Abstract
Emerging evidences suggest Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling pathway plays an important role in bone development and metabolism. Effects of JAK-STAT pathway on skeletal development are summarized based on skeletal phenotype of individual JAK and STAT gene knockout mouse. Furthermore, STAT3 has more profound effects on bone homeostasis compared with the other STATs. STAT3 mutation causes a disease called Job syndrome, most patients with which have associated craniofacial and skeletal features. Selective inactivation of STAT3 in osteoblasts decreases bone formation and skeletal responsiveness to mechanical loading. Future research includes investigating JAK-STAT signaling in osteoclasts and osteocytes.
Keywords: Janus kinase, signal transducer and activator of transcription, skeletal development, bone homeostasis, osteoblast, mechanotransduction
Introduction
The skeletal system is one of the most important systems found in the human body. It serves as the body’s structural support center, provides a framework for attachment of tissues, protects vital organs and helps direct the forces necessary for movement. Bones also act as a reservoir of calcium and phosphate, two compounds used for numerous metabolic processes.1 These essential functions lend to the importance of maintaining a healthy skeletal system. Skeleton develops and grows through a process called bone modeling,2 which leads to a net gain in bone mass and size. Bone homeostasis is maintained through bone remodeling in adult. Bone remodeling is a lifelong process and is achieved via a finely regulated balance between bone formation and resorption.3,4
All bones in the body continuously undergo remodeling, a process of removal of old bone and formation of new bone material.3,4 Usually bone formation matches bone resorption during this process for no net change in bone mass; however, osteoporosis is believed to be the result of a disparity between resorption and formation rates during remodeling.5 When bone is removed faster than it is replaced, bone mass, especially that of trabecular bone, decreases. Osteoporosis is a major disease characterized by a progressive decline in bone mineral density, which results in a dramatic increase in fracture risk for elderly individuals, especially in post-menopausal women.6 In order to prevent and treat osteoporosis, two main types of strategies have been applied: antiresorptive ones that slow the progression of bone resorption and anabolic ones that help stimulate bone formation.7
To ensure mechanical competence, bone tissue adapts its mass, architecture and mechanical properties to meet the prevailing dynamic loading environment. Bone’s adaptive response is regulated by the ability of resident bone cells to perceive and translate mechanical energy (e.g., from muscle contraction, substrate reaction forces, and gravity) into a cascade of structural and biochemical changes within the cells—a process known as mechanotransduction.8 The cellular products resulting from a transduced mechanical signal provide the signals necessary to fine tune—via bone resorption and/or formation—bone architecture and mass to adequately resist future loads.9,10 Osteoblasts are sensitive to mechanical environment and can generate anabolic response to mechanical stimulation. Therefore, exercise has been recommended as a useful way to improve bone strength. Many studies have shown exercise has effects on bone mass, the mineral content, size and shape. Numerous research studies demonstrate that exercise promotes and enhances the health of a person’s bones.
Understanding the underlying mechanisms of how bone remodeling is regulated and how bone response to mechanical stimulation will be helpful for discovering new targets to treat osteoporosis. Two major types of bone cells, osteoblasts and osteoclasts, which are responsible for bone formation and resorption, are directly involved in bone modeling and remodeling. Osteoblasts and osteoclasts are regulated, in part, by many local factors including cytokines in the bone microenvironment. Many cytokine signaling pathways have been found in bone cells, including the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway. The intent of this paper is to discuss the role of the JAK-STAT pathway in bone metabolism with the emphasis on STAT3.
The JAK-STAT pathway was originally identified as a receptor-activated pathway responsive primarily to interferon (IFN)-gamma and members of the interleukin-6 (IL-6) family. Members of the JAK family include JAK1, JAK2, JAK3 and Tyk2. All exhibit broad patterns of expression, except JAK3 whose expression is restricted to leukocytes. STAT proteins were originally discovered as latent cytoplasmic transcription factors two decades ago.11 There are seven known mammalian STAT proteins, STAT 1, 2, 3, 4, 5A, 5B, and 6.12 The STAT pathway can be activated via several means. Phosphorylation of STAT is initiated by cytokine binding to cell-surface cytokine receptors via the JAK-STAT pathway. The intracellular domains of many cytokine receptors, mainly IL-6 receptors, are physically associated with tyrosine kinases of the Janus kinase (JAK) family. IL-6 belongs to the IL-6 family (or gp130 family), which is composed of IL-6, IL-11, oncostatin M (OSM), leukemia inhibitory factor (LIF), cardiotrophin-1 (CT-1), and novel neurotrophin-1/B-cell stimulatory factor-3.13,14 They are pleiotropic cytokines, sharing the glycoprotein chain gp130 as a common signal transducer.13,15,16 The binding of IL-6 family cytokines to their receptors induces the homodimerization of gp130 and subsequently activates the gp130-associated JAKs.17 The activated JAKs phosphorylate the tyrosine residue of the YXXQ motif, leading to the recruitment through binding at the SH2 domain and tyrosine phosphorylation at the cytoplasmic tail of STATs. Activated STATs dimerize, enter the nucleus and regulate the transcription of various genes that regulate cell proliferation and differentiation in a cell-specific manner.18 In addition, JAKs can also be activated in response to distinct ligands that bind to G-protein-coupled receptors and stimulated STAT tyrosine phosphorylation. Furthermore, the STATs can be phosphorylated by growth factor binding to receptor tyrosine kinases, such as the epidermal growth factor (EGF) receptor and the platelet-derived growth factor (PDGF) receptor.
The JAK-STAT pathway plays a crucial role in the growth and differentiation of a variety of cell types. Although the significance of the JAK-STAT pathway in musculoskeletal system has not been well characterized, increasing evidence suggests that this pathway may be involved in regulation of bone homeostasis and bone response to mechanical loading. It is interesting to note that many cytokines that activate JAK-STAT signaling pathway affect osteoblast and osteoclast differentiation and proliferation. For example, the gp130 family of cytokines, including IL-6, IL-11, and oncostatin M, are expressed in osteoblasts and osteocytes and play an important role in osteoblast differentiation and bone formation.19 IL-6 and IL-11 in bone cells have been shown to increase with mechanical stimulation.20 Further, EGFR signaling suppresses osteoblast differentiation inhibits expression of master osteoblastic transcription factors Runx2 and Osterix.21 PDGF regulates bone formation and plays an important role in bone fracture repair and regeneration.22 Both EGFR and PDGFR signaling may be mediated by JAK-STAT pathway. However, the role of JAK-STAT signaling in bone homeostasis has not been extensively investigated despite the fact that almost all the JAKs and STATs are expressed in osteoblasts and osteoclasts.
JAK-STAT and Skeletal Development
The study of JAK and STAT knockout animals suggests that the JAK-STAT signaling pathway is important for skeletal development although not all the JAKs and STATs are equally essential for osteoblast and osteoclast biology. Table 1 summarizes the influence of JAKs and STATs on skeletal development from knockout animals.
Table 1. JAK-STAT and bone development.
| Deleted genes | Lethality | Bone phenotype |
|---|---|---|
|
JAKs |
|
|
| JAK126 |
Die perinatally |
Significantly small body mass. |
| JAK227,28 |
JAK2 knockout mice died of anemia at E12.5. |
Unknown. |
| JAK364,65 |
- |
JAK3−/− mice are born normally and develop in an indistinguishable manner from wild-type littermates and show no gross abnormality. |
| Tyk224,25 |
- |
TYK2-deficient mice are viable and fertile and show no obvious phenotype. |
|
STATs |
|
|
| STAT134-36 |
- |
STAT1−/− mice have higher bone density, but are indistinguishable from their normal counterparts on the basis of size, activity, or ability to reproduce. |
| STAT243 |
- |
The STAT2 null mice developed and bred normally in specific pathogen-free environment. |
| STAT338,60,62 |
STAT3−/− embryos die at E6.5–7.5 |
Selective inactivation of STAT3 in bone causes osteoporosis. |
| STAT442,45 |
- |
STAT4−/− mice are grossly indistinguishable from their wild-type littermates. |
| STAT5A/5B30-32 |
- |
The male and female STAT5A/5B double mutant mice, as well as the male STAT5B mutant mice are significantly smaller than their wild-type littermates. No effect is seen in the STAT5A mutant mice. |
| STAT641,44,46 | - | STAT6−/− mice are grossly indistinguishable from their littermate controls. |
JAK1, JAK2, and Tyk2 are expressed in bone cells and are involved in bone formation. For example, OSM is able to induce a rapid tyrosine phosphorylation of JAK1, JAK2, and Tyk2 in murine calvarial-derived osteoblastic cells.23 Among these 3 JAK family members, Tyk2 is not directly related to any developmental defects in skeleton because Tyk2-deficient mice are viable and fertile and show no obvious phenotype.24,25 JAK1 knockout mice die perinatally, but show a significantly small body mass in comparison with their littermates.26 Newborn JAK1-deficient mice weigh 40% less than their heterozygous or wild-type littermates. These data indicate bone growth delays without JAK1 in embryo, suggesting JAK1 is critical for skeletal development. Global JAK2 knockout mice die of anemia at E12.5 before bone formation starts.27,28 The study of mouse embryonic fibroblasts derived from JAK2 null embryos and their wild-type littermate controls demonstrates that JAK2 deficiency decouples growth hormone-receptor signaling from its downstream mediators, the STAT 5A and 5B.27,28 This implies that JAK2 may be also involved in bone development because the growth hormone essential for normal bone development. Unfortunately, no histological examination has been done on JAK1- or JAK2-deficient bone tissues so far. The underlying mechanism of how JAK1 and JAK2 affect signaling pathways in osteoblasts remains however to be investigated. One useful approach is to generate mice with selective inactivation of JAK2 in bone cells using the conditional JAK2 knockout mice and the study skeletal phenotype of these mice to further disclose detailed functions of JAK2 in osteoblasts and osteoclasts.29
All the STAT genes and proteins have been located in bone tissues. Among the seven STATs, only STAT5A and STAT5B knockout mice show obviously defective bone development.30-32 STAT5A and STAT5B are functionally quite pleiotropic. Biochemical and genetic studies have underscored the important role that STAT5A and STAT5B plays in directing a biological response to the IL-3 (IL-3, IL-5, and granulocyte-macrophage colony-stimulating factor), single-chain (e.g., growth hormone, prolactin, thrombopoietin, and erythropoietin), and γc (i.e., the IL-2, IL-7, IL-9, IL-15, and possibly IL-21) receptor families.33 The extensive sequence similarity between STAT5A and STAT5B (~96% amino acid identity) explains their functional redundancy. But, the responses to prolactin and growth hormone favor STAT5A and STAT5B, respectively. The male and female STAT5A/5B double mutant mice weighed 30–40% and 20–30% less than their wild-type littermates.31 Normal body mass is seen in the STAT5A mutant mice, while the male STAT5B mice are consistently 20–30% smaller than their wild-type littermates. These data show a delayed bone growth in STAT5A/5B mutant mice. The delayed skeletal development caused by STAT5A/5B mutation is consistent with IGF-1 function in bone. IGF-1 levels were significantly reduced in STAT5A/B mutant male relative to wild-type and female mice.31 The levels of serum IGF-1 were significantly reduced in male STAT5B mutant mice but not in the females, while the levels were normal in both male and female STAT5A mutant mice.
STAT1 and STAT3 influence skeletal development in different ways. STAT1 target genes appear to promote inflammation and antagonize proliferation. This contrasts the pro-proliferative and anti-inflammatory activities associated with STAT3. Thus, the ability of several cytokines to activate both STAT1 and STAT3 (e.g., members of the IFN-I and IL-6 families) may reflect an effort to achieve a more balanced response. The opposing effects of STAT1 and STAT3 exist in bone tissue.
Although STAT1−/− mice are indistinguishable from their normal controls,34,35 inactivation of STAT1 actually leads to an osteopetrotic bone (higher bone mass) phenotype in mouse when bone mineral content is measured.36 This can be explained by the inhibitory effect of STAT1 on Runx2 transcription in osteoblasts. Runx2 is the master transcription factor of osteoblast differentiation. Consistent with the higher bone mass in STAT1-deficient mice, inactivation STAT1 can accelerate fracture repair.37 These data suggest that STAT1 negatively regulates bone formation in vivo.
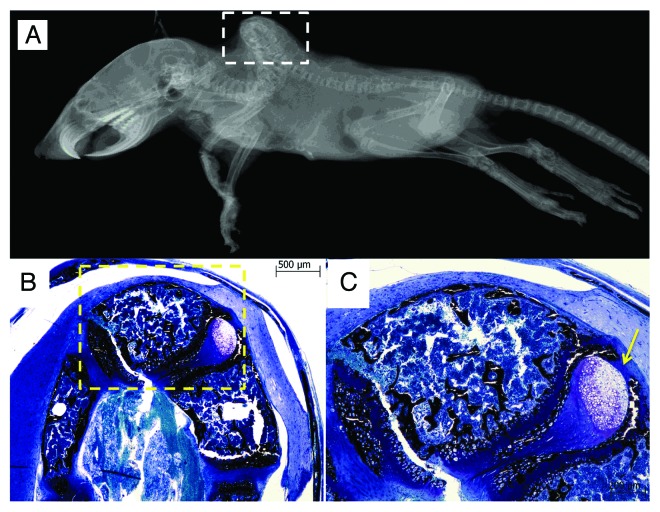
STAT3 is essential for early embryonic development because targeted disruption of the mouse STAT3 gene leads to embryonic lethality at 6.5–7.5 d.38 Tissue-specific deletion of STAT3 has demonstrated that STAT3 plays a crucial role in a variety of biological functions, including cell growth, suppression of apoptosis, and cell motility. STAT3 is a key signal transduction protein that mediates signaling by numerous cytokines, growth factors and oncoproteins.39 Upon stimulation, STAT3 is activated by tyrosine and serine phosphorylation, dimerizes, and translocates to the nucleus, where it binds specific gene-promoter sequences and induces gene expression. Genes that are regulated by activated STAT3 include antiapoptotic and proliferation-associated genes such as Bcl-xL, Bcl-2, Fas, Cyclin D1, Survivin, and c-Myc.40 Selective inactivation of STAT3 in osteoblasts causes smaller body mass and lower bone mass due to inhibition of bone formation, suggesting an important role of STAT3 in bone development. About 10% of those mice have severe spinal deformation. Histological examination of the deformed spine discloses abnormal morphology of the growth plate at the vertebral body with a delay in endochondral ossification (Fig. 1). Further, STAT3 signaling is also critical for maintaining bone homeostasis and this will be discussed later in the paper.
Figure 1. Kyphosis of a mouse with selective inactivation of STAT3 in osteoblasts/osteocytes occurs at the thoracic and lumbar regions in a radiograph (A). Histological examination at the spine (white square) shows deformity of vertebral bodies (B). Enlarged images of the histological section (yellow square) shows abnormally delayed endochondral ossicification as indication by the arrow (C). The histological section was stained with Von Kossa stain and bone tissues are stained black.
STAT2, STAT4, and STAT6 might not play a role in skeletal development because STAT2, STAT4, and STAT6 null mice are viable and fertile and show no overt phenotypic abnormalities.41-46 But, all of these three STATs may be involved in bone metabolism. For example, STAT2 may affect osteoblast indirectly via STAT1 since it heterodimerizes with STAT1 and the basal level of STAT1 is dependent on STAT2.43 STAT4 is essential for mediating responses to IL-12.42 A previous study has shown the inhibitory effect of IL-12 on macrophage colony stimulating factor/the receptor activator of nuclear factor-κB ligand-induced osteoclastogenesis.47 STAT6 is linked to inhibitory effect of IL-4 and IL-13 on osteoclast differentiation and osteoprotegerin production.48 However, STAT4 and STAT6 may play more critical role in arthritis, compared with other STATs, because STAT4 and STAT6 are induced by macrophage in synovial tissues from arthritis patients,49 which may contribute to Th1-mediated inflammation in arthritis.
STAT3 and Bone Homeostasis
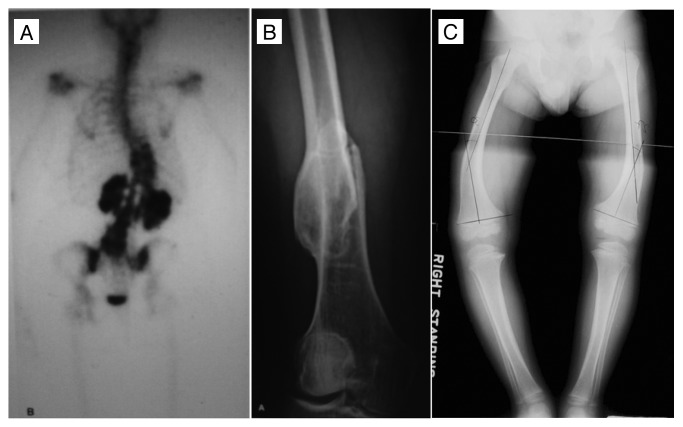
Among all the STATs, STAT3 is probably the most important transcription factor mediating intracellular signaling in osteoblasts and osteoclasts. In humans, STAT3 mutations reduce bone mass and increase incidence of minimal trauma fractures. In 2007 two independent groups of scientists from the US and Japan identified STAT3 mutation as the cause of a rare immunodeficiency disorder, Job syndrome, also known as hyperimmunoglobulin E syndrome (HIES).50,51 These two groups have identified multiple point mutations in STAT3, most of which are located in the DNA binding domain.50,51 Job syndrome, which was first reported in 1966,52 is an autosomal dominant multisystem disorder characterized by recurrent skin and pulmonary infections and extremely elevated levels of IgE in serum. Most of patients with Job syndrome have associated craniofacial and skeletal features (Fig. 2).53 Common findings among patients are recurrent fractures (in 57% of patients), hyperextensible joints (in 68% of patients), and scoliosis (in 76% of patients 16 y of age or older).54 Reduced bone mineral density is found in 64% of children and 54% of adults with HIES. Bone histology shows increased osteoclasts in spite of bisphosphonate treatment and osteomalacia.50 These clinical studies indicate that STAT3 mutation increases osteoclast number and bone resorption, suggesting that STAT3 play a role in osteoclast recruitment and activity. In addition, recurrent pathologic fractures occur most commonly in the long bones and ribs. The fractures do not occur at the same sites of typical osteoporosis in postmenopausal females. These findings indicate that STAT3 mutation has a detrimental effect on bone quality despite bone mineral content, which is likely related to mineralization. Overall, these clinical findings suggest the STAT3 mutations cause an increased bone resorption and abnormal bone formation.
Figure 2. Scoliosis (A), minimal trauma pathological fracture at femur (B) and rickets (C) in HIES patients. Adapted from Primary Immunodeficiency Diseases: A Molecular and Cellular Approach, edited by Ochs, Smith and Puck. 2006.
STAT3 mediates anabolic signals in osteoblasts and regulates bone formation. Stromal/osteoblastic cells express receptors for all members of the cytokine subfamily, such as IL-6, IL-11, etc., that share the gp130 signal transducer.55 Activation of the JAK-STAT3 signal transduction pathway promotes osteoblast differentiation, as shown by increased alkaline phosphatase activity and osteocalcin expression in vitro.19,23 Disruption of gp130 leads to defects in cartilage and bone development and metabolism. Sims et al. analyzed two lines of gp130 knock-in mice (gp130ΔSTAT/ΔSTAT and gp130Y757F/Y757F).56 gp130ΔSTAT/ΔSTAT exhibited bone shortening as a result of reduced chondrocyte differentiation. In another study using gp130−/− mice, osteoblasts are fewer in number in trabecular bone and exhibit widespread abnormalities of differentiated function.57 Osteoblasts from those mice exhibit decreased alkaline phosphatase mRNA and protein, both in vivo and in cultured osteoblasts. Because gp130ΔSTAT/ΔSTAT and gp130−/− are defective for all signals dependent on the cytoplasmic tail of gp130, including STAT3 activation, these results suggested that STAT3 is required for chondrocyte differentiation and osteoblast differentiation and function.
More direct evidence showing the role of STAT3 in osteoblasts have been acquired from bone-selective STAT3 knockout (KO) mice, in which inactivation of STAT3 occurs in osteoblasts/osteocytes. These conditional STAT3 KO mice are generated by breeding the transgenic mice in which Cre recombinase cDNA is cloned downstream of a 3.6 or 2.3 kb fragment of the rat Col1a1 promoter (Col3.6-Cre and Col2.3-Cre, respectively)58 with a strain of floxed mice in which the two loxP sites flank exons 18–20 of the STAT3 gene.59 Mice engineered with bone selective inactivation of STAT3 in osteoblasts and osteocytes exhibited significantly lower bone mineral density (7–12%, P < 0.05) and reduced ultimate force (21–34%, P < 0.01) compared with their age-matched littermate controls. This is consistent with a previous report showing that the mice with osteoblast-specific disruption of the STAT3 gene via the Cre-LoxP recombination system using α1(I)-collagen promoter Cre transgenic mice60 show an osteoporotic phenotype because of a reduced bone formation rate. These data clearly indicate that the STAT3 signal in osteoblasts plays a pivotal role in bone formation in vivo.
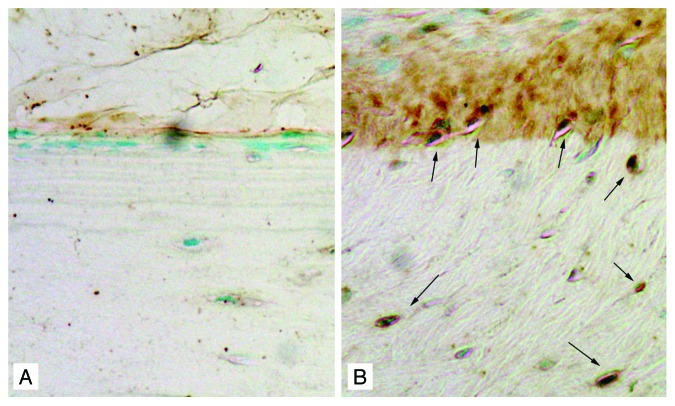
JAK-STAT pathway is involved in mechanotransduction. Mechanical Stretch activates JAK1, JAK2, Tyk2, STAT1, and STAT3 in rat cardiomyocytes.61 Our unpublished microarray data and immunohistochemical staining using a normal rat model have shown that STAT3 mRNA and protein levels markedly increase in response to mechanical loading (Fig. 3). In addition, as indicated by STAT3 phosphorylation in MC3T3-E1 osteoblastic cells, STAT3 activity significantly increases in response to fluid shear stress from 30 to 90 min.62 In order to further demonstrate the role that STAT3 plays in bone responsiveness to loading, the right ulnae of 16-week-old bone-specific STAT3 KO mice and the age-matched control mice were loaded with peak forces of 2.5 N and 2.75 N for female and male mice, respectively, at 2 Hz, 120 cycles/day for 3 consecutive days. The bone formation response was measured using histomorphometry. Mice with inactivation of STAT3 specific in bone were significantly less responsive to mechanical loading than the control mice as indicated by decreased relative mineralizing surface (rMS/BS, 47–59%, P < 0.05) and relative bone formation rate (rBFR/BS, 64–75%, P < 0.001).62 Bone responsiveness was equally decreased in mice in which STAT3 is inactivated either in early osteoblasts (Col3.6-Cre) or in mature osteoblasts (Col2.3-Cre). These data indicate loss-of-function mutations of STAT3 decrease the osteogenic response following mechanical loading.
Figure 3. Mechanical loading increase STAT3 expression in bone tissues. STAT3 positive osteoblasts at the periosteal surface of ulna and osteocytes in the cortex (indicated by arrows) were present in the loaded ulna (B) but not in the control ulna (A) by immunohistochemical staining with anti-STAT3 antibody, suggesting the involvement of STAT3 in mechanotransdcution.
STAT3 regulates cell respiration in mitochondria besides its action on gene transcription via binding specific gene-promoter sequences in nucleus. Without STAT3, for instance, an electron transport chain in mitochondria is inhibited leading to accumulation of reactive oxygen species (ROS) in osteoblasts.62 Clinical evidence suggests interplay between oxidative stress and age related bone loss. One reason is that oxidative stress antagonizes wnt signaling. Wnt molecules, on the other hand, have anabolic effect on bone formation. Indeed, ROS levels is higher in STAT3 deficient primary osteoblasts.62 It is known that mitochondrial activity is increased in response to physical activities. Therefore, high ROS level due to the lack of STAT3 in osteoblasts may antagonize bone anabolic factors and attenuate the mechanically induced new bone formation.
Future Direction
The future research includes identification of downstream genes affected by JAK-STAT pathway in osteoblasts. The STAT3 signaling upregulates the expression of receptor activator of nuclear factor κB ligand (RANKL) in osteoblasts, suggesting that the IL-6-gp130-STAT3 signal regulates the differentiation of osteoclast indirectly through the osteoblasts.63 However, the role of STAT3 in osteoclast activity in vivo has not been elucidated. This can be achieved using a conditional knockout moue model in which STAT3 inactivation occurs specifically in osteoclasts. In addition, osteocytes have been considered as the cells in bone tissue that sense the mechanical forces. All the member of IL-6 family have found in osteocytes. It is useful to find out if JAK-STAT signaling plays a role in sensing mechanical stress in bone. Further, it is of great interest to skeletal biologists understanding if the current anabolic and anti-resorptive drugs used to treat osteoporosis affect JAK-STAT signaling in osteoblasts and osteoclasts.
Glossary
Abbreviations:
- JAK
Janus kinase
- STAT
signal transducer and activator of transcription
- IL
interleukin
- IFN
interferon
- HIES
hyperimmunoglobulin E syndrome
- KO
knockout
- IGF
insulin like growth factor
- PDGF
platelet derived growth factor
- EGF
epidermal growth factor
- OSM
oncostatin M
- LIF
leukemia inhibitory factor
- CT-1
cardiotrophin-1
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/23930
References
- 1.Favus MJ. Primer on the metabolic bone diseases and disorders of mineral metabolism. Washington, DC: The American Society for Bone and Mineral Research, 2006. [Google Scholar]
- 2.Frost HM. Skeletal structural adaptations to mechanical usage (SATMU): 1. Redefining Wolff’s law: the bone modeling problem. Anat Rec. 1990;226:403–13. doi: 10.1002/ar.1092260402. [DOI] [PubMed] [Google Scholar]
- 3.Dempster DW. Anatomy and function of the adult skeleton. In: Favus MJ, ed. Primer on the metabolic bone diseases and disorders of mineral metabolism. Washington, DC: The American Society for Bone and Mineral Research, 2006:7-11. [Google Scholar]
- 4.Frost HM. Skeletal structural adaptations to mechanical usage (SATMU): 2. Redefining Wolff’s law: the remodeling problem. Anat Rec. 1990;226:414–22. doi: 10.1002/ar.1092260403. [DOI] [PubMed] [Google Scholar]
- 5.Recker R, Lappe J, Davies KM, Heaney R. Bone remodeling increases substantially in the years after menopause and remains increased in older osteoporosis patients. J Bone Miner Res. 2004;19:1628–33. doi: 10.1359/JBMR.040710. [DOI] [PubMed] [Google Scholar]
- 6.Looker AC, Orwoll ES, Johnston CC, Jr., Lindsay RL, Wahner HW, Dunn WL, et al. Prevalence of low femoral bone density in older U.S. adults from NHANES III. J Bone Miner Res. 1997;12:1761–8. doi: 10.1359/jbmr.1997.12.11.1761. [DOI] [PubMed] [Google Scholar]
- 7.Cusano NE, Bilezikian JP. Combination anabolic and antiresorptive therapy for osteoporosis. Endocrinol Metab Clin North Am. 2012;41:643–54. doi: 10.1016/j.ecl.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Duncan RL, Turner CH. Mechanotransduction and the functional response of bone to mechanical strain. Calcif Tissue Int. 1995;57:344–58. doi: 10.1007/BF00302070. [DOI] [PubMed] [Google Scholar]
- 9.Turner CH, Robling AG. Exercise as an anabolic stimulus for bone. Curr Pharm Des. 2004;10:2629–41. doi: 10.2174/1381612043383755. [DOI] [PubMed] [Google Scholar]
- 10.Turner CH, Warden SJ, Bellido T, Plotkin LI, Kumar N, Jasiuk I, et al. Mechanobiology of the skeleton. Sci Signal. 2009;2:pt3. doi: 10.1126/scisignal.268pt3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darnell JE., Jr. STATs and gene regulation. Science. 1997;277:1630–5. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 12.Jing N, Tweardy DJ. Targeting Stat3 in cancer therapy. Anticancer Drugs. 2005;16:601–7. doi: 10.1097/00001813-200507000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Heinrich PC, Behrmann I, Müller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998;334:297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senaldi G, Varnum BC, Sarmiento U, Starnes C, Lile J, Scully S, et al. Novel neurotrophin-1/B cell-stimulating factor-3: a cytokine of the IL-6 family. Proc Natl Acad Sci U S A. 1999;96:11458–63. doi: 10.1073/pnas.96.20.11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Müller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kishimoto T, Akira S, Narazaki M, Taga T. Interleukin-6 family of cytokines and gp130. Blood. 1995;86:1243–54. [PubMed] [Google Scholar]
- 17.Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19:2548–56. doi: 10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- 18.Ishihara K, Hirano T. Molecular basis of the cell specificity of cytokine action. Biochim Biophys Acta. 2002;1592:281–96. doi: 10.1016/S0167-4889(02)00321-X. [DOI] [PubMed] [Google Scholar]
- 19.Bellido T, Borba VZ, Roberson P, Manolagas SC. Activation of the Janus kinase/STAT (signal transducer and activator of transcription) signal transduction pathway by interleukin-6-type cytokines promotes osteoblast differentiation. Endocrinology. 1997;138:3666–76. doi: 10.1210/en.138.9.3666. [DOI] [PubMed] [Google Scholar]
- 20.Kido S, Kuriwaka-Kido R, Imamura T, Ito Y, Inoue D, Matsumoto T. Mechanical stress induces Interleukin-11 expression to stimulate osteoblast differentiation. Bone. 2009;45:1125–32. doi: 10.1016/j.bone.2009.07.087. [DOI] [PubMed] [Google Scholar]
- 21.Zhu J, Shimizu E, Zhang X, Partridge NC, Qin L. EGFR signaling suppresses osteoblast differentiation and inhibits expression of master osteoblastic transcription factors Runx2 and Osterix. J Cell Biochem. 2011;112:1749–60. doi: 10.1002/jcb.23094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollinger JO, Hart CE, Hirsch SN, Lynch S, Friedlaender GE. Recombinant human platelet-derived growth factor: biology and clinical applications. J Bone Joint Surg Am. 2008;90(Suppl 1):48–54. doi: 10.2106/JBJS.G.01231. [DOI] [PubMed] [Google Scholar]
- 23.Levy JB, Schindler C, Raz R, Levy DE, Baron R, Horowitz MC. Activation of the JAK-STAT signal transduction pathway by oncostatin-M cultured human and mouse osteoblastic cells. Endocrinology. 1996;137:1159–65. doi: 10.1210/en.137.4.1159. [DOI] [PubMed] [Google Scholar]
- 24.Shimoda K, Kato K, Aoki K, Matsuda T, Miyamoto A, Shibamori M, et al. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity. 2000;13:561–71. doi: 10.1016/S1074-7613(00)00055-8. [DOI] [PubMed] [Google Scholar]
- 25.Karaghiosoff M, Neubauer H, Lassnig C, Kovarik P, Schindler H, Pircher H, et al. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity. 2000;13:549–60. doi: 10.1016/S1074-7613(00)00054-6. [DOI] [PubMed] [Google Scholar]
- 26.Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, et al. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93:373–83. doi: 10.1016/S0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 27.Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, et al. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–95. doi: 10.1016/S0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 28.Neubauer H, Cumano A, Müller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/S0092-8674(00)81168-X. [DOI] [PubMed] [Google Scholar]
- 29.Krempler A, Qi Y, Triplett AA, Zhu J, Rui H, Wagner KU. Generation of a conditional knockout allele for the Janus kinase 2 (Jak2) gene in mice. Genesis. 2004;40:52–7. doi: 10.1002/gene.20063. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–86. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 31.Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, et al. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–50. doi: 10.1016/S0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 32.Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, et al. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci U S A. 1997;94:7239–44. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282:20059–63. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 34.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–42. doi: 10.1016/S0092-8674(00)81288-X. [DOI] [PubMed] [Google Scholar]
- 35.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–50. doi: 10.1016/S0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 36.Kim S, Koga T, Isobe M, Kern BE, Yokochi T, Chin YE, et al. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev. 2003;17:1979–91. doi: 10.1101/gad.1119303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tajima K, Takaishi H, Takito J, Tohmonda T, Yoda M, Ota N, et al. Inhibition of STAT1 accelerates bone fracture healing. J Orthop Res. 2010;28:937–41. doi: 10.1002/jor.21086. [DOI] [PubMed] [Google Scholar]
- 38.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A. 1997;94:3801–4. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nadiminty N, Lou W, Lee SO, Lin X, Trump DL, Gao AC. Stat3 activation of NF-kappaB p100 processing involves CBP/p300-mediated acetylation. Proc Natl Acad Sci U S A. 2006;103:7264–9. doi: 10.1073/pnas.0509808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bromberg J. Stat proteins and oncogenesis. J Clin Invest. 2002;109:1139–42. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–9. doi: 10.1016/S1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 42.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–7. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 43.Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/S1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 44.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, et al. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–30. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 45.Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–4. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 46.Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–3. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 47.Nagata N, Kitaura H, Yoshida N, Nakayama K. Inhibition of RANKL-induced osteoclast formation in mouse bone marrow cells by IL-12: involvement of IFN-gamma possibly induced from non-T cell population. Bone. 2003;33:721–32. doi: 10.1016/S8756-3282(03)00213-8. [DOI] [PubMed] [Google Scholar]
- 48.Yamada A, Takami M, Kawawa T, Yasuhara R, Zhao B, Mochizuki A, et al. Interleukin-4 inhibition of osteoclast differentiation is stronger than that of interleukin-13 and they are equivalent for induction of osteoprotegerin production from osteoblasts. Immunology. 2007;120:573–9. doi: 10.1111/j.1365-2567.2006.02538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker JG, Ahern MJ, Coleman M, Weedon H, Papangelis V, Beroukas D, et al. Expression of Jak3, STAT1, STAT4, and STAT6 in inflammatory arthritis: unique Jak3 and STAT4 expression in dendritic cells in seropositive rheumatoid arthritis. Ann Rheum Dis. 2006;65:149–56. doi: 10.1136/ard.2005.037929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–19. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 51.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–62. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 52.Davis SD, Schaller J, Wedgwood RJ. Job’s Syndrome. Recurrent, “cold”, staphylococcal abscesses. Lancet. 1966;287:1013–5. doi: 10.1016/S0140-6736(66)90119-X. [DOI] [PubMed] [Google Scholar]
- 53.Grimbacher B, Puck JM, Holland SM. Hyper-IgE Recurrent Infections Syndrome. In: Ochs HD, Smith CIE, Puck JM, eds. Primary Immunodeficiency Diseases: A Molecular & Cellular Approach. New York City, New York: Oxford University Press, Inc., 2007:496-504. [Google Scholar]
- 54.Grimbacher B, Holland SM, Gallin JI, Greenberg F, Hill SC, Malech HL, et al. Hyper-IgE syndrome with recurrent infections--an autosomal dominant multisystem disorder. N Engl J Med. 1999;340:692–702. doi: 10.1056/NEJM199903043400904. [DOI] [PubMed] [Google Scholar]
- 55.Bellido T, Stahl N, Farruggella TJ, Borba V, Yancopoulos GD, Manolagas SC. Detection of receptors for interleukin-6, interleukin-11, leukemia inhibitory factor, oncostatin M, and ciliary neurotrophic factor in bone marrow stromal/osteoblastic cells. J Clin Invest. 1996;97:431–7. doi: 10.1172/JCI118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sims NA, Jenkins BJ, Quinn JM, Nakamura A, Glatt M, Gillespie MT, et al. Glycoprotein 130 regulates bone turnover and bone size by distinct downstream signaling pathways. J Clin Invest. 2004;113:379–89. doi: 10.1172/JCI19872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shin HI, Divieti P, Sims NA, Kobayashi T, Miao D, Karaplis AC, et al. Gp130-mediated signaling is necessary for normal osteoblastic function in vivo and in vitro. Endocrinology. 2004;145:1376–85. doi: 10.1210/en.2003-0839. [DOI] [PubMed] [Google Scholar]
- 58.Liu F, Woitge HW, Braut A, Kronenberg MS, Lichtler AC, Mina M, et al. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int J Dev Biol. 2004;48:645–53. doi: 10.1387/ijdb.041816fl. [DOI] [PubMed] [Google Scholar]
- 59.Welte T, Zhang SS, Wang T, Zhang Z, Hesslein DG, Yin Z, et al. STAT3 deletion during hematopoiesis causes Crohn’s disease-like pathogenesis and lethality: a critical role of STAT3 in innate immunity. Proc Natl Acad Sci U S A. 2003;100:1879–84. doi: 10.1073/pnas.0237137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Itoh S, Udagawa N, Takahashi N, Yoshitake F, Narita H, Ebisu S, et al. A critical role for interleukin-6 family-mediated Stat3 activation in osteoblast differentiation and bone formation. Bone. 2006;39:505–12. doi: 10.1016/j.bone.2006.02.074. [DOI] [PubMed] [Google Scholar]
- 61.Pan J, Fukuda K, Saito M, Matsuzaki J, Kodama H, Sano M, et al. Mechanical stretch activates the JAK/STAT pathway in rat cardiomyocytes. Circ Res. 1999;84:1127–36. doi: 10.1161/01.RES.84.10.1127. [DOI] [PubMed] [Google Scholar]
- 62.Zhou H, Newnum AB, Martin JR, Li P, Nelson MT, Moh A, et al. Osteoblast/osteocyte-specific inactivation of Stat3 decreases load-driven bone formation and accumulates reactive oxygen species. Bone. 2011;49:404–11. doi: 10.1016/j.bone.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 63.O’Brien CA, Gubrij I, Lin SC, Saylors RL, Manolagas SC. STAT3 activation in stromal/osteoblastic cells is required for induction of the receptor activator of NF-kappaB ligand and stimulation of osteoclastogenesis by gp130-utilizing cytokines or interleukin-1 but not 1,25-dihydroxyvitamin D3 or parathyroid hormone. J Biol Chem. 1999;274:19301–8. doi: 10.1074/jbc.274.27.19301. [DOI] [PubMed] [Google Scholar]
- 64.Park SY, Saijo K, Takahashi T, Osawa M, Arase H, Hirayama N, et al. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3:771–82. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 65.Nosaka T, van Deursen JM, Tripp RA, Thierfelder WE, Witthuhn BA, McMickle AP, et al. Defective lymphoid development in mice lacking Jak3. Science. 1995;270:800–2. doi: 10.1126/science.270.5237.800. [DOI] [PubMed] [Google Scholar]