Abstract
Living organisms possess biological clocks that resonate with environmental cycles in light, temperature, and food availability. Recently, circadian oscillations in the redox state of peroxiredoxin have been described as an additional non-transcriptional timekeeping mechanism. Of note, this redox cycle is conserved in both prokaryotes and eukaryotes. How the classical “transcription-translation feedback loop” model and this redox oscillation are related is still poorly understood. In this minireview, we describe the most recent evidence pointing to cross-talk between the circadian clock and the redox status of the cell.
Keywords: Circadian, Circadian Clock, Circadian Rhythms, Hydrogen Peroxide, Peroxiredoxin, Redox, Redox Regulation, Redox Signaling, Thioredoxin, Thioredoxin Reductase
Introduction
The integration of biological clocks into cellular physiology has represented an important evolutionary advantage for multicellular and unicellular organisms, allowing them to anticipate and adapt to cyclical changes in environmental cues such as light, temperature, and food availability (1). The advantage conferred by resonating with environmental cycles has been technically challenging to demonstrate. However, pioneering experiments have shown that coordination with light/dark cycles can improve fitness in bacteria, flies, and plants (2–5).
In mammals, the timing system is composed of a series of biological clocks organized in a hierarchical manner. The main clock, also known as the “master pacemaker,” resides in the paired suprachiasmatic nuclei (SCN)2 of the hypothalamus, which receive and process light signals to achieve synchronization with the external environment. Through the release of hormones and neuropeptides, the SCN coordinate several other clocks distributed in different tissues and organs. These peripheral clocks in turn generate local self-sustained circadian rhythms (from Latin circa diem, about a day) of physiological processes to control tissue-specific functions (6–8).
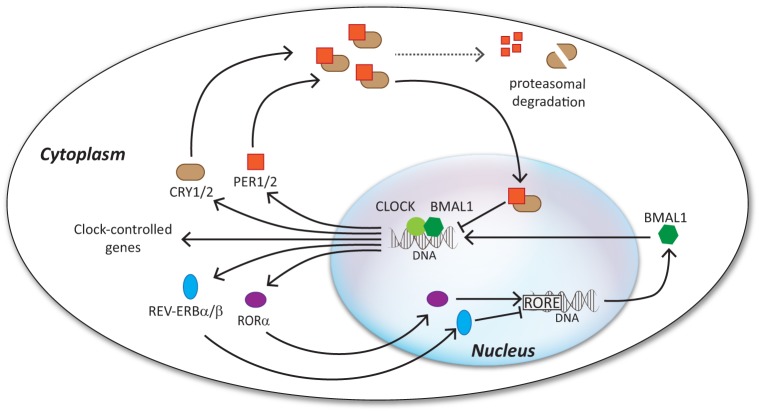
The first insights into the molecular mechanism of cellular rhythmicity came from relatively recent studies in Drosophila and Neurospora crassa. These studies showed that rhythmic oscillations in the expression of clock-controlled genes are generated by transcription-translation feedback loops (TTFLs) and that they are necessary to coordinate behavioral rhythmicity (9, 10). Similar timekeeping logic was later described in other organisms, although with different genes involved and different levels of complexity in the transcriptional circuits (11, 12). In mammals, for example, two positive activators, CLOCK (circadian locomotor output cycles kaput) and BMAL1 (brain and muscle Arnt-like protein 1), initiate transcription of the Period1/2 (Per1/2), Cryptochrome1/2 (Cry1/2), Rorα (retinoic acid receptor-related orphan receptor α), and Rev-erbα/β genes. When the level of expression of PER and CRY proteins reaches a particular threshold, they translocate into the nucleus and inhibit the transcriptional activity of the CLOCK-BMAL1 heterodimer, thereby blocking their own transcription. An additional loop is created by the REV-ERBα/β and RORα proteins, which instead repress or activate transcription of the Bmal1 gene, respectively (Fig. 1) (13). This classical model based on transcription has been slightly revisited in light of new data showing that proteasomal degradation, epigenetic modulation of gene expression, and post-translational modifications of mRNA play a key role in keeping rhythmicity (11, 14–16). For example, the turnover of PER and CRY is controlled by phosphorylation-mediated ubiquitination processes (17–21).
FIGURE 1.
Mammalian TTFL. The mammalian clock is sustained by a series of feedback loops involving several genes and the proteins that they encode. The two positive activators, CLOCK and BMAL1, initiate the transcription of the clock genes Per1/2, Cry1/2, Rorα, and Rev-erbα/β. PER1/2 and CRY1/2 proteins accumulate, dimerize, and translocate into the nucleus, where they bind to the CLOCK-BMAL1 dimer, thereby inhibiting its transcriptional activity. Eventually, proteasomal degradation of PER1/2 and CRY1/2 relieves the transcriptional repression on the CLOCK-BMAL1 complex, and the cycle can restart again. An additional loop involves the nuclear receptors RORα and REV-ERBα/β, which activate and repress the transcription of Bmal1, respectively. RORE, retinoic acid receptor-related orphan receptor response element.
Although conserved in many organisms, the TTFL cannot be considered as a universal building block for circadian clocks (11). For instance, the yeast Saccharomyces cerevisiae and the worm Caenorhabditis elegans show circadian rhythms but do not express the classical “clock genes” (22–24). Also, the cyanobacterium Synechococcus elongatus and the filamentous fungus N. crassa tend to favor protein phosphorylation as their basic timing mechanism (25, 26). Very recently, biochemical oscillations of the redox state of the protein peroxiredoxin (Prx) have been described as an additional timekeeping mechanism conserved in both eukaryotes and prokaryotes (12, 27, 28). These findings have thus revealed an intriguing link between the redox status of the cell and circadian clocks. We will discuss what we know about clock-relevant redox control systems and the reciprocal regulation between the redox state of the cells and circadian clocks.
Oxidative State and Redox Control Systems
The cellular redox environment is determined by the balance between the generation of oxidants and free radicals and the level of reducing agents. The most common oxidants are the reactive oxygen species (ROS), which are generated by intracellular enzymes during metabolic reactions. Some examples include superoxide anion (O2⨪), hydroxyl radical (HO•), and hydrogen peroxide (H2O2). To avoid oxidative damage, cells have adopted several detoxification strategies. Non-enzymatic mechanisms involve the synthesis of antioxidant molecules such as ascorbate, tocopherols (including vitamin E), and retinol (vitamin A). Enzymatic mechanisms include proteins such as superoxide dismutase, which catalyzes the dismutation of superoxide into oxygen and hydrogen peroxide, and catalase, which mediates the decomposition of hydrogen peroxide to water and oxygen. Additional redox buffering systems are provided by oxidize/reduced GSH and oxidized/reduced thioredoxin (Trx) (Fig. 2).
FIGURE 2.
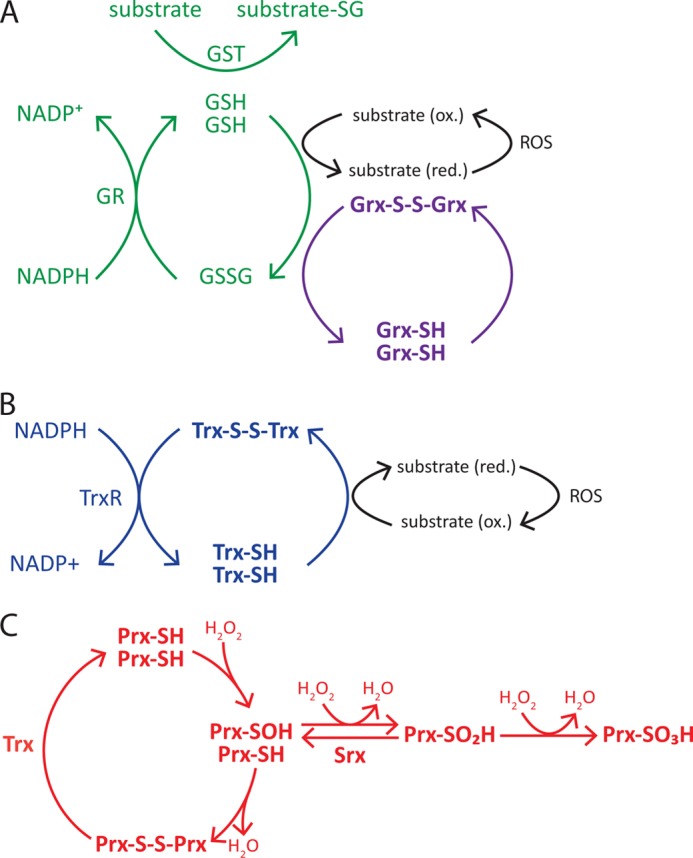
Redox systems. Shown is a schematic representation of the cellular redox systems and main antioxidant enzymes. A, GSH-Grx system. B, Trx system. C, Prx system. substrate-SG, glutathionylated substrate; GR, GSH reductase; ox., oxidized; red., reduced; TrxR, Trx reductase.
GSH is a low molecular weight antioxidant involved in the reduction of disulfide bonds and in the reduction of hydroperoxides by GSH peroxidases. Oxidized GSH (the disulfide GSSG) is potentially dangerous for the cell (29, 30), but it is normally reduced to GSH by GSH reductases via an NADPH-dependent reaction. Disulfide bridges in proteins are also reduced by glutaredoxins (Grxs), which rely on GSH for their non-enzymatic regeneration. GSH can also be conjugated to Cys residues on proteins by GST in a process called glutathionylation, which protects proteins from oxidation (31, 32). Similar to Grx, Trx proteins facilitate the reduction of several proteins by cysteine thiol-disulfide exchange. Oxidized Trxs are eventually reduced by Trx reductases via NADPH-dependent reactions. Among these antioxidant systems, Prxs have recently emerged has key players in the control of circadian rhythms.
Prx Cyclical Oxidation as the Prototype for Redox-regulated Cytosolic Clocks
Prxs are a highly conserved family of antioxidant proteins classified as class 1-Cys and class 2-Cys depending on the number of Cys residues involved in catalysis. In their catalytic site, Prxs contain a “peroxidatic” Cys residue that can be oxidized to a sulfenic acid (Cys-SOH) by an incoming peroxide (Fig. 2). In class 2-Cys Prxs after oxidation, this residue reacts with a “resolving” Cys residue to form an intermolecular (typical and atypical class 2-Cys) or intramolecular (atypical class 2-Cys) disulfide bond, which is eventually reduced by Trx. Typical class 2-Cys Prxs can undergo further oxidation (termed hyperoxidation), which generates sulfinic (Cys-SO2H) and sulfonic (Cys-SO3H) acid forms of the catalytic cysteine (33–35). The sulfinic acid form is catalytically inactive but can be reactivated by sulfiredoxin (Srx) through an ATP-dependent reduction reaction (36). In contrast, the sulfonic acid form is irreversibly oxidized, and its physiological occurrence is controversial (37, 38).
It has been recently demonstrated that Prxs follow circadian cycles of oxidation. In a recent study, in fact, the levels of dimeric Prx-SO2/3H were shown to oscillate with a period of 24 h with peaks of hyperoxidation at 12 h (circadian time) (27). Strikingly, these oscillations were demonstrated to occur in RBCs, which do not possess DNA, showing that Prx oscillations occur even in the absence of gene transcription (27). Oscillations in Prx have also been found in the small protist Ostreococcus tauri, which, contrary to RBCs, possesses an endogenous clock driven by transcription and translation of recognized plant clock genes (39). Importantly, oscillations in Prx could be detected also when this organism was shifted into a dark environment, a condition under which gene transcription of O. tauri is known to stop (28). In addition, when the organism was brought back to light, the clock did not reset, suggesting that a mechanism must be in place to keep track of time even in the absence of gene transcription. These studies therefore show that Prx redox cycling events could be an important mechanism for timekeeping.
Of note, circadian oxidation of Prx has been found not only in eukaryotes (including algae, fungi, flies, worms, and mammals) but also in archaea and bacteria (12, 24, 27, 28), suggesting that these oscillations might have been integrated early in evolution and likely coevolved with differing TTFLs in each organism. A key unanswered question is what determines Prx oscillations. Srx, which reduces the inactive sulfinic acid form into the active sulfenic acid form, might indeed account for these oscillations. However, some organisms that display oscillations in Prx do not express Srx homologs (i.e. C. elegans and N. crassa), suggesting that other mechanisms might be in place.
Given the highly conserved redox component of circadian oscillations, it is an important goal to now understand the relationship between the classical TTFL and Prx oscillations (12). Interestingly, when the transcriptional machinery is disrupted (e.g. in behavioral arrhythmic Drosophila mutants or in N. crassa mutants exhibiting a lengthened period), Prx oscillations are perturbed in phase, suggesting that gene transcription is not necessary but is related to cellular metabolic cycles. Along the same lines, when the Prx clock system is abolished, as occurs in mutants of S. elongatus and Arabidopsis thaliana deficient in well annotated 2-Cys Prx genes, circadian rhythms of clock genes persist with the same period as in control organisms, but are perturbed in either phase or amplitude (12). Taken together, these studies show that TTFL and Prx cycles are intertwined but potentially autonomous components of the circadian system. These results also raise the possibility that the redox status of the cell fluctuates and that these oscillations have critical and as yet incompletely understood biological consequences.
The Reciprocal Relationship between the Redox State and Circadian System
Initial hints that redox metabolism might be linked to the circadian clock were provided by work done by Rutter et al. (40) in which the ratio between oxidized and reduced forms of NAD and NADP cofactors was shown to regulate the DNA-binding activity of the CLOCK/NPAS2 (neuronal PAS domain protein 2)-BMAL1 heterodimer. However, these studies were purely biochemical, based solely on the use of purified recombinant proteins, and used concentrations of reactants much higher than is seen physiologically, making their wider interpretation difficult, especially in an in vivo context. More recently, in vivo oscillations in the redox state of FAD and NADPH have been described in organotypic slices of SCN (41). This study demonstrated that the redox state of SCN oscillates in a self-sustained fashion and that these oscillations contribute to determining the excitability of SCN neurons via non-transcriptional regulation of potassium channels. However, the connection between the transcriptional clock and redox oscillations in this tissue requires further investigation. Whether redox fluctuations are an output of circadian rhythms or whether they can act as input, or indeed both, is still under intense investigation.
In favor of a mechanistic link between redox fluctuations and the regulation of gene expression, studies in zebrafish demonstrated that changes in redox state actively control the expression of light-dependent genes. Light, which is the key entraining stimulus in this organism, generates H2O2, which in turn regulates the expression of the clock genes zCry1 and zPer2. Interestingly, oscillations in the mRNA levels of these genes are paralleled by antiphasic oscillations in mRNA and the activity of catalase (42), suggesting that this enzyme is involved in the control of H2O2-mediated circadian gene expression. Recently, LdpA (light-dependent period A), a component of the cyanobacterial circadian clock, was proposed to act as a redox sensor and to be used by the clock to adjust the period length (43). LdpA contains iron-sulfur centers and can sense the redox state of the cell, which correlates with the amount of light (high light correlates with a reduced redox state, whereas low light is associated with an oxidized redox state). Interestingly, on the basis of the light conditions, LdpA modulates the levels of CikA and KaiA, the latter of which is a key component of the central oscillator (44), thereby affecting the period length. Furthermore, cyanobacteria exposed to high light conditions show short periods, whereas cyanobacteria exposed to low light conditions display long periods. Finally, the effects of altered ROS and the circadian clock have also been observed in N. crassa (45, 46) and in the cyanobacterium Microcystis aeruginosa (47), in which H2O2 has been shown to impact on the daily expression pattern of clock genes as well as clock-controlled genes, including those involved in coordinating photosynthesis. These results clearly show that fluctuations in the redox state of the cells have an impact on the expression of clock-related genes in multiple diverse systems.
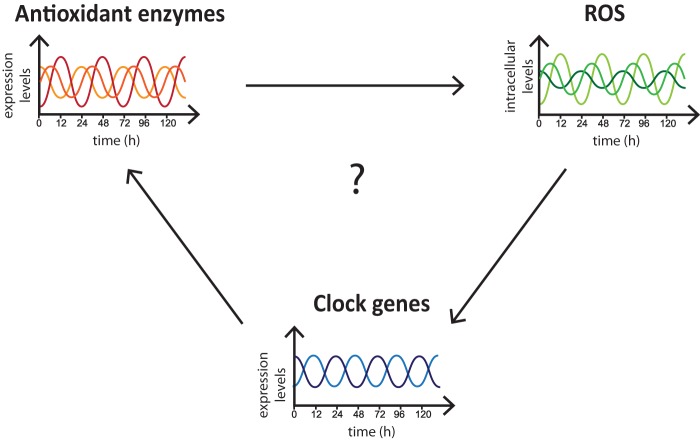
This scenario is further complicated by the finding that clock genes can in turn regulate the expression of antioxidant enzymes, thus providing an important and novel feedback loop (Fig. 3). For instance, in A. thaliana, the circadian clock coordinates ROS homeostasis and ROS-responsive genes, and H2O2 production and scavenging exhibit diurnal rhythms (48). Importantly, mutations in the core clock regulator CCA1 (circadian clock-associated 1) or in other components of the TTFL affect this time of the day specific pattern. In addition, it was observed that ROS can feed back to affect the transcription of clock-regulated genes. The importance of this cross-talk has been underlined in Drosophila melanogaster, in which the per gene has been shown to be essential for maintaining antioxidant defense. Indeed, flies exposed to H2O2 show daily mortality rhythms and are more susceptible during the late light phase. Mutation in the per gene abolishes this time of the day sensitivity and renders flies more susceptible to oxidative stress in general (49). Bmal1−/− mice show higher accumulation of ROS in several tissues compared with wild-type animals. This impairment in ROS homeostasis correlates with early aging and age-dependent pathologies. These data again suggest a connection between the circadian clock and redox homeostasis (50). More recently, the circadian system has been shown to also modulate the pathways involved in production and utilization of GSH (51). Wild-type Canton S flies show daily rhythms in the mRNA levels of glutamate-cysteine ligase, the rate-limiting enzyme in GSH biosynthesis, and GSTD1, which utilizes GSH in cellular detoxification. Importantly, mutants lacking the clock genes per and cyc show no rhythms in the expression of these proteins, underlying the link between GSH metabolism and the circadian system.
FIGURE 3.
Cross-talk between the circadian clock and redox homeostasis. The circadian clock and the redox state of the cell are interconnected. The expression level and activity of antioxidant enzymes determine the levels of intracellular ROS, which have been shown to impinge on the expression pattern of clock genes. In addition, some antioxidant enzymes have been shown to follow a circadian pattern of expression, suggesting that the clock system can regulate redox homeostasis.
Compartmentalization of Oxidative State and Redox Signaling: Future Perspectives
An emerging feature of redox signaling is its spatial and temporal compartmentalization. Recent developments highlight that different ROS signaling and redox buffering systems are spatially segregated and can have unique compartmentalized functions (Fig. 4) (52–55). For example, pools of mitochondrial, cytosolic, and nuclear GSH are separated within cells, and the trafficking of GSH, from the cytosol to the mitochondrial intramembrane space, is tightly regulated by porins in their membranes (56). Importantly, the maintenance of localized redox states is critical for cell function. Mitochondria-specific depletion of GSH makes mitochondria more sensitive to oxidative damage (57), whereas overexpression of the mitochondrial glutaredoxin Grx2 protects against oxidative stress to prevent apoptosis (58). The nuclear redox state is similarly pivotal for the activation of several redox-regulated transcription factors such as CLOCK and NPAS2 (40), NF-κB (59), Nrf2 (nuclear factor (erythroid-derived 2)-like 2) (60), and Rev-erbβ (61).
FIGURE 4.
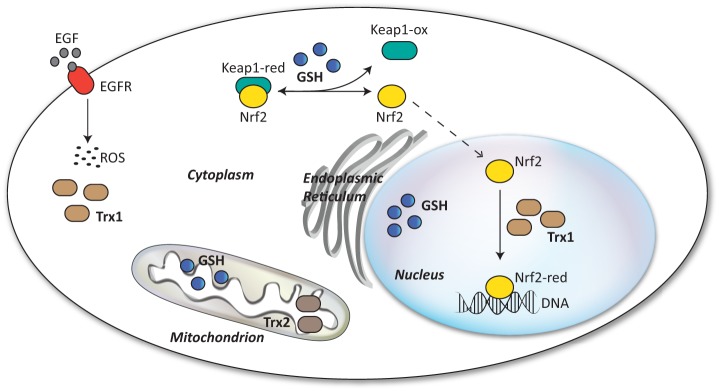
Compartmentalization of redox systems. Redox systems are compartmentalized, and pools of antioxidant enzymes are distributed differently in the cell. Pools of GSH and Trx have been described in the cytosolic, mitochondrial, and nuclear compartments. The cytosolic pool of Trx1 has been shown to limit ROS generated upon EGF receptor (EGFR) activation. The nuclear translocation of Nrf2 is regulated by a redox switch controlled by GSH and Keap1 oxidation, whereas its DNA-binding activity is regulated by a nuclear pool of Trx1. ox, oxidized; red, reduced.
Although evidence suggests that ROS are bona fide signaling molecules, some skepticism has been raised because of their high reactivity and low substrate specificity. However, there is evidence of tight coupling of ROS generators to the activity of antioxidant buffering systems and to specific targets, which would explain how the specificity of ROS signaling is brought about (62–64). In the adrenal gland, for example, H2O2 is involved in a feedback control loop to regulate corticosteroid synthesis (65). In the last phase of adrenocorticotropic hormone-induced steroidogenesis, cholesterol is imported in mitochondria, where cytochrome P450 enzymes catalyze the oxidative cleavage of its side chain. As a byproduct of their activity, cytochromes generate H2O2, which is eliminated by Prx3. During the catalytic cycle, Prx3 can occasionally be inactivated by hyperoxidation. Its activity is normally reverted by Srx. However, when corticosteroid synthesis increases, so does H2O2, and Srx activity is no longer sufficient to reduce and reactivate Prx3. This causes a further increase in H2O2 levels and the overflow of H2O2 in the cytosol. This last event triggers a signaling cascade involving p38 MAPK, which eventually inhibits corticosteroid synthesis. Of note, the levels of inactivated Prx3, activated p38 MAPK, and Srx exhibit circadian oscillations. In addition, tissue-specific ablation of Srx results in suppression of the adrenal circadian rhythms of corticosterone production, suggesting that Prx hyperoxidation, corticosteroid synthesis, and the circadian clock are interconnected.
Interestingly, oxidative signals can cause selective oxidation of specific redox couples. For example, EGF-mediated ROS signaling selectively oxidizes the cytosolic pool of Trx1 but not the mitochondrial pool of Trx2 (Fig. 4) (66, 67), suggesting that these pools are independently regulated. Furthermore, one of the major transcription factors activated by oxidative stress, Nrf2 can be differentially activated by redox signals: its translocation is promoted by a redox switch of Keap1 (Kelch-like ECH-associated protein 1), which is controlled by GSH, whereas its nuclear activity is under the control of Trx1 (Fig. 4) (60).
It is tempting to speculate that different redox systems are strategically located within the cell not only to protect substrates from excessive oxidation but also to regulate specific signaling pathways. In addition, different redox couples might act in concert to specifically modulate the response to ROS signals in proximity of key redox-sensitive proteins. Determining how this compartmentalized nature of cellular redox systems links to the clockwork will be critical to fully understand how the cell en masse keeps daily time. We believe that this will be an exciting area of investigation in the next few years.
Conclusions
Substantial evidence highlights the capability of living organisms to resonate with environmental cycles, which confers an evolutionary advantage because perturbing the clockwork reduces fitness. However, the biological mechanisms underlying the regulation of circadian rhythms are still elusive in the light of new insights coming from redox biology. In the post-genomic era, the dominance of gene regulation at the heart of circadian rhythms needs to be reconciled with mounting evidence demonstrating the importance of redox cycles and post-transcriptional/post-translational modifications (68).
It now appears that control of ROS signaling is deeply intertwined in the circadian clock system. Disruption of circadian rhythms in humans has been linked to several diseases such as breast cancer, obesity, diabetes, sleep disorders, and neurodegenerative diseases (69). Given the role of ROS in human pathophysiology, it is tempting to speculate that some of the pathologies associated with the deregulation of clock signaling are partially caused by alteration in redox signaling and possibly their compartmentalized nature. Thus, we propose that the understanding of how localized ROS production affects the activity of oscillators within cells will have important consequences for the development of dedicated therapies aimed at restoring aberrant signaling.
This work was supported by Wellcome Trust Grant 083643/Z/07/Z, European Research Council Starting Grant 281348 (MetaCLOCK), the EMBO Young Investigators Programme, the Medical Research Council (MRC) Centre for Obesity and Related Metabolic Disorders, and the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre. This is the sixth article in the Thematic Minireview Series on Redox-active Protein Modifications and Signaling.
- SCN
- suprachiasmatic nuclei
- TTFL
- transcription-translation feedback loop
- Prx
- peroxiredoxin
- ROS
- reactive oxygen species
- Trx
- thioredoxin
- Grx
- glutaredoxin
- Srx
- sulfiredoxin.
REFERENCES
- 1. Reddy A. B., O'Neill J. S. (2010) Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 20, 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ouyang Y., Andersson C. R., Kondo T., Golden S. S., Johnson C. H. (1998) Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 95, 8660–8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Woelfle M. A., Ouyang Y., Phanvijhitsiri K., Johnson C. H. (2004) The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr. Biol. 14, 1481–1486 [DOI] [PubMed] [Google Scholar]
- 4. Dodd A. N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J. M., Millar A. J., Webb A. A. (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633 [DOI] [PubMed] [Google Scholar]
- 5. Beaver L. M., Gvakharia B. O., Vollintine T. S., Hege D. M., Stanewsky R., Giebultowicz J. M. (2002) Loss of circadian clock function decreases reproductive fitness in males of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 99, 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balsalobre A., Damiola F., Schibler U. (1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937 [DOI] [PubMed] [Google Scholar]
- 7. Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. (2004) Circadian gene expression in individual fibroblasts. Cell 119, 693–705 [DOI] [PubMed] [Google Scholar]
- 8. Welsh D. K., Yoo S. H., Liu A. C., Takahashi J. S., Kay S. A. (2004) Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 14, 2289–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hardin P. E., Hall J. C., Rosbash M. (1990) Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343, 536–540 [DOI] [PubMed] [Google Scholar]
- 10. Aronson B. D., Johnson K. A., Loros J. J., Dunlap J. C. (1994) Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science 263, 1578–1584 [DOI] [PubMed] [Google Scholar]
- 11. Lakin-Thomas P. L. (2006) Transcriptional feedback oscillators: maybe, maybe not … J. Biol. Rhythms 21, 83–92 [DOI] [PubMed] [Google Scholar]
- 12. Edgar R. S., Green E. W., Zhao Y., van Ooijen G., Olmedo M., Qin X., Xu Y., Pan M., Valekunja U. K., Feeney K. A., Maywood E. S., Hastings M. H., Baliga N. S., Merrow M., Millar A. J., Johnson C. H., Kyriacou C. P., O'Neill J. S., Reddy A. B. (2012) Peroxiredoxins are conserved markers of circadian rhythms. Nature 485, 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guillaumond F., Dardente H., Giguère V., Cermakian N. (2005) Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms 20, 391–403 [DOI] [PubMed] [Google Scholar]
- 14. Lowrey P. L., Takahashi J. S. (2011) Genetics of circadian rhythms in mammalian model organisms. Adv. Genet. 74, 175–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mehra A., Baker C. L., Loros J. J., Dunlap J. C. (2009) Post-translational modifications in circadian rhythms. Trends Biochem. Sci. 34, 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng D., Lazar M. (2012) Clocks, metabolism, and the epigenome. Mol. Cell 47, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eide E. J., Woolf M. F., Kang H., Woolf P., Hurst W., Camacho F., Vielhaber E. L., Giovanni A., Virshup D. M. (2005) Control of mammalian circadian rhythm by CKI-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 25, 2795–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Siepka S. M., Yoo S. H., Park J., Song W., Kumar V., Hu Y., Lee C., Takahashi J. S. (2007) Circadian mutant Overtime reveals F-box protein FBXL3 regulation of Cryptochrome and Period gene expression. Cell 129, 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Busino L., Bassermann F., Maiolica A., Lee C., Nolan P. M., Godinho S. I., Draetta G. F., Pagano M. (2007) SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316, 900–904 [DOI] [PubMed] [Google Scholar]
- 20. Godinho S. I., Maywood E. S., Shaw L., Tucci V., Barnard A. R., Busino L., Pagano M., Kendall R., Quwailid M. M., Romero M. R., O'Neill J., Chesham J. E., Brooker D., Lalanne Z., Hastings M. H., Nolan P. M. (2007) The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316, 897–900 [DOI] [PubMed] [Google Scholar]
- 21. Yoo S. H., Mohawk J. A., Siepka S. M., Shan Y., Huh S. K., Hong H. K., Kornblum I., Kumar V., Koike N., Xu M., Nussbaum J., Liu X., Chen Z., Chen Z. J., Green C. B., Takahashi J. S. (2013) Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152, 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van der Linden A. M., Beverly M., Kadener S., Rodriguez J., Wasserman S., Rosbash M., Sengupta P. (2010) Genome-wide analysis of light- and temperature-entrained circadian transcripts in Caenorhabditis elegans. PLoS Biol. 8, e1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eelderink-Chen Z., Mazzotta G., Sturre M., Bosman J., Roenneberg T., Merrow M. (2010) A circadian clock in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 107, 2043–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Olmedo M., O'Neill J. S., Edgar R. S., Valekunja U. K., Reddy A. B., Merrow M. (2012) Circadian regulation of olfaction and an evolutionarily conserved, nontranscriptional marker in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 109, 20479–20484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vitalini M. W., de Paula R. M., Goldsmith C. S., Jones C. A., Borkovich K. A., Bell-Pedersen D. (2007) Circadian rhythmicity mediated by temporal regulation of the activity of p38 MAPK. Proc. Natl. Acad. Sci. U.S.A. 104, 18223–18228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakajima M., Imai K., Ito H., Nishiwaki T., Murayama Y., Iwasaki H., Oyama T., Kondo T. (2005) Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308, 414–415 [DOI] [PubMed] [Google Scholar]
- 27. O'Neill J. S., Reddy A. B. (2011) Circadian clocks in human red blood cells. Nature 469, 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O'Neill J. S., van Ooijen G., Dixon L. E., Troein C., Corellou F., Bouget F. Y., Reddy A. B., Millar A. J. (2011) Circadian rhythms persist without transcription in a eukaryote. Nature 469, 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Filomeni G., Rotilio G., Ciriolo M. R. (2003) Glutathione disulfide induces apoptosis in U937 cells by a redox-mediated p38 MAP kinase pathway. FASEB J. 17, 64–66 [DOI] [PubMed] [Google Scholar]
- 30. Park H. A., Khanna S., Rink C., Gnyawali S., Roy S., Sen C. K. (2009) Glutathione disulfide induces neural cell death via a 12-lipoxygenase pathway. Cell Death Differ. 16, 1167–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghezzi P. (2005) Review regulation of protein function by glutathionylation. Free Radic. Res. 39, 573–580 [DOI] [PubMed] [Google Scholar]
- 32. Dalle-Donne I., Rossi R., Colombo G., Giustarini D., Milzani A. (2009) Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem. Sci. 34, 85–96 [DOI] [PubMed] [Google Scholar]
- 33. Hall A., Karplus P. A., Poole L. B. (2009) Typical 2-Cys peroxiredoxins–structures, mechanisms and functions. FEBS J. 276, 2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wood Z. A., Poole L. B., Hantgan R. R., Karplus P. A. (2002) Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry 41, 5493–5504 [DOI] [PubMed] [Google Scholar]
- 35. Wood Z. A., Poole L. B., Karplus P. A. (2003) Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300, 650–653 [DOI] [PubMed] [Google Scholar]
- 36. Rhee S. G., Jeong W., Chang T. S., Woo H. A. (2007) Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int. Suppl. 106, S3–S8 [DOI] [PubMed] [Google Scholar]
- 37. Yang K. S., Kang S. W., Woo H. A., Hwang S. C., Chae H. Z., Kim K., Rhee S. G. (2002) Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J. Biol. Chem. 277, 38029–38036 [DOI] [PubMed] [Google Scholar]
- 38. Lim J. C., Choi H. I., Park Y. S., Nam H. W., Woo H. A., Kwon K. S., Kim Y. S., Rhee S. G., Kim K., Chae H. Z. (2008) Irreversible oxidation of the active-site cysteine of peroxiredoxin to cysteine sulfonic acid for enhanced molecular chaperone activity. J. Biol. Chem. 283, 28873–28880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Corellou F., Schwartz C., Motta J. P., Djouani-Tahri B., Sanchez F., Bouget F., Corellou F., Schwartz C., Motta J., Djouani-Tahri E. B., Sanchez F., Bouget F. Y. (2009) Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote Ostreococcus. Plant Cell 21, 3436–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rutter J., Reick M., Wu L. C., McKnight S. L. (2001) Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293, 510–514 [DOI] [PubMed] [Google Scholar]
- 41. Wang T. A., Yu Y. V., Govindaiah G., Ye X., Artinian L., Coleman T. P., Sweedler J. V., Cox C. L., Gillette M. U. (2012) Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science 337, 839–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hirayama J., Cho S., Sassone-Corsi P. (2007) Circadian control by the reduction/oxidation pathway: catalase represses light-dependent clock gene expression in the zebrafish. Proc. Natl. Acad. Sci. U.S.A. 104, 15747–15752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ivleva N. B., Bramlett M. R., Lindahl P. A., Golden S. S. (2005) LdpA: a component of the circadian clock senses redox state of the cell. EMBO J. 24, 1202–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ishiura M., Kutsuna S., Aoki S., Iwasaki H., Andersson C. R., Tanabe A., Golden S. S., Johnson C. H., Kondo T. (1998) Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523 [DOI] [PubMed] [Google Scholar]
- 45. Yoshida Y., Iigusa H., Wang N., Hasunuma K. (2011) Cross-talk between the cellular redox state and the circadian system in Neurospora. PLoS ONE 6, e28227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gyöngyösi N., Nagy D., Makara K., Ella K., Káldi K. (2013) Reactive oxygen species can modulate circadian phase and period in Neurospora crassa. Free Radic. Biol. Med. 58, 134–143 [DOI] [PubMed] [Google Scholar]
- 47. Qian H., Hu B., Yu S., Pan X., Wu T., Fu Z. (2012) The effects of hydrogen peroxide on the circadian rhythms of Microcystis aeruginosa. PLoS ONE 7, e33347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lai A. G., Doherty C. J., Mueller-Roeber B., Kay S. A., Schippers J. H., Dijkwel P. P. (2012) CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. U.S.A. 109, 17129–17134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Krishnan N., Davis A. J., Giebultowicz J. M. (2008) Circadian regulation of response to oxidative stress in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 374, 299–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kondratov R. V., Kondratova A. A., Gorbacheva V. Y., Vykhovanets O. V., Antoch M. P. (2006) Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 20, 1868–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Beaver L. M., Klichko V. I., Chow E. S., Kotwica-Rolinska J., Williamson M., Orr W. C., Radyuk S. N., Giebultowicz J. M. (2012) Circadian regulation of glutathione levels and biosynthesis in Drosophila melanogaster. PLoS ONE 7, e50454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Go Y. M., Jones D. P. (2008) Redox compartmentalization in eukaryotic cells. Biochim. Biophys. Acta 1780, 1273–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pani G., Bedogni B., Colavitti R., Anzevino R., Borrello S., Galeotti T. (2001) Cell compartmentalization in redox signaling. IUBMB Life 52, 7–16 [DOI] [PubMed] [Google Scholar]
- 54. Smith C. (1996) Compartmentation of glutathione: implications for the study of toxicity and disease. Toxicol. Appl. Pharmacol. 140, 1–12 [DOI] [PubMed] [Google Scholar]
- 55. Mishina N. M., Tyurin-Kuzmin P. A., Markvicheva K. N., Vorotnikov A. V., Tkachuk V. A., Laketa V., Schultz C., Lukyanov S., Belousov V. V. (2011) Does cellular hydrogen peroxide diffuse or act locally? Antioxid. Redox Signal. 14, 1–7 [DOI] [PubMed] [Google Scholar]
- 56. Kojer K., Bien M., Gangel H., Morgan B., Dick T. P., Riemer J. (2012) Glutathione redox potential in the mitochondrial intermembrane space is linked to the cytosol and impacts the Mia40 redox state. EMBO J. 31, 3169–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Marí M., Morales A., Colell A., García-Ruiz C., Kaplowitz N., Fernández-Checa J. (2013) Mitochondrial glutathione: features, regulation and role in disease. Biochim. Biophys. Acta 1830, 3317–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Enoksson M., Fernandes A. P., Prast S., Lillig C. H., Holmgren A., Orrenius S. (2005) Overexpression of glutaredoxin 2 attenuates apoptosis by preventing cytochrome c release. Biochem. Biophys. Res. Commun. 327, 774–779 [DOI] [PubMed] [Google Scholar]
- 59. Hirota K., Murata M., Sachi Y., Nakamura H., Takeuchi J., Mori K., Yodoi J. (1999) Distinct roles of thioredoxin in the cytoplasm and in the nucleus. A two-step mechanism of redox regulation of transcription factor NF-κB. J. Biol. Chem. 274, 27891–27897 [DOI] [PubMed] [Google Scholar]
- 60. Hansen J. M., Watson W. H., Jones D. P. (2004) Compartmentation of Nrf-2 redox control. Regulation of cytoplasmic activation by glutathione and DNA binding by thioredoxin-1. Toxicol. Sci. 82, 308–317 [DOI] [PubMed] [Google Scholar]
- 61. Gupta N., Ragsdale S. (2011) Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor Rev-erbβ. J. Biol. Chem. 286, 4392–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu R. F., Xu Y. C., Ma Z., Nwariaku F. E., Sarosi G. A., Jr., Terada L. S. (2005) Subcellular targeting of oxidants during endothelial cell migration. J. Cell Biol. 171, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ushio-Fukai M. (2006) Localizing NADPH oxidase-derived ROS. Sci. STKE 2006, re8. [DOI] [PubMed] [Google Scholar]
- 64. Hilenski L. L., Clempus R. E., Quinn M. T., Lambeth J. D., Griendling K. K. (2004) Distinct subcellular localizations of Nox1 and Nox4 in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 24, 677–683 [DOI] [PubMed] [Google Scholar]
- 65. Kil I. S., Lee S. K., Ryu K. W., Woo H. A., Hu M. C., Bae S. H., Rhee S. G. (2012) Feedback control of adrenal steroidogenesis via H2O2-dependent, reversible inactivation of peroxiredoxin III in mitochondria. Mol. Cell 46, 584–594 [DOI] [PubMed] [Google Scholar]
- 66. Watson W. H., Jones D. P. (2003) Oxidation of nuclear thioredoxin during oxidative stress. FEBS Lett. 543, 144–147 [DOI] [PubMed] [Google Scholar]
- 67. Halvey P. J., Watson W. H., Hansen J. M., Go Y. M., Samali A., Jones D. P. (2005) Compartmental oxidation of thiol-disulphide redox couples during epidermal growth factor signalling. Biochem. J. 386, 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Reddy A. B., Karp N. A., Maywood E. S., Sage E. A., Deery M., O'Neill J. S., Wong G. K., Chesham J., Odell M., Lilley K. S., Kyriacou C. P., Hastings M. H. (2006) Circadian orchestration of the hepatic proteome. Curr. Biol. 16, 1107–1115 [DOI] [PubMed] [Google Scholar]
- 69. Hastings M. H., Reddy A. B., Maywood E. S. (2003) A clockwork web: circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 4, 649–661 [DOI] [PubMed] [Google Scholar]