Abstract
The conservation of signaling cascades between humans and Drosophila, over more than 500 million years of evolutionary time, means that the genetic tractability of the fly can be used to its full advantage to understand the functional requirements for JAK-STAT pathway signaling across species. Here we review the background to how the pathway was first identified and the first characterization of JAK-STAT pathway phenotypes in the Drosophila system, highlighting the molecular, functional, and disease-related conservation of the pathway.
Identification of the Drosophila JAK-STAT Pathway
The molecular characterization of the JAK-STAT pathway has recently celebrated its 20th birthday, a milestone that marks the discovery of the tyrosine kinases TYK2 and JAK1 and the STAT1 and STAT2 transcription factors as the key factors underlying the cellular response to type I interferons.1 The initial description of the first JAK-STAT pathway components sparked a flurry of activity in multiple labs, which rapidly characterized a wide range of ligands, receptors, four JAKs, and seven STATs present in vertebrate cells. Collectively these factors signal through fundamentally similar mechanisms; a JAK-STAT pathway that now serves as a textbook example of how extracellular ligands can act at the cell surface to modulate nuclear gene expression.2-4
Hopscotch—the fly JAK
In contrast to the key role played by the Drosophila system in identifying many other key cellular signaling cascades, the initial steps leading to the identification of the Drosophila JAK-STAT pathway components lagged pioneering work being undertaken in vertebrate cell-based systems. However, the ball was set rolling in 1994 when the Perrimon lab cloned a novel gene termed Hopscotch (Hop), which encodes a maternally supplied protein required for the patterning of the embryonic cuticle (for an example of the LOF phenotype see Fig. 1) and the proliferation of diploid imaginal cells.5,6 Cloning of Hop identified it as a 1177 amino acid non-receptor tyrosine kinase, expressed throughout development, with a kinase domain, sharing 39% identity with JAK1, JAK2, and Tyk2, and an overall identity of 27% to JAK2. While not necessarily apparent at the time, the identification of JAK and the characteristic segmentation phenotype associated with pathway mutants represented a key insight and the first step on the path toward identifying the rest of the pathway.
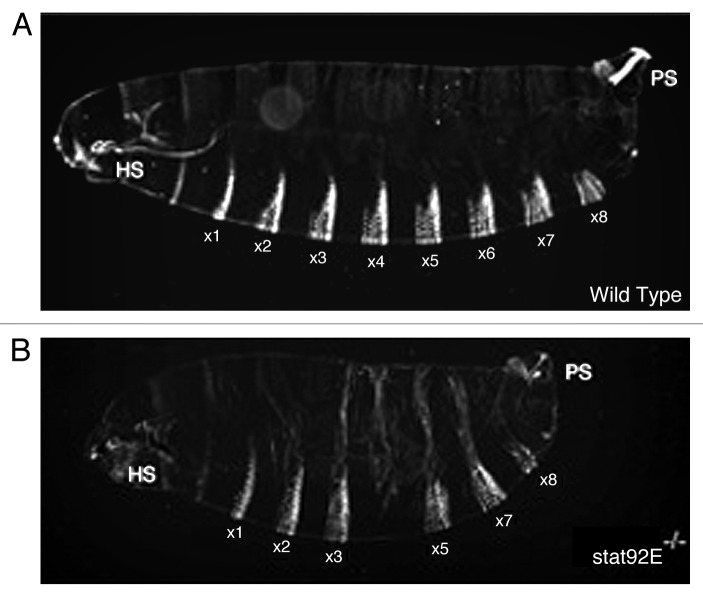
Figure 1. The embryonic cuticle phenotype associated with wild-type (A) and loss of JAK-STAT pathway activity (B)—in this case involving the removal of maternally contributed STAT92E. Head skeleton (HS) and posterior spiracles (PS) are both disrupted in pathway mutants. In addition, of the abdominal segments 1 to 8 (a1 to a8) a4 and a5 are missing and a8 reduced in the stat92E mutant. Figure reproduced with permission from reference 49.
The Drosophila STAT
Encouraged by the presence of at least one JAK kinase in Drosophila, and following a similar biochemical approach to that successfully employed in vertebrate cells, STAT-like activities were also soon demonstrated in vanadate/peroxide stimulated Drosophila S2 cells.7 This activity was identified on the basis of its ability to bind to a gamma-interferon response region containing a consensus STAT1 binding sequence. Molecular cloning and characterization of Drosophila STAT92E (then termed marelle and D-stat) was subsequently announced in two back-to-back Cell papers in 1996.8,9 Following up on the biochemical approach, the Darnell lab used a low stringency-PCR to clone Drosophila STAT, a transcription factor sharing its overall domain structure, and 33% overall amino acid identity, with human STAT5. They showed that STAT92E can be phosphorylated on a tyrosine residue conserved in vertebrate STATs and is able to bind to a consensus palindromic DNA sequence which is present in the promoter of the pair-rule gene evenskipped (eve)9—an insight into the segmentation phenotypes characteristic of fly JAK-STAT pathway mutants.10 By contrast, the parallel identification of STAT92E in the Perrimon lab followed the more traditional genetic approach involving a large-scale screen for autosomal lethal mutations associated with specific maternal-effect phenotypes.11 This screen identified P-element insertions in the STAT92E locus. In addition to allowing the rapid cloning of the STAT92E coding region, the insertional mutation also gave a segmentation phenotype (Fig. 1) very similar to that associated with maternal loss of Hop and a stripe-specific disruption in the expression of runt and eve. Epistatic analysis showed that STAT92E genetically interacts with weak Hop mutants in a manner consistent with STAT92E representing a bona fide downstream component of the pathway.8
The Outstretched/Unpaired family of ligands
Although originally named on the basis of a regulatory allele that produces an adult outstretched wing phenotype,12 amorphic alleles of the unpaired (upd) locus were first described in the Nobel Prize-winning saturation screen for loci affecting embryonic segmentation.13 Showing an atypical gap gene phenotype (described below and similar to Fig. 1A), which was later to be associated with loss of Hop and STAT92E, upd alleles were subsequently analyzed by generating mosaic mutant embryos. In this elegant example of genetic analysis, Gergen and Wieschaus showed that the protein encoded by the upd locus is likely to encode a diffusible factor that acts before gastrulation to influence the action of other genes.14 Based on the embryonic segmentation phenotype, as well as the conclusions of the genetic analysis,15 the Perrimon lab went on to characterize and clone the Upd locus. In addition to interacting genetically with hop mutants, Upd protein was found to be a dynamically expressed, secreted and glycosylated protein that interacts with the ECM and is able to activate both Hop and STAT92E in cell based systems.16-18 While much attention was focused on Upd as the “primary” JAK-STAT pathway ligand, publication of the Drosophila genome19 showed that two additional homologous genes are located in the genomic region surrounding Upd. Of these, upd2 is expressed in the same pattern as upd and encodes a secreted molecule that does not appear to be ECM associated, and thus has the potential to act systemically in vivo.17 While upd2 mutants are viable, expression of Upd2 can partially rescue the lethality of the updYM55 nonsense mutation17 and has recently been shown to play a pivotal role in nutrient sensing and insulin release.20 The third Drosophila JAK-STAT ligand, Upd3, is also secreted and able to interact with the ECM;21 however, its expression is more restricted with roles in the response to septic injury22 and the differentiation of lamellocytes in the developing lymph gland23 having been described.
The search for a receptor
Despite the identification of Hop in 1994, STAT92E in 1996, and the first characterization of Upd ligands in 1998, the identity of the pathway receptor remained unclear for some years. Indeed, even the publication of the Drosophila genome in 2000 and knowledge of vertebrate receptors such as GP130 and IL6R, searches based on sequence similarities were unable to identify the “missing” receptor. However, once again the embryonic segmentation phenotype, characteristic of JAK-STAT pathway mutants, gave the clue. In the Hombria lab, partial cloning of a protein with Fibronectin type III domains, whose loss affected embryonic posterior spiracles (structures also affected in hop and stat92E mutants), prompted the generation of germline clones, giving the loss of A5 cuticle phenotype.24 Biochemical characterization of the Domeless (Dome) locus showed that it encoded a transmembrane protein with homology to GP130 that is absolutely required for the expression of JAK-STAT pathway target genes.24 A later, biochemical identification of Dome by the Hou lab, confirmed these findings and demonstrated physical interactions of Dome with STAT92E and its requirement for Upd-induced phosphorylation of pathway components.25 Strikingly, while vertebrate systems contain many receptors, the three JAK-STAT pathway ligands present in flies, all signal through the same receptor.
This, however, turned out not to be the full story following the analysis of a gene lying immediately adjacent to dome that had originally been described as encoding a short Dome-like protein lacking an intracellular tail.26 In two recent reports, this locus has been shown to act as a negative regulator of pathway signaling both by RNAi screening and in vivo.23,27 Termed latran/eye transformer, this co-receptor contains a putative cytokine binding motif and can be co-immunoprecipitated with Dome and Hop, while its knockdown in S2 cells leads to increased STAT92E phosphorylation and transcriptional activity.27 The Crozatier group extended analysis of Latran to the lymph gland where its upregulation is a specific response to infestation by the parasitic wasp Leptopilina boulardi. They showed that downregulation of JAK-STAT pathway activity within the lymph gland is not only mediated by increased Latran expression, but also that this decrease is an essential for the differentiation of the lamellocyte blood cell lineage (see the review by Morin-Poulard et al., in this issue). Given the multitude of mechanisms by which the pathway can be downregulated (described below), it is intriguing that a negative co-receptor has evolved to regulate a single differentiation event in one tissue—it will be intriguing to see if Latran ultimately turns out to have, as yet, undiscovered secrets.
Negative pathway regulators
Following the cloning and initial characterization of the core Drosophila JAK-STAT pathway components, identification of potential regulators based on homology to the vertebrate pathway followed rapidly. This included the discovery of a Drosophila PIAS homolog as a negative modulator of STAT92E and a regulator of chromosomal integrity28,29 and the identification of three Drosophila SOCS family members (reviewed in ref. 30). Of these, SOCS36E has been best characterized both as a transcriptional target of the pathway, and as a potent negative regulator of not only JAK-STAT, but also EGFR, pathways.31-34
Finally, the tyrosine phosphatase, Ptp61F, was one of the last components to be discovered; identified as a potent negative regulator of JAK-STAT pathway signaling by two independent genome-wide RNAi screens,18,35 Ptp61F nonetheless represents the least well characterized regulator. Indeed, the existence of multiple splice forms and differing results has led to both STAT92E and the Hop/Dome receptor complex being proposed as substrates.31-34 Indeed, given that Dome, Hop, and STAT92E are all tyrosine phosphorylated, it is likely that other phosphatases targeting each component are functioning in vivo. More research into the dynamics of phosphatase activity, the identity of Ptp61F substrates, the potential role of the Drosophila SHP2 homolog, Corkscrew, and the validity of other phosphatases identified by RNAi screens all remain to be determined.
Drosophila JAK-STAT Phenotypes
The phenotypes and genetic interactions associated with JAK-STAT signaling form a key aspect of the reviews in this issue. While the cloning of the Drosophila JAK, STAT, ligand, and receptor may have lagged progress in vertebrate systems, the developmental genetic analysis available in the Drosophila system has made it one of the most powerful systems in which to examine the phenotypes associated with pathway disruption. Indeed, some of the first pathway phenotypes to be defined emerged from studies in the fruit fly.
Outstretched wings and small eyes
Drosophila geneticists, working over 80 years ago, are likely to have been the first scientists to observe and describe a JAK-STAT pathway phenotype. Named on the basis of their viable loss of function phenotypes, flies with outstretched wing posture and small eye phenotypes were collected and described.12 Only since the development of suitable genetic tools have these been shown to be viable hypomorphic alleles of the upd locus. Indeed, only very recently has the outstretched wing phenotype of adult wings held almost at 90° to the main body axis been shown to be caused by a defect in the development of the adult wing hinge.36 Characterization of outstretched alleles, show the loss of hinge structures in the dorsal adult wing, to result from the loss of one of five upd expression domain within the wing imaginal disc. Given the co-localization of the missing upd expression domain within a fold that also lies close to the fate mapped position of the missing hinge structure, it seems likely that the basis of this phenotype has now been solved.
While no molecular or gene expression changes associated with the small eye phenotype of the original upd alleles have been described, a requirement for the JAK-STAT pathway in the proliferation of cells within the developing eye imaginal disc likely provides an explanation for the effect.37 Consistent with this, the original characterization of Hop noted that “all larval diploid imaginal tissues are reduced in size, thus implying a zygotic role for hop in cellular proliferation”.6
Atypical gap-gene phenotypes
Described some years later,13 the embryonic segmentation phenotype associated with the loss of Upd, and indeed all core JAK-STAT pathway components, was a key factor in the identification of several pathway components. The defect is most readily characterized by the deletion of the fifth abdominal denticle belt (a5) and the posterior/mid-ventral portion of the fourth abdominal denticle belt (a4). Loss of pathway activity also leads to defects in the thoracic segments, the head skeleton, a8 and tail regions including the posterior spiracles (Fig. 1). Fusions of the sixth and seventh abdominal segments are also occasionally observed. Although clear in retrospect, the variable expressivity of the phenotype and the nature of the phenotype, which falls outside the broad “maternal, gap, pair-rule, or segment polarity” categorizations, lead to initial confusion and the characterization of upd alleles as representing an “atypical gap gene”.13 Although the expression of gap genes appears normal in Hop mutants, stripe-specific defects in the expression patterns of the pair-rule genes even-skipped (eve), runt, and fushi tarazu (ftz) are found.6,10 While not characterized within the promoters controlling every missing stripe, in the case of eve, JAK-STAT pathway activity acts as a transcriptional activator via a pair of STAT92E binding sites identified within a 500 bp eve stripe 3 promoter region.9 As expected, STAT92E is able to bind to both sites in in vitro mobility shift assays and their mutation in a reporter construct is sufficient to ablate stripe 3 eve expression.
While it seems likely that similar STAT92E binding activities control runt and ftz the precise underpinnings of other defects in the head skeleton and thoracic segments remain less clear. However, insights into the roles of JAK-STAT signaling in the posterior spiracles is certainly making progress with new findings into the cell biology underlying the formation of these structures a subject of a dedicated review in this issue.
Hemocyte overproliferation
While loss-of-function mutations in JAK-STAT pathway components provide valuable information about the developmental processes that require STAT92E, insights gained from gain-of-function mutations have provided intriguing insights into mechanisms relevant to human disease. Initially identified on the basis of a dominant temperature sensitive melanotic tumor phenotype,38,39 cloning of the mutation identified the lesion as an amino acid substitution within Hop which generates a potent gain-of-function effect.40 Phenotypically, this gain-of-function allele, termed HopTumorous lethal (HopTuml), results in the over-proliferation of hemocytes within the developing larva and the inappropriate differentiation of these hemocytes toward the lamellocyte lineage—a blood cell type that normally specializes in the encapsulation of parasitic wasp eggs. When present in excess, and in the absence of their normal targets, lamellocytes, in HopTuml backgrounds, encapsulate one another and rapidly form large melanized cell masses (Fig. 2). Furthermore, this phenotype is not an oddity of this one allele, but is also recapitulated by a second gain-of-function Hop allele, HopT42. HopT42 contains a substitution in a conserved residue present in the JH2 regulatory domain,41 a substitution that, then when mirrored in JAK2, is also sufficient to activate the vertebrate homolog.42
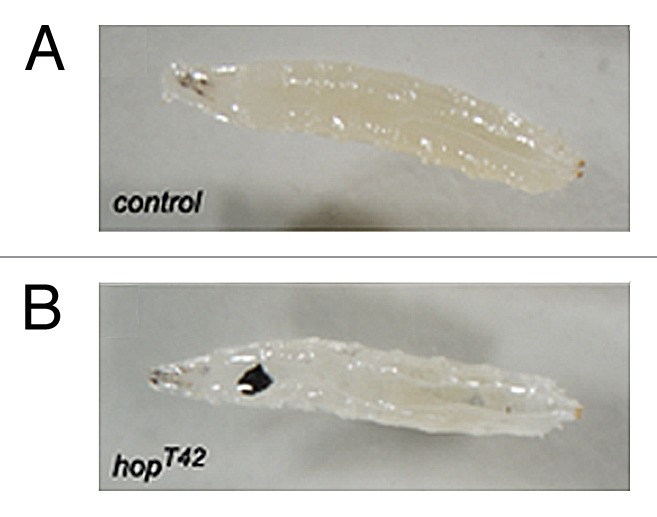
Figure 2. Third instar larvae either wild-type for the Hop locus (A) or carrying one copy of the gain-of-function HopT42 allele (B). Larvae with activated Hop alleles have increased numbers of circulating hemocytes which inappropriately differentiate into lamellocytes which then frequently form black melanized tumors (visible in [B]). See text for details.
Although multiple human leukemias and myelomas have long been associated with constitutive activation of the JAK-STAT pathway,43 the key significance of the HopTuml and HopT42 phenotypes was highlighted by the 2005 discovery of the human JAK2 V617F mutations.44-46 Reported essentially simultaneously by three groups, discovery of human JAK2 V617F substitutions as causative for the majority of myeloproliferative neoplasias has revolutionized the JAK-STAT field. The V617F substitution, also located within the JH2 domain of JAK2, results in constitutive activation of the molecule and the massive uncontrolled over proliferation of blood cells. The parallels between gain-of-function Hop alleles, and HopT42 in particular, with human JAK2 V617F, is indeed striking, and suggests that evolutionary conservation of the JAK-STAT pathway extends not just to the molecules, or even the normal functions such as hematopoiesis, but also to the gain-of-function disease states they can cause. Indeed, while the vertebrate field search for mechanisms to inhibit JAK activity in human disease, Drosophila has once again been used to raise the bar providing fundamental insights into non-canonical aspects of JAK-STAT pathway signaling arising from the study of HopTuml alleles47,48—a further subject of a review in this issue.
Summary
While not originally discovered or characterized in Drosophila, the rapid identification of a complete intact, low complexity ligand/receptor/JAK-STAT pathway in this genetically tractable and developmentally characterized model system, has provided a foundation for a broad and dynamic field. This special review issue summarizes some of the latest discoveries in the field and highlights how simple animal models remain at the forefront of research into the fundamentals of biology and human disease. We do indeed live in interesting times.

About Dr Martin Zeidler. Dr Zeidler received a BSc from the University of Sussex and undertook his PhD research at the European Molecular Biology Laboratory (EMBL) in Heidelberg Germany. Working on the developmental genetics of Drosophila, he then moved to the laboratory of Prof Norbert Perrimon at Harvard Medical School where he first started studying the roles of JAK-STAT pathway signaling in the fruit fly. Founding his own lab at the Max Planck Institute for Biophysical Chemistry in Göttingen, Germany, he was involved in genetic and genome-scale RNAi screens for regulators of JAK-STAT pathway activity. Moving to the University of Sheffield in 2006, he holds a Cancer Research UK Senior Cancer Research Fellowship and works within the MRC Centre for Developmental and Biomedical Genetics trying to work out what all the JAK-STAT regulatory genes actually do.

About Dr Nina Bausek. Dr Nina Bausek received an MSc from the University of Vienna, Austria. She was selected for the joint PhD program of the Institute of Molecular Pathology (IMP) and University of Vienna, Austria, studying the avian zona pellucida. Changing countries and model organism, she joined the lab of Dr Helen White-Cooper at the Department of Zoology, Oxford University to work on spermatogenesis-specific genes in Drosophila. Appreciating the scientific advantages of the fruit fly, she moved to the lab of Dr Martin Zeidler at the University of Sheffield, where she is funded by Cancer Research UK to specifically work on JAK-STAT downstream targets.
Acknowledgments
The authors wish to thank Stephen Brown, Norbert Perrimon, Katie Fisher, and other members of the Zeidler lab for their discussions and helpful comments. NB is supported by a Cancer Research-UK Senior Cancer Research fellowship held by MZ.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/25353
References
- 1.Stark GR, Darnell JE., Jr. The JAK-STAT pathway at twenty. Immunity. 2012;36:503–14. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohr A, Chatain N, Domoszlai T, Rinis N, Sommerauer M, Vogt M, et al. Dynamics and non-canonical aspects of JAK/STAT signalling. Eur J Cell Biol. 2012;91:524–32. doi: 10.1016/j.ejcb.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Harrison DA. The Jak/STAT pathway. Cold Spring Harb Perspect Biol. 2012;4:a011205. doi: 10.1101/cshperspect.a011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–16. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- 5.Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 1994;8:300–12. doi: 10.1101/gad.8.3.300. [DOI] [PubMed] [Google Scholar]
- 6.Perrimon N, Mahowald AP. l(1)hopscotch, A larval-pupal zygotic lethal with a specific maternal effect on segmentation in Drosophila. Dev Biol. 1986;118:28–41. doi: 10.1016/0012-1606(86)90070-9. [DOI] [PubMed] [Google Scholar]
- 7.Sweitzer SM, Calvo S, Kraus MH, Finbloom DS, Larner AC. Characterization of a Stat-like DNA binding activity in Drosophila melanogaster. J Biol Chem. 1995;270:16510–3. doi: 10.1074/jbc.270.28.16510. [DOI] [PubMed] [Google Scholar]
- 8.Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–9. doi: 10.1016/S0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- 9.Yan R, Small S, Desplan C, Dearolf CR, Darnell JEJ., Jr. Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–30. doi: 10.1016/S0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- 10.Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev Biol. 1996;175:314–24. doi: 10.1006/dbio.1996.0117. [DOI] [PubMed] [Google Scholar]
- 11.Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–9. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller HJ. Types of visible variations induced by X-rays in Drosophila. J Genet. 1930;22:299–334. doi: 10.1007/BF02984195. [DOI] [Google Scholar]
- 13.Wieschaus E, Nusslein-Volhard C, Jurgens G. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. III. Zygotic loci on the X-chromosome and fourth chromosome. Rouxs Arch Dev Biol. 1984;193:296–307. doi: 10.1007/BF00848158. [DOI] [PubMed] [Google Scholar]
- 14.Gergen JP, Wieschaus EF. Localized requirements for gene activity in segmentation of Drosophila embryos: Analysis of armadillo, fused, giant and unpaired mutations in mosaic embryos. Rouxs Arch Dev Biol. 1986;195:49–62. doi: 10.1007/BF00444041. [DOI] [PubMed] [Google Scholar]
- 15.Eberl DF, Perkins LA, Engelstein M, Hilliker AJ, Perrimon N. Genetic and developmental analysis of polytene section 17 of the X chromosome of Drosophila melanogaster. Genetics. 1992;130:569–83. doi: 10.1093/genetics/130.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–63. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hombría JC, Brown S, Häder S, Zeidler MP. Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Dev Biol. 2005;288:420–33. doi: 10.1016/j.ydbio.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 18.Müller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–5. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- 19.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 20.Rajan A, Perrimon N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell. 2012;151:123–37. doi: 10.1016/j.cell.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright VM, Vogt KL, Smythe E, Zeidler MP. Differential activities of the Drosophila JAK/STAT pathway ligands Upd, Upd2 and Upd3. Cell Signal. 2011;23:920–7. doi: 10.1016/j.cellsig.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev Cell. 2003;5:441–50. doi: 10.1016/S1534-5807(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 23.Makki R, Meister M, Pennetier D, Ubeda JM, Braun A, Daburon V, et al. A short receptor downregulates JAK/STAT signalling to control the Drosophila cellular immune response. PLoS Biol. 2010;8:e1000441. doi: 10.1371/journal.pbio.1000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown S, Hu N, Hombría JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. 2001;11:1700–5. doi: 10.1016/S0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- 25.Chen HW, Chen X, Oh SW, Marinissen MJ, Gutkind JS, Hou SX. mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 2002;16:388–98. doi: 10.1101/gad.955202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hombría JC, Brown S. The fertile field of Drosophila Jak/STAT signalling. Curr Biol. 2002;12:R569–75. doi: 10.1016/S0960-9822(02)01057-6. [DOI] [PubMed] [Google Scholar]
- 27.Kallio J, Myllymäki H, Grönholm J, Armstrong M, Vanha-aho LM, Mäkinen L, et al. Eye transformer is a negative regulator of Drosophila JAK/STAT signaling. FASEB J. 2010;24:4467–79. doi: 10.1096/fj.10-162784. [DOI] [PubMed] [Google Scholar]
- 28.Betz A, Lampen N, Martinek S, Young MW, Darnell JEJ., Jr. A Drosophila PIAS homologue negatively regulates stat92E. Proc Natl Acad Sci U S A. 2001;98:9563–8. doi: 10.1073/pnas.171302098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hari KL, Cook KR, Karpen GH. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 2001;15:1334–48. doi: 10.1101/gad.877901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stec WJ, Zeidler MP. Drosophila SOCS Proteins. J Signal Transduct. 2011;2011:894510. doi: 10.1155/2011/894510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callus BA, Mathey-Prevot B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21:4812–21. doi: 10.1038/sj.onc.1205618. [DOI] [PubMed] [Google Scholar]
- 32.Karsten P, Häder S, Zeidler MP. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech Dev. 2002;117:343–6. doi: 10.1016/S0925-4773(02)00216-2. [DOI] [PubMed] [Google Scholar]
- 33.Almudi I, Stocker H, Hafen E, Corominas M, Serras F. SOCS36E specifically interferes with Sevenless signaling during Drosophila eye development. Dev Biol. 2009;326:212–23. doi: 10.1016/j.ydbio.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 34.Herranz H, Hong X, Hung NT, Voorhoeve PM, Cohen SM. Oncogenic cooperation between SOCS family proteins and EGFR identified using a Drosophila epithelial transformation model. Genes Dev. 2012;26:1602–11. doi: 10.1101/gad.192021.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 2005;19:1861–70. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnstone K, Wells RE, Strutt D, Zeidler MP. Localised Jak/STAT pathway activation is required for Drosophila wing hinge development. PLoS One. 2013;8:e65076. doi: 10.1371/journal.pone.0065076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bach EA, Vincent S, Zeidler MP, Perrimon N. A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics. 2003;165:1149–66. doi: 10.1093/genetics/165.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corwin HO, Hanratty WP. Characterization of a unique lethal tumorous mutation in Drosophila. Mol Gen Genet. 1976;144:345–7. doi: 10.1007/BF00341734. [DOI] [PubMed] [Google Scholar]
- 39.Hanratty WP, Ryerse JS. A genetic melanotic neoplasm of Drosophila melanogaster. Dev Biol. 1981;83:238–49. doi: 10.1016/0012-1606(81)90470-X. [DOI] [PubMed] [Google Scholar]
- 40.Luo H, Hanratty WP, Dearolf CR. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. 1995;14:1412–20. doi: 10.1002/j.1460-2075.1995.tb07127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol. 2011;18:971–6. doi: 10.1038/nsmb.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo H, Rose P, Barber D, Hanratty WP, Lee S, Roberts TM, et al. Mutation in the Jak kinase JH2 domain hyperactivates Drosophila and mammalian Jak-Stat pathways. Mol Cell Biol. 1997;17:1562–71. doi: 10.1128/mcb.17.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward AC, Touw I, Yoshimura A. The Jak-Stat pathway in normal and perturbed hematopoiesis. Blood. 2000;95:19–29. [PubMed] [Google Scholar]
- 44.James C, Ugo V, Le Couédic JP, Staerk J, Delhommeau F, Lacout C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–8. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 45.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–90. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 46.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, et al. Cancer Genome Project Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–61. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 47.Shi S, Calhoun HC, Xia F, Li J, Le L, Li WX. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet. 2006;38:1071–6. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li WX. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol. 2008;18:545–51. doi: 10.1016/j.tcb.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müller P, Boutros M, Zeidler MP. Identification of JAK/STAT pathway regulators--insights from RNAi screens. Semin Cell Dev Biol. 2008;19:360–9. doi: 10.1016/j.semcdb.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]