) and ACTIN (actin). The SVMPs were the most abundant toxins in all of the venoms, particularly in the B. atrox, R. alternatus, R. cotiara and B. jararaca venoms, in which class P-III was notably the predominant toxin. PLA2s predominated in the B. jararacussu venom and was found in significant amounts in the B. neuwiedi venom. A significant contribution of C-type lectin-like proteins was also detected in the B. jararaca, R. alternatus and B. atrox venoms, whereas the SVSPs and LAAOs were almost equally distributed in all of the venoms. One interesting fact was the significant contribution of C-type (true) lectins in the B. jararacussu (8.8%) and B. neuwiedi (3.5%) venoms, in parallel with its absence (<1%) in the other venoms (Figure 1). Comparing these data with previous venomics studies [16], [18]–[20], [23], the major venom protein families as SVMPs, PLA2s and SVSPs were detected in our study in equivalent proportions. However, shotgun nanoESI-LTQ/Orbitrap allowed the detection in all venoms tested of some proteins not yet described as PDIEST, ECTONT, PLB and GLUTCYC. Also, NGF, detected here in all venoms, and HYALU, present in B. atrox, B. jararaca, R. alternatus and R. cotiara venoms, were previously detected in transcriptomes of B. jararacussu and Bothropoides pauloensis, respectively [19], [30], but not in their venomes. Five spectra identified as actin were detected in R. cotiara venom shotgun and due to the high sensitivity of the method, may derive from a minor contamination of the venom with venom gland cells. The most striking difference was the presence of significant amounts of LAAO, CLECL and CLEC spectra detected in our samples, compared to the previous venomics studies. Proteomics by shotgun nanoESI-LTQ/Orbitrap is based on a whole venom digestion by trypsin and the peptide mixture is then fractionated and analyzed in a high sensitive detection system. This approach may bias peptides with higher ionizable efficiency, but all protein families will be represented in the original mixture at the same proportions as they are present on venoms and the bias due to ionization efficiency will be the same for similar peptides present on venoms from different species. Thus, this method is appropriate for comparative studies, allowing the simultaneous analysis of different venoms, under exactly the same conditions. On the other hand, the traditional venomics [5] includes one step in which proteins are quantified and selected after SDS-PAGE separation, according to their staining by Coomassie blue. After trypsinization of selected bands, peptide detection and protein identification will also depend on peptide ionizable efficiency. It is well known that proteins present in venom mixtures in low proportions are hardly detectable by SDS-PAGE as some other venom proteins may be weakly stained. These proteins would be neglected in total protein detection and also when calculating their proportional participation in venom composition. The differences in protein separation methods and sensitivity of detection systems could explain the higher participation of some protein families described in our study when compared to the traditional venomics.
Figure 1. Protein family distribution for the venoms of the three different snake genera, as determined using a shotgun proteomics approach.
Each venom sample was prepared in duplicate, and the MS analyses were performed in triplicate for each venom sample replicate (a total of six MS analyses per venom). The data represent the mean of the normalized total spectral count distributed as follows: 1,891 (B. atrox); 1,727 (B. jararacussu); 2,719 (B. jararaca); 2,287 (B. neuwiedi); 1,252 (R. alternatus) and 1,767 (R. cotiara). The following were identified: SVMP-I, SVMP-II and SVMP–III (snake venom metalloproteinase - classes P-I, P-II and P-III); PLA2 (phospholipase A2); SVSP (snake venom serine proteinase); CLEC (C-type lectin); CLECL (C-type lectin-like); LAAO (L-amino acid oxidase); NGF (nerve growth factor); HYALU (hyaluronidase); VEGF (vascular endothelial growth factor); CRISP (cysteine-rich secretory protein); PDIEST (phosphodiesterase 1); ECTONT (ecto-5′-nucleotidase); PLB (phospholipase B); GLUTCYC (glutaminyl cyclase) and ACTIN (actin).
The venoms were also compared according to the elution profile from reverse-phase C-18 columns. To compare our findings with the previous data from B. atrox, B. cotiara and B. neuwiedi venomics studies [16], [19], [20], C-18 reverse-phase chromatography protocols using similar columns, buffer systems and elution conditions were used to fractionate the venoms. Figure 2 shows the chromatographic profile of the venoms from the six species selected for this study. As expected, the venoms presented comparable chromatographic profiles to those reported in the referenced studies. According to these previous studies, the major protein families were eluted as follows: disintegrins at approximately 50–60 min [19], [20]; basic PLA2s at approximately 110–120 min [19]; P-I SVMPs, some D-49 PLA2s and SVSPs between 120 and 160 min [18]–[20] and P-III SVMPs predominating after 160 min [18]–[20]. Using these data as references, P-III SVMPs appeared to be the most abundant antigens in the chromatograms of the B. atrox, R. alternatus, R. cotiara, B. jararaca and B. neuwiedi venoms, whereas several different peaks in the region corresponding to P-I SVMPs and SVSPs were detected. These observations are consistent with our venomic analysis results shown in Figure 1 and with previous proteomic studies in which P-III SVMPs comprised more than 50% of B. atrox venom [16], [18], approximately 50% of R. alternatus venom [23], approximately 70% of R. cotiara venom [20] and approximately 25.9% of B. neuwiedi venom [19]. SVMPs were also reported to comprise 53.1% of B. jararaca venom gland toxin transcripts [31]. The B. jararacussu venom was the most distinct venom in this group, showing a predominant peak in the PLA2 region and a low abundance of SVMPs, which is consistent with the literature showing a high expression of PLA2 in B. jararacussu venom glands and representing 35% of the total transcripts, followed by only 16% SVMPs and 2% SVSPs [30]. The marked difference in B. jararacussu venom compared to the other Bothrops species was previously reported [60], and a K-49 myotoxin yield of 25% from the crude venom was purified and considered to be the predominant antigen of the B. jararacussu venom [61].
Figure 2. Comparison of the elution profiles of venoms from snakes classified in different genera.
Samples containing 5 mg of crude lyophilized venom from Bothrops atrox, Bothrops jararacussu, Bothropoides jararaca, Bothropoides neuwiedi, Rhinocerophis alternatus and Rhinocerophis cotiara, species maintained at Instituto Butantan herpetarium, were applied to a Vydac C-18 column (4.6×250 mm, 10-µm particle size) coupled to an Agilent 1100 HPLC system. The fractions were eluted at 1 mL/min, with a gradient of 0.1% TFA in water (solution A) and 0.1% TFA in acetonitrile (solution B) (5% B for 10 min, followed by 5–15% B over 20 min, 15–45% B over 120 min, 45–70% B over 20 min and 70–100% B over 10 min). The separations were monitored at 214 nm.
According to the independent parameters used to compare the venoms, in Bothrops, the B. jararacussu profile was very different from that of B. atrox, showing a higher content of phospholipase A2 and a smaller amount of the class P-III metalloproteinase (SVMP) group, as detected either by proteomics or by the elution profile of the native proteins. Within the Bothropoides genus, major differences were observed by proteomics, such as the higher content of CLECL and P-III SVMP in B. jararaca and PI and PII SVMPs, PLA2 and CLEC in B. neuwiedi. The venoms were more similar within the Rhinocerophis genus, particularly when comparing the elution profile of the native proteins, though a higher contribution of CLECL was found in R. alternatus, and higher contents of L-amino acid oxidase and serine proteinase were detected in the R. cotiara venom using the proteomics approach. However, the distribution of B. atrox venom components was very similar to that of R. alternatus by both methods. Furthermore, the pattern observed for B. neuwiedi was closer to that of B. jararacussu venom due to the presence of higher levels of PLA2 and CLEC (Figures 1 and 2). Thus, apparently, venom composition was not related to the phylogenetic position of the snakes.
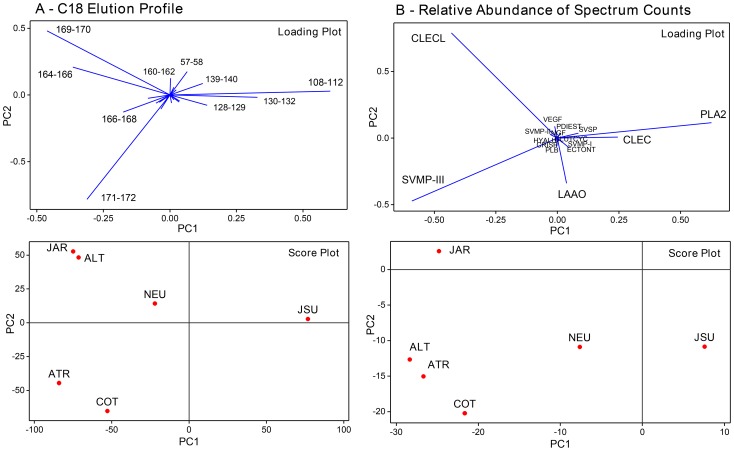
In order to statistically demonstrate these differences, the normalized values of the venom composition obtained by the total spectrum counts of each protein family, and the mAU values of the major peaks eluted in different volumes during the C-18 chromatography, were used as variables to cluster the venoms of snake species. A Principal Component Analysis (PCA) was also carried out in order to understand the key toxins responsible for the venom clustering. The resulting dendrograms and loading and score plots of the PCA are shown in Figures 3 and 4, respectively. Clustering according to the C-18 elution profile shows a strong similarity between R. alternatus and B. jararaca venoms. B. atrox and R. cotiara venoms also show similar elution profile, but different than R. alternatus and B. jararaca venoms, forming, therefore, two different clusters. On the other hand, B. neuwiedi and B. jararacussu venoms reveal lower similarity with the two former clusters, with B. jararacussu having the most distinct features (Figure 3). In the PCA, shown in Figure 4A, components with most prominent loadings that contributed to venom clusterization are the fractions eluted after 160 min with the highest negative values of PC1 (Fraction 164–166: PC1 = −0.365, PC2 = 0.209; Fraction 166–168: PC1 = −0.175, PC2 = −0.128; Fraction 169–170: PC1 = −0.461, PC2 = 0.481; Fraction 171–172: PC1 = −0.311, PC2 = −0.783). These fractions were characterized mostly as class P-III SVMPs in other studies [18]–[20] and reacted with MAJar-3 monoclonal antibodies in this study (see below). Fractions with the highest PC1 positive values were eluted between 108–112 min (PC1 = 0.630, PC2 = 0.029), recognized as PLA2s in previous studies [19], and fractions between 130–132 min (PC1 = 0.330, PC2 = −0.018), characterized as class P-I SVMP in the venom of adult B. atrox from El Paují (Orinoquia, Venezuela) that underwent ontogenetic variation [16]. With respect to proteomic data, B. atrox and R. alternatus venoms were the most closely related, and distances to this group increased gradually for R. cotiara, B. jararaca, B. neuwiedi and B. jararacussu venoms. The clustering of B. atrox and R. alternatus venoms is related to high values of CLECL and P-III SVMPs, which are the proteins with most prominent loadings (CLECL: PC1 = −0.431, PC2 = 0.789, P-III SVMPs: PC1 = −0.592, PC2 = −0.472), and low values of PLA2 and CLEC, also with significant loadings (PLA2: PC1 = 0.245, CLEC : PC1 = 0.245). R. cotiara venom shows similar pattern with respect to P-III SVMP, PLA2 and CLEC, but low values of CLECL and high values of LAAO (PC1 = 0.037, PC2 = −0.339). On the other hand, B. jararaca venom reveals low values of LAAO and large values of CLECL. B. neuwiedi and B. jararacussu venoms present an opposite pattern, with high values of PLA2 and CLEC and low values of PIII-SVMP (Figure 4 B).
Figure 3. Venom clustering according to toxin composition.
The venoms from Bothrops atrox (ATR), Bothrops jararacussu (JSU), Bothropoides jararaca (JAR), Bothropoides neuwiedi (NEU), Rhinocerophis alternatus (ALT) and Rhinocerophis cotiara (COT) were classified according to their protein composition by hierarchical clustering of the observations, including as a variable the normalized maximal mAU at 214 nm in defined elution intervals of C-18 reverse-phase chromatography (Panel A) or normalized total spectral counts of each protein group, as evaluated by shotgun mass spectrometry (Panel B). The procedure used an agglomerative hierarchical method linked by the minimum Euclidean distance between an item in one cluster and an item in another cluster (nearest neighbor) using the Minitab 16 software.
Figure 4. Principal Component Analysis relative to toxin composition.
Loading (top) and score (bottom) plots of the principal components 1 and 2 of the venoms from Bothrops atrox (ATR), Bothrops jararacussu (JSU), Bothropoides jararaca (JAR), Bothropoides neuwiedi (NEU), Rhinocerophis alternatus (ALT) and Rhinocerophis cotiara (COT) according to their protein composition including as variables the normalized maximal mAU at 214 nm in defined elution intervals of C-18 reverse-phase chromatography (Panel A), or the normalized total spectral counts of each protein group, as evaluated by shotgun mass spectrometry (Panel B). The Principal Component Analysis was based on the covariance matrix and all calculations were carried out in the software Minitab 16.
The dendrograms and PCAs obtained using the distinct sets of variables do not coincide, as they were based in distinct parameters. The number of total spectral counts of a given protein is not necessarily related to its mAU 214; moreover, chromatographic fractions represent mixtures of protein families treated as independent variables in the cluster corresponding to the proteomic data. In spite of these differences, both sets of variables indicate that the distribution of venoms is not related to the phylogenetic position of the snakes. It is important to note that a more comprehensive study using venoms from a larger number of species, quantitative assays for isolated components and also complete sequences of venom proteins would be essential to a definitive support of the lack of connection referred to above. Nevertheless, our data are supported by the literature. Taken together, the clusterization and PCA analysis indicate a polarization among the venoms. According to significant PC1 loadings, B. atrox, R. alternatus, R. cotiara and B. jararaca venoms are clearly opposite to B. jararacussu venom, the former group with prominent negative PC1 values of class P-III SVMPs, while B. jararacussu venom shows a polarization towards the presence of PLA2s and class P-I SVMPs. The same toxin polarization has been indicated to venoms from snakes that conserved the paedomorphic characteristics in their venoms (first group) and venoms of snakes whose venom underwent ontogenetic variation (in our study, B. jararacussu venom) [13], [16], [18], [38], [39]. Interestingly, B. neuwiedi venom was grouped closer to B. jararacussu in the cluster analysis, but showed smaller negative PC1 scores, in opposition to B. jararacussu venom. According to the distances, B. neuwiedi venom apparently conserved the paedomorphic phenotype, but may be suffering a transition to the ontogenetic changes observed in B. jararacussu or B. atrox from Colombia.
Correlations between phylogeny and venom composition have been appointed in the literature [36], [37]. Nevertheless, differences in composition of venoms from snakes belonging to the same genera are also present in the literature [62]–[64]. In a recent study, Gibbs et al. [65] found no evidence for significant phylogenetic signal in venom variation of Sistrurus spp, suggesting that diet variation may play a more important role in molding the venom composition. A remarkable variation in venom composition and toxicity was reported for rattlesnakes from Crotalus viridis/oreganus complex [66] and Crotalus durissus and Crotalus simius in Central and South American species [67]. In the latter, differences were related to the conservation of the newborn characteristics of Central American rattlesnake, C. simus, in the South American species and sub-species of C. durissus, a typical example of paedomorphism [67]. These examples are also found in snakes of the Bothrops complex. Tashima et al. [20] reported significant differences in venom composition between two species closely related, R. cotiara and R. fonsecai. A paedomorphic characteristic was also conserved along the dispersion of B. atrox from Central America to the Brazilian Amazon [16], including in the population used in this study. The conservation of the paedomorphic characteristics in B. atrox accounted for the concentration of class P-III SVMPs, which greatly contributes to the overall toxicity of Bothrops venoms [35]. Paedomorphic characteristics were not conserved in B. jararacussu venom, which has predominance of enzymatically inactive myotoxic PLA2s [60] and therefore, presents lower toxicity compared to B. atrox venom. The difference in composition and toxicity of B. atrox and B. jararacussu venoms argues in favor that the gain in toxicity was favorable in B. atrox due to its smaller size. According to this hypothesis, paedomorphic characteristic would not be essential to B. jararacussu snake that is very large and capable of inoculating large amount of venoms in mammalian preys.
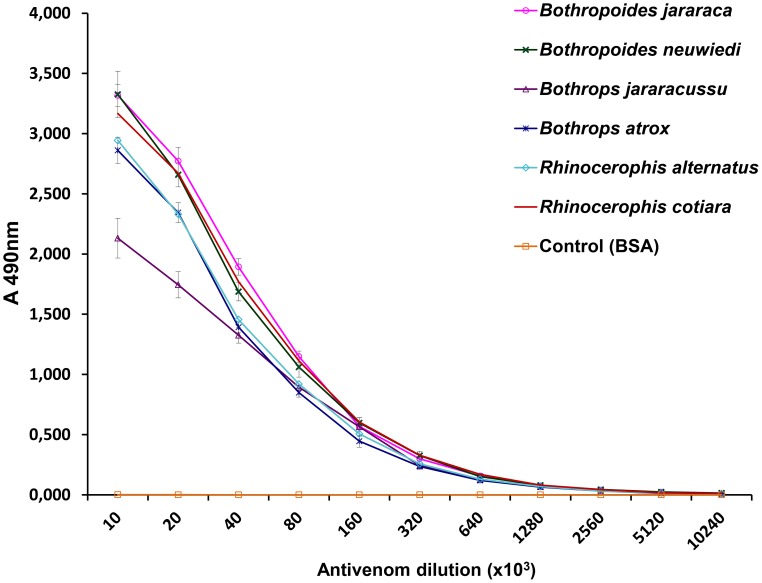
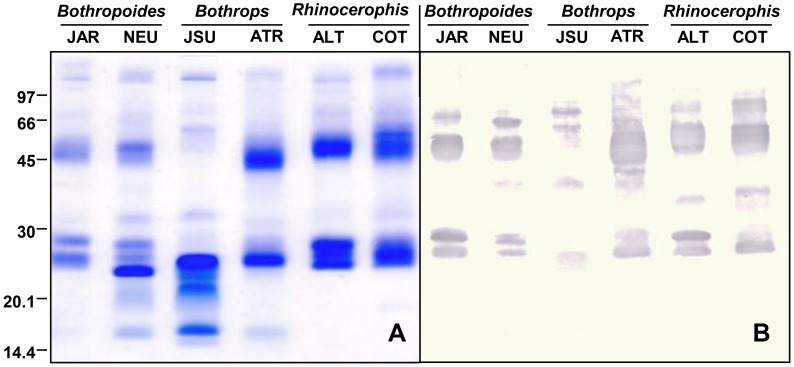
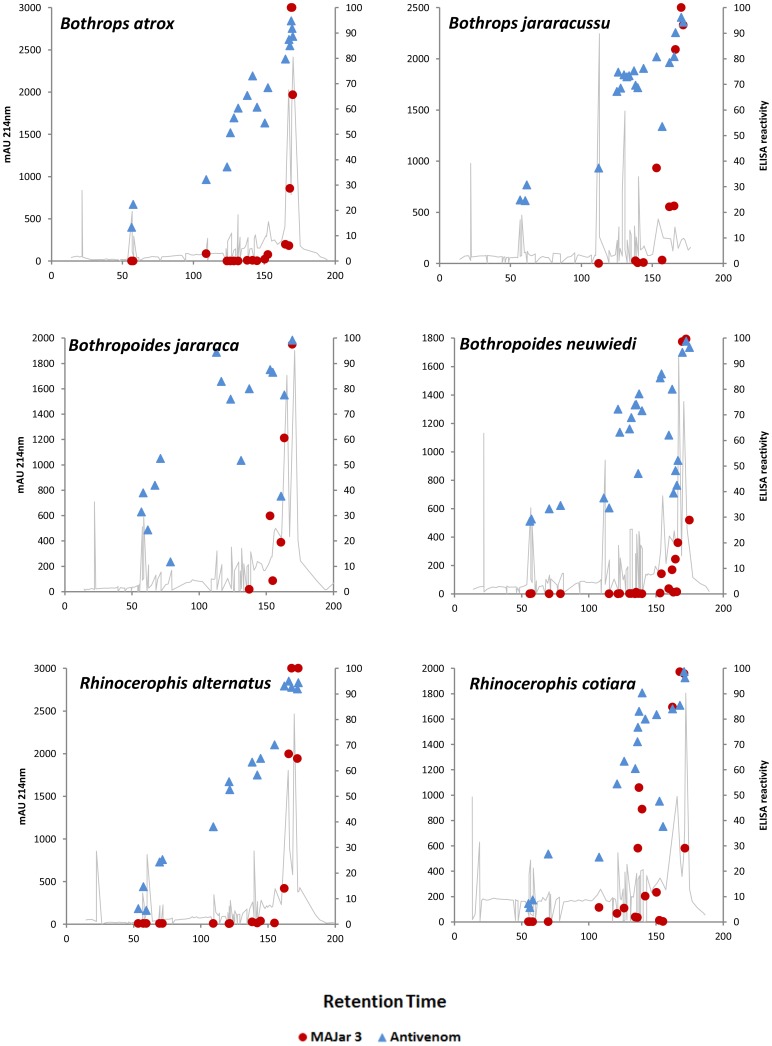
Our next approach was to evaluate the reactivity of the whole venoms and their isolated fractions with antivenoms. Figure 5 shows the titration curves of the antibothropic serum (SAB) in ELISA plates coated with equal amounts of each venom. The SAB antibody titers were the same, regardless of the antigen used, and they corresponded to a dilution of 640,000. The only differences among the venoms were the values obtained for the 10,000 and 20,000 dilutions of SAB against the B. jararacussu venom, which were significantly lower than comparing with other venoms. These dilutions reflect the zone at which the antigen concentration is the limiting factor, and differences in antibody binding may reflect the lower amount of reactive antigens in B. jararacussu venom, highlighting the antigenic relevance of P-III SVMPs. Indeed, the region correspondent to bands of approximately 50 kDa, which is the approximate molecular mass of P-III SVMPs, were less intense in the B. jararacussu venom electrophoresis than others (Figure 6A). SAB preferentially recognized bands of approximately 50 kDa by western blotting (Figure 6B), confirming the higher immunogenicity of SVMPs class P-III. Bands between 20 and 30 kDa, with masses corresponding to SVSPs and P-I SVMPs, were also recognized by SAB (Figure 6B). The SAB reactivity with each fraction from reverse-phase chromatography was also assessed and compared to the reactivity of a monoclonal antibody, MAJar-3, which recognizes the disintegrin domain of P-III SVMPs [51]. In Figure 7, we demonstrate the strong reactivity of the monoclonal antibody with the fractions eluted after 160 minutes (in all chromatograms), confirming that these fractions correspond to P-III SVMPs. The same fractions were the most SAB-reactive antigens in all venoms, regardless of whether these venoms were included in the immunization pool used to prepare the SAB antivenom. Even for the B. jararacussu venom, with a low abundance of SVMPs, the fractions eluted after 160 minutes were the most reactive. Intermediate levels of reactivity were detected with the fractions eluted between 120 and 160 minutes, with very limited reactivity for some, particularly the venoms of B. atrox and B. alternatus, suggesting a lower antigenicity of P-I SVMPs and SVSPs in relation to the SAB antivenom. Interestingly, three small peaks collected from the R. cotiara venom at approximately 140 minutes were strongly reactive with SAB and also with MAJar-3, suggesting the presence of P-III SVMPs in this venom, with distinct structural features and elution profiles. Despite the inclusion of B. jararacussu and B. neuwiedi venoms in the immunization pool, the reactivity of SAB with their fractions (showing PLA2 elution characteristics) from 100 to 110 minutes was moderate. The fractions eluted prior to 100 minutes in all of the chromatograms were poorly recognized by SAB. In other publications, fractions that eluted before 100 min under similar chromatographic conditions corresponded to disintegrins [19], [20], vasoactive peptides [19] or DC fragments of SVMPs [20].
Figure 5. Comparison of ELISA titration curves of Bothrops antivenom with venom from snakes classified in different genera.
Samples containing 100 µL whole venom (10 µg/mL) were used to coat maxisorb microplates (Nunc), which were incubated with crescent dilutions of SAB (starting from 1∶10,000), followed by incubation with anti-horse IgG labeled with peroxidase (1∶2,000). The reactions were developed with ortho-phenylenediamine/H2O2 as the enzyme substrate, and the products were detected at 490 nm. The experiments were performed in duplicate in three independent experiments, and the results are expressed as the mean ± sd of the six OD values.
Figure 6. Comparison of electrophoretic profile (A) and Bothrops antivenom antigenic reactivity (B) of venoms from snakes classified in different genera.
Samples containing 10 µg Bothropoides jararaca (JAR), Bothropoides neuwiedi (NEU), Bothrops atrox (ATR), Bothrops jararacussu(JSU), Rhinocerophis alternatus (ALT) and Rhinocerophis cotiara (COT) venoms were fractionated by SDS-PAGE (12.5% acrylamide gels) under non-reducing conditions and were either stained with Coomassie blue (A) or transferred to nitrocellulose membranes, which were then incubated with SAB (1∶1,000) as the primary antibody and peroxidase-labeled goat anti-horse IgG (1∶1,000). The reactive bands were detected by incubation with 4-chloro-α-naphthol and H2O2 (B). The numbers at the left indicate the mobility of the molecular mass markers in kDa. These results represent three independent runs.
Figure 7. ELISA reactivity with Bothrops antivenom and MAJar-3 monoclonal antibody of fractions collected from chromatograms of venom from snakes classified in different genera.
Samples containing 100 µL 1-µg/mL fractions collected at the elution times represented in the chromatograms were used to coat maxisorb microplates (Nunc), which were incubated with SAB (1∶1,000) or a monoclonal antibody against jararhagin (class P-III SVMP) MAJar-3 (1∶50), followed by incubation with anti-horse IgG (1∶2,000) or anti-mouse IgG (1∶1,000) labeled with peroxidase. The reactions were developed with ortho-phenylenediamine/H2O2 as the enzyme substrate, and the products were detected at 490 nm. The ELISA reactivity was calculated as % reactivity, taking as 100% the maximal OD value obtained in each of three independent experiments performed in duplicate.
Interestingly, despite the different methods used in this study, our results are comparable to those of Núñez et al. [18] and Calvete et al. [16], who showed the complete immunoprecipitation of PIII-SVMPs, to a minor extent of SVSPs and DC-fragments, and limited immunoreactivity towards PLA2 molecules and PI-SVMPs by antivenomics of B. atrox venom with commercial antivenoms. Using antivenomics of B. asper venom and commercial antivenoms, Gutiérrez et al. [9] also showed complete immunodepletion of P-III SVMPs and partial depletion of PLA2s, some serine proteinases, and P-I SVMPs. Correa-Neto et al. [26] approached the same issue by immunomics where the western blots of 2D-gel electrophoresed venoms revealed that antiserum against B. jararacussu venom showed higher reactivity to SVMPs and weaker reactivity towards SVSPs and PLA2s, and anti-jararaca serum preferentially recognized SVMPs and SVSPs among other antigens. Both of these sera failed to recognize low-molecular weight proteins [26]. Comparing the different methods, antivenomics is the best choice for a detailed study, since identifications of non-depleted proteins will show exactly the antigens that are partially immunodepleted or non-reactive with the antivenom. However, the method used here has the advantage to allow simultaneous tests of different venoms, at exactly the same conditions, and gives comparable results to antivenomics, thus is appropriate for comparative studies.
Important conclusions arise from these results. It becomes clear that P-III SVMPs are the predominant antigens in the venom of snakes from the Bothrops complex. Moreover, at least among the Bothrops, SVMPs are cross-reactive antigens that are equally recognized in venoms, regardless of their inclusion in the immunization pool. This is a good indication for antivenom efficacy, as P-III SVMPs are also abundant in most of these venoms and are related to the important symptoms of local and systemic envenomings, such as hemorrhage, the activation of coagulation factors, the inhibition of platelet aggregation and the activation of several factors that lead to local symptoms [35]. Interestingly, P-III and P-I SVMPs share similar catalytic domains and catalytic properties [68], which are involved in most of the toxic activities of SVMPs. Therefore, it is very intriguing that P-I SVMPs are less recognized by the antivenoms than are P-III SVMPs and raises some concerns about the neutralization efficacy of those activities related to the catalytic domain of these molecules. This observation suggests different interpretations: the most immunogenic epitopes of SVMPs may be located within the disintegrin-like or cysteine-rich domains; or catalytic domains of P-III SVMPs are more immunogenic than catalytic domains of P-I SVMPs. For instance, high hemorrhagic activity and the inhibition of platelet aggregation are typical for P-III SVMPs and depend upon disintegrin-like/cysteine-rich domains [69], [70], yet P-I SVMPs are able to induce local reactions [71] and activate coagulation factors [72], which are important effects of snake bites.
SVSPs and PLA2s are important toxins involved in the coagulopathy and local effects, respectively, of patients bitten by snakes of the Bothrops complex. Thus, the limited reactivity of SAB with these fractions should be addressed. Most SVSPs are thrombin-like enzymes involved in the blood coagulation disturbances induced by venom [33], and this symptom is easily controlled in patients after antivenom administration [73], suggesting that the presence of anti-SVSP antibodies in SAB is appropriate to neutralize the activity. However, PLA2s are generally myotoxic or pro-inflammatory [34], and these symptoms are not well neutralized by antivenoms. In the case of SVSPs, it appears that the low levels of antibodies present in SAB are sufficient to neutralize the systemic effects of SVSPs after intravenous administration. In contrast, this does not appear to be the case for the neutralization of the local effects of envenomings induced by PLA2s or P-I SVMPs. This lack of efficacy could most likely be dependent upon antivenom biodisponibility at the site of the lesion rather than on the potency of an antivenom against the myotoxic or dermonecrotic components of the venom [74] or the antibody titer against the toxins inducing the local effects.
Another important point observed in this study was the limited reactivity of antivenom with disintegrins and the DC fragments of SVMPs, which are recognized as inhibitors of platelet aggregation [69], [75], and its reactivity with vasoactive peptides. Although they are not presently considered major toxins correlated with the symptoms of envenomings, the additive or synergistic role of these small toxins in snake bite disorders cannot be ruled out. These low molecular mass peptides are known to be weakly immunogenic; however, in antivenomics studies, at least DC fragments and disintegrins were depleted from B. atrox [18] and B. asper [9] venoms by commercial antivenoms. Nevertheless, the presence of antibodies against such classes of low molecular size toxins in antivenoms should be regarded with more attention.
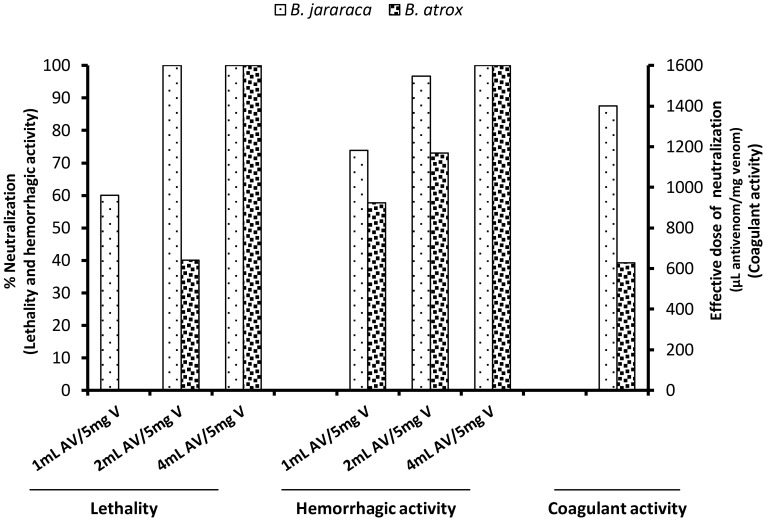
The next step of this study was to evaluate the SAB neutralization efficacy of the lethality, hemorrhagic and coagulant activities of B. atrox venom in comparison to B. jararaca venom. For these experiments, we used venoms from snakes collected in a region where many accidents are reported. The accepted potency of SAB efficacy, calculated as the volume necessary to neutralize the lethality of standard B. jararaca venom, is 1 mL antivenom/5 mg venom. This value was used as a reference to design the neutralization protocols used in this study, whereby this proportion was sufficient to protect more than 50% of mice from the challenge with 3 LD50 doses of B. jararaca venom (105 µg). However, neutralization of the 3 LD50 doses of B. atrox venom (225 µg) was achieved only when the proportion of 2 mL antivenom/5 mg venom was used (Figure 8). Most of the standard protocols to assess antivenom potency use a fixed LD50 value to challenge experimental mice. Therefore, this is also the reference assay used to compare the antivenom efficacy against different venoms. However, it is important to consider that LD50 values are variable among venoms and reflect the toxic activity of each toxin and their synergistic effect to induce death. Additionally, in most tests, the mice are challenged with pre-incubated mixtures of venoms and antivenoms, and, in these reactions, toxins are neutralized or cleared from the solution on a molar concentration basis rather than according to the neutralization of activity. This fact may explain why several previous studies reported that some venoms with higher LD50 values, such as B. atrox and B. jararacussu, are neutralized with higher concentrations of commercial antivenoms.
Figure 8. Neutralizing ability of Bothrops antivenom (SAB) against the major toxic activities of Bothropoides jararaca and Bothrops atrox venoms.
In the neutralization assays, the Bothrops atrox venom was pooled from 8 adult snakes collected in FLONA Tapajós, Santarém, Pará, Brazil. For the neutralization of lethality and hemorrhagic activity, doses of B. jararaca or B. atrox venoms were pre-incubated with SAB at ratios of 1, 2 or 4 times the SAB volume required to neutralize an equal amount of reference venom according to the manufacturer. To assess hemorrhage, 10 µg was incubated and injected intradermically in the dorsum of a group of 5 mice. The results show the % neutralization of the mean values, taken as 100% activity, of the data obtained after injection with venom incubated with saline. For the neutralization of lethal activity, 3 LD50 doses of B. jararaca (105 µg) or B. atrox (225 µg) venom were incubated, and the mixtures were injected intraperitoneally into groups of 5 mice; lethality was recorded over a period of 48 hours. The results represent the values obtained in 3 independent experiments and are expressed as % neutralization, considering the number of live/dead mice after this period. To assess the neutralization of coagulant activity, a constant amount of venom (2 times the minimum coagulant concentrations) was incubated with several dilutions of antivenom; the mixture was added to 100 µl bovine plasma, and the clotting times were recorded using a model ST4 mechanical coagulometer (Diagnostica Stago). The neutralization was expressed as the effective dose (ED), defined as the antivenom/venom ratio at which the clotting time was increased threefold when compared to the clotting time of plasma incubated with venom alone.
Similar findings were observed in our study regarding the neutralization of the coagulant activity of B. atrox and B. jararaca venoms. In this case, the B. atrox venom was more coagulant (minimal coagulant concentration in plasma: 10.8 µg/mL) than the B. jararaca venom (minimal coagulant concentration in plasma: 35.6 µg/mL), and higher concentrations of B. jararaca venom were used in the assays. The SAB neutralized the coagulating activity of both venoms; in this case, however, lower amounts of antivenoms were needed to neutralize the B. atrox activity (ED = 627 µL antivenom/mg venom), whereas B. jararaca venom neutralization required a higher antivenom concentration (ED = 1400 µL antivenom/mg venom), as shown in Figure 8.
The hemorrhagic activity was comparable between the venoms, and the ratio of 1 mL antivenom/5 mg venom neutralized more than 50% of the hemorrhage induced by both venoms (Figure 8). Taken together, these data suggest that SAB is efficient in neutralizing the most important effects of B. atrox venom despite the phylogenetic distance of the snake and the fact that the venom is not included in the immunization pool used to produce SAB. There are previously published papers in the literature suggesting the need to include B. atrox venom for horse immunization [76]–[78]. However, our data showing the opposite are supported by a previous study in which SAB immunodepleted the venom proteins from B. atrox populations exhibiting the paedomorphic venom phenotype, the same pattern found in specimens collected in Pará State, Brazil [16]. Moreover, our present data are supported by other pre-clinical assessments showing neutralization of the toxic activities of venoms not included in immunization protocols [79], [80] and by a clinical trial for the treatment of snake bite patients clinically classified as mild and moderate in Pará State (Brazil) demonstrated that the efficacy of a conventional antivenom (SAB) was comparable to the efficacy of an experimental antivenom prepared through horse immunization with B. atrox venom [81].
Recently, the understanding of venom composition by venomics [5], [6] and tests of the efficacy of antivenoms by antivenomics [8], [9], [11] have been extremely important approaches in order to achieve efficient antivenoms [82]–[84]. In this work, we approached this issue by a multifaceted comparative study of venoms from six species of snakes of distinct phylogenetic clades of Bothrops complex. Important differences were observed in venom composition of the snakes from Bothrops complex, mainly for B. jararacussu venom. However, these differences showed no apparent relationship with the phylogeny of the snakes. In this regard, although the taxonomy of this group is still under revision, the toxins present in the venoms are similar, in agreement with previous molecular data showing that the ancestral genes encoding Bothrops major toxin families were already present before the differentiation of the Bothrops species [85], [86]. As a result, the antivenom reacted similarly with toxins from the same protein family, as SVMPs, SVSPs or PLA2s, regardless of the snake phylogeny or the presence of the venom in the immunization pool used for antivenom production. Thus, we confirm previous data of antivenomics and suggest that it is possible to obtain pan-specific and efficient antivenoms to Bothrops snakes through immunization with venoms from a few species of snakes, if immunogenicity and antigenicity of the distinct protein classes of toxins are considered.
Funding Statement
This study was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES – Grant 063/2010 – Edital Toxinologia), Fundação de Amparo à Pesquisa no Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Programa Estratégico de Apoio à Pesquisa em Saúde (PAPES VI/FIOCRUZ). LFS, CAN and SSO are recipients of a student fellowship from CAPES; JLB, JAPJ and ILS are recipients of a student fellowship from FAPESP and PSP is recipient of a student fellowship from CNPq. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO (2007) Rabies and envenomings. A neglected public health issue. Available: http://whqlibdoc.who.int/publications/2007/9789241563482_eng.pdf
- 2. Kasturiratne A, Wickremasinghe AR, de Silva N, Gunawardena NK, Pathmeswaran A, et al. (2008) The global burden of snakebite: a literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med 5: e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ministério-da-Saúde (2001) Manual de diagnóstico e tratamento de acidentes por animais peçonhentos. 2nd ed. Brasília: Fundação Nacional de Saúde.
- 4.Brazil V (1918) Do envenenamento ofídico e seu tratamento. Collectanea de Trabalhos (1901–1917) - Instituto Butantan. São Paulo: Typographia do Diário Official. pp. 31–52.
- 5. Calvete JJ, Juárez P, Sanz L (2007) Snake venomics. Strategy and applications. J Mass Spectrom 42: 1405–1414. [DOI] [PubMed] [Google Scholar]
- 6. Calvete JJ (2013) Snake venomics: From the inventory of toxins to biology. Toxicon doi:10.1016/j.toxicon.2013.03.020 [DOI] [PubMed] [Google Scholar]
- 7. Calvete JJ (2011) Proteomic tools against the neglected pathology of snake bite envenoming. Expert Rev Proteomics 8: 739–758. [DOI] [PubMed] [Google Scholar]
- 8. Calvete JJ, Sanz L, Angulo Y, Lomonte B, Gutiérrez JM (2009) Venoms, venomics, antivenomics. FEBS Lett 583: 1736–1743. [DOI] [PubMed] [Google Scholar]
- 9. Gutiérrez JM, Lomonte B, León G, Alape-Girón A, Flores-Díaz M, et al. (2009) Snake venomics and antivenomics: Proteomic tools in the design and control of antivenoms for the treatment of snakebite envenoming. J Proteomics 72: 165–182. [DOI] [PubMed] [Google Scholar]
- 10. Gutiérrez JM, León G, Lomonte B, Angulo Y (2011) Antivenoms for snakebite envenomings. Inflamm Allergy Drug Targets 10: 369–380. [DOI] [PubMed] [Google Scholar]
- 11. Pla D, Gutiérrez JM, Calvete JJ (2012) Second generation snake antivenomics: comparing immunoaffinity and immunodepletion protocols. Toxicon 60: 688–699. [DOI] [PubMed] [Google Scholar]
- 12. Calvete JJ, Cid P, Sanz L, Segura A, Villalta M, et al. (2010) Antivenomic assessment of the immunological reactivity of EchiTAb-Plus-ICP, an antivenom for the treatment of snakebite envenoming in sub-Saharan Africa. Am J Trop Med Hyg 82: 1194–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alape-Girón A, Sanz L, Escolano J, Flores-Díaz M, Madrigal M, et al. (2008) Snake venomics of the lancehead pitviper Bothrops asper: geographic, individual, and ontogenetic variations. J Proteome Res 7: 3556–3571. [DOI] [PubMed] [Google Scholar]
- 14. Alape-Girón A, Flores-Díaz M, Sanz L, Madrigal M, Escolano J, et al. (2009) Studies on the venom proteome of Bothrops asper: perspectives and applications. Toxicon 54: 938–948. [DOI] [PubMed] [Google Scholar]
- 15. Calvete JJ, Borges A, Segura A, Flores-Díaz M, Alape-Girón A, et al. (2009) Snake venomics and antivenomics of Bothrops colombiensis, a medically important pitviper of the Bothrops atrox-asper complex endemic to Venezuela: Contributing to its taxonomy and snakebite management. J Proteomics 72: 227–240. [DOI] [PubMed] [Google Scholar]
- 16. Calvete JJ, Sanz L, Pérez A, Borges A, Vargas AM, et al. (2011) Snake population venomics and antivenomics of Bothrops atrox: Paedomorphism along its transamazonian dispersal and implications of geographic venom variability on snakebite management. J Proteomics 74: 510–527. [DOI] [PubMed] [Google Scholar]
- 17. Gutiérrez JM, Sanz L, Escolano J, Fernández J, Lomonte B, et al. (2008) Snake venomics of the Lesser Antillean pit vipers Bothrops caribbaeus and Bothrops lanceolatus: correlation with toxicological activities and immunoreactivity of a heterologous antivenom. J Proteome Res 7: 4396–4408. [DOI] [PubMed] [Google Scholar]
- 18. Núñez V, Cid P, Sanz L, De La Torre P, Angulo Y, et al. (2009) Snake venomics and antivenomics of Bothrops atrox venoms from Colombia and the Amazon regions of Brazil, Perú and Ecuador suggest the occurrence of geographic variation of venom phenotype by a trend towards paedomorphism. J Proteomics 73: 57–78. [DOI] [PubMed] [Google Scholar]
- 19. Rodrigues RS, Boldrini-França J, Fonseca FP, de la Torre P, Henrique-Silva F, et al. (2012) Combined snake venomics and venom gland transcriptomic analysis of Bothropoides pauloensis. J Proteomics 75: 2707–2720. [DOI] [PubMed] [Google Scholar]
- 20. Tashima AK, Sanz L, Camargo AC, Serrano SM, Calvete JJ (2008) Snake venomics of the Brazilian pitvipers Bothrops cotiara and Bothrops fonsecai. Identification of taxonomy markers. J Proteomics 71: 473–485. [DOI] [PubMed] [Google Scholar]
- 21. Fox JW, Ma L, Nelson K, Sherman NE, Serrano SM (2006) Comparison of indirect and direct approaches using ion-trap and Fourier transform ion cyclotron resonance mass spectrometry for exploring viperid venom proteomes. Toxicon 47: 700–714. [DOI] [PubMed] [Google Scholar]
- 22. Baramova EN, Shannon JD, Bjarnason JB, Fox JW (1989) Degradation of extracellular matrix proteins by hemorrhagic metalloproteinases. Archives of Biochemistry and Biophysics 275: 63–71. [DOI] [PubMed] [Google Scholar]
- 23. Ohler M, Georgieva D, Seifert J, von Bergen M, Arni RK, et al. (2010) The Venomics of Bothrops alternatus is a Pool of Acidic Proteins with Predominant Hemorrhagic and Coagulopathic Activities. Journal of Proteome Research 9: 2422–2437. [DOI] [PubMed] [Google Scholar]
- 24. Valente RH, Guimarães PR, Junqueira M, Neves-Ferreira AG, Soares MR, et al. (2009) Bothrops insularis venomics: a proteomic analysis supported by transcriptomic-generated sequence data. J Proteomics 72: 241–255. [DOI] [PubMed] [Google Scholar]
- 25. Zelanis A, Tashima AK, Pinto AF, Leme AF, Stuginski DR, et al. (2011) Bothrops jararaca venom proteome rearrangement upon neonate to adult transition. Proteomics 11: 4218–4228. [DOI] [PubMed] [Google Scholar]
- 26. Correa-Netto C, Teixeira-Araujo R, Aguiar AS, Melgarejo AR, De-Simone SG, et al. (2010) Immunome and venome of Bothrops jararacussu: a proteomic approach to study the molecular immunology of snake toxins. Toxicon 55: 1222–1235. [DOI] [PubMed] [Google Scholar]
- 27. Kohlhoff M, Borges MH, Yarleque A, Cabezas C, Richardson M, et al. (2012) Exploring the proteomes of the venoms of the Peruvian pit vipers Bothrops atrox, B. barnetti and B. pictus. J Proteomics 75: 2181–2195. [DOI] [PubMed] [Google Scholar]
- 28. Cardoso KC, Da Silva MJ, Costa GG, Torres TT, Del Bem LE, et al. (2010) A transcriptomic analysis of gene expression in the venom gland of the snake Bothrops alternatus (urutu). BMC Genomics 11: 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neiva M, Arraes FB, de Souza JV, Rádis-Baptista G, Prieto da Silva AR, et al. (2009) Transcriptome analysis of the Amazonian viper Bothrops atrox venom gland using expressed sequence tags (ESTs). Toxicon 53: 427–436. [DOI] [PubMed] [Google Scholar]
- 30. Kashima S, Roberto PG, Soares AM, Astolfi-Filho S, Pereira JO, et al. (2004) Analysis of Bothrops jararacussu venomous gland transcriptome focusing on structural and functional aspects: I–gene expression profile of highly expressed phospholipases A2. Biochimie 86: 211–219. [DOI] [PubMed] [Google Scholar]
- 31. Cidade DA, Simão TA, Dávila AM, Wagner G, Junqueira-de-Azevedo IL, et al. (2006) Bothrops jararaca venom gland transcriptome: analysis of the gene expression pattern. Toxicon 48: 437–461. [DOI] [PubMed] [Google Scholar]
- 32. Durban J, Juárez P, Angulo Y, Lomonte B, Flores-Diaz M, et al. (2011) Profiling the venom gland transcriptomes of Costa Rican snakes by 454 pyrosequencing. BMC Genomics 12: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Serrano SM, Maroun RC (2005) Snake venom serine proteinases: sequence homology vs. substrate specificity, a paradox to be solved. Toxicon 45: 1115–1132. [DOI] [PubMed] [Google Scholar]
- 34. Gutiérrez JM, Lomonte B (2013) Phospholipases A(2): Unveiling the secrets of a functionally versatile group of snake venom toxins. Toxicon 62: 27–39. [DOI] [PubMed] [Google Scholar]
- 35. Moura-da-Silva AM, Butera D, Tanjoni I (2007) Importance of snake venom metalloproteinases in cell biology: Effects on platelets, inflammatory and endothelial cells. Current Pharmaceutical Design 13: 2893–2905. [DOI] [PubMed] [Google Scholar]
- 36.Tu AT (1977) Venoms: Chemistry and Molecular Biology. New York, USA: John Wiley.
- 37.Tu AT (1991) Reptile Venoms and Toxins. Handbook of Natural Toxins. New York: Marcel Dekker, Inc.
- 38. Guercio RA, Shevchenko A, Lopez-Lozano JL, Paba J, Sousa MV, et al. (2006) Ontogenetic variations in the venom proteome of the Amazonian snake Bothrops atrox. Proteome Sci 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saldarriaga MM, Otero R, Núñez V, Toro MF, Díaz A, et al. (2003) Ontogenetic variability of Bothrops atrox and Bothrops asper snake venoms from Colombia. Toxicon 42: 405–411. [DOI] [PubMed] [Google Scholar]
- 40. Menezes MC, Furtado MF, Travaglia-Cardoso SR, Camargo AC, Serrano SM (2006) Sex-based individual variation of snake venom proteome among eighteen Bothrops jararaca siblings. Toxicon 47: 304–312. [DOI] [PubMed] [Google Scholar]
- 41. Gibbs HL, Sanz L, Calvete JJ (2009) Snake population venomics: proteomics-based analyses of individual variation reveals significant gene regulation effects on venom protein expression in Sistrurus rattlesnakes. J Mol Evol 68: 113–125. [DOI] [PubMed] [Google Scholar]
- 42. Daltry JC, Wüster W, Thorpe RS (1996) Diet and snake venom evolution. Nature 379: 537–540. [DOI] [PubMed] [Google Scholar]
- 43. Gibbs HL, Mackessy SP (2009) Functional basis of a molecular adaptation: prey-specific toxic effects of venom from Sistrurus rattlesnakes. Toxicon 53: 672–679. [DOI] [PubMed] [Google Scholar]
- 44. Barlow A, Pook CE, Harrison RA, Wüster W (2009) Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc Biol Sci 276: 2443–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Calvete JJ, Marcinkiewicz C, Monleon D, Esteve V, Celda B, et al. (2005) Snake venom disintegrins: evolution of structure and function. Toxicon 45: 1063–1074. [DOI] [PubMed] [Google Scholar]
- 46. Moura-da-Silva AM, Theakston RDG, Crampton JM (1996) Evolution of Disintegrin Cysteine-rich and mammalian Matrix-degrading Metalloproteinases: Gene duplication and divergence of a common ancestor rather than convergent evolution. Journal of Molecular Evolution 43: 263–269. [DOI] [PubMed] [Google Scholar]
- 47. Carrasco PA, Mattoni CI, Leynaud GC, Scrocchi GJ (2012) Morphology, phylogeny and taxonomy of South American bothropoid pit vipers (Serpentes, Viperidae). Zoologica Scripta 41: 109–124. [Google Scholar]
- 48.Wüster W, Salomão MG, Quijada-Mascarenhas JA, Thorpe RS (2002) Origins and evolution of the South American pitvipers fauna: evidence from mitocondrial DNA sequence analysis. In: Shuett W, Höggren M, Douglas ME, Greene HW, editors. Biology of the vipers. Eagle Mountain, UT: Eagle Mountain Publishing. pp. 111–129.
- 49. Castoe TA, Parkinson CL (2006) Bayesian mixed models and the phylogeny of pitvipers (Viperidae: Serpentes). Mol Phylogenet Evol 39: 91–110. [DOI] [PubMed] [Google Scholar]
- 50. Fenwick AM, Gutberlet RL Jr, Evans JA, Parkinson CL (2009) Morphological and molecular evidence for phylogeny and classification of South American pitvipers, genera Bothrops, Bothriopsis, and Bothrocophias (Serpentes: Viperidae). Zoological Journal of the Linnean Society 156: 617–640. [Google Scholar]
- 51. Tanjoni I, Butera D, Bento L, Della-Casa MS, Marques-Porto R, et al. (2003) Snake venom metalloproteinases: structure/function relationships studies using monoclonal antibodies. Toxicon 42: 801–808. [DOI] [PubMed] [Google Scholar]
- 52. Tanjoni I, Butera D, Spencer PJ, Takehara HA, Fernandes I, et al. (2003) Phylogenetic conservation of a snake venom metalloproteinase epitope recognized by a monoclonal antibody that neutralizes hemorrhagic activity. Toxicon 42: 809–816. [DOI] [PubMed] [Google Scholar]
- 53. Cunha Bastos VL, Salles JB, Valente RH, León IR, Perales J, et al. (2007) Cytosolic glutathione peroxidase from liver of pacu (Piaractus mesopotamicus), a hypoxia-tolerant fish of the Pantanal. Biochimie 89: 1332–1342. [DOI] [PubMed] [Google Scholar]
- 54. McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, et al. (2004) MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom 18: 2162–2168. [DOI] [PubMed] [Google Scholar]
- 55. Xu T, Venable JD, Park SK, Cociorva D (2006) ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Molecular and Cellular Proteomics 5: S–174. [Google Scholar]
- 56. Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392. [DOI] [PubMed] [Google Scholar]
- 57. Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658. [DOI] [PubMed] [Google Scholar]
- 58. Camey KU, Velarde DT, Sanchez EF (2002) Pharmacological characterization and neutralization of the venoms used in the production of Bothropic antivenom in Brazil. Toxicon 40: 501–509. [DOI] [PubMed] [Google Scholar]
- 59. Gené JA, Roy A, Rojas G, Gutiérrez JM, Cerdas L (1989) Comparative study on coagulant, defibrinating, fibrinolytic and fibrinogenolytic activities of Costa Rican crotaline snake venoms and their neutralization by a polyvalent antivenom. Toxicon 27: 841–848. [DOI] [PubMed] [Google Scholar]
- 60. Moura-da-Silva AM, Cardoso DF, Tanizaki MM (1990) Differences in distribution of myotoxic proteins in venoms from different Bothrops species. Toxicon 28: 1293–1301. [DOI] [PubMed] [Google Scholar]
- 61. Moura-da-Silva AM, Desmond H, Laing G, Theakston RDG (1991) Isolation and Comparison of Myotoxins Isolated From Venoms of Different Species of Bothrops Snakes. Toxicon 29: 713–723. [DOI] [PubMed] [Google Scholar]
- 62.Mackessy SP (2008) Venom composition in rattlesnakes: trends and biological significance. In: Hayes WK, Beaman KR, Cardwell MD, Bush SP, editors. The Biology of Rattlesnakes. Loma Linda, CA, USA : Loma Linda University Press. pp. 495–510.
- 63. Angulo Y, Escolano J, Lomonte B, Gutiérrez JM, Sanz L, et al. (2008) Snake venomics of Central American pitvipers: clues for rationalizing the distinct envenomation profiles of Atropoides nummifer and Atropoides picadoi. J Proteome Res 7: 708–719. [DOI] [PubMed] [Google Scholar]
- 64. Fernández J, Lomonte B, Sanz L, Angulo Y, Gutiérrez JM, et al. (2010) Snake venomics of Bothriechis nigroviridis reveals extreme variability among palm pitviper venoms: different evolutionary solutions for the same trophic purpose. J Proteome Res 9: 4234–4241. [DOI] [PubMed] [Google Scholar]
- 65. Gibbs HL, Sanz L, Sovic MG, Calvete JJ (2013) Phylogeny-Based Comparative Analysis of Venom Proteome Variation in a Clade of Rattlesnakes (Sistrurus sp.). PLoS One 8: e67220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mackessy SP (2010) Evolutionary trends in venom composition in the western rattlesnakes (Crotalus viridis sensu lato): toxicity vs. tenderizers. Toxicon 55: 1463–1474. [DOI] [PubMed] [Google Scholar]
- 67. Calvete JJ, Sanz L, Cid P, de la Torre P, Flores-Díaz M, et al. (2010) Snake venomics of the Central American rattlesnake Crotalus simus and the South American Crotalus durissus complex points to neurotoxicity as an adaptive paedomorphic trend along Crotalus dispersal in South America. J Proteome Res 9: 528–544. [DOI] [PubMed] [Google Scholar]
- 68. Fox JW, Serrano SMT (2008) Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. Febs Journal 275: 3016–3030. [DOI] [PubMed] [Google Scholar]
- 69. Serrano SM, Kim J, Wang D, Dragulev B, Shannon JD, et al. (2006) The cysteine-rich domain of snake venom metalloproteinases is a ligand for von Willebrand factor A domains: role in substrate targeting. Journal of Biological Chemistry 281: 39746–39756. [DOI] [PubMed] [Google Scholar]
- 70. Baldo C, Jamora C, Yamanouye N, Zorn TM, Moura-da-Silva AM (2010) Mechanisms of Vascular Damage by Hemorrhagic Snake Venom Metalloproteinases: Tissue Distribution and In Situ Hydrolysis. Plos Neglected Tropical Diseases 4: e727 doi:10.1371/journal.pntd.0000727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gutierrez JM, Rucavado A (2000) Snake venom metalloproteinases: Their role in the pathogenesis of local tissue damage. Biochimie 82: 841–850. [DOI] [PubMed] [Google Scholar]
- 72. Modesto JC, Junqueira-de-Azevedo IL, Neves-Ferreira AG, Fritzen M, Oliva ML, et al. (2005) Insularinase A, a prothrombin activator from Bothrops insularis venom, is a metalloprotease derived from a gene encoding protease and disintegrin domains. Biol Chem 386: 589–600. [DOI] [PubMed] [Google Scholar]
- 73. Cardoso JL, Fan HW, França FO, Jorge MT, Leite RP, et al. (1993) Randomized comparative trial of three antivenoms in the treatment of envenoming by lance-headed vipers (Bothrops jararaca) in São Paulo, Brazil. Quarterly Journal of Medicine 86: 315–325. [PubMed] [Google Scholar]
- 74. Gutierrez JM, Leon G, Rojas G, Lomonte B, Rucavado A, et al. (1998) Neutralization of local tissue damage induced by Bothrops asper (terciopelo) snake venom. Toxicon 36: 1529–1538. [DOI] [PubMed] [Google Scholar]
- 75. McLane MA, Marcinkiewicz C, Vijay-Kumar S, Wierzbicka-Patynowski I, Niewiarowski S (1998) Viper venom disintegrins and related molecules. Proceedings of the Society for Experimental Biology and Medicine 219: 109–119. [DOI] [PubMed] [Google Scholar]
- 76. Furtado MeF, Cardoso ST, Soares OE, Pereira AP, Fernandes DS, et al. (2010) Antigenic cross-reactivity and immunogenicity of Bothrops venoms from snakes of the Amazon region. Toxicon 55: 881–887. [DOI] [PubMed] [Google Scholar]
- 77. Queiroz GP, Pessoa LA, Portaro FC, Furtado MeF, Tambourgi DV (2008) Interspecific variation in venom composition and toxicity of Brazilian snakes from Bothrops genus. Toxicon 52: 842–851. [DOI] [PubMed] [Google Scholar]
- 78. Muniz EG, Maria WS, Estevão-Costa MI, Buhrnheim P, Chávez-Olórtegui C (2000) Neutralizing potency of horse antibothropic Brazilian antivenom against Bothrops snake venoms from the Amazonian rain forest. Toxicon 38: 1859–1863. [DOI] [PubMed] [Google Scholar]
- 79. Segura A, Castillo MC, Núñez V, Yarlequé A, Gonçalves LR, et al. (2010) Preclinical assessment of the neutralizing capacity of antivenoms produced in six Latin American countries against medically-relevant Bothrops snake venoms. Toxicon 56: 980–989. [DOI] [PubMed] [Google Scholar]
- 80. Saravia P, Rojas E, Escalante T, Arce V, Chaves E, et al. (2001) The venom of Bothrops asper from Guatemala: toxic activities and neutralization by antivenoms. Toxicon 39: 401–405. [DOI] [PubMed] [Google Scholar]
- 81. Pardal PPO, Souza SM, Monteiro MRCC, Fan HW, Cardoso JLC, et al. (2004) Clinical trial of two antivenoms for the treatment of Bothrops and Lachesis bites in the north eastern Amazon region of Brazil. Transactions of the Royal Society of Tropical Medicine and Hygiene 98: 28–42. [DOI] [PubMed] [Google Scholar]
- 82. Calvete JJ (2010) Antivenomics and venom phenotyping: A marriage of convenience to address the performance and range of clinical use of antivenoms. Toxicon 56: 1284–1291. [DOI] [PubMed] [Google Scholar]
- 83. Calvete JJ, Domont GB (2011) Omic technologies to fight the neglect. J Proteomics 74: 1483–1484. [DOI] [PubMed] [Google Scholar]
- 84. Gutiérrez JM, Solano G, Pla D, Herrera M, Segura A, et al. (2013) Assessing the preclinical efficacy of antivenoms: From the lethality neutralization assay to antivenomics. Toxicon 69: 168–179. [DOI] [PubMed] [Google Scholar]
- 85. Casewell NR, Wagstaff SC, Harrison RA, Renjifo C, Wüster W (2011) Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol Biol Evol 28: 2637–2649. [DOI] [PubMed] [Google Scholar]
- 86. Fry BG, Vidal N, van der Weerd L, Kochva E, Renjifo C (2009) Evolution and diversification of the Toxicofera reptile venom system. J Proteomics 72: 127–136. [DOI] [PubMed] [Google Scholar]