Abstract
Post-translational S-glutathionylation occurs through the reversible addition of a proximal donor of glutathione to thiolate anions of cysteines in target proteins, where the modification alters molecular mass, charge, and structure/function and/or prevents degradation from sulfhydryl overoxidation or proteolysis. Catalysis of both the forward (glutathione S-transferase P) and reverse (glutaredoxin) reactions creates a functional cycle that can also regulate certain protein functional clusters, including those involved in redox-dependent cell signaling events. For translational application, S-glutathionylated serum proteins may be useful as biomarkers in individuals (who may also have polymorphic expression of glutathione S-transferase P) exposed to agents that cause oxidative or nitrosative stress.
Keywords: Cysteine-mediated Cross-linking, Glutathione, Glutathionylation, Peroxiredoxin, Serpin, Glutathione S-Transferase, Kinase Signaling, Nitric Oxide, Nitrosylation, Peroxidases
Introduction
S-Glutathionylation targets cysteines in a basic environment (low pKa) perhaps in close three-dimensional proximity to Arg, His, or Lys residues. Reports of reversible S-glutathionylation emerged as early as 1985 (1), but in concert with understanding the importance of reactive oxygen/nitrogen species (ROS/RNS)2 as second messengers, publications have increased substantially over the past 10 years. ROS/RNS signaling operates through a set of post-translational protein modifications that are discrete, site-specific, and reversible. Certain proteins undergo reversible chemical changes in response to altered localized redox potential. Among the most susceptible redox-sensitive targets are thiol (–SH) groups on cysteines. Signaling events are facilitated through redox-active proteins when one or more cysteines can exist as reactive thiolate anions. These cysteines are more nucleophilic and become susceptible to attack by GSH.
Oxygen-based metabolism is arguably the most efficient and evolved method for producing energy from nutrients, but ROS as byproducts of ATP generation provide physiological challenges. There are >200,000 cysteines encoded by the human genome, making cysteine one of the least commonly used amino acids (2). This sparing usage does not imply restricted functionality but instead indicates evolutionary importance particularly because organism cysteine content correlates with degree of biological complexity (3). Sulfur has unique characteristics (variable valence, nucleophilicity, and capacity to exist in different oxidation states) that make it intrinsically useful in a biologically oxidative environment. The major non-protein source of cysteine in cells is GSH (γ-glutamylcysteinylglycine), where the sulfhydryl can be deprotonated to a thiolate anion. Reported pKa values for the thiol group of GSH range from 8.7 (4) to 9.25 (5), with variation contingent upon the measurement methodology. Balance between oxidation and reduction reactions plays an essential role in numerous signaling cascades, including those associated with proliferation, inflammatory responses, apoptosis, and senescence. Post-translational modifications of cysteines can directly influence these processes. The flexibility of the thiol group of cysteine predicates that a number of post-translational modifications can occur (Table 1). As a response to ROS/RNS, S-glutathionylation is an established modification. The basal levels of S-glutathionylated proteins in resting cells are at least an order of magnitude lower than those in cells under oxidative stress (6). Quantitative analysis of the relationship between cysteine modifications and the corresponding functional changes in various cellular contexts is needed to define the frequency of S-glutathionylation events versus other cysteinyl modifications. Bioinformatic approaches may prove useful in predicting the susceptibility of individual cysteines to S-glutathionylation (7). Furthermore, S-nitrosylation may precede and initiate S-glutathionylation (8–10).
TABLE 1.
Possible post-translational modifications of protein-associated cysteine residues
| Group added to Cys residue | Notes |
|---|---|
| Thiol, disulfide, thiolate, sulfenic, sulfinic, or sulfonic acids | These occur following different degrees of ROS exposure, causing different extents of oxidation. |
| S-Cysteinylation | This occurs in ROS, where a disulfide bond converts l-Cys to S- (l-cysteinyl)-l-Cys. Donors can be cysteine or GSSG. |
| S-Nitrosylation | This involves NO-mediated formation of S-nitrosothiol and is reversible and sometimes short-lived. Consequences are important in cell signaling events. |
| S-Glutathionylation | This is a consequence of GS− addition, with both protective and signaling consequences. |
| S-Sulfhydration | This is a physiological post-translational modification caused by hydrogen sulfide, with subsequent effects on cell signaling pathways. |
| S-Isoprenylation, S-farnesylation, S-geranylgeranylation, and S-palmitoylation | These involve lipidation through addition of hydrophobic residues. Frequently, the affected protein has modified affinity for other proteins. Although particularly relevant in cell membranes, a number of enzymes are influenced by the process. |
Thiol Modifications Caused by ROS
ROS include oxygen-centered radicals (singlet oxygen (1O2), hydroxyl radical (•OH), superoxide anion radical (O2⨪, SO), and peroxyl radical (R-O2⨪)) as well as hydrogen (and other organic) peroxide (H2O2, ROOH) and ozone (O3). Protein cysteines can occur as surface residues generally exposed to an aqueous environment, as is predominant in mitochondria (11), or buried inside globular domains. Either could be catalytically or structurally significant.
Regardless of their origin, neither 1O2 nor •OH can be considered as a specific protein thiol oxidant. Contributory factors to this include their short half-life (∼10−9 s) (12) and high nonspecific reactivities with nearly diffusion-controlled rate constants (13). SO can oxidize thiols to thiyl radicals at a rate constant of ∼103 m−1 s−1 at pH 7.4 (14). Because the rate constant for cytosolic and mitochondrial superoxide dismutases is ∼107–109 m−1 s−1 (15) and that for spontaneous dismutation of SO is ∼105 m−1 s−1 (16), protein thiol oxidation by SO is too slow to be significant. Both spontaneous and catalyzed dismutations of SO result in an accumulation of H2O2. Generally, the intracellular levels of H2O2 are controlled by catalase (an enzyme with a high turnover rate) (17), GSH peroxidases 1–4 (18), and peroxiredoxins (19). Site-specifically, H2O2 levels can be elevated enough to cause thiol oxidation. Thus, H2O2 and ROOH specifically oxidize thiols through a two-electron nucleophilic substitution reaction (13). This reaction requires dissociation (deprotonation) of the thiol to a thiolate anion (RS−) to reach the required nucleophilicity (13). The pKa of protein cysteines approximates 8.5 (4). Thus, only a small proportion of protein thiols are deprotonated at physiological pH (pH 7.0–7.4). Cysteines within a basic environment (e.g. three-dimensionally surrounded by Arg, Lys, or His residues) have higher reaction rates with H2O2 primarily as a consequence of greater deprotonation. One result of this oxidation is formation of cysteine sulfenic acid, which is highly reactive with the GSH sulfhydryl and can facilitate protein S-glutathionylation (20). Fig. 1A provides an outline of some of these reactions.
FIGURE 1.
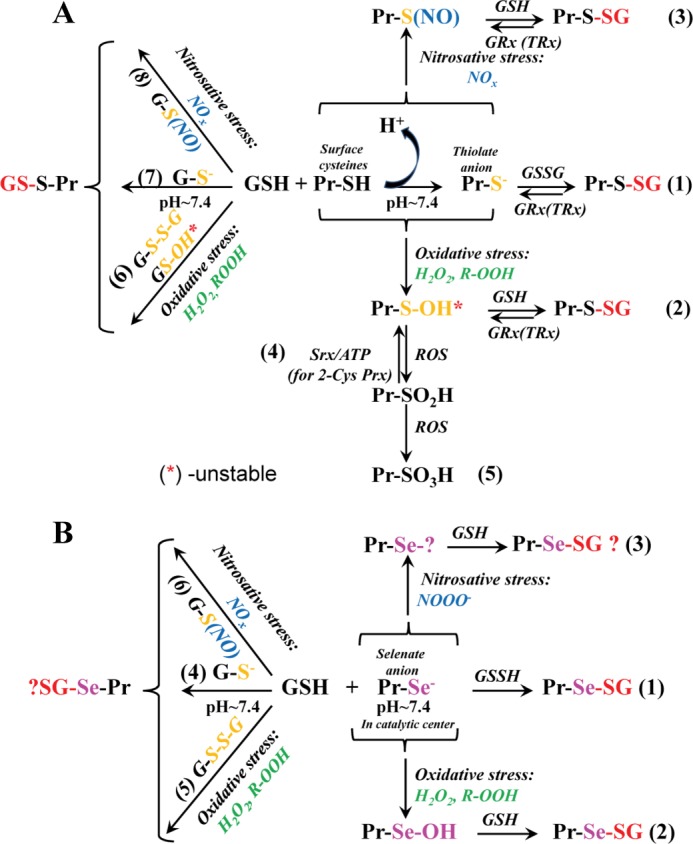
Chemical basis for the biological S-glutathionylation of regular Cys (A) and Sec (B). A, surface protein cysteines at physiological pH are not reactive because of high pKa values. In a basic microenvironment, the thiol deprotonates to a thiolate anion, which attacks the sulfhydryl of intracellular GSSG, with the subsequent formation of disulfide (pathway 1). Under oxidative (pathway 2) or nitrosative (pathway 3) stress, a surface protein (Pr) sulfhydryl becomes activated through cysteine sulfenate or nitro-S-cysteine formation and then undergoes S-glutathionylation. Deglutathionylation of glutathionylated cysteines is predominately catalyzed by Grx. Site-specifically, thioredoxin (TRx) can also catalyze deglutathionylation. Sulfiredoxin (Srx) catalyzes the reduction of a protein cysteine sulfinic acid into sulfenate for 2-Cys peroxiredoxins (Prx; pathway 4). Generally, overoxidation of protein cysteines is terminal (pathway 5). Similar to protein thiols, the cysteine of GSH can be activated through pathways 6–8, resulting in protein S-glutathionylation. The relative abundance of intracellular GSH influences the probability and specificity of particular reactions (see text). B, for proteins containing Sec (GSH peroxidases, etc.), its low pKa results in the formation of a selenate anion at physiological pH, permanently activated (pathway 1). S-Glutathionylation of protein Sec residues is affected by ROS/RNS (pathways 2 and 3), but the mechanism of the latter is unclear. S-Glutathionylation of Sec could be further accelerated by activation of GSH (pathways 4–6), but the mechanism of such is not yet defined.
GSH is in most cells at millimolar levels (21). The reasons for the nonspecific reactivity of GSH with 1O2 and •OH are the same as for protein thiols (see above). Reaction of GSH with SO usually generates GSSG and an intermediate thiyl radical (GS•) and thiolate anion (GS−) (22). Because of the high concentrations of GSH and the instability of GSH sulfenate as an intermediate, the reaction of GSH with H2O2 (ROOH) also generates GSSG (22). GSH reductase reduces intracellular GSSG to GSH using NADPH as a source of electrons (23). A high cellular content of GSH could in principle result in site-specific accumulation of GSSG and thiol-disulfide exchange-driven S-glutathionylation of proteins.
Thiol Modification by RNS
RNS include moieties such as nitric oxide (NO) and NO-derived compounds, nitroxyl anion (NO−, HNO), nitrosonium cation (NO+), higher oxides of nitrogen (N2O, NO2, N2O3), peroxynitrite (ONOO−/ONOOH), S-nitrosothiols (RSNO), and dinitrosyl iron complexes ((RS−)2Fe+(NO+)2 … (−SR)2)−) (24).
NO is a reactive molecule generated intracellularly by NOS that may nitrosylate protein thiols directly, through reaction with molecular oxygen (generating N2O3 with high affinity for thiols) (25) or with SO radicals (generating peroxynitrite) (26). Generally, the half-lives of nitrosylated cysteines are short as a consequence of spontaneous denitrosylation by excess GSH (generating GSNO). However, for a few proteins, S-nitrosylation may be stabilized by conformation-mediated inaccessibility for GSH (27). Glutathione S-transferase P (GSTP) in the presence of excess GSH can be S-glutathionylated, providing another (albeit indirect) pathway of NO signaling. The reaction of peroxynitrite with thiols can generate thiyl radicals or sulfenates (13), with the implication that the forward reaction of S-glutathionylation might be independent of GSH/GSSG ratios.
A primary role for glutaredoxin (Grx) in deglutathionylation is premised on the nucleophilic attack of the cysteine thiolate anion on a protein-GSH mixed disulfide (28). There are reports that Grx can catalyze GSSG-dependent S-glutathionylation of proteins such as actin, GAPDH, and protein-tyrosine phosphatase 1B by an alternative mechanism involving •OH-mediated GSH thiol radical generation under hypoxia (29). Considering the high reactivity and nonspecificity of •OH-mediated reactions, the overall contribution of this mechanism of catalysis remains to be established.
In the presence of O2, NO may react with the sulfhydryl of GSH to produce (likely through intermediate(s) of higher nitrogen oxides) GSNO, which can serve as a physiological scavenger of NOx species (30). GSNO can react with GSH, yielding GSSG, nitrite, nitrous oxide, and ammonia (31), products that may contribute to protein nitrosylation. Protein thiol transnitrosylation by GSNO (28) is regulated by expression of GSNO reductase (32). High intracellular GSH concentrations can shift the equilibrium of the reaction with NO to GSSG, but during nitrosative stress, certain cell compartments can harbor high levels of GSNO. Under such conditions, GSNO may contribute to protein nitrosylation and/or S-glutathionylation (9, 10). The specificity of GSNO-mediated post-translational modifications depends on the microenvironment of the target cysteine within protein tertiary and quaternary structures.
Can Selenium Substitute for Sulfur?
Selenium and sulfur are in the same group in the periodic table and share common properties, including participation in oxidation/reduction reactions. Selenium is found in the form of selenocysteine (Sec; the 21st amino acid), coded by the UGA codon in 25 proteins (33). From an evolutionary standpoint, recoding the UGA stop codon into the Sec sense codon requires commitment of biological energy, implying that it fulfills some critical biological role that sulfur-containing cysteines do not. Selenol groups are substantially more acidic than thiols, with pKa values of ∼5.3 (34). At physiological pH, selenol is completely deprotonated and exists as a selenate anion (RSe−). Selenyl radicals are less oxidizing than thiyl radicals and are more easily produced (35). Generally, Sec is less stable than Cys (22), and native selenoproteins do not necessarily have higher activities than native sulfur proteins (36). Selenols (as selenate anions) are better nucleophiles than thiols and become oxidized by H2O2 (ROOH) to selenic acid (RSeOH). Selenic acid is unstable and reacts with GSH, forming a selenenyl sulfide adduct (glutathionylated selenenyl). A second molecule of GSH reduces this selenenyl sulfide bond to selenol and generates GSSG (37). These reactions occur in the catalytic center of GSH peroxidases and may require a more hydrophobic environment. Oxidations of surface protein selenols and their subsequent reactivity with GSH have not been demonstrated, and overoxidation of Sec to seleninic or selenonic acid has not been reported (38).
GSH peroxidase can be inactivated by peroxynitrite (39), and stop-flow experiments have monitored its reaction with selenate anions (E-Se−) of Sec in the enzyme active site. This reaction can transform the selenate anion into a form of selenium that is difficult to reduce back to selenol (39). This reaction occurs in the specific hydrophobic environment of the enzyme catalytic center. To date, there are no reports that RNS may mediate the reactivity of Sec residues with GSH. Fig. 1B summarizes some of these reactions. In summary, selenium and sulfur have not evolved to perform identical antioxidant functions within the cell.
S-Glutathionylation as a Cycle
The forward reaction of S-glutathionylation can be either spontaneous or catalyzed by GSTP. At physiological pH, GSTP binds GSH and lowers the pKa of the thiol, producing a thiolate anion (GS−) at the active site (40). This catalyzes the forward reaction, a specific example of which is provided by the reactivation of peroxiredoxin-6 (41). Reversal of S-glutathionylation may be impacted by enzymes such as thioredoxin (42), Grx (43), and sulfiredoxin (44). Although studies have confirmed a role for Grx in protein deglutathionylation (45), Grx may also facilitate GS radical recombination with a protein thiyl radical and catalyze S-glutathionylation via stabilization of the GS• thiyl radical as an enzyme disulfide anion radical intermediate (Grx1-SSG⨪) (46). Reversal of S-glutathionylation is contingent upon the extent and duration of the initial stress, removal of which usually permits deglutathionylation to follow a half-life of ∼2–3 h (47). Relatively high levels of ROS would give precedence to the protective function, masking critical thiols from further irreversible oxidation. Whereas sulfinates or sulfonates frequently lead to proteolysis, S-glutathionylated residues can maintain dormancy during stress.
Relative to the proteome, the number of proteins susceptible to S-glutathionylation is not large. Proteomic techniques reveal a high number of putative S-glutathionylated proteins in unicellular organisms such as Plasmodium falciparum and Chlamydomonas reinhardtii (48, 49). A literature review reveals some clustering of function and includes enzymes with catalytically important cysteines (in particular, those involved with protein folding and stability, NO regulation, and redox homeostasis); cytoskeletal proteins; signaling proteins (particularly kinases and phosphatases); transcription factors; Ras proteins; protein folding and degradation; ion channels, calcium pumps, and binding proteins (involved in calcium homeostasis); and energy metabolism and glycolysis. Within these categories, there are some notable examples of functional alterations associated with the modification: cytoskeletal restructuring during cell growth or motility is impacted by S-glutathionylation, particularly of actin (50). Some studies report inhibition of enzymatic activity/protein function by S-glutathionylation, e.g. GAPDH (51), eNOS (52, 53), the heteromeric Kir4.1-Kir5.1 channel (54), low molecular weight tyrosine phosphatase (55), MAPK phosphatase 1 (56), mitochondrial thymidine kinase 2 (57), Na,K-ATPase (58), and protein-disulfide isomerase (59). The position of cysteine is important in determining the impact of the modification. For instance, carbonic anhydrase III can be inhibited or activated depending on the cysteine involved (60), whereas the inositol 1,4,5-triphosphate receptor (61) and Rac2 (62) are each activated by S-glutathionylation. The canonical phosphorylation/dephosphorylation pathways are cyclical, flexibly controlling signaling events as a response to external or internal stimuli. A significant number of kinases and phosphatases are subject to secondary regulation by S-glutathionylation, providing secondary control of critical pathways following stimuli that cause homeostatic changes in ROS/RNS. Additional detailed examples are discussed in recent reviews (63, 64).
One emerging example merits emphasis. Fas (CD95, Apo-1) is a constitutively expressed member of the TNF family of death receptors with roles in apoptosis signaling. Additional regulatory roles of Fas, including immune cell activation and proliferation, have been identified (65). Fas is a substrate for S-glutathionylation (Fas-SSG) at Cys-294, sustained via caspase-dependent degradation of Grx1 (66). Fas-SSG enhances Fas recruitment into lipid rafts and promotes FasL binding, the formation of death-inducing signaling complexes, and caspase activation, resulting in amplification of cell death pathways (66). Distinct subcellular localization of Fas exists, with oxidative conditions causing the majority of Fas-SSG to be found in the endoplasmic reticulum (ER). Moreover, stimulation with FasL causes protein/protein interactions between Fas, ERp57 (a member of the protein-disulfide isomerase family), and GSTP. Genetic or pharmacologic diminution of ERp57 and GSTP1 decreases FasL-induced S-glutathionylation of Fas, producing lowered death-inducing signaling complex formation and enhanced cell survival. Increased expression of ERp57 and GSTP1 enhances the kinetics of translocation of Fas-SSG from the ER to the cytosol/plasma membrane, enhances caspase activation, and promotes cell death. As a disease correlate, interactions between Fas, ERp57, and GSTP1 and levels of Fas-SSG are increased in murine pulmonary fibrosis, which depends on functional Fas, implying that oxidative processing of Fas and its subsequent S-glutathionylation may provide an important ligand regulatory switch to control the apoptotic signal (67). Furthermore, S-glutathionylation of the GTPase Ras does not affect Ras activity unless it is modified by GSH through a radical-mediated mechanism, which promotes activation through nucleotide dissociation and GTP binding. Binding of GSSG through disulfide exchange or via Ras oxidation by a non-radical oxidant does not impact Ras activation (68).
Translational Applications for Protein S-Glutathionylation
Human response to ROS/RNS is influenced by numerous variables. For example, GSTP polymorphisms arise from nucleotide transitions that change codon 105 from Ile to Val and codon 114 from Ala to Val, generating four possible GSTP1 alleles: wild-type GSTP1-1A (Ile-105/Ala-114), GSTP1-1B (Val-105/Ala-114), GSTP1-1C (Val-105/Val-114), and GSTP1-1D (Ile-105/Val-114). Population frequencies of 0.65 (GSTP1-1A), 0.262 (GSTP1-1B), and 0.068 (GSTP1-1C) occur in Caucasians (69). The I105V and A114B substitutions do not alter GSH binding affinity but cause steric changes at the substrate-binding site of the enzyme (70). What impact GSTP polymorphisms may have in establishing risk factors for response to ROS/RNS is not clear. One example is provided by the hierarchical effectiveness of polymorphic variants of GSTP in regulating the peroxidase activity of Prdx6 (71). Prdx6 is a dual-functioning enzyme that detoxifies lipid peroxides particularly in biological membranes with a peroxidase function activated by GSTP. Activation of Prdx6 depends on its heterodimerization and subsequent S-glutathionylation by GSH-loaded GSTP (41). The catalytic Cys-47 residue is buried in the hydrophobic globular core region of Prdx6. Following reduction of peroxides, this cysteine is oxidized to a sulfenate, and GSTP participates in overcoming the accessibility barrier for GSH to reach this hydrophobic region and reactivate Prdx6. In silico modeling or deletion studies suggest that the site of closest contact between Prdx6 and the GSTP monomer is at the interface of their interaction at residues 115–124 for GSTP and residues 163–169 for Prdx6. Presumably, the combination of differences in molecular volume (Ala, 69Å3; Val, 120Å3; and Ile, 204Å3) and hydrophobicity in the side chains impacts the affinity of GSTP for Prdx6. The affinities of GSTP1-1A (or GSTP1-1C) for Prdx6 are higher (KD ∼ 51–57 nm) than those of GSTP1-1B (or GSTP1-1D) (KD ∼ 101–94 nm) (71). These values determine proximity between the catalytic Cys-47 sulfenate of Prdx6 and activated GSH (thiolate) bound to the GSTP allelic variant and, consequently, the efficiency with which the sulfenate at Cys-47 is S-glutathionylated and reduced (i.e. activated) (71). The peroxidase activities of cells transiently transfected with GSTP1-1A (or GSTP1-1C) are higher than those transfected with GSTP1-1B (or GSTP1-1D), and catalytically inactive Y7F GSTP does not support activation of Prdx6 (71). These observations carry the implication that human polymorphisms of GSTP differentially mediate activation of Prdx6 and that, contingent upon their GSTP genotype, individuals will have potential differences in mounting an antioxidant response, particularly impacting protection of cell membranes against lipid peroxidation. Polymorphic variants of GSTP may also potentially impact the forward reaction in S-glutathionylation of other client protein clusters (72, 73). Large-scale epidemiology studies will be required to analyze the fidelity of the relationship between GSTP polymorphisms and susceptibility to ROS/RNS.
Because altered S-glutathionylation patterns occur in diseases characterized by redox deregulation (e.g. cardiovascular, lung, inflammatory, and neurodegenerative diseases and cancer), there is opportunity to use them as biomarkers. S-Glutathionylated hemoglobin and actin have been investigated as biomarkers in patients with Friedreich ataxia, diabetes, hyperlipidemia, and uremia associated with hemodialysis or peritoneal dialysis (74–76). In hypertension, oxidative stress-induced S-glutathionylation of eNOS acts as a switch providing redox regulation of cell signaling and vascular functions (52, 53). S-Glutathionylation of the tumor suppressor p53 has been found in human cancers, and functional inactivation of p53 by S-glutathionylation during ROS/RNS inhibits DNA binding in cancer cells (77). As such, the extent of p53 S-glutathionylation in cancers and normal tissues exposed to chronic ROS may define a relationship between redox control of p53 and disease progression and/or therapeutic response.
Redox homeostasis and thiol regulation have been linked to hematopoiesis and the regulation of bone marrow stem cell proliferation. As a tissue, the marrow environment is separated into distinct niches characterized by adhesive molecules, cytokines, chemokines, growth factors, Ca2+, and O2 gradients that can define how hematopoietic stem cells (HSCs) and hematopoietic progenitor cells (HPCs) migrate, proliferate, and differentiate (78). Given that ROS and H2O2 levels impact HSC quiescence (79) and that recent data link HSC location in the distal endosteum and ROS-induced DNA damage (80), it does not seem unreasonable to propose that a redox gradient might influence HSC migration and differentiation. S-Glutathionylation of ER chaperone proteins regulates their function and the induction of the unfolded response (59, 81), which has roles in hematopoiesis and T and B lymphocyte differentiation (82). These types of relationships provide evidence that niche thiol status and chaperone protein S-glutathionylation may be involved in hematopoietic events. Interrogation of these pathways may be a productive avenue in determining how redox-altering myeloproliferative therapeutics may function (83).
In patients with ischemic heart disease and heart failure, superoxide dismutase mimetics or low doses of H2O2 have been used in clinical trials with bone marrow progenitor cell therapies (84, 85). Achieving effective therapeutic indices in such studies would benefit from the identification of specific biomarkers that might define the parameters that monitor either efficacy or toxicity. In this vein, accumulating evidence supports a role for serine protease inhibitor (serpins) family members in myeloproliferation and HPC mobilization (86, 87). The down-regulation of serpins A1 and A3 during cell mobilization influences the marrow microenvironment and migratory behavior of HPCs. Furthermore, radiation causes serpin A1 up-regulation in bone marrow and has been linked to inhibition of HSC and HPC mobilization (87). Serpin A1/A3 enzymatic activity is regulated by modifications of key redox-active cysteines (88). S-Glutathionylation of serpin A1 results in conformational changes that weaken its affinity for its target protease, thereby suppressing proteolytic activity. Serpins A1 and A3 in both mouse and human plasma are S-glutathionylated in a time- and dose-dependent manner when treated with a redox-modulating agent that mitigates myeloproliferation (89). In addition to regulating serpin enzymatic activity, S-glutathionylation of serpin family members may also influence myeloproliferation. As such, serpin S-glutathionylation may predict marrow response to myeloproliferative agents. Inhibition of disulfide bond formation in the redox-sensitive serpin bomapin directly affects the responsiveness of myeloid progenitor cells to their microenvironment (90). Further evaluation of protein S-glutathionylation in a therapeutic setting may provide the sort of biomarkers that are important in drug development and optimization of treatment strategies.
Techniques for Measuring S-Glutathionylation
Although methodology is improving, there are still challenges in enhancing sensitivity of detection of S-glutathionylation. Problems include the generally low levels of cellular S-glutathionylated proteins and the labile nature of protein-SSG bonds. With a putative proteome of >20,000, there is a relatively limited number of proteins (hundreds) subject to modification. Recent technical advances have facilitated enhanced sensitivity of detection, and there are informative reviews (64, 91, 92). The following methodologies are used for detection and/or quantification of S-glutathionylation.
Essentially two “direct” or “indirect” approaches have been used to detect the protein-SSG bond. The direct method is based on the use of radiolabeled GSH (93). Cells are preincubated with cycloheximide to block protein synthesis, incubated with [35S]cysteine to radiolabel GSH, exposed to ROS, and compared with control cells (92). After removal of free radiolabeled GSH, radiolabeled S-glutathionylated proteins are detected and quantified by radiography. When coupled with two-dimensional gel and mass spectrometry, the site of the modification can be identified. One advantage of this approach is the in vivo relevance, but this method requires cycloheximide, likely perturbing cell physiology. Methods based on biotinylated GSH or membrane-permeable biotinylated reduced GSH ethyl esters have been reported (94). Tagging GSH with biotin allows detection of purified S-glutathionylated proteins through avidin-conjugated agarose beads followed by immunoblotting (anti-biotin antibodies) or avidin-based affinity chromatography. A prevalent method is to use commercial anti-GSH antibodies directed against the GS moiety bound to an unknown antigenic protein at an unknown site. The low sensitivity of this antibody imparts a limited detection potential, and many of the proteins identified are abundant, e.g. actin (95), tubulin (96), and HSP70 (93). Immunoprecipitation followed by immunoblotting with the anti-GSH antibodies can enhance detection (89).
Indirect techniques require reduction of S-glutathionylated proteins by Grx (6) followed by labeling the reduced protein thiol with iodoacetyl or maleimide. Derivatives of these compounds with additional mass (97), fluorescence (98), or biotin derivatives (99) in combination with LC-MS/MS, fluorescent microscopy, or immunoblotting enable labeling and monitoring of changes in redox state. Proteins are initially alkylated by N-ethylmaleimide to block free thiols; after reduction by Grx, these newly accessible thiols react with derivatives of iodoacetyl and maleimide groups for further application. Rapid fluorometric methods to quantify S-glutathionylation were developed by using GSH derivatized with 2,3-naphthalenedicarboxaldehyde (100).
Concluding Remarks and Perspectives
Cysteine is a highly malleable amino acid susceptible to many types of post-translational modifications. S-Glutathionylation serves both to protect and modify structure/function. The cyclical nature of the S-glutathionylation process allows regulation of signaling pathways to occur during variations in ROS/RNS homeostasis. So far, the number of proteins susceptible to S-glutathionylation is not large (hundreds) compared with the proteome, but improved sensitivity of detection will likely lead to the unearthing of further candidates. There is a nascent opportunity to consider what impact protein S-glutathionylation may have in the cause/effect relationship of a number of human pathologies. Indeed, there are already early indications of examples in which candidate proteins may be biomarkers of exposure/response to ROS/RNS. Polymorphisms in GSTP imply that the forward reaction for S-glutathionylation may be subject to population differences, again raising the possibility of a link with human disease.
This work was supported, in whole or in part, by National Institutes of Health Grants CA08660 and CA117259 and National Center for Research Resources (NCRR) Grant P20 RR024485-COBRE in Oxidants, Redox Balance, and Stress Signaling. This work was also supported by the South Carolina Centers of Excellence Program, by the Drug Metabolism and Clinical Pharmacology Shared Resource of Hollings Cancer Center of the Medical University of South Carolina, and by Swedish Research Council Grant 524-2011-6998 (to J. Z.). This work was conducted in a facility constructed with support from National Institutes of Health Grant C06 RR015455 from the NCRR Extramural Research Facilities Program. This is the fifth article in the Thematic Minireview Series on Redox-active Protein Modifications and Signaling.
- ROS
- reactive oxygen species
- RNS
- reactive nitrogen species
- GSTP
- glutathione S-transferase P
- Grx
- glutaredoxin
- Sec
- selenocysteine
- ER
- endoplasmic reticulum
- HSC
- hematopoietic stem cell
- HPC
- hematopoietic progenitor cell.
REFERENCES
- 1. Ziegler D. M. (1985) Role of reversible oxidation-reduction of enzyme thiol-disulfides in metabolic regulation. Annu. Rev. Biochem. 54, 305–329 [DOI] [PubMed] [Google Scholar]
- 2. Jones D. P. (2008) Radical-free biology of oxidative stress. Am. J. Physiol. Cell Physiol. 295, C849–C868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miseta A., Csutora P. (2000) Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol. Biol. Evol. 17, 1232–1239 [DOI] [PubMed] [Google Scholar]
- 4. Srinivasan U., Mieyal P. A., Mieyal J. J. (1997) pH profiles indicative of rate-limiting nucleophilic displacement in thioltransferase catalysis. Biochemistry 36, 3199–3206 [DOI] [PubMed] [Google Scholar]
- 5. Tajc S. G., Tolbert B. S., Basavappa R., Miller B. L. (2004) Direct determination of thiol pKa by isothermal titration microcalorimetry. J. Am. Chem. Soc. 126, 10508–10509 [DOI] [PubMed] [Google Scholar]
- 6. Lind C., Gerdes R., Hamnell Y., Schuppe-Koistinen I., von Löwenhielm H. B., Holmgren A., Cotgreave I. A. (2002) Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch. Biochem. Biophys. 406, 229–240 [DOI] [PubMed] [Google Scholar]
- 7. Sun C., Shi Z. Z., Zhou X., Chen L., Zhao X. M. (2013) Prediction of S-glutathionylation sites based on protein sequences. PLoS ONE 8, e55512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Townsend D. M., Findlay V. J., Fazilev F., Ogle M., Fraser J., Saavedra J. E., Ji X., Keefer L. K., Tew K. D. (2006) A glutathione S-transferase π-activated prodrug causes kinase activation concurrent with S-glutathionylation of proteins. Mol. Pharmacol. 69, 501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giustarini D., Milzani A., Aldini G., Carini M., Rossi R., Dalle-Donne I. (2005) S-Nitrosation versus S-glutathionylation of protein sulfhydryl groups by S-nitrosoglutathione. Antioxid. Redox Signal. 7, 930–939 [DOI] [PubMed] [Google Scholar]
- 10. Coles S. J., Easton P., Sharrod H., Hutson S. M., Hancock J., Patel V. B., Conway M. E. (2009) S-Nitrosoglutathione inactivation of the mitochondrial and cytosolic BCAT proteins: S-nitrosation and S-thiolation. Biochemistry 48, 645–656 [DOI] [PubMed] [Google Scholar]
- 11. Requejo R., Hurd T. R., Costa N. J., Murphy M. P. (2010) Cysteine residues exposed on protein surfaces are the dominant intramitochondrial thiol and may protect against oxidative damage. FEBS J. 277, 1465–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roots R., Okada S. (1975) Estimation of life times and diffusion distances of radicals involved in x-ray-induced DNA strand breaks of killing of mammalian cells. Radiat. Res. 64, 306–320 [PubMed] [Google Scholar]
- 13. Forman H. J., Maiorino M., Ursini F. (2010) Signaling functions of reactive oxygen species. Biochemistry 49, 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Winterbourn C. C., Metodiewa D. (1995) Reaction of superoxide with glutathione and other thiols. Methods Enzymol. 251, 81–86 [DOI] [PubMed] [Google Scholar]
- 15. Forman H. J., Fridovich I. (1973) Superoxide dismutase: a comparison of rate constants. Arch. Biochem. Biophys. 158, 396–400 [DOI] [PubMed] [Google Scholar]
- 16. Fridovich I. (1983) Superoxide radical: an endogenous toxicant. Annu. Rev. Pharmacol. Toxicol. 23, 239–257 [DOI] [PubMed] [Google Scholar]
- 17. Chelikani P., Fita I., Loewen P. C. (2004) Diversity of structures and properties among catalases. Cell. Mol. Life Sci. 61, 192–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arthur J. R. (2000) The glutathione peroxidases. Cell. Mol. Life Sci. 57, 1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hofmann B., Hecht H. J., Flohé L. (2002) Peroxiredoxins. Biol. Chem. 383, 347–364 [DOI] [PubMed] [Google Scholar]
- 20. Forman H. J., Fukuto J. M., Torres M. (2004) Redox signaling: thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am. J. Physiol. Cell Physiol. 287, C246–C256 [DOI] [PubMed] [Google Scholar]
- 21. Reed D. J. (1990) Glutathione: toxicological implications. Annu. Rev. Pharmacol. Toxicol. 30, 603–631 [DOI] [PubMed] [Google Scholar]
- 22. Winterbourn C. C., Metodiewa D. (1999) Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic. Biol. Med. 27, 322–328 [DOI] [PubMed] [Google Scholar]
- 23. Worthington D. J., Rosemeyer M. A. (1976) Glutathione reductase from human erythrocytes. Catalytic properties and aggregation. Eur. J. Biochem. 67, 231–238 [DOI] [PubMed] [Google Scholar]
- 24. Martínez M. C., Andriantsitohaina R. (2009) Reactive nitrogen species: molecular mechanisms and potential significance in health and disease. Antioxid. Redox Signal. 11, 669–702 [DOI] [PubMed] [Google Scholar]
- 25. Stamler J. S., Singel D. J., Loscalzo J. (1992) Biochemistry of nitric oxide and its redox-activated forms. Science 258, 1898–1902 [DOI] [PubMed] [Google Scholar]
- 26. West M. B., Hill B. G., Xuan Y. T., Bhatnagar A. (2006) Protein glutathiolation by nitric oxide: an intracellular mechanism regulating redox protein modification. FASEB J. 20, 1715–1717 [DOI] [PubMed] [Google Scholar]
- 27. Paige J. S., Xu G., Stancevic B., Jaffrey S. R. (2008) Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem. Biol. 15, 1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allen E. M., Mieyal J. J. (2012) Protein-thiol oxidation and cell death: regulatory role of glutaredoxins. Antioxid. Redox Signal. 17, 1748–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qanungo S., Starke D. W., Pai H. V., Mieyal J. J., Nieminen A. L. (2007) Glutathione supplementation potentiates hypoxic apoptosis by S-glutathionylation of p65-NFκB. J. Biol. Chem. 282, 18427–18436 [DOI] [PubMed] [Google Scholar]
- 30. Wink D. A., Nims R. W., Darbyshire J. F., Christodoulou D., Hanbauer I., Cox G. W., Laval F., Laval J., Cook J. A., Krishna M. C., DeGraff W. G., Mitchell J. B. (1994) Reaction kinetics for nitrosation of cysteine and glutathione in aerobic nitric oxide solutions at neutral pH. Insights into the fate and physiological effects of intermediates generated in the NO/O2 reaction. Chem. Res. Toxicol. 7, 519–525 [DOI] [PubMed] [Google Scholar]
- 31. Singh S. P., Wishnok J. S., Keshive M., Deen W. M., Tannenbaum S. R. (1996) The chemistry of the S-nitrosoglutathione/glutathione system. Proc. Natl. Acad. Sci. U.S.A. 93, 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu L., Hausladen A., Zeng M., Que L., Heitman J., Stamler J. S. (2001) A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410, 490–494 [DOI] [PubMed] [Google Scholar]
- 33. Kryukov G. V., Castellano S., Novoselov S. V., Lobanov A. V., Zehtab O., Guigó R., Gladyshev V. N. (2003) Characterization of mammalian selenoproteomes. Science 300, 1439–1443 [DOI] [PubMed] [Google Scholar]
- 34. Huber R. E., Criddle R. S. (1967) Comparison of the chemical properties of selenocysteine and selenocystine with their sulfur analogs. Arch. Biochem. Biophys. 122, 164–173 [DOI] [PubMed] [Google Scholar]
- 35. Nauser T., Dockheer S., Kissner R., Koppenol W. H. (2006) Catalysis of electron transfer by selenocysteine. Biochemistry 45, 6038–6043 [DOI] [PubMed] [Google Scholar]
- 36. Nauser T., Steinmann D., Koppenol W. H. (2012) Why do proteins use selenocysteine instead of cysteine? Amino Acids 42, 39–44 [DOI] [PubMed] [Google Scholar]
- 37. Sarma B. K., Mugesh G. (2005) Glutathione peroxidase (GPx)-like antioxidant activity of the organoselenium drug ebselen: unexpected complications with thiol exchange reactions. J. Am. Chem. Soc. 127, 11477–11485 [DOI] [PubMed] [Google Scholar]
- 38. Reeves M. A., Hoffmann P. R. (2009) The human selenoproteome: recent insights into functions and regulation. Cell. Mol. Life Sci. 66, 2457–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Padmaja S., Squadrito G. L., Pryor W. A. (1998) Inactivation of glutathione peroxidase by peroxynitrite. Arch. Biochem. Biophys. 349, 1–6 [DOI] [PubMed] [Google Scholar]
- 40. Graminski G. F., Kubo Y., Armstrong R. N. (1989) Spectroscopic and kinetic evidence for the thiolate anion of glutathione at the active site of glutathione S-transferase. Biochemistry 28, 3562–3568 [DOI] [PubMed] [Google Scholar]
- 41. Manevich Y., Feinstein S. I., Fisher A. B. (2004) Activation of the antioxidant enzyme 1-Cys peroxiredoxin requires glutathionylation mediated by heterodimerization with πGST. Proc. Natl. Acad. Sci. U.S.A. 101, 3780–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arnér E. S., Holmgren A. (2000) Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267, 6102–6109 [DOI] [PubMed] [Google Scholar]
- 43. Fernandes A. P., Holmgren A. (2004) Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox Signal. 6, 63–74 [DOI] [PubMed] [Google Scholar]
- 44. Findlay V. J., Townsend D. M., Morris T. E., Fraser J. P., He L., Tew K. D. (2006) A novel role for human sulfiredoxin in the reversal of glutathionylation. Cancer Res. 66, 6800–6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reynaert N. L., van der Vliet A., Guala A. S., McGovern T., Hristova M., Pantano C., Heintz N. H., Heim J., Ho Y. S., Matthews D. E., Wouters E. F., Janssen-Heininger Y. M. (2006) Dynamic redox control of NF-κB through glutaredoxin-regulated S-glutathionylation of inhibitory κB kinase β. Proc. Natl. Acad. Sci. U.S.A. 103, 13086–13091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Starke D. W., Chock P. B., Mieyal J. J. (2003) Glutathione-thiyl radical scavenging and transferase properties of human glutaredoxin (thioltransferase). Potential role in redox signal transduction. J. Biol. Chem. 278, 14607–14613 [DOI] [PubMed] [Google Scholar]
- 47. Townsend D. M., Manevich Y., He L., Hutchens S., Pazoles C. J., Tew K. D. (2009) Novel role for glutathione S-transferase π. Regulator of protein S-glutathionylation following oxidative and nitrosative stress. J. Biol. Chem. 284, 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kehr S., Jortzik E., Delahunty C., Yates J. R., 3rd, Rahlfs S., Becker K. (2011) Protein S-glutathionylation in malaria parasites. Antioxid. Redox Signal. 15, 2855–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zaffagnini M., Bedhomme M., Groni H., Marchand C. H., Puppo C., Gontero B., Cassier-Chauvat C., Decottignies P., Lemaire S. D. (2012) Mol. Cell. Proteomics 11, M111.014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bowers R. R., Manevich Y., Townsend D. M., Tew K. D. (2012) Sulfiredoxin redox-sensitive interaction with S100A4 and non-muscle myosin IIA regulates cancer cell motility. Biochemistry 51, 7740–7754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mohr S., Hallak H., de Boitte A., Lapetina E. G., Brüne B. (1999) Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 274, 9427–9430 [DOI] [PubMed] [Google Scholar]
- 52. Chen C. A., Wang T. Y., Varadharaj S., Reyes L. A., Hemann C., Talukder M. A., Chen Y. R., Druhan L. J., Zweier J. L. (2010) S-Glutathionylation uncouples eNOS and regulates its cellular and vascular function. Nature 468, 1115–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Manevich Y., Townsend D. M., Hutchens S., Tew K. D. (2010) Diazeniumdiolate mediated nitrosative stress alters nitric oxide homeostasis through intracellular calcium and S-glutathionylation of nitric oxide synthetase. PLoS ONE 5, e14151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jin X., Yu L., Wu Y., Zhang S., Shi Z., Chen X., Yang Y., Zhang X., Jiang C. (2012) S-Glutathionylation underscores the modulation of the heteromeric Kir4.1-Kir5.1 channel in oxidative stress. J. Physiol. 590, 5335–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Abdelsaid M. A., El-Remessy A. B. (2012) S-Glutathionylation of LMW-PTP regulates VEGF-mediated FAK activation and endothelial cell migration. J. Cell Sci. 125, 4751–4760 [DOI] [PubMed] [Google Scholar]
- 56. Kim H. S., Ullevig S. L., Zamora D., Lee C. F., Asmis R. (2012) Redox regulation of MAPK phosphatase 1 controls monocyte migration and macrophage recruitment. Proc. Natl. Acad. Sci. U.S.A. 109, E2803–E2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sun R., Eriksson S., Wang L. (2012) Oxidative stress induced S-glutathionylation and proteolytic degradation of mitochondrial thymidine kinase 2. J. Biol. Chem. 287, 24304–24312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Petrushanko I. Y., Yakushev S., Mitkevich V. A., Kamanina Y. V., Ziganshin R. H., Meng X., Anashkina A. A., Makhro A., Lopina O. D., Gassmann M., Makarov A. A., Bogdanova A. (2012) S-Glutathionylation of the Na,K-ATPase catalytic α subunit is a determinant of the enzyme redox sensitivity. J. Biol. Chem. 287, 32195–32205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xiong Y., Manevich Y., Tew K. D., Townsend D. M. (2012) S-Glutathionylation of protein disulfide isomerase regulates estrogen receptor α stability and function. Int. J. Cell Biol. 2012, 273549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cabiscol E., Levine R. L. (1996) The phosphatase activity of carbonic anhydrase III is reversibly regulated by glutathiolation. Proc. Natl. Acad. Sci. U.S.A. 93, 4170–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lock J. T., Sinkins W. G., Schilling W. P. (2012) Protein S-glutathionylation enhances Ca2+-induced Ca2+ release via the IP3 receptor in cultured aortic endothelial cells. J. Physiol. 590, 3431–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kil I. S., Shin S. W., Park J. W. (2012) S-Glutathionylation regulates GTP-binding of Rac2. Biochem. Biophys. Res. Commun. 425, 892–896 [DOI] [PubMed] [Google Scholar]
- 63. Tew K. D., Manevich Y., Grek C., Xiong Y., Uys J., Townsend D. M. (2011) The role of glutathione S-transferase P in signaling pathways and S-glutathionylation in cancer. Free Radic. Biol. Med. 51, 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xiong Y., Uys J. D., Tew K. D., Townsend D. M. (2011) S-Glutathionylation: from molecular mechanisms to health outcomes. Antioxid. Redox Signal. 15, 233–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Feig C., Peter M. E. (2007) How apoptosis got the immune system in shape. Eur. J. Immunol. 37, S61–S70 [DOI] [PubMed] [Google Scholar]
- 66. Anathy V., Aesif S. W., Guala A. S., Havermans M., Reynaert N. L., Ho Y. S., Budd R. C., Janssen-Heininger Y. M. (2009) Redox amplification of apoptosis by caspase-dependent cleavage of glutaredoxin 1 and S-glutathionylation of Fas. J. Cell Biol. 184, 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Anathy V., Roberson E., Cunniff B., Nolin J. D., Hoffman S., Spiess P., Guala A. S., Lahue K. G., Goldman D., Flemer S., van der Vliet A., Heintz N. H., Budd R. C., Tew K. D., Janssen-Heininger Y. M. (2012) Oxidative processing of latent Fas in the endoplasmic reticulum controls the strength of apoptosis. Mol. Cell. Biol. 32, 3464–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hobbs G. A., Bonini M. G., Gunawardena H. P., Chen X., Campbell S. L. (2013) Glutathiolated Ras: characterization and implications for Ras activation. Free Radic. Biol. Med. 57, 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Harris M. J., Coggan M., Langton L., Wilson S. R., Board P. G. (1998) Polymorphism of the Pi class glutathione S-transferase in normal populations and cancer patients. Pharmacogenetics 8, 27–31 [DOI] [PubMed] [Google Scholar]
- 70. Zimniak P., Nanduri B., Pikuła S., Bandorowicz-Pikuła J., Singhal S. S., Srivastava S. K., Awasthi S., Awasthi Y. C. (1994) Naturally occurring human glutathione S-transferase GSTP1-1 isoforms with isoleucine and valine in position 104 differ in enzymic properties. Eur. J. Biochem. 224, 893–899 [DOI] [PubMed] [Google Scholar]
- 71. Manevich Y., Hutchens S., Tew K. D., Townsend D. M. (2013) Allelic variants of glutathione S-transferase P1-1 differentially mediate the peroxidase function of peroxiredoxin VI and alter membrane lipid peroxidation. Free Radic. Biol. Med. 54, 62–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hayes J. D., Pulford D. J. (1995) The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 30, 445–600 [DOI] [PubMed] [Google Scholar]
- 73. Townsend D. M. (2007) S-Glutathionylation: indicator of cell stress and regulator of the unfolded protein response. Mol. Interv. 7, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Giustarini D., Rossi R., Milzani A., Colombo R., Dalle-Donne I. (2004) S-Glutathionylation: from redox regulation of protein functions to human diseases. J. Cell. Mol. Med. 8, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Niwa T., Naito C., Mawjood A. H., Imai K. (2000) Increased glutathionyl hemoglobin in diabetes mellitus and hyperlipidemia demonstrated by liquid chromatography/electrospray ionization-mass spectrometry. Clin. Chem. 46, 82–88 [PubMed] [Google Scholar]
- 76. Takayama F., Tsutsui S., Horie M., Shimokata K., Niwa T. (2001) Glutathionyl hemoglobin in uremic patients undergoing hemodialysis and continuous ambulatory peritoneal dialysis. Kidney Int. Suppl. 78, S155–S158 [DOI] [PubMed] [Google Scholar]
- 77. Velu C. S., Niture S. K., Doneanu C. E., Pattabiraman N., Srivenugopal K. S. (2007) Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry 46, 7765–7780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Iwasaki H., Suda T. (2009) Cancer stem cells and their niche. Cancer Sci. 100, 1166–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jang Y. Y., Sharkis S. J. (2007) A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood 110, 3056–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chambers S. M., Shaw C. A., Gatza C., Fisk C. J., Donehower L. A., Goodell M. A. (2007) Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 5, e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Townsend D. M., Manevich Y., He L., Xiong Y., Bowers R. R., Jr., Hutchens S., Tew K. D. (2009) Nitrosative stress-induced S-glutathionylation of protein disulfide isomerase leads to activation of the unfolded protein response. Cancer Res. 69, 7626–7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Brunsing R., Omori S. A., Weber F., Bicknell A., Friend L., Rickert R., Niwa M. (2008) B- and T-cell development both involve activity of the unfolded protein response pathway. J. Biol. Chem. 283, 17954–17961 [DOI] [PubMed] [Google Scholar]
- 83. Grek C. L., Townsend D. M., Tew K. D. (2011) The impact of redox and thiol status on the bone marrow: pharmacological intervention strategies. Pharmacol. Ther. 129, 172–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ceradini D. J., Yao D., Grogan R. H., Callaghan M. J., Edelstein D., Brownlee M., Gurtner G. C. (2008) Decreasing intracellular superoxide corrects defective ischemia-induced new vessel formation in diabetic mice. J. Biol. Chem. 283, 10930–10938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kubo M., Li T. S., Suzuki R., Ohshima M., Qin S. L., Hamano K. (2007) Short-term pretreatment with low-dose hydrogen peroxide enhances the efficacy of bone marrow cells for therapeutic angiogenesis. Am. J. Physiol. Heart Circ. Physiol. 292, H2582–H2588 [DOI] [PubMed] [Google Scholar]
- 86. Winkler I. G., Hendy J., Coughlin P., Horvath A., Lévesque J. P. (2005) Serine protease inhibitors serpina1 and serpina3 are down-regulated in bone marrow during hematopoietic progenitor mobilization. J. Exp. Med. 201, 1077–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. van Pel M., van Os R., Velders G. A., Hagoort H., Heegaard P. M., Lindley I. J., Willemze R., Fibbe W. E. (2006) Serpina1 is a potent inhibitor of IL-8-induced hematopoietic stem cell mobilization. Proc. Natl. Acad. Sci. U.S.A. 103, 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Griffiths S. W., King J., Cooney C. L. (2002) The reactivity and oxidation pathway of cysteine 232 in recombinant human α1-antitrypsin. J. Biol. Chem. 277, 25486–25492 [DOI] [PubMed] [Google Scholar]
- 89. Grek C. L., Townsend D. M., Uys J. D., Manevich Y., Coker W. J., 3rd, Pazoles C. J., Tew K. D. (2012) S-Glutathionylated serine proteinase inhibitors as plasma biomarkers in assessing response to redox-modulating drugs. Cancer Res. 72, 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Przygodzka P., Ramstedt B., Tengel T., Larsson G., Wilczynska M. (2010) Bomapin is a redox-sensitive nuclear serpin that affects responsiveness of myeloid progenitor cells to growth environment. BMC Cell Biol. 11, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Mieyal J. J., Gallogly M. M., Qanungo S., Sabens E. A., Shelton M. D. (2008) Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid. Redox Signal. 10, 1941–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gao X. H., Bedhomme M., Veyel D., Zaffagnini M., Lemaire S. D. (2009) Methods for analysis of protein glutathionylation and their application to photosynthetic organisms. Mol. Plant 2, 218–235 [DOI] [PubMed] [Google Scholar]
- 93. Michelet L., Zaffagnini M., Vanacker H., Le Maréchal P., Marchand C., Schroda M., Lemaire S. D., Decottignies P. (2008) In vivo targets of S-thiolation in Chlamydomonas reinhardtii. J. Biol. Chem. 283, 21571–21578 [DOI] [PubMed] [Google Scholar]
- 94. Yang Y., Shi W., Cui N., Wu Z., Jiang C. (2010) Oxidative stress inhibits vascular KATP channels by S-glutathionylation. J. Biol. Chem. 285, 38641–38648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Pizarro G. O., Ogut O. (2009) Impact of actin glutathionylation on the actomyosin-S1 ATPase. Biochemistry 48, 7533–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Landino L. M., Moynihan K. L., Todd J. V., Kennett K. L. (2004) Modulation of the redox state of tubulin by the glutathione/glutaredoxin reductase system. Biochem. Biophys. Res. Commun. 314, 555–560 [DOI] [PubMed] [Google Scholar]
- 97. Go Y. M., Pohl J., Jones D. P. (2009) Quantification of redox conditions in the nucleus. Methods Mol. Biol. 464, 303–317 [DOI] [PubMed] [Google Scholar]
- 98. Zhang C., Rodriguez C., Circu M. L., Aw T. Y., Feng J. (2011) S-Glutathionyl quantification in the attomole range using glutaredoxin-3-catalyzed cysteine derivatization and capillary gel electrophoresis with laser-induced fluorescence detection. Anal. Bioanal. Chem. 401, 2165–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hamnell-Pamment Y., Lind C., Palmberg C., Bergman T., Cotgreave I. A. (2005) Determination of site-specificity of S-glutathionylated cellular proteins. Biochem. Biophys. Res. Commun. 332, 362–369 [DOI] [PubMed] [Google Scholar]
- 100. Menon D., Board P. G. (2013) A fluorometric method to quantify protein glutathionylation using glutathione derivatization with 2,3-naphthalenedicarboxaldehyde. Anal. Biochem. 433, 132–136 [DOI] [PubMed] [Google Scholar]


