Abstract
C. elegans PAQR-2 is homologous to the insulin-sensitizing adiponectin receptors in mammals, and essential for adaptation to growth at 15°C, a low but usually acceptable temperature for this organism. By screening for novel paqr-2 suppressors, we identified mutations in genes involved in phosphatidylcholine synthesis (cept-1, pcyt-1 and sams-1) and fatty acid metabolism (ech-7, hacd-1, mdt-15, nhr-49 and sbp-1). We then show genetic evidence that paqr-2, phosphatidylcholines, sbp-1 and Δ9-desaturases form a cold adaptation pathway that regulates the increase in unsaturated fatty acids necessary to retain membrane fluidity at low temperatures. This model is supported by the observations that the paqr-2 suppressors normalize the levels of saturated fatty acids, and that low concentrations of detergents that increase membrane fluidity can rescue the paqr-2 mutant.
Author Summary
Cold-blooded organisms such as insects, fish or worms must make physiological adjustments when the temperature in their environment decreases. One essential adaptive measure is to increase the fluidity of the cellular membranes that are made of fatty molecules and would tend to harden at low temperatures, just as butter would. In our study we identify genes that are regulated by PAQR-2, a membrane protein that we show to be essential for adjusting the membrane fluidity during cold adaptation in the nematode C. elegans. Interestingly, the genes influenced by PAQR-2 are all involved in fatty acid metabolism. We speculate that the human homologs of PAQR-2, which are receptors for the hormone adiponectin, may have similar functions.
Introduction
To function over a range of temperatures, poikilotherms must make adaptive adjustments to their physiology [1]–[4]. For example, adaptation to cold in Drosophila correlates with changes in the phospholipid composition of cellular membranes, favoring a higher abundance of shorter and/or unsaturated fatty acids to maintain membrane fluidity [5]–[8]. Another aspect of cold adaptation involves the activation of stress responses, including the inaptly named heat shock proteins [4], [9], [10]. On the whole however, little is known about the regulatory pathways, from receptors to effectors, that coordinate the well-documented physiological changes that poikilotherms make as they adapt to cold temperatures.
The C. elegans paqr-2 mutant is unable to adapt to growth at 15°C, which is close to the lowest temperature at which this species can be propagated [11], [12]. paqr-2 encodes a protein with seven transmembrane domains homologous to the mammalian adiponectin receptors, AdipoR1 and AdipoR2, that are insuling-sensitizing metabolic regulators of which the signaling pathways are not well understood [1]–[3], [13]–[15]. Like its mammalian homologs, paqr-2 is important for metabolic regulation: the paqr-2 mutant shows an abnormal fatty acid (FA) composition, an excess of lipid droplets when combined with a paqr-1 mutation, and is synthetic lethal with loss-of-function (lof) mutations in genes that promote FA turnover such as sbp-1 and nhr-49 [5], [12], which are C. elegans homologs of SREBP and HNF4 [16]–[19]. One important limitation of our earlier work on paqr-2 is that we used a candidate gene approach, mostly inspired from previous studies in mice, to try and identify genes that may interact with paqr-2. While often successful, such an approach makes it impossible to discover unexpected interactions. Here, we used an unbiased forward genetic approach in which we screened randomly mutagenized worm populations to isolate and then characterize mutations that suppress the cold adaptation defect of the paqr-2 mutant. This is a classical investigative strategy for which C. elegans is eminently suited [20]. Using this approach, we isolated nine paqr-2 suppressors and establish a novel regulatory pathway connecting paqr-2, phosphatidylcholine (PC) synthesis and the regulation of FA desaturation to increase membrane fluidity during cold adaptation.
Results
The paqr-2(tm3410) mutant
The paqr-2(tm3410) allele carries a deletion that eliminates the first transmembrane domain and introduces a premature stop codon prior to all remaining transmembrane domains [12] (Figure S1A). Consistently, antibodies raised against the C-terminus of PAQR-2 detect a band of ∼66 kDa that is absent in the paqr-2(tm3410) mutant but reappears when the wild-type paqr-2 gene is reintroduced as a transgene (Figure S1B). paqr-2(tm3410), later referred to simply as paqr-2, is therefore likely a null allele.
A paqr-2 suppressor screen
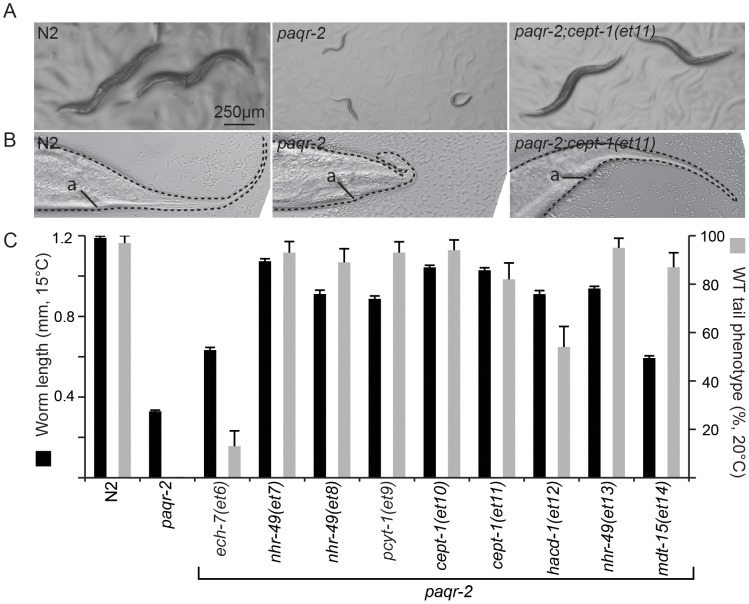
We mutagenized paqr-2 mutant worms and screened the F2 progeny for their ability to grow and reproduce at 15°C. A screen of ∼15 000 mutagenized haploid genomes led to the isolation of 9 paqr-2 suppressor mutants (alleles et6-et14; Figure 1A and C; Figure S2A) all of which also suppressed the characteristic paqr-2 withered tail tip phenotype (Figure 1B and C), as well as brood size and length defects at 20°C (one exception is the et6 allele which only slightly rescued the tail phenotype, did not rescue brood size and caused a slight reduction in length at 20°C; Figure S2B–C). The suppressor mutations therefore suppress multiple aspects of the paqr-2 mutant phenotype, rather than merely improving cold-adaptation.
Figure 1. Overview of the paqr-2 suppressor mutations.
(A) Wild-type (N2), paqr-2 mutants and paqr-2 mutant carrying the suppressor mutant allele et11 photographed after 144 hours of cultivation at 15°C. (B) Tail of wild-type (N2), paqr-2 mutant and paqr-2 mutant carrying the suppressor mutant allele et11 grown at 20°C. “a” indicates the position of the anus. (C) Quantification of the suppression of two paqr-2 phenotypes by the nine isolated suppressor alleles (et6–et14).
paqr-2 suppressors
To molecularly define the suppressor mutations, we used a strategy based on back-crossing and whole-genome sequencing [21], [22]: each suppressor was outcrossed 4 or 6 times to wild type and paqr-2 single mutants, and its genome was then sequenced. For each suppressor, mutation clusters present in the outcrossed strains defined a region of interest in which to look for the suppressor mutation. Candidate mutations were tested using three strategies: 1) Attempt to desuppress the paqr-2 phenotypes by introducing the wild-type version of the candidate gene into the suppressor strain; 2) Test whether available loss-of-function (lof) mutations or RNAi inhibition of the candidate gene suppress the paqr-2 phenotypes; and 3) When a gain-of-function (gof) mutation was considered, attempt to suppress the paqr-2 phenotypes by introducing the mutated version of the candidate gene as a transgene. Figures 2 and S3 show the experimental data confirming the identification of the paqr-2 suppressors, Figure 3 and Table 1 describe their molecular nature, and Figure 4 presents them in the form of a metabolic/genetic pathway.
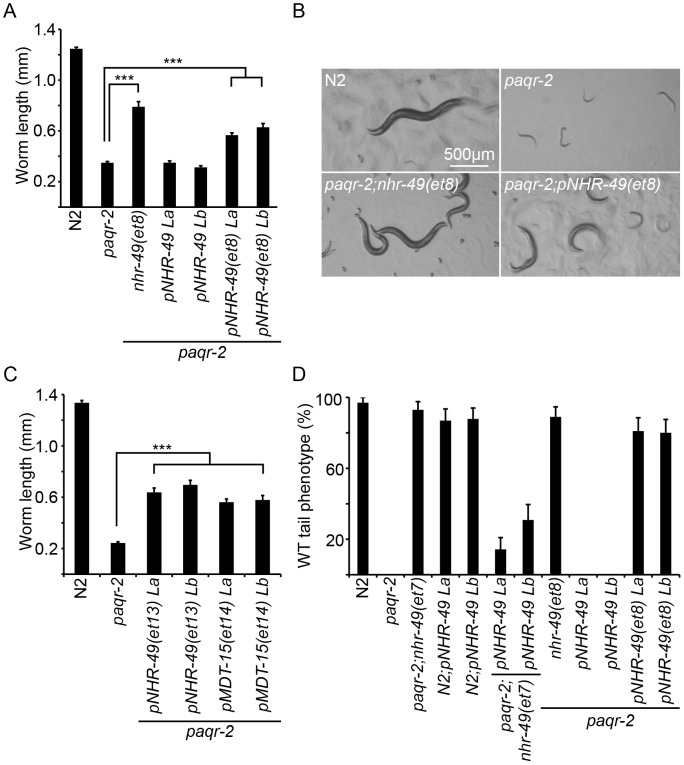
Figure 2. The paqr-2 suppressor mutations in nhr-49 and mdt-15 are gof alleles.
(A) Length of worms grown for 144 hours at 15°C. Results from two paqr-2 lines carrying the plasmids pNHR-49 and pNHR-49(et8) are shown; only strains carrying pNHR-49(et8) showed improved growth indicating that it is a gof allele. (B) Representative images from four of the genotypes used in panel A. (C) Providing nhr-49(et13) or mdt-15(et14) as transgenes suppress the 15°C growth defect of the paqr-2 mutant, indicating that they are gof alleles. The results from two lines are shown for each transgene. (D) Homozygosity for the nhr-49(et7) or nhr-49(et8) mutations suppress the paqr-2 withered tail tip phenotype. Providing nhr-49(et8) as a transgene also suppresses the paqr-2 the tail tip defect, again demonstrating that et8 is a gof allele. ***: p<0.001.
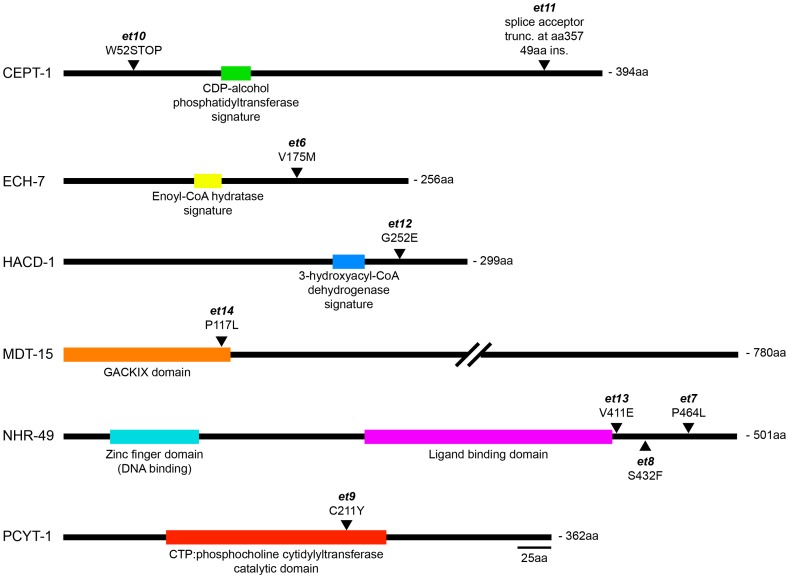
Figure 3. The novel paqr-2 suppressor mutations affect proteins that regulate lipid metabolism.
For each protein, the position and nature of the novel paqr-2 suppressor alleles are indicated as well as the position of important functional domains.
Table 1. Genes that can be mutated to suppress the paqr-2 cold adaptation defect.
| Gene | Allele/transgene | Type | Molecular description | Flanking sequence (25 bp, (+) strand) | Experimental evidence for paqr-2 supp ressive activity |
| aak-2 | ok524 | Lof | X: 408 bp deletion | L: agtacaccttctgacattttcatgR: tagaaatccaattaaagtaattgaa | Introduction of aak-2(ok524) partially suppresses the 15°C growth defect of paqr-2 [12] |
| cept-1 | et10 | lof | X: G12763193A; W52STOP | L: tttatcgatgtacgtttttcagatgR: tgggagtttgtgattacattatgcc | Introduction of WT cept-1 as a transgene in paqr-2;cept-1(et10) desuppresses the paqr-2 tail defect at 20°C and brood size at 15°C (Figure S3C–D) |
| cept-1 | et11 | lof | X: G12765379A, splice acceptor | L: gtatattttacttcatccactttcaR: atatactgttatacttctcttttca | Introduction of WT cept-1 as a transgene in paqr-2;cept-1(et11) desuppresses the paqr-2 tail defect (Figure S3D) |
| ech-7 | et6 | lof | I: C14368566T; V175M | L: tcaggaggcgaaggaagacggcctgR: tgagcaaggtgttcccggtgcagca | Injected ech-7 RNAi suppresses the 15°C growth defect of paqr-2 (Figure S3A) |
| hacd-1 | et12 | lof | V: G1461311A; G252E | L: ccaattgaattgtgtgactatgttgR: acttgatgttctgcagagcactttg | Introduction of hacd-1(ok2776) suppresses the 15°C growth defect of paqr-2. Introduction of WT hacd-1 as a trasgene in paqr-2;hacd-1(et12) desuppresses the paqr-2 tail defect (Figure S3B and D) |
| mdt-15 | et14 | gof | III: C5832666T; P117L | L: gctcctgtgcctccagatccacaacR: aacatcagctcaggcaagaaatcca | Introduction of mdt-15(et14) as a transgene suppresses the 15°C growth defect of paqr-2 (Figure 2C) |
| nhr-49 | et7 | gof | I: C9874044T; P464L | L: atgctctcagcaactcttgcagctcR: attggcaattcatcctctccaatca | Introduction of WT nhr-49 as a transgene in paqr-2;nhr-49(et7) desuppresses the paqr-2 tail defect (Figure 2D) |
| nhr-49 | et8 | gof | I: C9873765T; S432F | L: aatgatcattcgacggctccggtctR: tttacagcaacatctttcatctccg | Introduction of nhr-49(et8) as a transgene suppresses the tail and 15°C growth defect of paqr-2 (Figure 2A and D) |
| nhr-49 | et13 | gof | I: T9873702A; V411E | L: tccgtacataaatgatagcttcaggR: ggattctctgctctccgagttcatt | Introduction of nhr-49(et13) as a transgene suppresses the 15°C growth defect of paqr-2 (Figure 2C) |
| nhr-80 | tm1011 | lof | III: 446 bp deletion | L: tttattagaaaactcacacaatgctR: gtctgcggggacacattcgcgaatt | Introduction of nhr-80(tm1011) partially suppresses the 15°C growth defect of paqr-2 [12] |
| pcyt-1 | et9 | lof | X: G7579636A; C211Y | L: ggtgtctctacaagtgacgtcgtctR: cagaatcatccgtgattacgataag | Injected pcyt-1 RNAi partially suppresses the 15°C growth defect of paqr-2 (Figure S3A) |
| sams-1 | ok2946 | lof | X: 707 bp deletion | L: gtttcaatgtttttgttcaggattR:tcggaacttccccactcaccgacga | Introduction of sams-1(ok2946) suppresses the 15°C growth defect of paqr-2 (Figure S3E) |
| sbp-1 | epEx141 | Over exp. | sbp-1::GFP::SBP-1+rol-6(su1006) | Extra chromosomal array | Introduction of epEx141 suppresses the 15°C growth defect of paqr-2 (Figure S3F) |
The et6 to et14 alleles were isolated in this study. Genomic positions refer to Wormbase release WS200. The aak-2 and nhr-80 alleles were identified as paqr-2 suppressors in an earlier study [12].
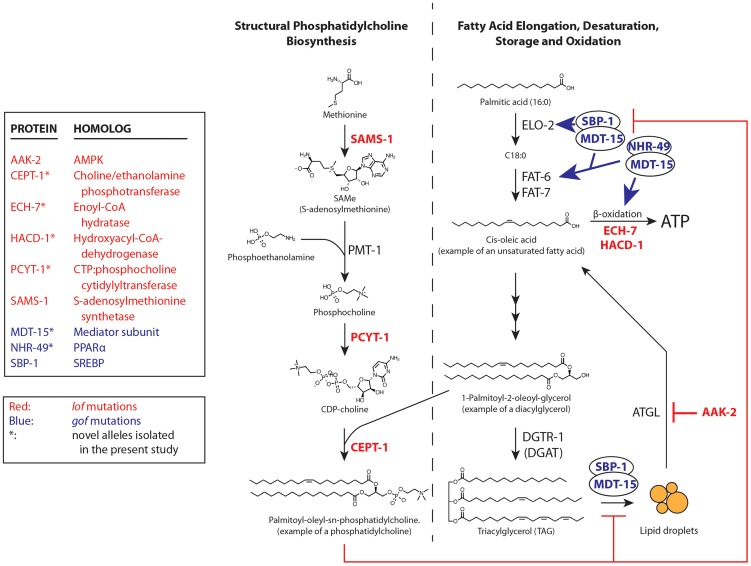
Figure 4. paqr-2 suppressors either inhibit phosphatidylcholine synthesis or promote fatty acid metabolism.
Red- and blue-labeled names indicate proteins where lof and gof alleles can suppress the paqr-2 phenotypes, respectively; for sbp-1 we used a multicopy transgene as a gof allele. The nhr-80(tm1011) lof allele also suppresses the paqr-2 cold adaptation defect [12]; it is omitted from the figure for clarity.
Briefly, three paqr-2 suppressors are gof alleles of nhr-49 (alleles et7, et8 and et13; Figure 2), and a fourth is a gof mutation in mdt-15 (allele et14; Figure 2C). nhr-49 encodes a nuclear hormone receptor homologous to the mammalian HNF4 and with activities that are sometimes compared to that of PPARα, while mdt-15 encodes a subunit of Mediator, an RNA polymerase II transcription co-regulator, that interacts with the NHR-49 and SBP-1 transcription factors to promote FA desaturation, elongation and β-oxidation [17], [23]–[25]. Several lines of evidence support the conclusion that these paqr-2 suppressors are gof alleles: 1) They act in a dominant fashion when introduced as multicopy transgenes into paqr-2 mutant worms (Figure 2); 2) The nhr-49(et8) acts as a dominant allele in a standard genetics assay whereby worms heterozygous for this allele but homozygous for the paqr-2 mutation were scored for growth at 15°C (data: 59.5% of F2 progeny from paqr-2;nhr-49(et8)/+ mothers grew into adults at 15°C, which matches the 62.5% expectation if nhr-49(et8) is dominant rather than the 37.5% if it was recessive; n = 341); and 3) The nhr-49(et8) gof allele suppresses paqr-2 phenotypes while the nhr-49(gk405) lof allele is synthetic lethal with paqr-2, and these two alleles have opposite effects on fatty acid composition and fat-7 regulation (see below).
Two other paqr-2 suppressor mutations are lof alleles of the genes ech-7 (allele et6; Figure S3A) and hacd-1 (allele et12; Figure S3B and D). These genes encode the worm homologs of enoyl-CoA hydratase and of hydroxyacyl-CoA dehydrogenase, which perform consecutive reactions during FA β-oxidation in mitochondria.
The remaining three paqr-2 suppressor mutations are lof alleles of pcyt-1 (allele et9; Figure S3A) and cept-1 (alleles et10 and et11; Figure S3C–D), the worm homologs of CTP:phosphocholine cytidylyltransferase and of choline/ethanolamine phosphotransferase, which are enzymes important for PC synthesis [19]. While our experimental evidence show that pcyt-1(et9) is a lof allele, it may be a hypomorph rather than a null allele since homozygosity for the pcyt-1(ok547) deletion allele is reported in Wormbase to cause lethality. Note however that the pcyt-1(ok547) allele is unpublished and may not have been thoroughly outcrossed.
The newly isolated paqr-2 suppressors, as well as the previously identified partial suppressors aak-2(ok524) and nhr-80(tm1011) [12], are all involved in PC synthesis and FA metabolism. Building on these findings, we also tested other genes involved in these processes and found that a sams-1 lof mutation can suppress the cold adaptation defect of paqr-2 mutants (Figure S3E). sams-1 encodes S-adenosylmethionine synthetase that converts methionine to S-adenosylmethionine, the methyl donor during phosphocholine synthesis [19]. Separately, we found that an overexpression transgene of sbp-1 can rescue the paqr-2 cold adaptation defect (Figure S3F). sbp-1 is an activator of the Δ9 desaturases fat-5, fat-6 and fat-7 that also regulates lipid storage [23], [26]. In summary, the paqr-2 suppressor mutations affect genes of two basic metabolic pathways (Figure 4): 1) PC biosynthesis: sams-1, pcyt-1 and cept-1; and 2) FA metabolism: aak-2, ech-7, hacd-1, mdt-15, nhr-49, nhr-80 and sbp-1. Importantly, these two pathways are functionally related: low levels of PCs are associated with activation of sbp-1/SREBP in metazoans [19].
paqr-2 mutants accumulate saturated fatty acids
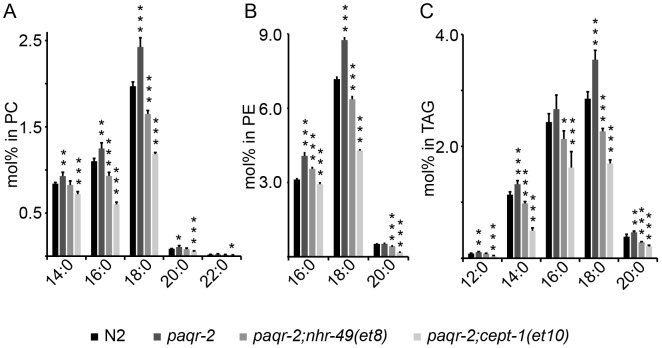
Mechanistically, the identity of the paqr-2 suppressors strongly implicates this transmembrane receptor as a regulator of FA metabolism during cold adaptation and tail tip maintenance. We hypothesized that a common outcome of all the paqr-2 suppressor mutations is an increase in unsaturated fatty acids. To test this, we determined the FA composition of PCs, phosphatidylethanolamines (PEs) and triacylglycerols (TAGs) in synchronized wild-type, paqr-2 mutants, and paqr-2 mutants carrying suppressor mutations. An analysis of 5 biological replicates per genotype showed that 35 of 98 PC species and 19 of 82 PE species are present at significantly elevated levels in the paqr-2 mutant, and that most of the elevated species carry either one (29/54) or two (10/54) saturated FAs (Tables S1, S2). Similarly, 9 of 13 TAGs that contain two or three saturated FAs were significantly increased in the paqr-2 mutant (Table S3). Measuring directly the relative abundance of individual FA chains in each of the three lipid types studied showed that saturated even-length FAs are almost all significantly increased in the paqr-2 mutant and decreased in the paqr-2;nhr-49(et8) and paqr-2;cept-1(et10) double mutants (Figure 5; Table S4). It is evident that the paqr-2 mutant has an abnormally high abundance of saturated FAs in all lipid types assayed, and that suppression of the paqr-2 phenotypes is associated with an increased abundance of unsaturated FAs.
Figure 5. Fatty acid composition defects in the paqr-2 mutant.
The paqr-2 mutant has an excess of saturated even-chained FAs in PCs (A), PEs (B) and TAGs (C), and these defects are corrected or overcompensated by the nhr-49(et8) and cept-1(et10) suppressor mutations. *: p<0.05; **: p<0.01; ***: p<0.001.
To better describe the consequences of the nhr-49(et8) and cept-1(et10) mutations, we also analyzed the lipid profiles of these single mutants in comparison with those of wild-type and paqr-2 mutant worms (Tables S5, S6, S7, S8). The nhr-49(gk405) null mutant was also included in this analysis. The results show that the nhr-49(et8) and nhr-49(gk405) alleles often have opposite effects on the degree of fatty acid desaturation. For example, in PCs the et8 and gk405 alleles went 18 times in opposite directions compared to N2 control worms, and only 4 times in the same direction, the trend being a decrease in the proportion of saturated FAs in the et8 mutant. The effect was also found in the composition of PEs where et8 and gk405 went 25 times in opposite directions and 14 times in the same direction. In general, nhr-49(et8) had much less even-chained saturated FAs (16∶0, 18∶0 and 20∶0) than nhr-49(gk405), and much more 18∶2 and 20∶2 in all three lipid classes. These results are consistent with the two alleles having opposite effects on the activity of nhr-49, with et8 causing a decrease of saturated fatty acids and gk405 causing an increase in saturated FAs. As shown in Tables S5, S6, S7, S8, the cept-1(et10) mutant is also very enriched in unsaturated FAs, especially in PEs and TAGs, and showed the most depletion of TAGs with no or just a single unsaturated bond. In conclusion, the nhr-49(et8) and nhr-49(gk405) behave opposite in many lipid measurements, as expected from a gain-of-function vs loss-of-function comparison, and both nhr-49(et8) and cept-1(et10) tend to have lower levels of saturated FAs, consistent with the hypothesis that they boost the activity of Δ9-desaturases.
Intriguingly, paqr-2 suppressors normalized the overall PE/PC ratio, which is slightly elevated in the paqr-2 mutant at 20°C (Figure S4A). This was true even for the cept-1 lof alleles, which would be expected to lower PC levels, hence raise the PE/PC ratio. This result suggests the possibility that the paqr-2 mutant may compensate for an inability to upregulate FA unsaturation by increasing the PE/PC ratio, perhaps by regulating the insertion of dietary lipid types into membranes or by regulating their turnover. Modulating the PE/PC ratio has important effects on membrane dynamics since PEs are bilayer-destabilizing phospholipids [1]. Of the single mutants nhr-49(et8), cept-1(et10) and nhr-49(gk405), only the cept-1 mutant had a PE/PC ratio that differed significantly from wild-type, being elevated as expected from a lof allele of cept-1 given the role of this gene in PC biosynthesis (Figure S4B).
The paqr-2 suppressors do not act via paqr-1
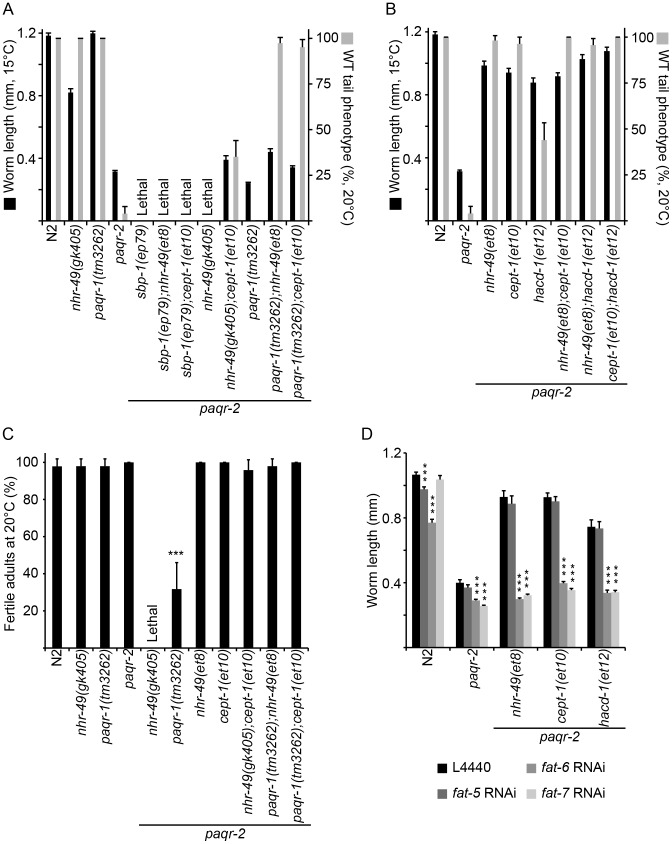
paqr-1 shows sequence homology to paqr-2, and although the paqr-1 mutant has neither cold adaptation or tail tip morphology defects, there is some functional overlap between the two genes [12]. It was therefore important to explore the possibility that the paqr-2 suppressor mutations act by boosting the activity of paqr-1. This is not the case: the nhr-49(et8) and cept-1(et10) mutations still partially suppress the growth defect at 15°C and completely suppress the abnormal tail phenotype of the paqr-2 mutant at 20°C even when paqr-1 is mutated (Figure 6A). Similarly, nhr-49(et8) and cept-1(et10) can completely suppress the grave fertility defect of paqr-1;paqr-2 at 20°C (Figure 6C). paqr-1 is therefore not required for the suppression of paqr-2 phenotypes.
Figure 6. paqr-1 is not required for paqr-2 suppression while sbp-1 and Δ9-desaturases are especially important.
(A) and (B) Length of worms after cultivation of L1s at 15°C for 144 hours. Note that paqr-1 is not essential for the ability of nhr-49(et8) or cept-1(et10) to suppress the tail phenotype at 20°C. Also, the paqr-2;nhr-49(gk405) synthetic lethality is suppressed by cept-1(et10), while the paqr-2 sbp-1(ep79) synthetic lethality is not. (C) Fertility at 20°C. Note that the nhr-49(et8) and cept-1(et10) alleles are able to restore high fertility to the paqr-1;paqr-2 double mutants, indicating again that their effects are independent of paqr-1. Also, note that cept-1(et10) completely suppresses the synthetic lethality of paqr-2;nhr-49(gk405), demonstrating that et10 does not act via nhr-49. (D) The paqr-2 suppressor effects of nhr-49(et8), cept-1(et10) and hacd-1(et12) are abolished when fat-6 or fat-7 are inhibited by RNAi while the control L4440 RNAi vector or RNAi against fat-5 has no effect.
Lowered PC synthesis suppresses the paqr-2 cold adaptation defect by promoting fatty acid desaturation
Genetic interaction experiments suggest that nhr-49(et8) and cept-(et10) likely act on the same target to suppress the paqr-2 phenotypes. One evidence is the fact that the triple mutant paqr-2;nhr-49(et8);cept-1(et10) is viable and grows as well at 15°C as either the paqr-2;nhr-49(et8) or paqr-2;cept-1(et10) double mutants (Figure 6B). This is consistent with at least two interpretations: 1) the gof mutation nhr-49(et8) and the lof mutation cept-1(et10) act on a common target; or 2) nhr-49(et8) acts downstream of cept-1(et10). However, we can reject the second of these hypotheses using the nhr-49(gk405) null allele since the double mutant paqr-2;nhr-49(gk405) is lethal while the triple mutant paqr-2;nhr-49(gk405); cept-1(et10) is quite viable (Figure 6A and C). Since nhr-49 is a well-established promoter of fatty acid desaturation, we tentatively conclude that this is also the essential outcome of the cept-1(et10) mutation. We previously showed that a lof mutation in nhr-80, another nuclear hormone receptor that can activate Δ9-desaturases, partially suppresses the paqr-2 cold adaptation defect, although this suppression is not nearly as effective as that caused by the nhr-49 gof alleles described here [12]. The observations that a lof allele of nhr-80 and a gof allele of nhr-49 can both suppress paqr-2 mutant phenotypes while the lof nhr-49(gk405) is synthetic lethal with the paqr-2 mutation are at first glance surprising given the documented overlap in function between nhr-80 and nhr-49, for example as activators of Δ9-desaturases. There must therefore be important functional differences between the two genes. NHR-49 has many interaction partners, including NHR-80, and it is possible that the nhr-80 lof mutation suppresses the paqr-2 phenotypes either by allowing more interaction of NHR-49 with its other partners, or by causing the enhanced activation of compensatory pathways, such as SBP-1, to restore desaturase activation. Indeed, nhr-49 and nhr-80 mutants are known to have distinct metabolic profiles, with nhr-49 being involved in a wider range of lipid homeostasis pathways [18], [27].
Walker and co-workers have shown that lower PC levels lead to activation of sbp-1/SREBP, and that this regulation is conserved among metazoans [19]. Here, sbp-1 appears to be particularly important for the survival of paqr-2 mutants: the sbp-1(ep79) partial loss of function allele is lethal in combination with paqr-2 [12] (Figure 6A). Furthermore, and in contrast to the paqr-2;nhr-49(gk405) synthetic lethality, the paqr-2;sbp-1(ep79) lethality cannot be suppressed by cept-1(et10) nor, incidentally, by nhr-49(et8) (Figure 6A). These results are consistent with cept-1 acting through sbp-1, and with sbp-1 being more important than nhr-49 for the survival of paqr-2 mutants.
If lof mutations in enzymes of the PC synthesis pathway suppress the cold adaptation defect of the paqr-2 mutant by increasing Δ9-desaturase activity via sbp-1, then inhibiting the desaturases should abolish the paqr-2 suppression. This is indeed the case: RNAi against fat-6 or fat-7 completely desuppresses the 15°C growth of paqr-2;cept-1(et10) double mutants (Figure 6D). Furthermore, inhibiting fat-6 or fat-7 also abolishes the paqr-2 suppression effects of nhr-49(et8) and hacd-1(et12) (Figure 6D), suggesting that the effects of all three suppressor mutations converge on FA desaturation. Note that in practice we cannot rely on our RNAi results to distinguish the relative importance of fat-6 and fat-7 since these show extensive sequence homology such that the RNAi against either gene probably results in inhibition of both genes.
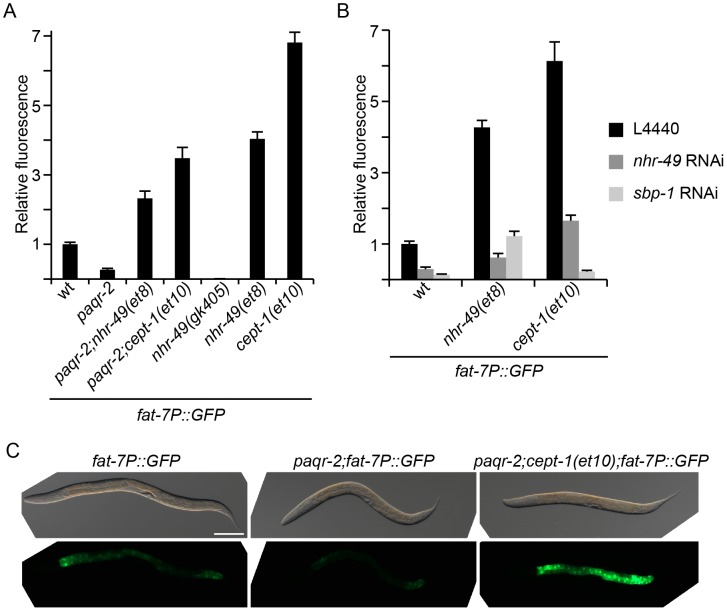
The desaturase fat-7 is regulated by paqr-2 and its suppressors
Our observations so far suggest that paqr-2 normally participates in cold adaptation by regulating the activity of Δ9-desaturases via sbp-1 (via changes in PC levels) or nhr-49. We investigated the possible regulation of Δ9-desaturases by paqr-2 using a previously published fat-7 promoter GFP reporter [19]. As anticipated, the paqr-2 mutant exhibits a marked decrease in fat-7 expression (Figure 7A and C). Furthermore, the nhr-49(et8) and cept-1(et10) alleles show a dramatic increase in fat-7 expression either as single mutants or, importantly, when combined with the paqr-2 mutation. The nhr-49(gk405) has very low fat-7 expression levels, consistent with its lack of activity. These results clearly show that paqr-2 is a regulator of fat-7, and suggest that mutations that can compensate for the lack of paqr-2 do so, at least in part, by boosting the activity of fat-7. Additionally, by using RNAi to separately knock down nhr-49 or sbp-1, we made the interesting observation that the nhr-49(et8) allele depends at least partially on sbp-1 to cause an increase in fat-7 expression, and that, conversely, cept-1(et10) depends on both nhr-49 and sbp-1 to cause an increase in fat-7 expression. This suggests that there is considerable overlap or even mutual dependency between the nhr-49 and cept-1 pathways that regulate fat-7.
Figure 7. The fat-7 desaturase is regulated by paqr-2 and its suppressors.
(A) The paqr-2 mutant has decreased expression of fat-7, while strains carrying the paqr-2 suppressors nhr-49(et8) or cept-1(et10) show a marked increase in fat-7 expression when compared to control worms (wt). (B) The nhr-49(et8) and cept-1(et10) alleles are dependent on both nhr-49 and sbp-1 for the upregulation of fat-7. (C) Representative images of worms from panel (A). Note the strong GFP signal in the paqr-2;cept-1(et10) double mutant, especially compared to the low levels of the paqr-2 single mutant. Scale bar is 100 µm. All strains in this figure carry the fat-7 promoter GFP reporter in integrated form.
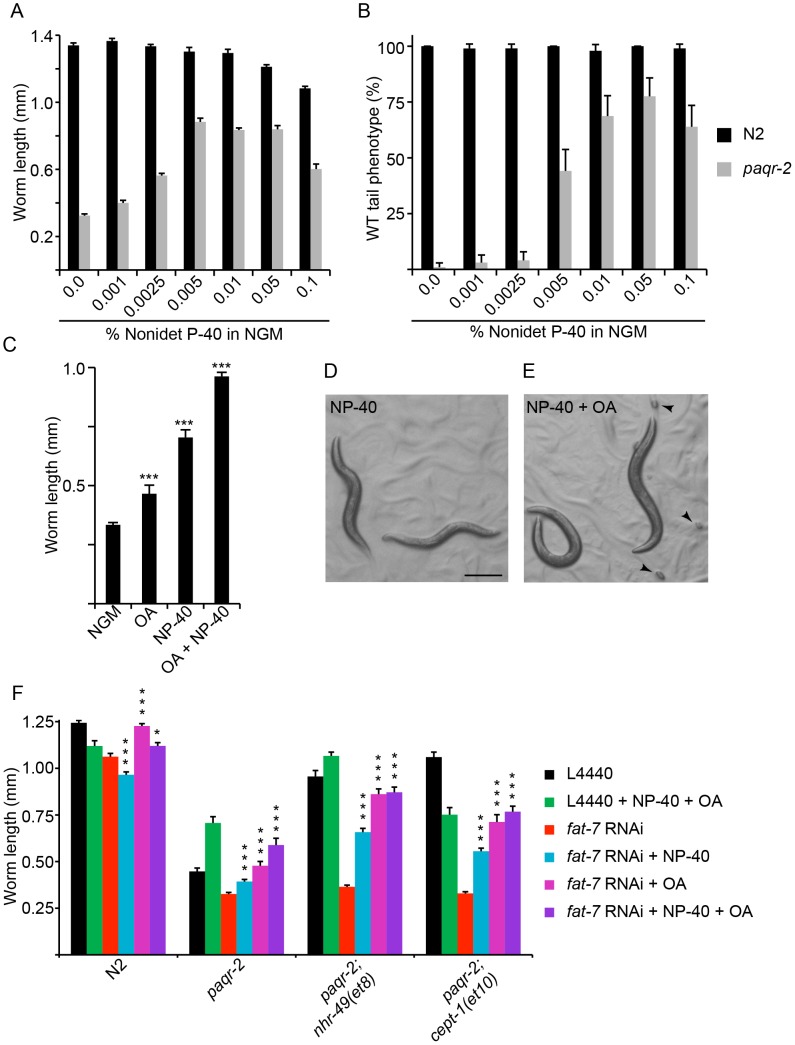
Detergents can suppress the paqr-2 mutant phenotypes
The results described so far suggest that the paqr-2 mutant cannot adapt to 15°C because of a failure to activate Δ9 desaturases, hence to increase membrane fluidity. Furthermore, since the tail tip morphology defect is correlated with the 15°C growth arrest in most experiments, it is likely that this phenotype also is due to a defect in desaturase regulation. We were intrigued by the possibility of rescuing the paqr-2 mutant by using detergents at concentrations expected to increase membrane fluidity [28], [29]. Strikingly, we found that small amounts of Nonidet P-40 or Triton X-100 are excellent at rescuing the tail tip morphology at 20°C and the growth at 15°C (Figure 8A–B and Figure S5A–B). The detergents alone did not however provide a full paqr-2 rescue at 15°C: treated worms grew well, but produced no progeny. It seems likely that the detergents cannot fulfill all biological functions that desaturated FAs have beyond their role in improving membrane fluidity, including possible signaling functions. We therefore tested oleic acid, a monounsaturated FA, as a more biologically natural way of compensating for the reduced levels of unsaturated FAs in the paqr-2 mutant. On its own, oleic acid produced only a marginal growth rescue at 15°C (Figure 8C and Figure S5C–D). However, the combination of oleic acid and Nonidet P-40 caused complete rescue, allowing the treated paqr-2 worms to grow and reproduce (Figure 8C–E). Indeed, the combination of 1 mM oleic acid and 0.05% Nonidet P-40 even suppressed the synthetic lethality of paqr-2 sbp-1(ep79) worms, allowing double homozygous worm progeny of a paqr-2 sbp-1(ep79)/+ mother to grow into adults and establish a stable line. Finally, we show that Nonidet P-40 alone, or in combination with oleic acid, suppresses the paqr-2 growth defect at 15°C even when fat-7 is inhibited by RNAi (Figure 8F). In other words, exogenously providing either a mild detergent or a desaturated fatty acid can relieve the paqr-2 mutant from its dependency on increased Δ9-desaturases for growth at a low temperature. These are striking results given our earlier observation that the paqr-2 suppressor mutations are dependent on the Δ9-desaturases in order to allow growth at 15°C (Figure 6D), and again support the hypothesis that failing to regulate membrane fluidity is a key defect of the paqr-2 mutant at 15°C or of the paqr-2 sbp-1 double mutant at 20°C.
Figure 8. Detergents and oleic acid can suppress the paqr-2 mutant phenotypes.
Low concentrations of Nonidet P-40 (NP-40; A–B) suppress the paqr-2 growth defect at 15°C and the tail tip defect at 20°C. (C) 1 mM oleic acid (OA) slightly improves growth of paqr-2 mutants at 15°C, but not as well as 0.1% Nonidet P-40 (NP-40) or the combination of both. paqr-2 mutants treated with 0.1% NP-40 alone grow into sterile adults (D), and adding also 1 mM oleic acid (OA) restores fertility (E; arrowheads indicate eggs). (F) RNAi against fat-7 inhibits the paqr-2 suppression by nhr-49(et8) and cept-(et10) but even these double mutants can grow at 15°C when NP-40 or OA are provided exogenously either alone or in combination. Scale bar in D represents 250 µm. *:p<0.05; ***: p<0.001.
Discussion
The role of paqr-2 during cold adaptation
We have shown that mutations in either of two metabolic pathways can suppress the paqr-2 phenotypes: 1) mutations that reduce phosphatidylcholine synthesis; and 2) mutations that affect FA metabolism. Mutations in either pathway suppress the paqr-2 phenotypes by augmenting FA desaturation either directly (gof alleles of nhr-49 or mdt-15, or sbp-1 overexpression), or indirectly by reducing PC levels hence upregulating sbp-1 (lof alleles of sams-1, pcyt-1 or cept-1), or by increasing the exposure of FA to Δ9 desaturases via increased release from TAGs (lof allele of aak-2) or inhibition of β-oxidation (lof alleles of ech-7 or hacd-1). Increasing the relative abundance of unsaturated FAs in biological membranes increases their fluidity [1]–[3], and this is likely the essential effect of paqr-2 during cold adaptation.
Several evidences suggest that the pathway outlined in Figure 4 is indeed regulated by paqr-2, rather than being a parallel pathway important for cold adaptation. Firstly, all nine paqr-2 suppressors isolated in our non-biased screen were part of that pathway. Secondly, all nine suppressors also suppressed a very specific paqr-2 morphological phenotype that was not screened for, namely the withered tail tip. Thirdly, the convergence of the identified pathway onto Δ9-desaturases is consistent with their established importance during cold adaptation [5], [7]. Fourthly, both the cold adaptation and tail phenotypes of paqr-2 were corrected by the use of low concentrations of detergents expected to mimic the effects of increasing fatty acid desaturation on membrane fluidity. The present study therefore suggests that paqr-2 is a transmembrane protein essential for the regulation of Δ9 desaturases during cold adaptation via the modulation of PC levels, hence sbp-1. However, the direct point of interaction between PAQR-2 and its target pathway is not immediately apparent, but one interesting hypothesis is that PAQR-2 acts on FA metabolism by regulating PC abundance via an associated phospholipase activity. Indeed, PAQR-2 belongs to a large and diverse protein family of membrane hydrolases that includes Per1, a yeast GPI-phospholipase A2 [9].
Others have previously correlated an increase in unsaturated FAs with adaptation to 15°C growth in C. elegans [11], and with cold adaptation in many other organisms [1]–[3]. A role for Δ9 desaturases for growth at low temperatures in C. elegans has also been documented previously, and it is particularly striking that fat-6;fat-7 double mutants exhibit a temperature sensitivity similar to the paqr-2 mutant, growing well at 20°C and poorly at 15°C [5]. Here we add a novel level of regulation by linking paqr-2, PCs, sbp-1 and Δ9 desaturases into a regulatory axis controlling the increase in unsaturated FAs during cold adaptation. At present we do not know whether regulating the activity of Δ9 desaturases is the only essential function of paqr-2 during cold adaptation.
Our finding that paqr-2 dependent increases in FA unsaturation is an essential component of the adaptation to 15°C in C. elegans may seem to conflict with Murray et al. who previously estimated that FA unsaturation changes contribute only about 15% of the cold adaptation response [16]. However, it is important to keep in mind that Murray et al. were concerned with survival of C. elegans at the acutely lethal temperature of 0°C while we are concerned here with adaptive changes at a low but physiologically acceptable temperature.
Evolutionary considerations
Could the paqr-2 homologs play a role during cold adaptation even in homeotherms? Increased serum adiponectin levels have been observed in rats kept at 4°C for 24 hours, with adiponectin mRNA levels becoming elevated in brown adipose tissue (BAT) [30]. Also, human subjects wearing a 10°C liquid-conditioned suit for two hours have shown a near-doubling of circulating adiponectin levels [31]. Furthermore, the AdipoRs are expressed in temperature-sensitive neurons in the brain [32], and adiponectin itself has sequence homology with hibernation-associated plasma proteins in Asian chipmunks and woodchucks [30], [33], which again suggests that it may be part of a protein family that functions during cold adaptation in mammals.
Materials and Methods
C. elegans cultivation, strains and transgenes
Maintenance of worms were performed as described elsewhere [34]. The wild type reference strain was the C. elegans Bristol variety strain, N2. Unless otherwise stated, strains were obtained from the C. elegans Genetics Center (CGC; MN, USA) and cultivated at 20°C.
Western blot
Crowded plates of various C. elegans strains were harvested and washed twice in water before lysis by boiling in SDS sample loading buffer. Twenty micrograms of total protein was loaded in each lane, electrophoresed on a SDS polyacrylamide gel, and transferred to nitrocellulose. For Western blot detection of the PAQR-2 protein, a rabbit antiserum was generated by immunizing a rabbit against the peptide PLNVRDWTPADVGL corresponding to the C-terminal amino acids. Crude serum was used at a dilution of 1∶1000. Blotto (5% skim milk powder in TBST) was used for blocking. Antibody dilutions and washes were carried out in TBST. A goat anti-rabbit HRP-conjugated secondary antibody (GE) was used at a final concentration of 1∶2 500 to detect bound primary antibody. Detection of bound antibody was performed using an ECL Detection Kit, as per the manufacturer's instruction (Thermo Scientific).
Mutagenesis and screen for paqr-2 suppressors
paqr-2(tm3410) worms were mutagenized for 4 hours by incubation in the presence of 0.05 M ethyl methane sulfonate (EMS) according to the standard protocol [34]. The worms were then washed and placed on a culture dish. Two hours later, vigorous hermaphrodite L4 animals were transferred to new culture plates. Five days later, F1 progeny were bleached, washed and their eggs allowed to hatch overnight in M9 (22 mM KH2PO4, 42 mM Na2HPO4, 85.5 mM NaCl and 1 mM MgSO4). The resulting L1 larvae were transferred to new plates, cultivated at 15°C, then screened from day 4 to day 6 to identify paqr-2 suppressors able to reproduce at that temperature, which were picked to new plates for further analysis. Presence of the paqr-2(tm3410) mutant allele in each suppressor was confirmed using a PCR assay [12].
The isolated suppressor alleles, named et6 to et14 were outcrossed 4-to-6 times prior to whole genome sequencing (see below), and 10 times prior to their phenotypic characterization or use in the experiments presented here. Outcrossing was done by mating wild-type N2 males to a suppressor, then crossing the male progeny to paqr-2 single mutant worms (themselves previously outcrossed 10 times to wild-type worms); progeny from this cross were picked to individual plates and kept at 20°C then screened for homozygozity for paqr-2 using PCR, followed by testing their F2 progeny for ability to grow at 15°C. Five such cycles were carried out amounting to ten outcrosses (five to N2 worms and five to paqr-2(tm3410) worms).
Whole genome sequencing
The genomes of suppressor mutants that had been outcrossed 4 or 6 times were sequenced to a depth of 25–40x as previously described [22]. The sequencing results were analyzed using the MAPQGene software to produce tables listing all differences between the reference N2 genome and that of the mutants, and sort these differences by criteria such as non-coding substitutions, termination mutations, splice-site mutations, etc. [35]. For each suppressor mutant, one or two hot spots, i.e. small genomic area containing several mutations, were identified, which is in accordance to previous reports [21]. Mutations in the hot spot that were still retained after 10 outcrosses were considered candidate paqr-2 suppressors and tested experimentally as described in the text.
Plasmids
The pNHR-49 plasmids (WT, et8 and et13) were all constructed by amplification of the nhr-49 promoter, gene and UTR using primers 5′-atccttactggacgccgtctaca-3′ and 5′-gtagtacagtaaccaactttcccgaagt-3′ with the corresponding genotype as template. The resulting ∼6.6 kb PCR products were cloned into pCR-XL-TOPO (Invitrogen).
The pCEPT-1 plasmid was constructed by amplification of the cept-1 promoter, gene and UTR using primers 5′-gagtttcgataaagtatagcttgtcacga-3′ and 5′- ggttaagcttgttttgtgtaatgcgtagt-3′ and a mixture of N2 worms as template. The resulting ∼6.6 kb PCR product was cloned into pCR-XL-TOPO.
The pHACD-1 plasmid was constructed by amplification of the hacd-1 promoter, gene and UTR using primers 5′-gaatagttaacaagcgctcagtcatga-3′ and 5′-acagtgtcgccacttcaggactact-3′ and a mixture of N2 worms as template. The resulting ∼4.6 kb PCR product was cloned into pCR-XL-TOPO.
The pMDT-15(et14) plasmid was constructed by amplification of the mdt-15 promoter, gene and UTR using primers 5′-gaaattgctcatctatcgggtct-3′ and 5′-atcgcaggaatcggattcgta-3′ and a mixture of paqr-2;mdt-15(et14) worms as template. The resulting ∼7.7 kb PCR product was cloned into pCR-XL-TOPO.
Generation of transgenic animals
Plasmids were prepared with a Qiagen miniprep kit (Qiagen) and used with the following concentrations: pRF4 (rol-6) of 50 ng/µl, test plasmids of 5–10 ng/µl, and pBSKS (Stratagene) of 40–45 ng/µl to a total of 100 ng/µl.
15°C growth assays
For confirmation of suppressor alleles or assay of double/triple mutants using the paqr-2 cold adaptation phenotype at 15°C, synchronized L1s were placed on plates and incubated at 15°C for 5 or 6 days. Pictures were taken using a Zeiss Axiophot microscope and worm lengths measured in ImageJ [36].
Brood size assay
The brood size experiment confirming cept-1(et10) was performed essentially as described [12]. All strains were incubated at 20°C from L1 to 1 day adults before transfer to 15°C.
paqr-2 tail tip phenotype assay
For confirmation of suppressor alleles using the withered tail tip phenotype, L4s were picked and scored 24 h later as young adults. n>100 for all tail experiments.
RNAi feeding
All strains were grown on control L4440 RNAi bacteria for one generation at 20°C, then synchronized and L1s placed onto assay RNAi, incubated at 15°C and scored on day 6. Feeding RNAi clones were from the Ahringer RNAi library.
RNAi injections
Template for in vitro transcription was made by a PCR reaction using a T7 primer (5′-cgtaatacgactcactatag-3′) and feeding RNAi clones as template. In vitro transcription was performed using the Riboprobe System-T7 (Promega) and the dsRNAi was purified and injected into paqr-2 worms. Injected worms were allowed to lay eggs at 20°C and the eggs were transferred to 15°C for scoring of worm length 6 days later. The ech-7 and pcyt-1 RNAi clones were from the ORF-RNAi library.
Quantification of fat-7::GFP expression
The pfat-7::GFP (rtIs30) carrying strain HA1842 has been described elsewhere [19] and was a kind gift from Amy K. Walker. Quantitative measurements of the GFP intensity was performed on synchronized L4s using the image processing program ImageJ [36]. Presence of the rtIs30 array in the nhr-49(gk405) mutant was assayed by PCR reactions against GFP.
Lipidomics
Samples were composed of synchronized L4 larvae (one 15 cm diameter plate/sample). Worms were washed 3 times with M9, pelleted and stored at −80°C until analysis. For lipid extraction, the pellet was sonicated for 10 minutes in methanol and then extracted according to published methods [37]. Internal standards were added in the chloroform phase during the extraction. Lipid extracts were evaporated and reconstituted in chloroform∶methanol [1∶2] with 5 mM ammonium acetate. This solution was infused directly (shotgun approach) into a QTRAP 5500 mass spectrometer (ABSciex, Toronto, Canada) equipped with a Nanomate Triversa (Advion Bioscience, Ithaca, NY) as described previously [38]. Phospholipids were measured using multiple precursor ion scanning [39], [40] and triacylglycerols were measured using neutral loss scanning [41]. The data was evaluated using the LipidProfiler software [39].
Detergents and oleic acid
Detergent or oleic acid plates were made by adding the appropriate amount of 10% Nonidet P-40 substitute, 10% Triton X-100 or 0.5 M oleic acid stock solutions to NGM. Synchronized L1s were placed on detergent plates, incubated at 15°C for 6 days and scored for worm length.
Statistics
Error bars for worm length measurements show the standard error of the mean, and t-tests were used to identify significant differences between worm lengths. Error bars for the frequency of the tail tip defect show the 95% confidence interval determined using Z-tests.
Supporting Information
Structure of the paqr-2 gene and characterization of the tm3410 allele. (A) Structure of the paqr-2 transcript (top) and PAQR-2 protein, with the deleted regions in the tm3410 allele indicated by the red underlines. (B) Western blot showing the PAQR-2 band at ∼66 kDa, which is absent in the paqr-2(tm3410) mutant but recovered when a paqr-2 transgene is reintroduced. Bracketed bands in (B) indicate degradation PAQR-2 products in the transgenic animals, and other bands are due to non-specific binding of the antibody.
(TIF)
Fertility, brood size and growth rate of the paqr-2 suppressors. (A) Percentage of L1s that grow into fertile adults at three different temperatures. Note that all suppressor mutations permit reproductive growth of the paqr-2 mutant at 15°C. (B) Total self-brood size at 20°C. Note that all suppressor mutations dramatically improve self-brood size, except for ech-7(et6). (C) Length of worms grown from the L1 stage at 20°C for different amounts of time. Note that all suppressor mutations improve the growth of the paqr-2 mutant, except again for ech-7(et6). The figure legend in C also applies to panel A. *: p<0.05; **: p<0.01; ***: p<0.001.
(TIF)
Experimental tests confirming the identity of paqr-2 suppressor mutations. (A) Injected RNAi against pcyt-1 or ech-7 can suppress the paqr-2 phenotype, suggesting that the paqr-2 suppressors et6 and et9 are lof alleles of these genes. (B) The hacd-1(ok2776) deletion allele is as effective at suppressing the paqr-2 15°C growth defect as the hacd-1(et12) allele, confirming that et12 is also a lof allele. (C) Providing wild-type cept-1 as a transgene desuppresses the self-brood size defect in paqr-2;cept-1(et10), indicating that cept-1(et10) is a lof allele. (D) Homozygosity for the cept-1(et10), cept-1(et11) or hacd-1(et12) mutation suppresses the tail defect in paqr-2 worms, but this phenotype is desuppressed by introducing wild-type cept-1 or hacd-1 transgenes in the paqr-2;cept-1(et10) and paqr-2;cept-1(et11), or paqr-2;hacd-1(et12) strains, respectively. This indicates that et10, et11 and et12 are lof alelles. (E) and (F) The sams-1(ok2946) deletion allele and the sbp-1 overexpression transgene epEx141 can also suppress the paqr-2 15°C growth defect, respectively. ***: p<0.001.
(TIF)
Analysis of PE/PC ratios. (A) The PE/PC ratio is elevated in the paqr-2 mutant and is partially normalized by the nhr-49(et8) or cept-1(et10) mutations. (B) In a separate experiment, again the PE/PC ratio is elevated in the paqr-2 mutant, unaffected in the nhr-49(et8) or nhr-49(gk405) mutants, and elevated in the cept-1(et10) mutant. The average amounts of lipids recovered per mg of protein from 5 samples for each genotype are also indicated. There is a fair amount of variation in the lipid extraction, as evidenced by the larger error bars for the amount of lipids. However, this did not affect the relative recovery of PEs and PCs, as evidenced by the small error bars for the PE/PC ratios.
(TIF)
The non-ionic detergent Triton X-100 and oleic acid can independently suppress paqr-2 phenotypes. (A) Inclusion of 0.05–0.2% Triton X-100 in the culture plates allows the paqr-2 mutant to grow at 15°C. (B) Including 0.05% Triton X-100 in culture plates allows the paqr-2 mutant to develop and maintain normal tail tips at 20°C. (C) and (D) Inclusion of 0.5–2 mM oleic acid in culture plates causes a slight but dose-dependent improvement in the growth of paqr-2 mutants at 15°C and in the quality of the tail morphology at 20°C, respectively.
(TIF)
PC composition (each row indicates the % of total PC that had the indicated number of carbon atoms and double bonds in its two FAs). Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests. Note: 17:Δ is cis-9,10-methylenehexadecanoic acid and 19:Δ is cis-11,12-methylene octadecanoic acid.
(XLSX)
PE composition (each row indicates the % of total PE that had the indicated number of carbon atoms and double bonds in its two FAs). Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests. Note: 17:Δ is cis-9,10-methylenehexadecanoic acid and 19:Δ is cis-11,12-methylene octadecanoic acid.
(XLSX)
TAG composition (each row indicates the % of total TAG that had the indicated number of carbon atoms and double bonds in its FAs). Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests.
(XLSX)
FA composition of PCs, PEs and TAGs. Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests. Note: 17:Δ is cis-9,10-methylenehexadecanoic acid and 19:Δ is cis-11,12-methylene octadecanoic acid.
(XLSX)
PC composition (each row indicates the % of total PC that had the indicated number of carbon atoms and double bonds in its two FAs). Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests. Note: 17:Δ is cis-9,10-methylenehexadecanoic acid and 19:Δ is cis-11,12-methylene octadecanoic acid.
(XLSX)
PE composition (each row indicates the % of total PE that had the indicated number of carbon atoms and double bonds in its two FAs). Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests. Note: 17:Δ is cis-9,10-methylenehexadecanoic acid and 19:Δ is cis-11,12-methylene octadecanoic acid.
(XLSX)
TAG composition (each row indicates the % of total TAG that had the indicated number of carbon atoms and double bonds in its FAs). Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests.
(XLSX)
FA composition of PCs, PEs and TAGs. Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests. Note: 17:Δ is cis-9,10-methylenehexadecanoic acid and 19:Δ is cis-11,12-methylene octadecanoic acid.
(XLSX)
Acknowledgments
Some strains were provided by the Caenorhabditis Genetics Center (CGC). We thank Peter Carlsson for comments on the manuscript.
Funding Statement
This work was funded by the Swedish Research Council, Magnus Bergvalls Stiftelse, Diabetesfonden, the IngaBritt och Arne Lundbergs Research Foundation and Carl Tryggers Stiftelse. Some strains were provided by CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hazel JR (1995) Thermal adaptation in biological membranes: is homeoviscous adaptation the explanation? Annu Rev Physiol 57: 19–42. [DOI] [PubMed] [Google Scholar]
- 2. Guschina IA, Harwood JL (2006) Mechanisms of temperature adaptation in poikilotherms. FEBS Lett 580: 5477–5483 doi:10.1016/j.febslet.2006.06.066 [DOI] [PubMed] [Google Scholar]
- 3. Crockett EL (2008) The cold but not hard fats in ectotherms: consequences of lipid restructuring on susceptibility of biological membranes to peroxidation, a review. J Comp Physiol, B 178: 795–809 doi:10.1007/s00360-008-0275-7 [DOI] [PubMed] [Google Scholar]
- 4. Hayward SAL, Murray PA, Gracey AY, Cossins AR (2007) Beyond the lipid hypothesis: mechanisms underlying phenotypic plasticity in inducible cold tolerance. Adv Exp Med Biol 594: 132–142 doi:_10.1007/978-0-387-39975-1_12 [DOI] [PubMed] [Google Scholar]
- 5. Brock TJ, Browse J, Watts JL (2007) Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans . Genetics 176: 865–875 doi:10.1534/genetics.107.071860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ohtsu T, Kimura MT, Katagiri C (1998) How Drosophila species acquire cold tolerance–qualitative changes of phospholipids. Eur J Biochem 252: 608–611. [DOI] [PubMed] [Google Scholar]
- 7. Savory FR, Sait SM, Hope IA (2011) DAF-16 and Δ9 desaturase genes promote cold tolerance in long-lived Caenorhabditis elegans age-1 mutants. PLoS ONE 6: e24550 doi:10.1371/journal.pone.0024550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Overgaard J, Sørensen JG, Petersen SO, Loeschcke V, Holmstrup M (2005) Changes in membrane lipid composition following rapid cold hardening in Drosophila melanogaster. J Insect Physiol 51: 1173–1182 doi:10.1016/j.jinsphys.2005.06.007 [DOI] [PubMed] [Google Scholar]
- 9. Pei J, Millay DP, Olson EN, Grishin NV (2011) CREST–a large and diverse superfamily of putative transmembrane hydrolases. Biol Direct 6: 37 doi:10.1186/1745-6150-6-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colinet H, Lee SF, Hoffmann A (2010) Temporal expression of heat shock genes during cold stress and recovery from chill coma in adult Drosophila melanogaster . FEBS J 277: 174–185 doi:10.1111/j.1742-4658.2009.07470.x [DOI] [PubMed] [Google Scholar]
- 11. Tanaka T, Ikita K, Ashida T, Motoyama Y, Yamaguchi Y, et al. (1996) Effects of growth temperature on the fatty acid composition of the free-living nematode Caenorhabditis elegans . Lipids 31: 1173–1178. [DOI] [PubMed] [Google Scholar]
- 12. Svensson E, Olsen L, Mörck C, Brackmann C, Enejder A, et al. (2011) The Adiponectin Receptor Homologs in C. elegans Promote Energy Utilization and Homeostasis. PLoS ONE 6: e21343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, et al. (2003) Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 423: 762–769 doi:10.1038/nature01705 [DOI] [PubMed] [Google Scholar]
- 14. Yamauchi T, Nio Y, Maki T, Kobayashi M, Takazawa T, et al. (2007) Targeted disruption of AdipoR1 and AdipoR2 causes abrogation of adiponectin binding and metabolic actions. Nat Med 13: 332–339 doi:10.1038/nm1557 [DOI] [PubMed] [Google Scholar]
- 15. Holland WL, Miller RA, Wang ZV, Sun K, Barth BM, et al. (2011) Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nat Med 17: 55–63 doi:10.1038/nm.2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murray P, Hayward SAL, Govan GG, Gracey AY, Cossins AR (2007) An explicit test of the phospholipid saturation hypothesis of acquired cold tolerance in Caenorhabditis elegans . Proc Natl Acad Sci USA 104: 5489–5494 doi:10.1073/pnas.0609590104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR (2005) Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol 3: e53 doi:10.1371/journal.pbio.0030053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pathare PP, Lin A, Bornfeldt KE, Taubert S, van Gilst MR (2012) Coordinate regulation of lipid metabolism by novel nuclear receptor partnerships. PLoS Genet 8: e1002645 doi:10.1371/journal.pgen.1002645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walker A, Jacobs R, Watts J, Rottiers V (2011) A Conserved SREBP-1/Phosphatidylcholine Feedback Circuit Regulates Lipogenesis in Metazoans. Cell 147: 840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Z, Sherwood DR (2011) Dissection of genetic pathways in C. elegans. Methods Cell Biol 106: 113–157 doi:10.1016/B978-0-12-544172-8.00005-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zuryn S, Le Gras S, Jamet K, Jarriault S (2010) A strategy for direct mapping and identification of mutations by whole-genome sequencing. Genetics 186: 427–430 doi:10.1534/genetics.110.119230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sarin S, Prabhu S, O'Meara MM, Pe'er I, Hobert O (2008) Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat Methods 5: 865–867 doi:10.1038/nmeth.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun Z-Y, et al. (2006) An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442: 700–704 doi:10.1038/nature04942 [DOI] [PubMed] [Google Scholar]
- 24. Taubert S, van Gilst MR, Hansen M, Yamamoto KR (2006) A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans . Genes Dev 20: 1137–1149 doi:10.1101/gad.1395406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Atherton HJ, Jones OAH, Malik S, Miska EA, Griffin JL (2008) A comparative metabolomic study of NHR-49 in Caenorhabditis elegans and PPAR-alpha in the mouse. FEBS Lett 582: 1661–1666 doi:10.1016/j.febslet.2008.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McKay RM, McKay JP, Avery L, Graff JM (2003) C. elegans: a model for exploring the genetics of fat storage. Dev Cell 4: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brock TJ, Browse J, Watts JL (2006) Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet 2: e108 doi:10.1371/journal.pgen.0020108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henriksen JR, Andresen TL, Feldborg LN, Duelund L, Ipsen JH (2010) Understanding Detergent Effects on Lipid Membranes: A Model Study of Lysolipids. Biophysj 98: 2199–2205 doi:10.1016/j.bpj.2010.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ahyayauch H, Bennouna M, Alonso A, Goñi FM (2010) Detergent Effects on Membranes at Subsolubilizing Concentrations: Transmembrane Lipid Motion, Bilayer Permeabilization, and Vesicle Lysis/Reassembly Are Independent Phenomena. Langmuir 26: 7307–7313 doi:10.1021/la904194a [DOI] [PubMed] [Google Scholar]
- 30. Yoda M, Nakano Y, Tobe T, Shioda S, Choi-Miura NH, et al. (2001) Characterization of mouse GBP28 and its induction by exposure to cold. Int J Obes Relat Metab Disord 25: 75–83. [DOI] [PubMed] [Google Scholar]
- 31. Imbeault P, Dépault I, Haman F (2009) Cold exposure increases adiponectin levels in men. Metab Clin Exp 58: 552–559 doi:10.1016/j.metabol.2008.11.017 [DOI] [PubMed] [Google Scholar]
- 32. Klein I, Sanchez-Alavez M, Tabarean I, Schaefer J, Holmberg KH, et al. (2011) AdipoR1 and 2 are expressed on warm sensitive neurons of the hypothalamic preoptic area and contribute to central hyperthermic effects of adiponectin. Brain Res 1423: 1–9 doi:10.1016/j.brainres.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wong GW, Wang J, Hug C, Tsao T-S, Lodish HF (2004) A family of Acrp30/adiponectin structural and functional paralogs. Proc Natl Acad Sci USA 101: 10302–10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sulston JE, Hodgkin JA (1988) Methods. In: Wood WB, editor. The Nematode Caernorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. pp. 587–606.
- 35. Bigelow H, Doitsidou M, Sarin S, Hobert O (2009) MAQGene: software to facilitate C. elegans mutant genome sequence analysis. Nat Methods 6: 549 doi:10.1038/nmeth.f.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 doi:10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Folch J, Lees M, Sloane Stanley GH (1957) A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226: 497–509. [PubMed] [Google Scholar]
- 38. Jung HR, Sylvänne T, Koistinen KM, Tarasov K, Kauhanen D, et al. (2011) High throughput quantitative molecular lipidomics. Biochim Biophys Acta 1811: 925–934 doi:10.1016/j.bbalip.2011.06.025 [DOI] [PubMed] [Google Scholar]
- 39. Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, et al. (2009) Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proceedings of the National Academy of Sciences 106: 2136–2141 doi:10.1073/pnas.0811700106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ekroos K, Ejsing CS, Bahr U, Karas M, Simons K, et al. (2003) Charting molecular composition of phosphatidylcholines by fatty acid scanning and ion trap MS3 fragmentation. J Lipid Res 44: 2181–2192 doi:10.1194/jlr.D300020-JLR200 [DOI] [PubMed] [Google Scholar]
- 41. Murphy RC, James PF, McAnoy AM, Krank J, Duchoslav E, et al. (2007) Detection of the abundance of diacylglycerol and triacylglycerol molecular species in cells using neutral loss mass spectrometry. Anal Biochem 366: 59–70 doi:10.1016/j.ab.2007.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structure of the paqr-2 gene and characterization of the tm3410 allele. (A) Structure of the paqr-2 transcript (top) and PAQR-2 protein, with the deleted regions in the tm3410 allele indicated by the red underlines. (B) Western blot showing the PAQR-2 band at ∼66 kDa, which is absent in the paqr-2(tm3410) mutant but recovered when a paqr-2 transgene is reintroduced. Bracketed bands in (B) indicate degradation PAQR-2 products in the transgenic animals, and other bands are due to non-specific binding of the antibody.
(TIF)
Fertility, brood size and growth rate of the paqr-2 suppressors. (A) Percentage of L1s that grow into fertile adults at three different temperatures. Note that all suppressor mutations permit reproductive growth of the paqr-2 mutant at 15°C. (B) Total self-brood size at 20°C. Note that all suppressor mutations dramatically improve self-brood size, except for ech-7(et6). (C) Length of worms grown from the L1 stage at 20°C for different amounts of time. Note that all suppressor mutations improve the growth of the paqr-2 mutant, except again for ech-7(et6). The figure legend in C also applies to panel A. *: p<0.05; **: p<0.01; ***: p<0.001.
(TIF)
Experimental tests confirming the identity of paqr-2 suppressor mutations. (A) Injected RNAi against pcyt-1 or ech-7 can suppress the paqr-2 phenotype, suggesting that the paqr-2 suppressors et6 and et9 are lof alleles of these genes. (B) The hacd-1(ok2776) deletion allele is as effective at suppressing the paqr-2 15°C growth defect as the hacd-1(et12) allele, confirming that et12 is also a lof allele. (C) Providing wild-type cept-1 as a transgene desuppresses the self-brood size defect in paqr-2;cept-1(et10), indicating that cept-1(et10) is a lof allele. (D) Homozygosity for the cept-1(et10), cept-1(et11) or hacd-1(et12) mutation suppresses the tail defect in paqr-2 worms, but this phenotype is desuppressed by introducing wild-type cept-1 or hacd-1 transgenes in the paqr-2;cept-1(et10) and paqr-2;cept-1(et11), or paqr-2;hacd-1(et12) strains, respectively. This indicates that et10, et11 and et12 are lof alelles. (E) and (F) The sams-1(ok2946) deletion allele and the sbp-1 overexpression transgene epEx141 can also suppress the paqr-2 15°C growth defect, respectively. ***: p<0.001.
(TIF)
Analysis of PE/PC ratios. (A) The PE/PC ratio is elevated in the paqr-2 mutant and is partially normalized by the nhr-49(et8) or cept-1(et10) mutations. (B) In a separate experiment, again the PE/PC ratio is elevated in the paqr-2 mutant, unaffected in the nhr-49(et8) or nhr-49(gk405) mutants, and elevated in the cept-1(et10) mutant. The average amounts of lipids recovered per mg of protein from 5 samples for each genotype are also indicated. There is a fair amount of variation in the lipid extraction, as evidenced by the larger error bars for the amount of lipids. However, this did not affect the relative recovery of PEs and PCs, as evidenced by the small error bars for the PE/PC ratios.
(TIF)
The non-ionic detergent Triton X-100 and oleic acid can independently suppress paqr-2 phenotypes. (A) Inclusion of 0.05–0.2% Triton X-100 in the culture plates allows the paqr-2 mutant to grow at 15°C. (B) Including 0.05% Triton X-100 in culture plates allows the paqr-2 mutant to develop and maintain normal tail tips at 20°C. (C) and (D) Inclusion of 0.5–2 mM oleic acid in culture plates causes a slight but dose-dependent improvement in the growth of paqr-2 mutants at 15°C and in the quality of the tail morphology at 20°C, respectively.
(TIF)
PC composition (each row indicates the % of total PC that had the indicated number of carbon atoms and double bonds in its two FAs). Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests. Note: 17:Δ is cis-9,10-methylenehexadecanoic acid and 19:Δ is cis-11,12-methylene octadecanoic acid.
(XLSX)
PE composition (each row indicates the % of total PE that had the indicated number of carbon atoms and double bonds in its two FAs). Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests. Note: 17:Δ is cis-9,10-methylenehexadecanoic acid and 19:Δ is cis-11,12-methylene octadecanoic acid.
(XLSX)
TAG composition (each row indicates the % of total TAG that had the indicated number of carbon atoms and double bonds in its FAs). Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests.
(XLSX)
FA composition of PCs, PEs and TAGs. Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests. Note: 17:Δ is cis-9,10-methylenehexadecanoic acid and 19:Δ is cis-11,12-methylene octadecanoic acid.
(XLSX)
PC composition (each row indicates the % of total PC that had the indicated number of carbon atoms and double bonds in its two FAs). Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests. Note: 17:Δ is cis-9,10-methylenehexadecanoic acid and 19:Δ is cis-11,12-methylene octadecanoic acid.
(XLSX)
PE composition (each row indicates the % of total PE that had the indicated number of carbon atoms and double bonds in its two FAs). Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests. Note: 17:Δ is cis-9,10-methylenehexadecanoic acid and 19:Δ is cis-11,12-methylene octadecanoic acid.
(XLSX)
TAG composition (each row indicates the % of total TAG that had the indicated number of carbon atoms and double bonds in its FAs). Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests.
(XLSX)
FA composition of PCs, PEs and TAGs. Red and blue shadings indicate FAs that are significantly elevated or lowered, respectively, compared to the control N2 worms and based on t-tests. Note: 17:Δ is cis-9,10-methylenehexadecanoic acid and 19:Δ is cis-11,12-methylene octadecanoic acid.
(XLSX)