Abstract
Aberrant activation of the STAT3 transcription factor has been reported in a large group of tumors and a strong biological basis now defines this protein as an oncogenic driver. Consequently, STAT3 is considered to be a promising target in the field of cancer therapy. For its inhibition to result in a successful therapeutic approach, the definition of a target tumor population identified by specific and detectable alterations is critical. The canonical activation model of STAT3 relies on a constitutive phosphorylation on its 705 tyrosine site, resulting in its dimerization, nuclear translocation, and the consequent activation of cancer genes. Therefore, it is expected that tumors expressing this phosphorylated form are addicted to STAT3 and will be sensitive to existing drugs which are targeting this dimeric form. However, recent results have shown that STAT3 can function as an oncogene in the absence of this tyrosine phosphorylation. This indicates that different forms of the transcription factor also play an important role in tumor growth and chemotherapy resistance. This complicates the definition of STAT3 as an oncogene and as a potential prognosis and predictive biomarker. The obligation to target a defined tumor type implies that future clinical trials should use a precise definition of STAT3 activation. This will allow tumors addicted to this oncogene to be identified correctly, leading to a strong rationale for patient stratification.
Keywords: NFκB, STAT3, chemotherapy, resistance, targeted therapy
The identification of oncogenic signaling pathways that allow cell transformation, tumor growth and chemotherapy failure has greatly increased the knowledge of tumor cell biology, leading to a paradigm shift in cancer treatment. Initially performed with large populations and uncharacterized diseases, clinical trials have essentially defined chemotherapy treatments for the average patient. These trials are now moving to the field of personalized medicine and to the design of targeted therapies that should be applied to a molecularly defined tumor type and followed by predictive biomarkers.1-5 Clinical successes of these new approaches are illustrated by the targeting of the BCR-ABL fusion protein with Gleevec, of the HER2 receptor with herceptin or of the mutated form of the EGFR with erlotinib or gefitinib. Although much progress has been made in the identification of kinase or receptor inhibitors, the situation is more complex for oncogenic transcription factors such as Myc, E2F, NFκB, or STAT proteins. Transcription factors are attractive targets since they coordinate most of the oncogenic intracellular pathways. However, designing specific inhibitory drugs that can be used in a clinical setting is challenging in the absence of a reliable catalytic site. Nevertheless, recent results have demonstrated the feasability of targeting transcription factors in vivo, showing that Myc inhibition prevents lung tumor progression in vivo in mice.6
Among these potential targets, STAT3 proteins are cytoplasmic transcription factors that get phosphorylated upon activation, translocate into the nucleus and activate target genes. Besides its normal functions, this protein plays an important role in cellular transformation and tumorigenesis.7 In contrast to normal cells where its phosphorylation is transient, constitutive activation of STAT3 has been reported in several primary cancers and in many oncogene-transformed cells. Following the description of its importance in v-src signaling,8,9 one of the first pieces of evidence that STAT3 is directly oncogenic comes from reports that a constitutively-activated form induces cell transformation and tumors in mice.10 Cysteine substitutions at the 661 and 663 residues allow the formation of disulfite bridges and induce the constitutive activation of a STAT3 dimer. This protein transforms rodent fibroblasts and induces tumor growth in vivo. Equivalent modifications located within the SH2 domain (positions 657 and 658) have been recently detected in inflammatory hepatocellular adenomas. These mutants showed constitutive activity and Y705 phosphorylation in the absence of cytokine signaling.11 Similar results have been obtained in large granular lymphocytic leukemia using whole genome sequencing. In this disease, 40% of patients express a mutated form of STAT3 on its SH2 domain, and this mutation induces a constitutive phosphorylation of the transcription factor.12 Although it was initially believed that STAT3 activation only results from a constitutive phosphorylation, we might expect mutations to be discovered in other tumors.
Since most tumors express active forms of the transcription factor, STAT3 proteins are now considered to be some of the leading targets for cancer therapy.13 It is anticipated that the inhibition of this signaling pathway will be useful against a wide range of cancers. Accordingly, several inhibitors already used in clinic (such as drugs targeting the EGFR receptors, the Bcr-Abl fusion protein, or mTOR signaling for instance) are supposed to act at least in part through an indirect inactivation of the STAT3 pathway. STAT3 addiction of tumor cells has been largely demonstrated in cell lines or xenografts models, using direct or indirect inhibitors that target the oncogene or its upstream activating pathways. Several murine knockout models have confirmed that its inhibition prevents tumor growth when deleted in specific cells. For instance, the inactivation of the transcription factor in intestinal epithelial cells significantly prevents cancer progression in colitis-associated cancer.14 The specific knockdown of STAT3 in keratinocytes prevents skin tumor initiation in response to DMBA.15 Similarly, its downregulation inhibits anaplastic large cell lymphomas following inactivation by gene targeting or by antisense nucleotides.16 At the molecular level and depending on the cell type, this effect is generally explained by the inhibition of cancer cell growth, cell survival, invasion, and angiogenic signaling. Interestingly, STAT3 regulates growth in three dimensions, suggesting that its inhibition might also target the interaction between cancers cells and the microenvironment, probably through the downregulation of cytokine production and adhesion molecules.17-19
Altogether, it is now clear from these in vitro and in vivo models that most tumor cells are addicted to the STAT3 pathway and that a strong biological basis defines the oncogene as a driver candidate for the design of new targeted therapies.7 With this objective in mind, one should recall that unselected clinical trials that define treatments for the average patient have serious limitations. As a result, all clinical trial designs or analyses of cancer samples should use a precise definition of the STAT3 oncogene. This will allow a correct identification of tumors that are addicted to the transcription factor, leading to a strong rationale for patient stratification.
STAT3 activation is known to rely on the phosphorylation of a tyrosine residue (Y705) that induces its dimerization, nuclear translocation, and DNA binding. Hence, for many years, STAT3 activation has been described essentially through the analysis of this phosphorylation which has been shown to be necessary for the activation of cell cycle and survival genes such as Myc, cdc25A, survivin, or Bcl-xL.20-23 Consequently, the ability of STAT3 inhibitors to prevent cell cycle progression and to induce cell death has generally been correlated with the downregulation of the Y705 phosphorylation. Many convincing studies have described an important role of the STAT3 dimer in carcinogenesis, using either in vitro approaches or animal models. It has generally been concluded that tumors that do not express this Y705-phosphorylated form are not addicted to the STAT3 oncogene. Therefore, its activation on tumor samples has been mainly investigated through the detection of Y705 detection by immunochemistry, leading to the widespread conclusion that STAT3 is often activated in tumor samples and that this upregulation is correlated with poor prognosis and tumor failure. In the context of targeted therapies and the imperative patient stratification, this implies that future phase II clinical trials should be performed according to the prior detection of STAT3 Y705 phosphorylation on biopsies. STAT3 inhibitors, or drugs that are targeting this pathway, should not be used if the corresponding tumor does not express this tyrosine phosphorylated form of the oncogene. In addition, since STAT3 is known to play an important role in drug resistance,24 one can speculate that tumors expressing high levels of its Y705-phosphorylated form would be more resistant to genotoxic treatments. Therefore, the prior detection of this modification by immunochemistry on biopsies could also be useful in identifying tumors that are resistant to conventional chemotherapy.
These first studies have described what could be called now a canonical function of STAT3. Recent results have shown that its activity, either normal or oncogenic, does not rely solely on this Y705 phosphorylation. A second phosphorylation site located in the C-terminal domain (S727) is well known to play an important role in gene activation,25 probably through the interaction with transcriptional coactivators such as SRC, cdk9, or CBP.26,27 However, its functions are probably more complex since the S727 site is, for instance, phosphorylated in response to various cellular stresses in the absence of Y705 phosphorylation. These observations suggest that STAT3 might have different functions that do not rely on its canonical activation and that different forms of the protein might exist within the cell. Important results have recently shown that STAT3 can activate transcription in the absence of Y705 phosphorylation28 which illustrates this hypothesis. In this condition, STAT3 interacts with the NFκB subunits to induce specific cancer genes that the homodimer does not regulate by itself.29 The driving function and cooperations of the STAT3 and NFκB oncogenes have been clearly described in colorectal and breast cancers, showing in particular that these transcription factors activate a secretory phenotype that is necessary for tumor growth.14,19,30,31 Although it appears that the STAT3–NFκB interaction does not require the Y705 phosphorylation, it remains to be determined whether this association relies on the regulation of the S727 site. From a clinical point of view, this raises the question of the detection of the STAT3–NFκB complex on tumor samples if this enhances or defines aggressive tumors in the absence of Y705 phosphorylation. Proximity ligation assay can theoretically be used to detect interactions on frozen and FFPE tissues but this approach needs to be implemented by pathologists in clinical practice.
Recent results have extended these observations, showing that different oncogenic forms of STAT3 are present within cancer cells and that they might be specific to different oncogenic pathways. STAT3 phosphorylation on S727 has been described in chronic lymphocytic leukemia and is necessary for cell survival in the absence of Y705 phosphorylation.32 It has also been reported that Ras-mediated transformation is significantly reduced when STAT3 is mutated on its S727 residue.33 This was explained by a defect in the regulation of the electron transport chain and respiration.34 It is important to note that this new cytoplasmic function is observed in Ras-expressing cells but that v-src classically relies on the canonical Y705-phosphorylated form of the oncogene. Interestingly, the same differences have been reported in breast cancer. Using the MCF7 cell line, it has been shown that the survival of cancer initiating cells (using the side population as a criteria) relies on STAT3 S727 phosphorylation and on the mTOR kinase.35 This is not the case in basal-like cancer cells where the survival of CD44+CD24− breast cells depends on the opposite on the activation of STAT3 on Y705 by the JAK2 kinase.36 Consequently, a JAK2 inhibitor targets this CD44+CD24− cell population by reducing STAT3 Y705 phosphorylation. In light of these results, it is expected that drugs targeting the STAT3 pathway will be useful in inducing the death of resistant cancer stem cells. However, it seems important to use these compounds according to the activation of the mTOR-STAT3S727 or JAK2-STAT3Y705 pathways. Therefore, if phase II clinical trials are conducted to inactivate the STAT3 oncogene in breast cancer, different subtypes of this disease should not be treated with the same drugs. Different STAT3-targeting drugs may also be used to treat tumors expressing the Ras or v-src oncogenes. In addition, it will also interesting to determine if drugs targeting indirectly the STAT3 pathway, for instance through EGFR, Bcr-Abl, or mTOR inactivation, act by the downregulation of STAT3-Y705 pathway, of STAT3S727 signaling, or both. STAT3 knock-in cells reconstituted with the different mutants should be useful to investigate these questions.
Besides its importance as a new cancer target, STAT3 characterization might also be useful to identify tumors that are resistant to conventional genotoxic treatments.24 In response to irinotecan and oxaliplatin, the main treatments of colorectal cancer, STAT3 is phosphorylated on its S727 site, not on Y705, to induce the expression of DNA repair genes.37,38 Interestingly, STAT3 phosphorylation—on S694—has also been detected on a phospho-proteomic screen of ATM/ATR targets following DNA damage.39 In addition, genetic knockdown experiments performed in Drosophila have reported that STAT92E, when non phosphorylated, plays an important role in the protection against DNA damage through its interaction with HP1 and the regulation of heterochromatin formation.40,41 Thus, like STAT142,43 and STAT5,44,45 STAT3 is connected to DNA damage and repairs signaling pathways. Immunochemistry experiments performed in our laboratory have shown that a significant proportion of tumor cells are phosphorylated on this S727 site in colorectal tumors, before any chemotherapy treatment. We speculate that this is related to the genetic instability and DNA damage that occur early in this disease. Therefore, it will be interesting to determine if this S727 phosphorylation is useful to define tumors that have enhanced DNA repair capacities and are resistant to conventional genotoxic treatments.
Besides the STAT3 Y705 and S727 sites, several other post-translational modifications have been described and the transcription factor can be acetylated or methylated. Following cytokine treatment, STAT3 is acetylated by CBP on its K685 residue and inhibited by the HDAC1 and HDAC2 histone deacetylases.46 This acetylation does not affect the phosphorylation of STAT3 but is necessary for a stable dimer formation, for DNA binding and for the induction of genes such as cyclin D1, Myc, or Bcl-xL. This K685 acetylation has been detected in melanoma and colorectal cancer tissue using immunochemistry.47 Interestingly, its mutation decreased tumor growth in vivo and this effect was associated with promoter demethylation and with an enhanced expression of several tumor suppressor genes, including p53. This was explained by the interaction between STAT3 and DNMT1 and the consequent promoter methylations and gene silencing. From these results, it appears that the K685 site also plays an important role in cancer growth besides Y705 and S727, thus questioning whether this methylation defines the same oncogenic activities and target genes and whether it is correlated with the same prognosis as the Y705 phosphorylation.
Other results have shown that STAT3 is also dimethylated by Set9 on K140 and inactivated by the LSD1 demethylase.48 This modification reduces the phosphorylation of the transcription factor on Y705 and its DNA binding and transcriptional activities. Interestingly, the K140 dimethylation is a nuclear event that can be detected on promoters and that requires prior DNA binding and S727 phosphorylation. Importantly, the Set9-mediated regulation of the transcription factor only occurs on a subset of genes, suggesting that Set9 could regulate the specificity of STAT3 transcription activity. It will be interesting to determine whether this pathway is involved in the function of the oncogene, and whether it affects its regulation of cancer genes or the response to chemotherapy. As stated above, if the K140 dimethylation is detectable on tumor samples, it could be used to stratify the disease and it will be interesting to determine whether it overlaps with the information provided by the S727, Y705, and K685 staining.
Another important result of this study was that this modification was detected not only by western blot but also by chromatin immunoprecipitation (ChIP) experiments. Different phosphorylated forms of STAT3 can be detected by ChIP on specific promoters in response to DNA damage.37,38 It is important to point out that western blot results used to describe phosphorylation pathways might essentially characterize soluble proteins. Depending on the method, transcription factors which are bound to their DNA sequences are not always released by common extraction procedures. If one (wrongly, see above) assumes that the oncogenic functions of STAT3 are only related to its transcriptional activation of cancer genes, then chromatin immunoprecipitation experiments should be used to define its active oncogenic modifications. DNA-bound forms of the transcription factors might have different modifications patterns as compared with the soluble forms isolated from classical extracts. If this hypothesis is correct, the conventional lysis used for western blot might miss which active forms of the transcription factor are truly involved in the activation of cancer genes.
In light of this important information, the STAT3 paradigm has been modified and the Y705 phosphorylation should no longer be considered a unique marker of activation. The S727, K170, and K685 sites and other modifications yet to be discovered play an important role in STAT3 functions and, as such, are expected to affect its oncogenic functions. Although this remains to be fully demonstrated, one can speculate that STAT3 activates different transcriptional programs depending on its posttranslational modifications and specific partners. If this holds true, it is not obvious that all forms of STAT3 will have equivalent oncogenic activity, nor that all target genes will have the same impact on tumor progression. As a consequence, STAT3 activation on tumor samples should not only be investigated through Y705 detection by immunochemistry. It is not certain that the known correlations between this oncogene and bad prognosis will be identical once all modifications have been taken into account.
Surprisingly, recent publications have reported tumor suppressive functions for STAT3. As stated above, this oncogene is known to be constitutively activated and to play an important role in colorectal cancer, both using in vitro and in vivo models.14 Although STAT3 is necessary for early adenoma formation, its inactivation promotes late tumor progression in an APC+/− mouse model of colorectal cancer,49 which is surprising. In thyroid cancer, its activation is correlated with reduced metastasis, and its inactivation with enhanced tumor growth.50 This putative tumor suppressive function of STAT3 has previously been suggested on a molecular level. Although STAT3 is well known to induce cell cycle and survival genes, it also activates cell cycle inhibitors such as p21waf1, which is rather counterintuitive for an oncogene involved in the abnormal activation of cell cycle. This is generally explained by stating that results are cell type-specific, but these potential opposite functions could also be explained by different post-translational modifications. This would allow the formation of suppressive or oncogenic enhanceosomes, leading to the expression of specific genes involved in cell cycle arrest or on the opposite in abnormal proliferation.
This certainly complicates the STAT3 oncogenic field because one can speculate that different forms of the oncogene are present within cancer cells, that this might vary during the different stages of the disease or in response to treatment or as a consequence of tumor adaptation. In addition, it is also known that STAT3 phosphorylation is heterogeneous in tumor samples. The Y705-phosphorylated form of STAT3 is known to be highly expressed at the leading edge of the tumor,7 probably as a result of increased cytokine production and inflammation. In this case, drugs targeting the dimeric form of STAT3 are expected to be useful on the specific subpopulation located at the edge of the tumor but not necessarily at its core. However, we believe that this does not mean that the other regions of the tumor do not rely on STAT3. As stated above, the oncogene can interact with NFκB in the absence of Y705 phosphorylation or can eventually be modified on the S727, K170, or K685 sites. It is obvious that a drug targeting the dimer and its Y705 phosphorylation will probably be useless if a tumor depends on a STAT3 modification that does not imply its dimeric form.
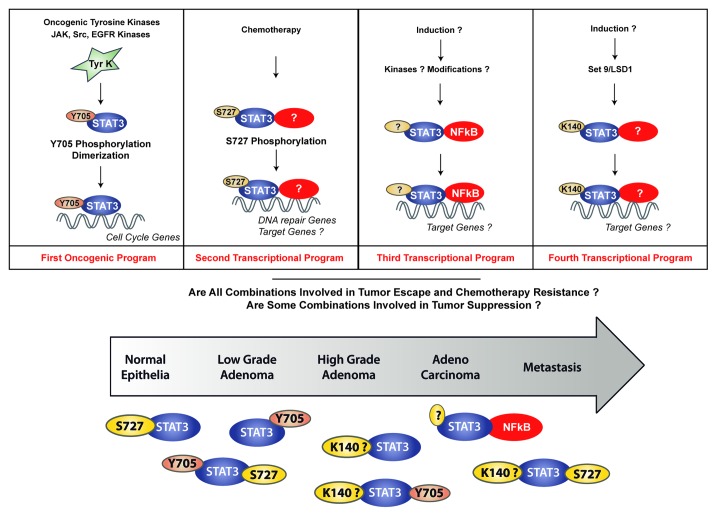
We believe that it is important that we now characterize the different forms of the transcription factor and to determine if they have the same partners, the same target genes and identical functions. If STAT3 inhibition is to result in a useful therapeutic approach, one of the critical steps will be the definition of a target population identified by its specific alterations. We might discover that some modifications have a different impact on its oncogenic functions and that different targeted therapies should be used, according to the prior detection of a specific form (see Fig. 1 for a proposed hypothesis). It will be also important to show whether these different activations are correlated with tumor stages and with the resistance to a particular chemotherapy. STAT3 inhibitors could be used both as targeted therapies and powerful agents used in combination with genotoxic treatments, provided that STAT3 detection can be standardized in a clinical context to define a resistant population.
Figure 1. A different oncogenic role for different STAT3 modifications?
Acknowledgments
Work in our laboratory is supported by grants from the Ligue Contre le Cancer (comité du Maine et Loire), the Canceropole Grand Ouest, the Region Pays de la Loire and the Agence Nationale de la Recherche. We thank Jill Hunter and Jacqueline Butterworth for their critical reading of the original text.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/24716
References
- 1.de Bono JS, Ashworth A. Translating cancer research into targeted therapeutics. Nature. 2010;467:543–9. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 2.Bernards R. It’s diagnostics, stupid. Cell. 2010;141:13–7. doi: 10.1016/j.cell.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 3.van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–70. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 4.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–52. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 5.Sawyers C. Targeted cancer therapy. Nature. 2004;432:294–7. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 6.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–83. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sansone P, Bromberg J. Targeting the interleukin-6/Jak/stat pathway in human malignancies. J Clin Oncol. 2012;30:1005–14. doi: 10.1200/JCO.2010.31.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JEJ., Jr. Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–8. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–52. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/S0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 11.Pilati C, Amessou M, Bihl MP, Balabaud C, Nhieu JT, Paradis V, et al. Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas. J Exp Med. 2011;208:1359–66. doi: 10.1084/jem.20110283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmäki H, Andersson EI, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366:1905–13. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 14.Bollrath J, Phesse TJ, von Burstin VA, Putoczki T, Bennecke M, Bateman T, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Chan KS, Sano S, Kiguchi K, Anders J, Komazawa N, Takeda J, et al. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J Clin Invest. 2004;114:720–8. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R, et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med. 2005;11:623–9. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- 17.Leslie K, Gao SP, Berishaj M, Podsypanina K, Ho H, Ivashkiv L, et al. Differential interleukin-6/Stat3 signaling as a function of cellular context mediates Ras-induced transformation. Breast Cancer Res. 2010;12:R80. doi: 10.1186/bcr2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bollrath J, Greten FR. IKK/NF-kappaB and STAT3 pathways: central signalling hubs in inflammation-mediated tumour promotion and metastasis. EMBO Rep. 2009;10:1314–9. doi: 10.1038/embor.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirogane T, Fukada T, Muller JM, Shima DT, Hibi M, Hirano T. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity. 1999;11:709–19. doi: 10.1016/S1074-7613(00)80145-4. [DOI] [PubMed] [Google Scholar]
- 21.Fukada T, Ohtani T, Yoshida Y, Shirogane T, Nishida K, Nakajima K, et al. STAT3 orchestrates contradictory signals in cytokine-induced G1 to S cell-cycle transition. EMBO J. 1998;17:6670–7. doi: 10.1093/emboj/17.22.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barré B, Vigneron A, Coqueret O. The STAT3 transcription factor is a target for the Myc and riboblastoma proteins on the Cdc25A promoter. J Biol Chem. 2005;280:15673–81. doi: 10.1074/jbc.M413203200. [DOI] [PubMed] [Google Scholar]
- 23.Barré B, Avril S, Coqueret O. Opposite regulation of myc and p21waf1 transcription by STAT3 proteins. J Biol Chem. 2003;278:2990–6. doi: 10.1074/jbc.M210422200. [DOI] [PubMed] [Google Scholar]
- 24.Barré B, Vigneron A, Perkins N, Roninson IB, Gamelin E, Coqueret O. The STAT3 oncogene as a predictive marker of drug resistance. Trends Mol Med. 2007;13:4–11. doi: 10.1016/j.molmed.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Wen Z, Zhong Z, Darnell JE., Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–50. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 26.Giraud S, Hurlstone A, Avril S, Coqueret O. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene. 2004;23:7391–8. doi: 10.1038/sj.onc.1207972. [DOI] [PubMed] [Google Scholar]
- 27.Giraud S, Bienvenu F, Avril S, Gascan H, Heery DM, Coqueret O. Functional interaction of STAT3 transcription factor with the coactivator NcoA/SRC1a. J Biol Chem. 2002;277:8004–11. doi: 10.1074/jbc.M111486200. [DOI] [PubMed] [Google Scholar]
- 28.Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, et al. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–47. [PubMed] [Google Scholar]
- 29.Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007;21:1396–408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–96. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hazan-Halevy I, Harris D, Liu Z, Liu J, Li P, Chen X, et al. STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells. Blood. 2010;115:2852–63. doi: 10.1182/blood-2009-10-230060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–6. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–7. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, Wulfkuhle J, Zhang H, Gu P, Yang Y, Deng J, et al. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci U S A. 2007;104:16158–63. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44⁺CD24⁻ stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121:2723–35. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Courapied S, Sellier H, de Carné Trécesson S, Vigneron A, Bernard AC, Gamelin E, et al. The cdk5 kinase regulates the STAT3 transcription factor to prevent DNA damage upon topoisomerase I inhibition. J Biol Chem. 2010;285:26765–78. doi: 10.1074/jbc.M109.092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vigneron A, Gamelin E, Coqueret O. The EGFR-STAT3 oncogenic pathway up-regulates the Eme1 endonuclease to reduce DNA damage after topoisomerase I inhibition. Cancer Res. 2008;68:815–25. doi: 10.1158/0008-5472.CAN-07-5115. [DOI] [PubMed] [Google Scholar]
- 39.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 40.Yan SJ, Lim SJ, Shi S, Dutta P, Li WX. Unphosphorylated STAT and heterochromatin protect genome stability. FASEB J. 2011;25:232–41. doi: 10.1096/fj.10-169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi S, Larson K, Guo D, Lim SJ, Dutta P, Yan SJ, et al. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol. 2008;10:489–96. doi: 10.1038/ncb1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Townsend PA, Cragg MS, Davidson SM, McCormick J, Barry S, Lawrence KM, et al. STAT-1 facilitates the ATM activated checkpoint pathway following DNA damage. J Cell Sci. 2005;118:1629–39. doi: 10.1242/jcs.01728. [DOI] [PubMed] [Google Scholar]
- 43.Thomas M, Finnegan CE, Rogers KM, Purcell JW, Trimble A, Johnston PG, et al. STAT1: a modulator of chemotherapy-induced apoptosis. Cancer Res. 2004;64:8357–64. doi: 10.1158/0008-5472.CAN-04-1864. [DOI] [PubMed] [Google Scholar]
- 44.Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO, et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8:795–806. doi: 10.1016/S1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 45.Slupianek A, Hoser G, Majsterek I, Bronisz A, Malecki M, Blasiak J, et al. Fusion tyrosine kinases induce drug resistance by stimulation of homology-dependent recombination repair, prolongation of G(2)/M phase, and protection from apoptosis. Mol Cell Biol. 2002;22:4189–201. doi: 10.1128/MCB.22.12.4189-4201.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–73. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 47.Lee H, Zhang P, Herrmann A, Yang C, Xin H, Wang Z, et al. Acetylated STAT3 is crucial for methylation of tumor-suppressor gene promoters and inhibition by resveratrol results in demethylation. Proc Natl Acad Sci U S A. 2012;109:7765–9. doi: 10.1073/pnas.1205132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci U S A. 2010;107:21499–504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Musteanu M, Blaas L, Mair M, Schlederer M, Bilban M, Tauber S, et al. Stat3 is a negative regulator of intestinal tumor progression in Apc(Min) mice. Gastroenterology. 2010;138:1003–11, e1-5. doi: 10.1053/j.gastro.2009.11.049. [DOI] [PubMed] [Google Scholar]
- 50.Couto JP, Daly L, Almeida A, Knauf JA, Fagin JA, Sobrinho-Simões M, et al. STAT3 negatively regulates thyroid tumorigenesis. Proc Natl Acad Sci U S A. 2012;109:E2361–70. doi: 10.1073/pnas.1201232109. [DOI] [PMC free article] [PubMed] [Google Scholar]