Abstract
Objective
The rs1333049, rs10757278 and rs4977574 are single nucleotide polymorphisms (SNPs) of chromosome 9p21 locus that are associated with prevalence of acute coronary syndromes (ACS). The rs1333049 SNP was also associated with cardiac outcome 6 months post ACS. No data concerning their association with long term prognosis after myocardial infarction is available. The aim of our study was to investigate the association of the 9p21.3 locus with 5-year overall mortality in patients with ST-elevation myocardial infarction (STEMI) treated invasively.
Materials and Methods
We performed a retrospective analysis of data collected prospectively in a registry of consecutive patients with STEMI treated with primary PCI. Genotyping was performed with a TaqMan method. The analyzed end-point was total 5-year mortality.
Results
The study group comprised 589 patients: 25.3% of females (n = 149), mean age 62.4±11.9 years, total 5-year mortality 16.6% (n = 98). When all the study group was analyzed, no significant differences in mortality were found between the genotypes. However, in high-risk patients (Grace risk score ≥155 points, n = 238), low-risk homozygotes had significantly better 5-year survival compared to other genotypes. The hazard ratio associated with high-risk genotype (high-risk homozygote or heterozygote) was: HR = 2.9 (95%CI 1.4–6.1) for the rs4977574 polymorphism, HR = 2.6 (1.25–5.3) for the rs1333049 one and HR = 2.35 (1.2–4.6) for the rs10757278 one (Cox proportional hazards model).
Conclusions
The 9p21.3 locus is associated with 5-year mortality in high-risk patients with STEMI. This finding, due to very high effect size, could potentially be applied into clinical practice, if appropriate methods are elaborated.
Introduction
Several genome-wide association studies have shown a strong association between the chromosomal locus 9p21.3 and coronary artery disease (CAD) or myocardial infarction [1]–[8]. The results were further replicated in large-scale case-control studies [9]–[10]. The same locus was reported to give significant genomic signal for other diseases, like type 2 diabetes [11]–[14], aortic or intracranial aneurysms, peripheral artery disease or cancers [15]–[17].
There are several single nucleotide polymorphisms (SNPs) of the 9p21.3 locus associated with cardiovascular diseases, however, the functional link remains hypothetical. None of the SNPs is located within a protein coding region. The 9p21.3 locus contains only a sequence for an antisense RNA (ANRIL, CDKN2BAS). The nearby genes are coding cyclin-dependent kinases 2B and 2A (CDKN2A and CDKN2B) or methylthioadenosine phosphorylase (MTAP). The CDKN2B is potentially involved in pathogenesis of atherosclerosis, while it is a downstream target for transforming growth factor beta [18]–[19]. The problem is that SNPs influencing CDKN2B expression do not affect CAD risk and vice versa [20].
There is a very strong evidence for association between the 9p21.3 locus and myocardial infarction (MI). However, data regarding its influence on further prognosis is equivocal. In the GRACE registry that was performed in Europe (n = 3247, patients with all forms of an acute coronary syndrome, 6 months of follow-up), C allele of the rs1333049 polymorphism was independently associated with recurrent myocardial infarction or cardiac death [21]. No association with outcome was found in the population of the Post-Myocardial Infarction study (New Zealand, n = 816, median follow-up 9 years) or in Han Chinese patients with first ST-segment elevation myocardial infarction (STEMI, n = 520, median follow-up 29 months) [22]–[23]. The discrepancies were not related to type of treatment. Han Chinese patients underwent invasive procedure, participants from New Zealand received fibrinolytics and in GRACE registry patients were enrolled irrespective of applied strategy.
There is a debate if genetic testing may become part of the risk assessment in MI patients. Some authors claim that the influence of particular polymorphisms on prognosis does not exceed 15–30%, thus making a whole genetic analysis useless for clinical risk assessment. Their opinions, however, are based on genome wide association studies that are especially biased to recognize markers with limited influence. This fact does not mean that particular polymorphisms would not be useful for special populations, where they could identify high risk patients.
In the present study we aimed to investigate the association of the 9p21.3 locus with 5-year all-cause mortality of patients with myocardial infarction. We tested 3 previously described SNPs (rs4977574, rs1333049, rs10757278) as markers of this locus. The further goal of the study was selecting and characterizing patients who could have the greatest benefit from genotyping.
Materials and Methods
Ethics statement
The study protocol was approved by Ethics Committee of Medical University of Bialystok. The study was performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki. Informed written consent has been obtained from the subjects.
We performed a retrospective analysis of data collected prospectively in a single center. It comprised Caucasian patients with STEMI, inhabitants of North-Eastern Poland, who were hospitalized in years 2001–2005 and survived first 48 hours after admission. The 48 hour survival threshold was implemented because of the time from blood sampling to potential result of genetic testing. In this subpopulation it would be of no benefit, but the very early mortality might affect final results. No additional exclusion criteria were introduced. All the patients underwent coronary angiography within 12 hours from symptoms onset. STEMI was diagnosed based on rise in troponin I concentration or creatine kinase –MB fraction activity accompanied by chest pain history and new ECG abnormalities (lasting >20 minutes ST-segment elevation or left bundle branch block). The analyzed data included patients' history, physical examination on admission, routine laboratory tests, echocardiography, results of coronary angiography and invasive treatment. MDRD (Modification of Diet in Renal Disease) formula was used to estimate creatinine clearance. Grace risk score was calculated retrospectively, based on previously described method [24]. Appropriate number of points were given for: age, heart rate, systolic blood pressure, creatinine plasma concentration, Killip class and cardiac arrest (all parameters assessed on admission). All patients were also scored for ST-segment deviation and elevated cardiac markers. Next, all subjects were divided into high-risk group (≥155 points) and non-high-risk group, according to previously evaluated clinical cut-off level [25]. All the patients were treated according to contemporary guidelines.
Blood samples were collected to EDTA tubes, treated with commercial DNA extraction kit (Blood Mini, A&A Biotechnology) and stored in −20 degrees Celsius. The SNPs (rs1333049, rs10757278, rs4977574) were assessed with a TaqMan SNP Genotyping Assay on the ABI 7500 real time PCR platform (Applied Biosystems), according to manufacturer's instructions. Ten percent of samples were genotyped twice (quality control requirements).
The analyzed end-point was 5-year all-cause mortality. Data concerning survival was retrieved from the local population registry run by a Government Office, assuring the most complete follow-up possible.
A statistical analysis was performed with STATISTICA 9.0 software. Distribution of variables was tested with Shapiro-Wilk test. Next, clinical parameters were compared between the genotypes with chi-2, t-Student or Mann-Whitney tests, as appropriate. Survival was compared with log-rank test. Univariate and multivariate analyses for 5-year survival were performed with Cox proportional hazards model. Variables with significant association with survival were included in a primary model of multivariate regression. In the case of the 3 SNPs, due to strong linkage between them, only the rs4977574 was included, because it showed the strongest effect, based on literature [2], [4], [5], [6]. The final model was selected in backward stepwise manner. Additionally, all SNPs in a univariate analysis as well as multivariate model were adjusted for severity of coronary artery disease (number of vessels with significant stenosis). Two-sided p value<0.05 was considered statistically significant. In the case of survival analysis (chi-2 and log-rank tests) multiple tests were performed, therefore p values were adjusted for Bonferroni correction (4 subgroup analyses). Due to very strong linkage between the 3 SNPs, we did not consider them for further multiple testing. The biostatistical parameters were calculated using ARLEQUIN v.3.0 software.
The study was designed to have a statistical power of at least 80 percent to detect a 66% percent relative risk increase in 5-year mortality of high-risk homozygotes compared to other genotypes. Assuming 18% overall mortality rate and percentage of high-risk homozygotes around 25% [2], the target of events would be achieved in a group of 570 patients. Estimation of sample sizes or effect sizes in survival functions were performed with chi-square test.
Results
The registry comprised 609 patients. Nine of them were lost to follow-up (1.5%) and genotype could not be determined in 11 remaining cases due to poor sample quality (1.8%). No genotyping discrepancies were observed in the samples genotyped twice. The final study group consisted of 589 patients: 25.3% of females (n = 149), mean age 62.4±11.9 years, TIMI 3 obtained in 92% of patients (n = 542).
Genotyping results are presented in table 1. The genotype frequency distributions showed no significant deviations from Hardy-Weinberg equilibrium (rs1333049 SNP: p = 0.12; rs10757278: p = 0.21; rs4977574: p = 0.17). The specific allele frequencies are comparable to previous reports (2, 4, 5). According to genetic localisation data, the SNPs rs1333049, rs10757278 and rs4977574 are closely linked. Consequently, pairwise comparison using the exact test disequilibrium analysis yielded departures from independence for all pairs of loci (p<0.0001, table 2). Therefore we chose the rs4977574 SNP with the strongest effect based on literature [2], [4], [5], [6] to present clinical characteristics of the study group (G allele carriers vs. AA homozygotes, table 3). No significant differences were observed between the genotypes.
Table 1. Percentages of specific genotypes and associated mortality rates.
| Polymorphism (risk allele) | rs1333049 (C) | rs10757278 (G) | rs4977574(G) | ||||||
| The whole study group (n = 589) | |||||||||
| Genotype | CC | CG | GG | GG | AG | AA | GG | AG | AA |
| Percentage (n) | 24.8 (146) | 46.7 (275) | 28.5 (168) | 24.2 (143) | 47.4 (279) | 28.3 (167) | 24.1 (142) | 47.0 (277) | 28.9 (170) |
| 5-year mortality (n) | 14.4 (21) | 20.0 (55) | 13.1 (22) | 15.4 (22) | 19.4 (54) | 13.2 (22) | 16.2 (23) | 19.5 (54) | 12.4 (21) |
p = 0.01;
p = 0.029;
p = 0.008;
p>0.05 compared to homozygotes; all p values adjusted for Bonferroni correction.
Table 2. Linkage disequilibrium of investigated SNPs.
| p | SNP1 | SNP2 | D′ | LD | r2 |
| <0.0001 | rs1333049 | rs10757278 | 0.88 | 0.21 | 0.77 |
| <0.0001 | rs1333049 | rs4977574 | 0.83 | 0.2 | 0.68 |
| <0.0001 | rs10757278 | rs4977574 | 0.89 | 0.22 | 0.78 |
Table 3. Baseline characteristics of the study group based on rs4977574 genotype (risk allele- G).
| Characteristic | Overall population N = 589 | rs4977574 G-allele carriers N = 419 | rs4977574 AA homozygotes N = 170 | P |
| Age (years) | 62.4 (12.0) | 63.0 (11.7) | 60.9 (12.6) | 0.43 |
| Female gender (%) | 25.3 (n = 149) | 25.3 (n = 106) | 25.3 (n = 43) | 0.99 |
| Body mass index (kg/m2) | 24.7 (3.7) | 24.5 (3.8) | 25.0 (3.5) | 0.6 |
| Hypertension (%) | 53.3 (n = 314) | 54.1 (n = 227) | 51.2 (n = 87) | 0.5 |
| Type 2 diabetes (%) | 22.1 (n = 130) | 23.4 (n = 98) | 18.8 (n = 32) | 0.22 |
| Previous myocardial infarction (%) | 10.9 (n = 64) | 11.5 (n = 48) | 9.4 (n = 16) | 0.47 |
| Systolic blood pressure (mmHg) | 138.7 (28.3) | 138.3 (29.8) | 139.8 (24.3) | 0.88 |
| Heart rate (beats/min) | 75.7 (17.8) | 76.2 (18.1) | 74.7 (16.9) | 0.26 |
| Killip class III or IV (%) | 4.1 (n = 24) | 4.0 (n = 17) | 4.1 (n = 7) | 0.97 |
| ST-elevation in anterior leads | 39.4 (n = 232) | 40.1 (n = 168) | 37.6 (n = 64) | 0.58 |
| TIMI flow grade 3 after procedure | 92.0 (n = 542) | 91.6 (n = 384) | 92.9 (n = 158) | 0.59 |
| Stent implantation (%) | 77.1 (n = 454) | 76.6 (n = 321) | 78.2 (n = 133) | 0.67 |
| No of vessels with significant stenosis | 1.7 (0.8) | 1.7 (0.8) | 1.8 (0.8) | 0.12 |
| eGFR (ml/min/1.73 m2) | 79.9 (23.3) | 79.1 (24.1) | 82.0 (21.2) | 0.51 |
| Haemoglobin on admission (mg/dl) | 13.05 (1.6) | 13.0 (1.6) | 13.1 (1.7) | 0.26 |
| Ejection fraction (%) | 45.9 (9.5) | 45.2 (9.0) | 45.9 (9.5) | 0.26 |
| Grace risk score | 149 (35) | 151 (35) | 145 (34) | 0.09 |
eGFR- estimated GFR, mean values with standard deviations are given, unless otherwise specified.
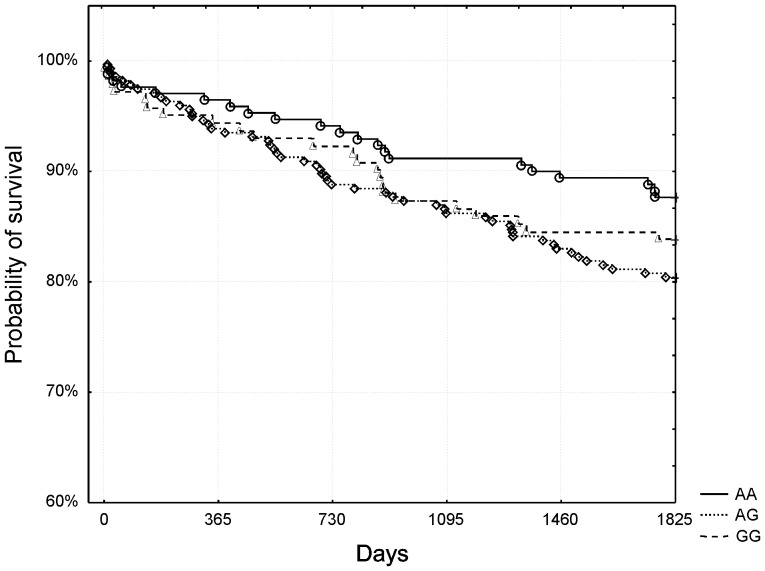
A 5-year follow-up with a median of 1950 days (minimum 1824, maximum 3378 days) was performed. At the cut-off point of 1825 days (5 years) 98 patients died (16.6%). When all the study group was analyzed, no significant differences in mortality were found between the genotypes (table 1). Figure 1 presents Kaplan-Meier surviving curves for specific genotypes of rs4977574 polymorphism and 5-year mortality (p = 0.6 after adjustment for Bonferroni correction, log-rank test).
Figure 1. The rs4977574 polymorphism and 5-year survival.
No significant differences were observed between the genotypes.
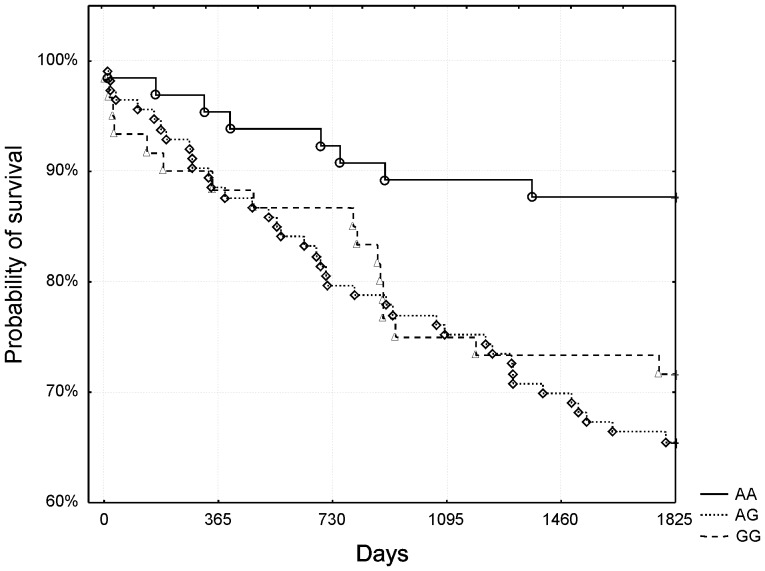
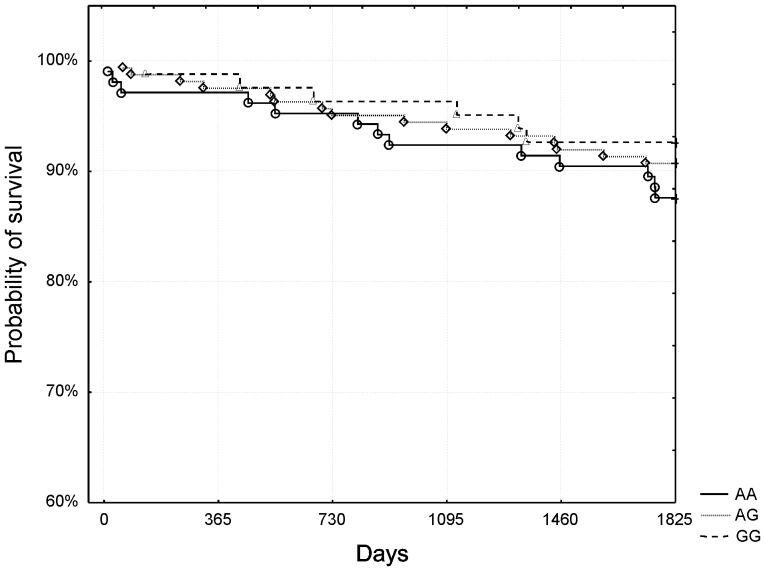
However, in the subgroup of high-risk patients (GRACE risk score ≥155, n = 238, 26.9% mortality [n = 64]), visual analysis of survival curves showed strikingly better survival of low-risk homozygotes compared to both other genotypes. The curves for heterozygotes and high-risk homozygotes had almost similar course and therefore were collapsed together for further analysis. Kaplan-Meier surviving curves for the subgroup of high-risk patients and rs4977574 polymorphism are shown in figure 2. The difference was statistically significant (p = 0.008 after adjustment for Bonferroni correction, log-rank test). No such trend was observed in low and medium-risk patients (p>0.99 after adjustment for Bonferroni correction, log-rank test, figure 3). Mortality rates for specific genotypes in the subgroup of high-risk patients are shown in table 1.
Figure 2. The rs4977574 polymorphism and 5-year survival - subgroup of high-risk patients (Grace risk score ≥155).
AA low-risk homozygotes had significantly higher probability of survival compared to other genotypes (p = 0.008 after adjustment for Bonferroni correction, log-rank test).
Figure 3. The rs4977574 polymorphism and 5-year survival – medium and low-risk patients (Grace risk score <155).
No significant differences were observed between the genotypes.
Table 4 presents results of Cox proportional hazards model for 5-year survival in a subgroup of high-risk patients. The hazard ratio associated with high-risk genotype (high-risk homozygote or heterozygote) was: HR = 2.9 (95%CI 1.4–6.1) for the rs4977574 polymorphism, HR = 2.6 (1.25–5.3) for the rs1333049 one and HR = 2.35 (1.2–4.6) for the rs10757278 one. In a univariate analysis, apart from 3 investigated polymorphism, significant association was found in the case of Killip class, ejection fraction and type 2 diabetes. In multivariate regression the rs4977574 polymorphism remained related to mortality together with ejection fraction and type 2 diabetes. After adjustment for severity of coronary artery disease all p values were still below 0.05.
Table 4. Univariate and multivariate analysis for 5-year mortality in a subgroup of high-risk patients (Grace risk score ≥155).
| Variable | Hazard ratio | 95% CI | p |
| Killip class | 1.4 | 1.09–1.7 | 0.01 |
| Ejection fraction (%) | 0.95 | 0.93–0.98 | 0.0002 |
| Type 2 diabetes | 2.1 | 1.3–3.5 | 0.006 |
| rs1333049 CC or CG genotype | 2.6 (2.3) | 1.25–5.3 (1.2–4.5) | 0.004 (0.014) |
| rs10757278 GG or AG genotype | 2.35 (2.2) | 1.2–4.6 (1.1–4.2) | 0.006 (0.017) |
| rs4977574 GG or AG genotype | 2.9 (2.65) | 1.4–6.1 (1.3–5.4) | 0.004 (0.006) |
| No of vessels with significant stenosis | 0.98 | 0.9–1.06 | 0.66 |
| Multivariate analysis | |||
| Ejection fraction (%) | 0.95 (0.96) | 0.93–0.98 (0.94–0.99) | 0.001 (0.01) |
| Type 2 diabetes | 1.9 (1.97) | 1.1–3.2 (1.2–3.3) | 0.014 (0.008) |
| rs4977574 GG or AG genotype | 2.7 (2.46) | 1.3–5.7 (1.2–5.0) | 0.009 (0.01) |
In the brackets values adjusted for severity of coronary artery disease (number of vessels with significant stenosis) are given.
Next, similar trends between genotypes and survival were found in patients with 3-vessel disease (n = 122, Table 1). Additional analysis was done using Cox proportional hazard model that showed following hazard ratios associated with high-risk genotype: HR = 3.65 (95%CI 1.3–10.5) for the rs4977574 polymorphism, HR = 2.6 (1.002–6.9) for the rs1333049 one and HR = 3.0 (1.03–8.5) for the rs10757278 one.
Discussion
The 9p21.3 locus showed a significant association with 5-year survival of high-risk patients with STEMI (GRACE risk score ≥155 points or 3-vessel disease). This phenomenon is probably related to an increased number of events in the chosen subgroups. On the other hand, the 9p21.3 locus may influence atherosclerotic plaque development, stability, and, in this way, survival [15]. Such effect would be more pronounced in patients with advanced coronary atherosclerosis. It was already reported that 9p21 locus is associated with mortality in patients with multivessel coronary artery disease [26]. Another patomechanism that needs to be considered is sudden cardiac death due to probable arrhythmia that was shown in a prospective cohort [27]. Based on multivariate analysis, 9p21 locus polymorphism added prognostic information to previously established risk factors like ejection fraction and type 2 diabetes.
This report supports results from the GRACE registry, that was also performed in European populations and showed association between the rs1333049 polymorphism and outcome within 6 months [21]. Our findings extend this effect to 5 years, however, they are limited to high-risk patients. Another difference is study population (patients with all forms of an acute coronary syndrome in the GRACE registry vs. the ones with STEMI). We observed remarkably high effect sizes for the association between high-risk genotypes and 5-year outcome. It makes these novel risk markers potentially more applicable in everyday practice. Furthermore, if only a particular subgroup of patients was to be genotyped, the method would be more cost-effective. The future interventional studies where additional therapeutic actions are applied in high-risk clinical and genetic profile would help translate this effect into clinical practice.
The results from European registries are not in concordance with studies performed in other populations [22], [23]. The effect sizes of specific genotypes may be strongly related to genetic background as well as environmental factors. Therefore it is essential to validate results of genetic studies in regional populations. In Han Chinese patients with STEMI no differences were found between the 9p21 locus genotypes in 2-year event-free survival (cardiac death, non-fatal MI, recurrent angina or heart failure requiring hospitalization) [23]. Similarly, patients from Post-Myocardial Infarction Study (New Zealand, 9-year follow-up) had genotype-dependent variation neither in total mortality nor in hospital admissions due to reinfarctions or heart failure [22].
There is an Italian study that investigates closely linked polymorphism: rs1333040 [28]. Included participants had early-onset myocardial infarction (<45 years) and underwent coronary angiography without index event coronary revascularization. During 10-year follow-up the genotype significantly affected risk of coronary revascularization, but no influence on cardiac death or recurrent myocardial infarctions was observed. In general, the rate of events was low due to early age (5.1%, n = 77) and possibly therefore no effect on survival was shown.
We have found no association between the 9p21.3 locus and either participants' age or severity of coronary artery disease, which is again consistent with the GRACE registry [21]. On the contrary, in the study from New Zealand high-risk homozygotes (rs1333049 SNP) were significantly younger, compared to other genotypes (60.6 vs. 62.8 years, p = 0.009) [22]. The report from the United States of America showed that the rs10757278 SNP influenced angiographic severity and progression of coronary artery disease [29]. Finally, in Chinese population the rs1333049 SNP contributed to severity of coronary artery disease, but only in diabetics [30]. The discrepancies might be again explained by different genetic effect sizes depending on a chosen population.
It is surprising that no significant deviation from Hardy-Weinberg equilibrium was shown in our population. However, the study was not designed to prove the association between the 3 SNPs and myocardial infarction. Such analysis would require remarkably larger sample sizes.
The number of patients investigated in this study was relatively low, but in our opinion it was large enough to search for associations of clinical importance. Very large study groups enable finding significant correlations of very small effect sizes that have no further meaning in everyday practice. The further limitation of the study is a retrospective type of analysis, however, the data was collected prospectively.
Conclusions
The 9p21.3 locus was associated with 5-year mortality in high-risk patients with STEMI. This finding, due to very high effect size, could potentially be applied into clinical practice. However, optimal set of risk genotypes and appropriate tools for this clinical setting are still to be identified and elaborated.
Funding Statement
This work was supported by National Science Center, Poland [N N 402 529139]. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, et al. (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, et al. (2007) Genome-wide association analysis of coronary artery disease. N Engl J Med 357: 443–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koch W, Türk S, Erl A, Hoppmann P, Pfeufer A, et al. (2011) The chromosome 9p21 region and myocardial infarction in a European population. Atherosclerosis 217: 220–6. [DOI] [PubMed] [Google Scholar]
- 4. Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, et al. (2007) A Common Variant on Chromosome 9p21 Affects the Risk of Myocardial Infarction. Science 316: 1491–93. [DOI] [PubMed] [Google Scholar]
- 5. Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, et al. (2009) Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy-number variants. Nat Genet 41: 334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O'Donnell CJ, Kavousi M, Smith AV, Kardia SL, Feitosa MF, et al. (2011) Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation 124: 2855–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takeuchi F, Yokota M, Yamamoto K, Nakashima E, Katsuya T, et al. (2012) Genome-wide association study of coronary artery disease in the Japanese. Eur J Hum Genet 20: 333–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee JY, Lee BS, Shin DJ, Woo Park K, Shin YA, et al. (2013) A genome-wide association study of a coronary artery disease risk variant. J Hum Genet 58: 120–6. [DOI] [PubMed] [Google Scholar]
- 9. Butterworth AS, Braund PS, Farrall M, Hardwick RJ, Saleheen D, et al. (2011) Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet 7: e1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scheffold T, Kullmann S, Huge A, Binner P, Ochs HR, et al. (2011) Six sequence variants on chromosome 9p21.3 are associated with a positive family history of myocardial infarction: a multicenter registry. BMC Cardiovasc Disord 11: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, et al. (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331–6. [DOI] [PubMed] [Google Scholar]
- 12. Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, et al. (2007) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316: 1336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, et al. (2007) A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cugino D, Gianfagna F, Santimone I, de Gaetano G, Donati MB, et al. (2012) Type 2 diabetes and polymorphisms on chromosome 9p21: A meta-analysis. Nutr Metab Cardiovasc Dis 22: 619–25. [DOI] [PubMed] [Google Scholar]
- 15. Nambi V, Boerwinkle E, Lawson K, Brautbar A, Chambless L, et al. (2012) The 9p21 genetic variant is additive to carotid intima media thickness and plaque in improving coronary heart disease risk prediction in white participants of the Atherosclerosis Risk in Communities (ARIC) Study. Atherosclerosis 222: 135–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, et al. (1994) Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet 8: 27–32. [DOI] [PubMed] [Google Scholar]
- 17. Cannon-Albright LA, Goldgar DE, Meyer LJ, Lewis CM, Anderson DE, et al. (1992) Assignment of a locus for familial melanoma, MLM, to chromosome 9p13–p22. Science 258: 1148–52. [DOI] [PubMed] [Google Scholar]
- 18. Kalinina N, Agrotis A, Antropova Y, Ilyinskaya O, Smirnov V, et al. (2004) Smad expression in human atherosclerotic lesions: evidence for impaired TGF-b/Smad signaling in smooth muscle cells of fibrofatty lesions. Arterioscler. Thromb Vasc Biol 24: 1391n6. [DOI] [PubMed] [Google Scholar]
- 19. Schmid M, Sen M, Rosenbach MD, Carrera CJ, Friedman H, et al. (2000) A methylthioadenosine phosphorylase (MTAP) fusion transcript identifies a new gene on chromosome 9p21 that is frequently deleted in cancer. Oncogene 19: 5747–54. [DOI] [PubMed] [Google Scholar]
- 20. Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, et al. (2010) Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS One 5: e10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buysschaert I, Carruthers KF, Dunbar DR, Peuteman G, Rietzschel E, et al. (2010) A variant at chromosome 9p21 is associated with recurrent myocardial infarction and cardiac death after acute coronary syndrome: The GRACE Genetics Study. Eur Heart J 31: 1132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ellis KL, Pilbrow AP, Frampton CM, Doughty RN, Whalley GA, et al. (2010) A common variant at chromosome 9P21.3 is associated with age of onset of coronary disease but not subsequent mortality. Circ Cardiovasc Genet 3: 286–93. [DOI] [PubMed] [Google Scholar]
- 23. Peng WH, Lu L, Zhang Q, Zhang RY, Wang LJ, et al. (2009) Chromosome 9p21 polymorphism is associated with myocardial infarction but not with clinical outcome in Han Chinese. Clin Chem Lab Med 47: 917–22. [DOI] [PubMed] [Google Scholar]
- 24. Granger CB, Goldberg RJ, Dabbous O, Pieper KS, Eagle KA, et al. (2003) Global Registry of Acute Coronary Events Investigators. Predictors of hospital mortality in the Global Registry of Acute Coronary Events. Arch Intern Med 163: 2345–2353. [DOI] [PubMed] [Google Scholar]
- 25.Global Registry of Acute Coronary Events website. Available: http://www.outcomes-umassmed.org/GRACE/grace_risk_table.aspx. Accessed: 25 Jul 2013.
- 26. Gioli-Pereira L, Santos PC, Ferreira NE, Hueb WA, Krieger JE, et al. (2012) Higher incidence of death in multi-vessel coronary artery disease patients associated with polymorphisms in chromosome 9p21. BMC Cardiovasc Disord 12: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Newton-Cheh C, Cook NR, VanDenburgh M, Rimm EB, Ridker PM, et al. (2009) A common variant at 9p21 is associated with sudden and arrhythmic cardiac death. Circulation 120: 2062–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ardissino D, Berzuini C, Merlini PA, Mannuccio Mannucci P, Surti A, et al. (2011) Investigators. Influence of 9p21.3 genetic variants on clinical and angiographic outcomes in early-onset myocardial infarction. J Am Coll Cardiol 58: 426–34. [DOI] [PubMed] [Google Scholar]
- 29. Patel RS, Su S, Neeland IJ, Ahuja A, Veledar E, et al. (2010) The chromosome 9p21 risk locus is associated with angiographic severity and progression of coronary artery disease. Eur Heart J 31: 3017–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang W, Peng WH, Lu L, Zhang RY, Zhang Q, et al. (2011) Polymorphism on chromosome 9p21.3 contributes to early-onset and severity of coronary artery disease in non-diabetic and type 2 diabetic patients. Chin Med J 124: 66–71. [DOI] [PubMed] [Google Scholar]