Background: myo-Inositol-3-phosphate synthase (MIPS) catalyzes the first step in de novo biosynthesis of inositol in eukaryotes.
Results: MIPS is a phosphoprotein. Phosphorylation regulates the activity of yeast and human MIPS.
Conclusion: Phosphorylation of MIPS is a novel regulatory mechanism of inositol biosynthesis.
Significance: This may explain the causes and consequences of perturbation of inositol metabolism implicated in human disorders.
Keywords: Inositol 1, Phospholipid Metabolism, Phosphorylation, Post-translational Modification, Yeast, myo-Inositol Phosphate Synthase, Phosphorylation Site Mutants, Valproate
Abstract
myo-Inositol-3-phosphate synthase (MIPS) plays a crucial role in inositol homeostasis. Transcription of the coding gene INO1 is highly regulated. However, regulation of the enzyme is not well defined. We previously showed that MIPS is indirectly inhibited by valproate, suggesting that the enzyme is post-translationally regulated. Using 32Pi labeling and phosphoamino acid analysis, we show that yeast MIPS is a phosphoprotein. Mass spectrometry analysis identified five phosphosites, three of which are conserved in the human MIPS. Analysis of phosphorylation-deficient and phosphomimetic site mutants indicated that the three conserved sites in yeast (Ser-184, Ser-296, and Ser-374) and humans (Ser-177, Ser-279, and Ser-357) affect MIPS activity. Both S296A and S296D yeast mutants and S177A and S177D human mutants exhibited decreased enzymatic activity, suggesting that a serine residue is critical at that location. The phosphomimetic mutations S184D (human S279D) and S374D (human S357D) but not the phosphodeficient mutations decreased activity, suggesting that phosphorylation of these two sites is inhibitory. The double mutation S184A/S374A caused an increase in MIPS activity, conferred a growth advantage, and partially rescued sensitivity to valproate. Our findings identify a novel mechanism of regulation of inositol synthesis by phosphorylation of MIPS.
Introduction
Inositol is a six-carbon cyclitol that is ubiquitous in biological systems. It is a precursor for inositol lipids and inositol phosphates that are required for various structural and functional processes, including membrane formation, signaling, and membrane trafficking (1, 2). Inositol plays a major role in the transcriptional regulation of hundreds of genes, including lipid biosynthesis genes that harbor the inositol-sensitive upstream activating sequence (UASINO)3 in their promoters (3). In addition, inositol plays a key role in osmoregulation. For example, kidney cells rely on myo-inositol along with sodium to offset the change in osmolarity generated by the loss of water molecules (4).
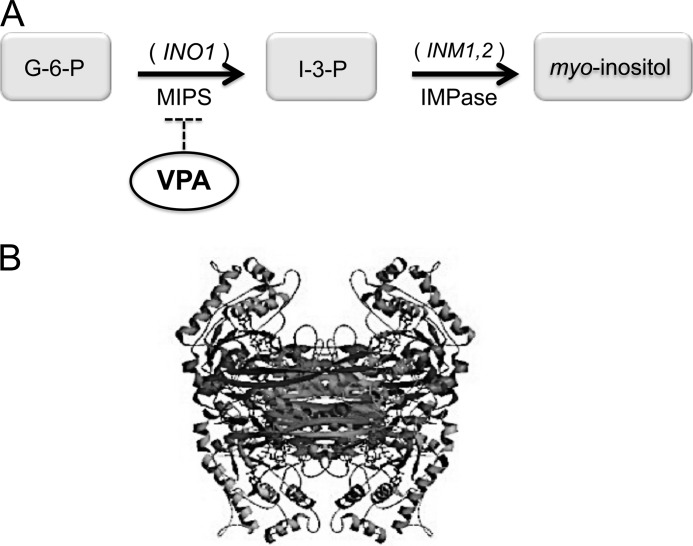
The enzyme responsible for the rate-limiting step of de novo synthesis of inositol is myo-inositol-3-phosphate synthase (MIPS), which catalyzes the conversion of glucose 6-phosphate (G-6-P) to inositol 3-phosphate (I-3-P) in the cytosol in a four-step catalytic reaction using NAD+ as a cofactor (Fig. 1A). MIPS has been isolated from bacteria (5), archaea (6), protozoa (7), plants (8), and animals (9, 10). In yeast, MIPS is encoded by the well characterized INO1 gene (11); in mammals, it is encoded by ISYNA1 and exists as multiple isoforms (12–14). The crystal structure of yeast MIPS shows that it is a homotetramer (Fig. 1B) (15, 16), whereas mammalian MIPS exists as a trimer (10). Each monomer has three major domains: a catalytic domain that binds the substrate glucose 6-phosphate, an NAD+-binding domain, and a central domain, consisting of the N and C termini, which stabilizes the two other domains (17). Sequence analysis showed that the enzyme is highly conserved, with remarkable conservation of the amino acid residues that are important for catalytic activity (18).
FIGURE 1.
MIPS catalyzes the rate-limiting step in inositol biosynthesis. A, MIPS, encoded by INO1, catalyzes the first and rate-limiting step in de novo biosynthesis of inositol. B, the three-dimensional structure of MIPS (Protein Data Bank code 1P1I) (16). G-6-P, glucose 6-phosphate; I-3-P, inositol 3-phosphate.
Regulation of yeast MIPS at the level of INO1 transcription has been extensively studied and well characterized (19–21). The INO1 gene is tightly regulated by inositol. When inositol is limiting, the transcription activators Ino2p and Ino4p form a dimer that binds to the UASINO, leading to the transcription of INO1. However, when inositol is available, the negative regulator Opi1p prevents activation by Ino2p and Ino4p and represses transcription of INO1 (22, 23).
The significance of maintaining inositol homeostasis for cell physiology is exemplified by the inositol-less death phenomenon, in which cells starved for inositol die within a few hours of starvation (24). Inositol depletion leads to various consequences (25). Growth in the absence of inositol elicits profound changes in lipid metabolism and activates numerous stress responses (26). Altered inositol levels in the brain have been associated with a variety of mental disorders, including bipolar disorder (27), Alzheimer disease (28, 29), and manic depressive psychosis (30). Thus, the ability of cells to maintain the right balance of inositol at all times undoubtedly necessitates a high level of regulation.
Although the regulation of inositol biosynthesis at the level of INO1 transcription has been extensively characterized, as discussed above, the regulation of MIPS activity has not been studied. The anticonvulsant drug valproate (VPA) induces depletion of inositol, and MIPS has been suggested as a possible target of the drug (31–33). Our initial finding that VPA leads to a decrease in inositol 3-phosphate in yeast (33) suggested that VPA inhibits MIPS activity. The subsequent observation that VPA-mediated inhibition was indirect (31) suggested that MIPS is regulated post-translationally. Phosphorylation controls the regulation and localization of numerous enzymes, many of which, like MIPS, are transcriptionally regulated by UASINO elements (34–36). To our knowledge, nothing is known about the post-translational regulation of MIPS.
Here we report for the first time that MIPS, from yeast and humans, is regulated by phosphorylation of at least three residues, one in the catalytic domain and two in the NAD+-binding domain, thus identifying a novel mechanism of regulation of inositol biosynthesis. These sites are conserved in yeast and human MIPS, suggesting that regulation by phosphorylation is a conserved regulatory mechanism. We also show that mutation of the two inhibitory phosphosites, Ser-184 and Ser-374, leads to decreased sensitivity to VPA, suggesting that VPA may inhibit MIPS activity as a consequence of phosphorylation of these two residues.
EXPERIMENTAL PROCEDURES
Materials
All chemicals used were reagent grade or better. Media components were from Difco, EMD Biochemicals, Sigma, or Fisher. Inositol, valproate, glucose 6-phosphate, NaIO4, Na2SO3, ascorbic acid, and ammonium molybdate were from Sigma or Fisher. Restriction enzymes, Pfu DNA polymerase, Phusion Hotstart DNA polymerase, and T4 DNA ligase were from Promega and New England Biolabs. The Wizard Plus plasmid DNA purification and DNA gel extraction kits were from Promega. Pureproteome magnetic beads and Centricon filters were from Millipore. Alkaline phosphatase was from New England Biolabs. The protease inhibitor mixture Complete Mini and the phosphatase inhibitor PhoSTOP were from Roche. ProBond nickel chelated resin and the purification columns were from Bio-Rad. Protein assay reagents and electrophoresis reagents were from Bio-Rad. PCR and sequencing primers were from IDT and Eurofins.
Strains, Media, and Growth Conditions
The Escherichia coli and Saccharomyces cerevisiae strains used in this work are listed in Table 1. Yeast was grown in synthetic minimal medium containing 2% glucose and lacking inositol unless otherwise stated. Media were supplemented with the amino acids histidine, leucine, methionine, uracil, and lysine using Difco standard concentrations. For selection of cells carrying specific plasmids, appropriate amino acids were omitted from the media. For overexpression purposes, induction medium contained 2% galactose and 1% raffinose with no glucose added. Where indicated, VPA was used at a concentration of 1 mm, and inositol was used at 75 μm. Yeast strains were grown at 30 °C. E. coli strain DH5α was used for plasmid maintenance and amplifications. Bacteria were grown at 37 °C in LB medium (0.5% yeast extract, 1% tryptone, 1% NaCl), supplemented with ampicillin (100 μg/ml) for selection purposes. For growth on plates, media were supplemented with 1.5 and 2% agar for E. coli and yeast, respectively. Growth in liquid cultures was monitored spectrophotometrically by measuring absorbance at 550 nm.
TABLE 1.
Strains used in this study
| E. coli | |
| DH5α | F−φ80dlacZΔm15Δ (lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rkmk+)phoA supE44I−thi-1gyrA96relA1 |
| S. cerevisiae | |
| BY4741 | MATa his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 |
| BY4741 ino1Δ | MATa his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, INO1::KanMX |
DNA Manipulations, PCR, and DNA Sequencing
Standard methods were followed for isolation of genomic DNA, plasmid purification, digestion with restriction endonucleases, and ligation. Transformation of bacterial and yeast cells was carried out using electroporation. All PCRs were optimized. DNA sequencing was carried out using the ACGT Inc. sequencing facility (Wheeling, IL).
Construction of Plasmids and Expression of Yeast and Human MIPS in S. cerevisiae
All plasmids and primers used in this work are listed in Tables 2 and 3, respectively. The INO1 gene was amplified from S. cerevisiae genomic DNA using the primer pair 5′-GTTGTCGGGTTCCTAATGTT-3′ and 5′-CAACAATCTCTCTTCGAATCT-3′ and tagged with His6 and an Xpress epitope on the N terminus using pRSETA as the source of the tag. The tagged INO1 was then subcloned into pYES6 containing the blasticidin marker. HindIII and XhoI were used to transfer the tagged INO1 into pRD015 to make the pRDINO1 construct, a high copy GAL1-driven plasmid containing a URA3 marker. All subsequent manipulations and mutagenesis experiments for the yeast MIPS were carried out using the pRDINO1 vector. All mutations were confirmed by sequencing. The low copy set of all mutants was prepared by transferring the mutated genes to the centromeric low copy vector p415-ADH purchased from ATCC. For human MIPS, the human INO1 gene (hINO1) was transferred from pRSETA-hINO1 (31) and cloned into pRD015. All subsequent experiments for the human MIPS, including mutagenesis, growth, and enzyme purification were carried out using pRDhINO1 and its derivatives.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristics | Source/Reference |
|---|---|---|
| pRSET A | E. coli expression vector | Invitrogen |
| pYES6-CT | Multicopy shuttle vector containing blasticidin marker | Invitrogen |
| pYES2-CT | Multicopy shuttle vector containing URA3 marker | Invitrogen |
| P415-ADH | Low copy shuttle vector containing LEU2 marker | ATCC®87374TM |
| pYES6-INO1 | Plasmid containing yeast INO1 | This study |
| pRD015 | Dual tag shuttle vector constructed using pYES2-CT as a backbone | This study |
| pRDINO1 | INO1 gene from pYES6-INO1 ligated into Hindlll/Xho1 site | This study |
| pRD-T48V | INO1T48V derivative of pRDINO1 | This study |
| pRD-S177A | INO1S177A derivative of pRDINO1 | This study |
| pRD-S184A | INO1S184A derivative of pRDINO1 | This study |
| pRD-S296A | INO1S296A derivative of pRDINO1 | This study |
| pRD-S374A | INO1S374A derivative of pRDINO1 | This study |
| pRD-S177D | INO1S177D derivative of pRDINO1 | This study |
| pRD-S184D | INO1S184D derivative of pRDINO1 | This study |
| pRD-S296D | INO1S296D derivative of pRDINO1 | This study |
| pRD-S374D | INO1S374D derivative of pRDINO1 | This study |
| pRD-S184A/S374A | INO1S184A/S374A derivative of pRDINO1 | This study |
| pADH-INO1 | INO1 gene from pRDINO1 | This study |
| pADH-T48V | INO1T48V derivative of pADH-INO1 | This study |
| pADH-S177A | INO1S177A derivative of pADH-INO1 | This study |
| pADH-S184A | INO1S184A derivative of pADH-INO1 | This study |
| pADH-S296A | INO1S296A derivative of pADH-INO1 | This study |
| pADH-S374A | INO1S374A derivative of pADH-INO1 | This study |
| pADH-S177D | INO1S177D derivative of pADH-INO1 | This study |
| pADH-S184D | INO1S184D derivative of pADH-INO1 | This study |
| pADH-S296D | INO1S296D derivative of pADH-INO1 | This study |
| pADH-S374D | INO1S374D derivative of pADH-INO1 | This study |
| pADH-S184A/S374A | INO1S184A/S374A derivative of pADH-INO1 | This study |
| pRSETA/hINO1 | Plasmid containing human MIP synthase cDNA | Ref. 31 |
| pRD-hINO1 | Human INO1 (ISYNA1) from pRSETA/hINO1 | This study |
| pADH-hINO1 | Human INO1 (ISYNA1) from pRD-hINO1 | This study |
| pRD-hS177A | hINO1S177A derivative of pRD-hINO1 | This study |
| pRD-hS177D | hINO1S177D derivative of pRD-hINO1 | This study |
| pRD-hS279A | hINO1S279A derivative of pRD-hINO1 | This study |
| pRD-hS279D | hINO1S279D derivative of pRD-hINO1 | This study |
| pRD-hS357A | hINO1S357A derivative of pRD-hINO1 | This study |
| pRD-hS357D | hINO1S357D derivative of pRD-hINO1 | This study |
TABLE 3.
Primers used for site-directed mutagenesis of yeast and human MIPS
| Primer | Sequence (5′–3′) |
|---|---|
| Yeast MIPS | |
| S184A F | CCTTGGTGAAGCCTCTTCCTGCCATTTACTACCCT |
| S184A R | AGGGTAGTAAATGGCAGGAAGAGGCTTCACCAAGG |
| S184D F | CCTTGGTGAAGCCTCTTCCTGACATTTACTACCCTGATTTCA |
| S184D R | TGAAATCAGGGTAGTAAATGTCAGGAAGAGGCTTCACCAAGG |
| S296A F | GTCCCCTATATTAATGGTGCACCGCAGAATACTTTTG |
| S296A R | CAAAAGTATTCTGCGGTGCACCATTAATATAGGGGAC |
| S296D F | CTATCTTGGAAGGTGTCCCCTATATTAATGGTGATCCGCAGAATACTTTTGTT |
| S296D R | AACAAAAGTATTCTGCGGATCACCATTAATATAGGGGACACCTTCCAAGATAG |
| S374A F | GTCTAAGGAGATTTCCAAAGCTTCTGTCATAGATGACATC |
| S374A R | GATGTCATCTATGACAGAAGCTTTGGAAATCTCCTTAGAC |
| S374D F | ATTTAGGTCTAAGGAGATTTCCAAAGATTCTGTCATAGATGACATCATCGC |
| S374D R | GCGATGATGTCATCTATGACAGAATCTTTGGAAATCTCCTTAGACCTAAAT |
| Human MIPS | |
| hS177A F | CGGCCCCGGCCTGCTGTTTACATCCC |
| hS177A R | GGGATGTAAACAGCAGGCCGGGGCCG |
| hS177D F | CCCTGCGGCCCCGGCCTGATGTTTACATCCC |
| hS177D R | GGGATGTAAACATCAGGCCGGGGCCGCAGGG |
| hS279A F | CCTTCCTCAATGGGGCTCCGCAGAACACC |
| hS279A R | GGTGTTCTGCGGAGCCCCATTGAGGAAGG |
| hS279D F | TGCCTTCCTCAATGGGGATCCGCAGAACACCCTG |
| hS279D R | CAGGGTGTTCTGCGGATCCCCATTGAGGAAGGCA |
| hS357A F | TAAGGAGGTGTCCAAGGCCAACGTGGTGGACGAC |
| hS357A R | GTCGTCCACCACGTTGGCCTTGGACACCTCCTTA |
| hS357D F | CTAAGGAGGTGTCCAAGGACAACGTGGTGGACGACA |
| hS357D R | TGTCGTCCACCACGTTGTCCTTGGACACCTCCTTAG |
| Sequencing primers | |
| GAL1 | TGCATAACCACTTTAACT |
| ADH | TCAAGCTATACCAAGCATACAATCA |
| Seq INO1 | TTAATGACACCATGGAAAACCTC |
| Seq hINO1 | CACCATTGAGCTCGGTCTG |
Site-directed Mutagenesis
Site mutations were constructed using a two-step PCR protocol developed for this study. For each mutation, two overlapping, non-phosphorylated primers were designed. Two PCRs were run simultaneously, using one primer in each reaction. pRDINO1 and pRDhINO1 were used as templates for yeast MIPS and human MIPS, respectively. The first PCR was run for 10 cycles. The two samples from the first PCR were then pooled together, and the PCR was resumed for another 20 cycles. After cooling, ligase was added to the reaction mix and incubated for 1 h at room temperature. Gel analysis was used to detect the presence of the amplified plasmid. The parent strands were digested using DpnI treatment for 2 h at 37 °C. Transformation of competent E. coli cells was carried out using electroporation. Selection was carried out on LB plates supplemented with ampicillin. Plasmids were purified, and all mutations were verified by sequencing. The constructs harboring the mutations were then used to transform yeast ino1Δ cells.
Purification of Recombinant MIPS
S. cerevisiae BY4741 ino1Δ mutant bearing the pRDINO1 construct (or its derivatives) was used to express the recombinant His6-tagged MIPS. Cells were grown at 30 °C in synthetic minimal medium lacking uracil. Galactose (2%) was used to induce overexpression of the recombinant protein. Cell extracts were prepared by disrupting cells with glass beads (0.5 mm diameter), vortexing for 30 min intermittently, keeping cells on ice. The disruption buffer contained 50 mm Tris-Cl (pH 7.4) and 300 mm NaCl. A mixture of protease and phosphatase inhibitors was added to the disruption buffer before breaking the cells. MIPS was purified using ProBond nickel-chelating resin to bind the protein, gently mixing at 4 °C for 1 h. The resin was washed twice with cold 20 mm and then 60 mm imidazole in Tris buffer (50 mm Tris-Cl (pH 7.4) and 300 mm NaCl). The protein was eluted with 300 mm imidazole in Tris buffer, dialyzed, concentrated, and resuspended in 50 mm Tris-Cl, 50 mm NaCl, 10 mm dithiothreitol (DTT). Protein concentration was determined by the Bradford method using bovine serum albumin as the standard.
MIPS Activity Assay
Purified MIPS enzyme activity was determined by the rapid colorimetric method of Barnett et al. (37) with minor modifications. Purified protein was suspended in the reaction buffer containing 100 mm Tris acetate (pH 8.0), 0.8 mm NAD, 2 mm DTT, and 14 mm NH4Cl. After the addition of 5 mm glucose 6-phosphate to a final volume of 150 μl, the reaction mixture was incubated for 1 h at 37 °C. The reaction was terminated by the addition of 50 μl of 20% (w/v) trichloroacetic acid and kept on ice for 10 min. The precipitated protein was removed by centrifugation. The supernatant (200 μl) was incubated with 200 μl of 0.2 m NaIO4 for 1 h at 37 °C. This was followed by the addition of 200 μl of 1 m Na2SO3 to remove excess NaIO4. For the measurement of phosphate, a 600-μl reagent mixture consisting of 240 μl of H2O, 120 μl of 2.5% ammonium molybdate, 120 μl of 10% ascorbic acid, and 120 μl of 6 n sulfuric acid was added to the reaction mix and incubated for 1 h at 37 °C. Absorbance was measured at 820 nm, and activity was determined by the amount of inorganic phosphate liberated. A unit of MIPS activity is defined as the amount of enzyme that catalyzes the formation of 1 nmol of product/min. Specific activity is defined as units/mg of protein.
Dephosphorylation of MIPS
Dephosphorylation reactions were performed at 37 °C in a total volume of 50 μl for 1 h. Ten μg of purified MIPS was gently mixed with nickel-coated magnetic beads for 1 h at 4 °C. The supernatant was aspirated, and the beads were resuspended in alkaline phosphatase buffer consisting of 50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 10 mm MgCl2, and 1 mm DTT. Alkaline phosphatase (10 units) was added, and the mixture was incubated at 37 °C for 45 min, after which orthovanadate was added to the reaction mix to a final concentration of 10 mm to inactivate the alkaline phosphatase. The beads were then collected and washed three times with Tris purification buffer in preparation for the MIPS assay.
In Vivo Phosphorylation of MIPS
Cells with recombinant MIPS were precultured in synthetic minimal medium, inoculated to A550 0.03 in 20 ml of phosphate-free medium to which 2 mCi of 32Pi was added, and grown to the early stationary phase with or without VPA. Cells were harvested by centrifugation, washed with water, and disrupted with glass beads in 100 μl of buffer containing 150 mm NaCl, 50 mm Tris-Cl (pH 7.5), 0.5% deoxycholate, 0.1% SDS, 1% Nonidet P-40, 10 mm NaF, 5 mm B-glycerophosphate, 1 mm sodium vanadate, and protease inhibitor mixture. The cell extracts were precleaned with 10 μl of protein G plus protein A-agarose for 1 h at 4 °C and incubated with 2 μg of anti-Xpress antibody, 50 μl of protein G plus protein A-agarose overnight at 4 °C. After washing with the same buffer, the MIPS·anti-Xpress antibody complex was dissociated by boiling for 5 min in protein sample buffer containing 2% SDS, 10% glycerol, 100 mm DTT, 60 mm Tris-Cl (pH 6.8), and 0.001% bromphenol blue. After a brief low speed centrifugation, proteins were analyzed by SDS-PAGE. Bands were visualized by Coomassie Blue staining, and 32Pi-labeled proteins were identified by autoradiography.
Phosphoamino acid Analysis
MIPS labeled with 32Pi was digested with 6 n HCl. The acid was evaporated, and the sample was dissolved in 10 μl of TLE buffer (pH 1.9) and mixed with 1 μg of cold phosphoserine, phosphothreonine, and phosphotyrosine standards. The phosphoamino acids were separated in two dimensions, by electrophoresis followed by thin layer chromatography. Standard phosphoamino acids were visualized by ninhydrin staining of the thin layer chromatography plate. Labeled phosphoamino acids were visualized by autoradiography.
Mass Spectrometry Analysis of MIPS Phosphorylation Sites
Purified protein samples were reduced with dithiothreitol, alkylated with iodoacetamide, and digested overnight with sequencing grade trypsin (Promega) or chymotrypsin (Roche Applied Science) at 37 οC. Peptides were desalted and separated by reverse phase chromatography before introduction into a linear ion trap mass spectrometer (LTQ-XL, Thermo Scientific). The top five peaks in the MS1 scan (400–1700 m/z) were sequentially selected for fragmentation by collision-induced dissociation (normalized collision energy = 30, activation Q = 0.25, activation time = 30 ms). Dynamic exclusion was turned on (if 2 hits in 5 s then excluded for 20 s, list size = 200). MS2 spectra were scored against a yeast FASTA protein database (NCBI; 6298 entries) using the SEQUEST algorithm (version 27, revision 13). Search parameters included 1.6 Da/1.0 Da parent/fragment ion tolerances; +57 on Cys fixed modification; +16 on Met and +80 on Ser/Thr/Tyr variable modifications; and up to 2 missed cleavages. Results were imported into Scaffold (version 3.5; Proteome Software), and MS2 spectra were reanalyzed against a subset database using X!Tandem (version 2007.01.01.1). Peptide probabilities were scored using the Peptide Prophet algorithm, and a ≥90% threshold was utilized. Localization probabilities of post-translational modifications were scored using Scaffold PTM (version 2.1.1; Proteome Software).
RESULTS
VPA Causes a Decrease in MIPS Activity
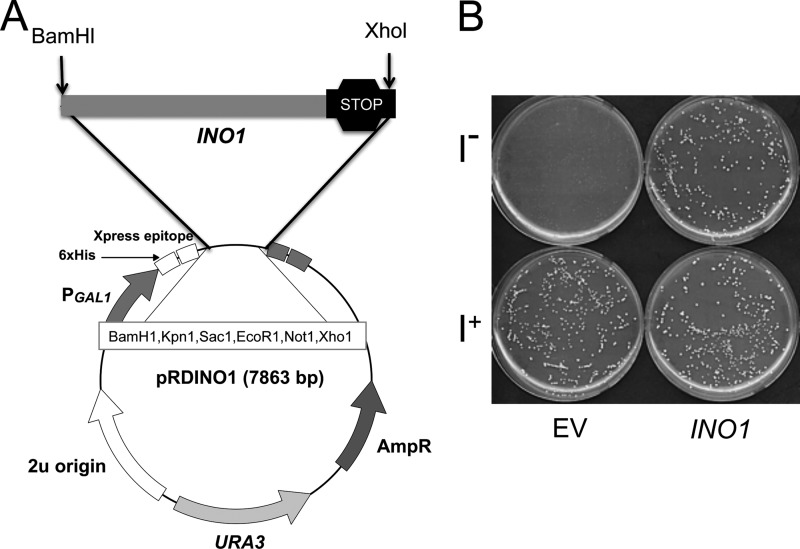
The INO1 coding sequence (1578 bp) was amplified from yeast genomic DNA, tagged with His6 and an Xpress epitope, and cloned under the GAL1 promoter in the expression vector pRD015 with URA3 as the selection marker (the construct referred to herein as pRDINO1) (Fig. 2A). To determine if the tagged MIPS is functional, pRDINO1 was transformed into ino1Δ cells that lack endogenous MIPS and therefore cannot grow independently in the absence of inositol. Transformants (ino1Δ-INO1) were selected on Ura− plates and then tested for growth in the absence of inositol (I−) in comparison with cells carrying the empty vector. The tagged MIPS rescued growth on I− medium (Fig. 2B).
FIGURE 2.
The tagged MIPS is functional and rescues growth of ino1Δ in inositol-deficient medium. A, map of the INO1 construct used for expression and purification of MIPS. B, ino1Δ cells transformed with the empty vector (pRD015) or with the vector carrying the tagged INO1 (pRDINO1) were precultured to the mid-logarithmic phase in synthetic medium (SM) supplemented with 75 μm inositol. Cells were washed twice with distilled H2O to remove residual inositol, diluted, and plated on selective SM with (I+) or without (I−) inositol supplementation.
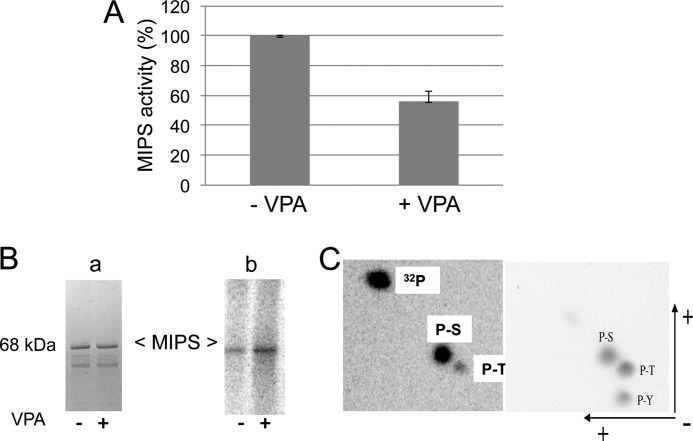
We have previously shown that cells grown in the presence of VPA exhibit a decrease in levels of inositol and inositol-3-phosphate (33) and that VPA does not directly inhibit MIPS (31). To test the possibility that VPA indirectly inhibits MIPS activity by bringing about a post-translational change in the enzyme, we expressed and purified MIPS from ino1Δ-INO1 cells grown to the mid-logarithmic phase and treated with 1 mm VPA for 3 h. MIPS purified from the VPA-treated cells showed a decrease in activity of about 40% compared with enzyme purified from untreated cells, suggesting that MIPS is regulated post-translationally (Fig. 3A).
FIGURE 3.
MIPS is a phosphoprotein. A, MIPS purified from VPA-treated cells showed decreased activity. ino1Δ cells harboring the pRDINO1 vector were grown to the mid-logarithmic phase in inositol-free SM supplemented with galactose and raffinose. 1 mm VPA was added to one culture, and cells were incubated for 3 h and then harvested. MIPS was extracted and purified, and activity was assayed using the method of Barnett et al. (37). B, ino1Δ cells harboring the tagged INO1 were resuspended in phosphate-free induction medium in the presence of 32Pi with or without VPA for 1 h and then harvested. Cell extracts were precleaned with protein G plus protein A-agarose and incubated overnight at 4 °C with anti-Xpress antibody and 50 μl of protein G plus protein A-agarose. After washing with buffer, the MIPS·anti-Xpress antibody-agarose complex was dissociated by boiling for 5 min and analyzed by SDS-PAGE. Bands were visualized by Coomassie Blue staining (a), and 32Pi-labeled MIPS was identified by phosphorimaging (b). Data represent three independent experiments. C, phosphoamino acid analysis of MIPS protein. 32Pi-Labeled MIPS was digested with 6 n HCl. The acid was evaporated, and the sample was dissolved in 10 μl of TLE buffer (pH 1.9) and mixed with 1 μg of cold phosphoserine, phosphothreonine, and phosphotyrosine standards. The phosphoamino acids were separated in two dimensions by electrophoresis followed by thin layer chromatography. Ninhydrin staining of the TLC plate (right) shows the migration of the standards. An autoradiogram (left) shows the 32Pi-labeled phosphoamino acids present in MIPS. The data are representative of three independent experiments. Error bars, S.E.
MIPS Is a Phosphoprotein
To determine if MIPS is post-translationally modified by phosphorylation, ino1Δ cells harboring the tagged INO1 were grown in the presence of 32Pi with or without VPA, and MIPS was precipitated with anti-Xpress antibody using protein A plus protein G-conjugated Sepharose. The 32P-labeled MIPS protein was resolved using SDS-PAGE and visualized with Coomassie Blue and autoradiography. MIPS purified from VPA-treated cells showed a higher degree of phosphorylation compared with MIPS purified from untreated cells (Fig. 3B). Phosphoamino acid analysis of the 32P-labeled protein indicated that MIPS was phosphorylated mostly at serine residues, although a faint label of threonine residues was also detected (Fig. 3C).
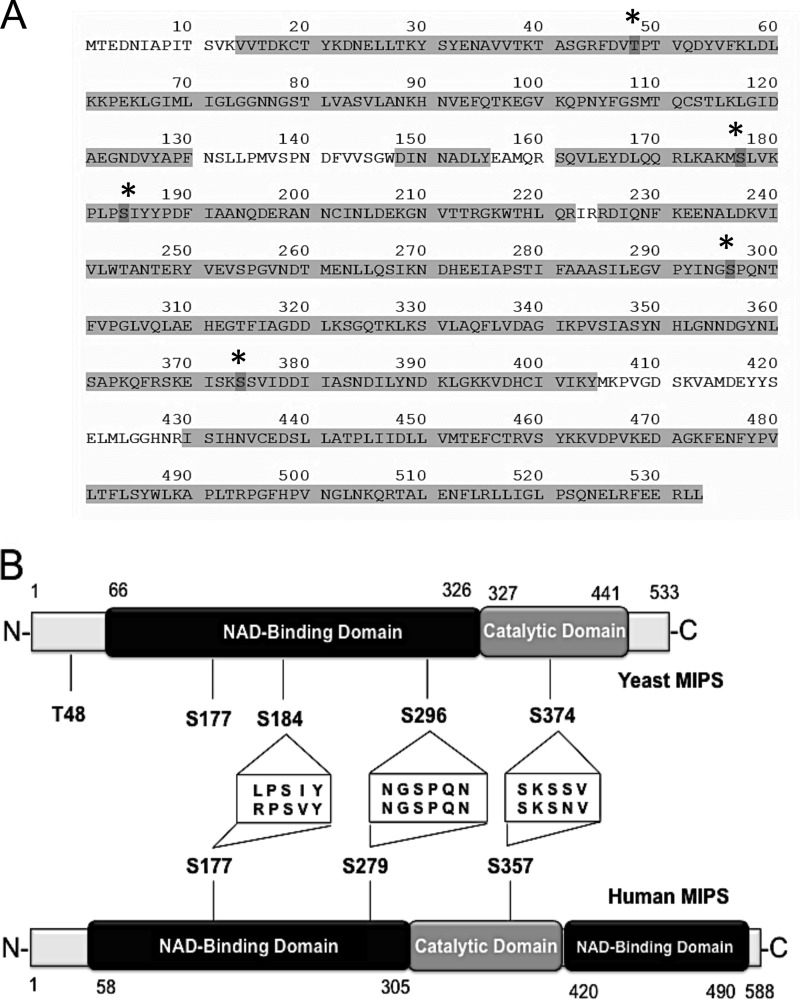
To identify the putative phosphorylated residues, MIPS was overexpressed and purified from ino1Δ-INO1 cells grown to the mid-logarithmic phase. Duplicate samples of control (no treatment) and VPA (1 mm)-treated cells were utilized. The protein was purified using ProBond resin, dialyzed, concentrated, and digested with either trypsin or chymotrypsin. The resulting peptides were separated by reverse phase chromatography and analyzed by tandem electrospray mass spectrometry. MIPS peptides and phosphopeptides were identified using an algorithm that compares MS/MS spectra against database sequences. A total sequence coverage of 88.4% was obtained with both enzymes (Fig. 4A).
FIGURE 4.
Domains and phosphosites of MIPS. A, protein sequence of yeast MIPS (NCBI accession NP_012382.2). Sequenced residues are highlighted (471/533 = 88.4% sequence coverage). Phosphosites sequenced in at least two of four samples analyzed are marked with an asterisk. B, a schematic diagram illustrating the domain structure of yeast and human MIPS and the position of the phosphorylation sites identified by mass spectrometry.
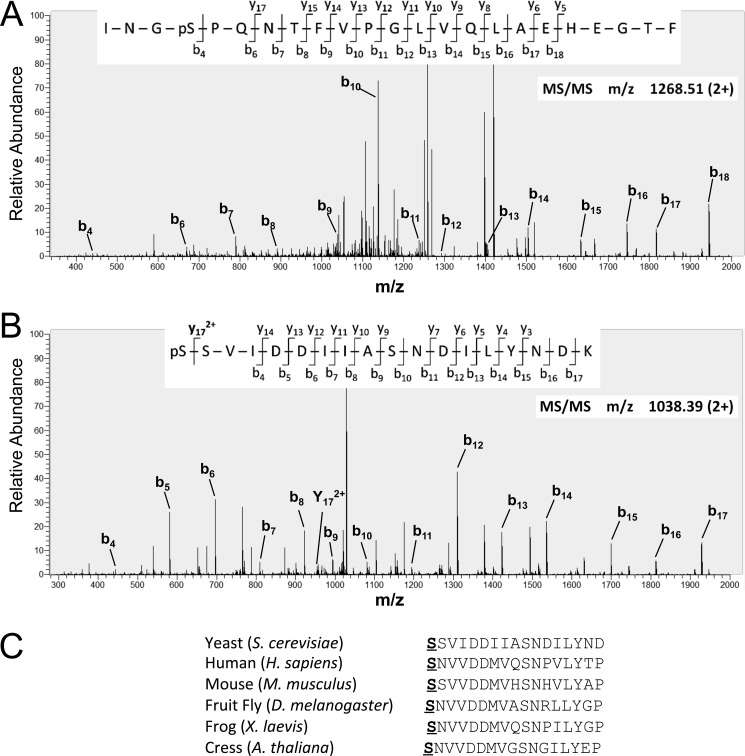
Five phosphosites were identified in four independent samples: Thr(P)-48 in the N-terminal domain; Ser(P)-177, Ser(P)-184, and Ser(P)-296 in the NAD+-binding domain; and Ser(P)-374 in the catalytic domain (Fig. 4B). For Ser(P)-48, Ser(P)-177, Ser(P)-184, and Ser(P)-296, unambiguous localization of the phosphosites was made on the basis of the corresponding MS/MS spectra. A representative fragmentation spectrum for the identification of Ser(P)-296 is shown in Fig. 5A. More complete MS/MS annotations for all phosphopeptide spectral identifications can be found in supplemental Fig. S1 and Table S1.
FIGURE 5.
MIPS is phosphorylated at five serine and threonine residues. A, representative MS/MS spectrum of the doubly charged MIPS phosphopeptide 293INGpSPQNTFVPGLVQLAEHEGTF315 (where pS represents phosphoserine) obtained after chymotryptic digestion. The phosphorylation site was unambiguously assigned to Ser(P)-296 based on the mass assignment of b4. Not all peaks are annotated. More complete MS/MS peak assignments for this spectrum and for Thr(P)-48, Ser(P)-177, Ser(P)-184, and Ser(P)-374 phosphopeptide spectra can be found in the supplemental material. B, the b4 fragment ion within the MS/MS spectrum of 374SSVIDDIIASNDILYNDK391 indicates that one of the N-terminal serine residues is phosphorylated. Two lines of evidence suggest that the phosphorylated residue is Ser(P)-374; the y172+ fragment ion indicates that Ser-375 is not phosphorylated, and the Ser-374 residue within yeast MIPS is highly conserved across several species (shown in C).
The termini of peptides do not yield fragmentation information as robustly as the central region; therefore, phosphosite determination for the 374SSVIDDIIASNDILYNDK391 phosphopeptide was not as direct. The b4–b9 ion series indicated that the phosphosite was either at Ser-374 or Ser-375 and not Ser-383 or Thr-388 (Fig. 5B). A doubly charged y17 ion was consistently seen in MS/MS spectra that matched this phosphopeptide. Although it has relatively low abundance, this fragment ion indicated that Ser-375 was not phosphorylated (Fig. 5B). Finally, the serine residue at position 374 of yeast MIPS is conserved across disparate species (Fig. 5C). This correlative evidence suggests that Ser-374 has a critical role for protein function.
Three Phosphosites Modulate Activity of Yeast MIPS
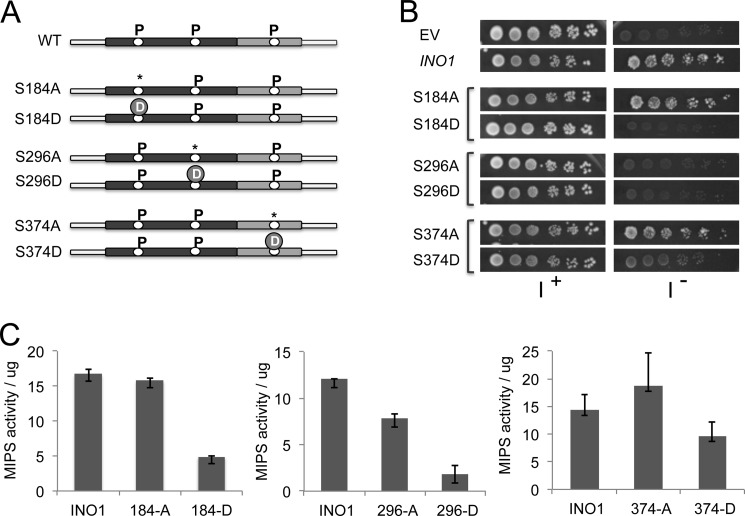
To determine which of the identified phosphorylation residues are functionally important for the activity of MIPS, two site mutants were constructed for each of the residues: 1) a phosphorylation-deficient mutant in which serine was changed to the unphosphorylatable alanine, or threonine was changed to valine and 2) a phosphomimetic mutant, in which serine was changed to the phosphorylation-mimicking aspartate (Fig. 6A). All of the mutations were confirmed by sequencing. Each mutation was constructed in two vectors: 1) a centromeric low expression vector, p415-ADH, driven by the ADH promoter, used for growth experiments and 2) a high expression vector, pRD015, driven by the GAL1 promoter, used for overexpression and purification of the enzyme. All constructs were transformed into ino1Δ cells. To assess the physiological effect of the mutations on growth, cells transformed with the empty vector, wild type INO1 gene, or mutated genes were grown on synthetic medium with or without inositol. When MIPS was expressed from the high copy vector, all of the site mutants supported growth of ino1Δ cells (data not shown). However, clear differences in growth were observed in ino1Δ cells transformed with the MIPS mutants on the low copy vector (Fig. 6B). Cells carrying the plasmid with wild type INO1 grew normally on inositol-deficient medium. Mutations in residues in the NAD-binding domain, Ser-184 and Ser-296, affected growth. Cells carrying S184A grew well, but S184D did not support growth, suggesting that phosphorylation of this residue inhibits the activity of MIPS and, hence, the synthesis of inositol. Both mutants S296A and S296D did not support growth on inositol-deficient medium, suggesting that a serine residue is essential at that position. Mutants of residue Ser-374 in the catalytic domain were also assessed. Cells carrying S374A grew well, but S374D did not support growth, suggesting that phosphorylation of this residue inhibits MIPS activity. Cells carrying mutations of Thr-48 and Ser-177 showed the same growth pattern as that of WT (data not shown), indicating that these sites are not critical or regulated by phosphorylation.
FIGURE 6.
Effect of site mutations of yeast MIPS on growth and enzyme activity. A, schematic representation of the mutated phosphorylation sites in the yeast MIPS protein. *, phosphodeficient mutation; D, phosphomimetic mutation. B, ino1Δ cells transformed with the empty vector pADH, vector carrying wild type yeast INO1, or vector carrying the indicated mutated INO1 gene were precultured in Leu− I+ medium, washed twice in distilled water to remove inositol, and spotted in a 10-fold dilution series on Leu− medium in the presence or absence of 75 μm inositol (I). Plates were incubated for 5 days at 30 °C. C, effect of site mutations on activity of yeast MIPS. Ino1Δ cells expressing the wild type (INO1) or mutated MIPS were grown at 30 °C to the exponential phase in selective SM supplemented with 75 μm inositol. MIPS was purified and assayed for activity using the method of Barnett et al. (37) as described under “Experimental Procedures,” and activity per μg of protein is indicated. The results are representative of three experiments. Values are mean ± S.E. (error bars).
To determine if the altered growth patterns seen in the mutants were due to altered MIPS activity, wild type and mutant MIPS were overexpressed and purified from ino1Δ cells grown to the late log phase in selective medium supplemented with inositol. Using equivalent amounts of purified protein from all mutants, MIPS activity was assayed using the method of Barnett et al. (37). The activity of MIPS from phosphorylation deficient mutants S184A and S374A was similar to or slightly greater than that of the wild type MIPS (Fig. 6C). In contrast, MIPS from the phosphomimetic mutants S184D and S374D showed a decrease in activity to about 30 and 60% of wild type levels, respectively. This suggests that phosphorylation of residues Ser-184 and Ser-374 has an inhibitory effect on the activity of MIPS. In contrast to Ser-184 and Ser-374, both mutations of residue Ser-296 (S296A and S296D), led to a decrease in MIPS activity (Fig. 6C), suggesting that a serine residue at this site is important for maintaining catalytic activity.
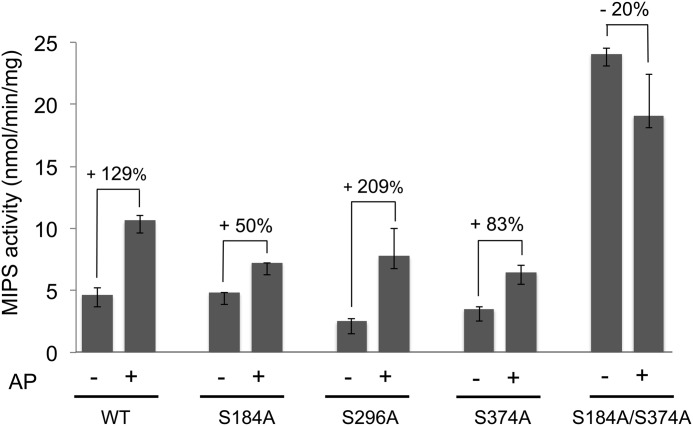
The effect of phosphorylation on MIPS activity was also addressed by dephosphorylating the protein using alkaline phosphatase. Dephosphorylation increased the activity of wild type MIPS by about 130% (Fig. 7). Dephosphorylation of the S184A and S374A mutants showed a smaller increase in activity (50 and 83% for S184A and S374A, respectively), suggesting that each of these putative inhibitory sites partially contributes to the overall inhibitory effect. Dephosphorylation of S296A caused more of an increase in activity than either S184A or S374A, suggesting that the two phosphosites Ser-184 and Ser-374 have a greater inhibitory effect than either site alone.
FIGURE 7.
Effect of dephosphorylation on MIPS activity. ino1Δ cells expressing wild type (WT) or mutated yeast MIPS were grown to the exponential phase in SM I+. MIPS was purified and mixed with nickel-coated magnetic beads and gently mixed for about 1 h at 4 °C. The beads were collected, rinsed, and resuspended in alkaline phosphatase (AP) buffer with or without alkaline phosphatase and incubated at 37 °C for 1 h. The beads were then washed three times with buffer to eliminate alkaline phosphatase and free phosphate. Activity of the bound MIPS was assayed as described under “Experimental Procedures.” The percentage change in activity after dephosphorylation is indicated. Values are mean of at least two experiments ± S.E. (error bars).
To examine the effect of a phosphorylated Ser-296, a double mutant S184A/S374A was constructed. Dephosphorylation of the double mutant did not significantly alter MIPS activity (Fig. 7), suggesting that under the physiological conditions in which the protein was purified, phosphorylation of Ser-296 was not significant.
Taken together, these results indicate that yeast MIPS is regulated by phosphorylation of at least three residues, two of which are inhibitory when phosphorylated (Ser-184 and Ser-374). The third residue, Ser-296, is strictly dependent on the presence of a serine at that position.
The Phosphorylation Sites are Conserved in Human MIPS
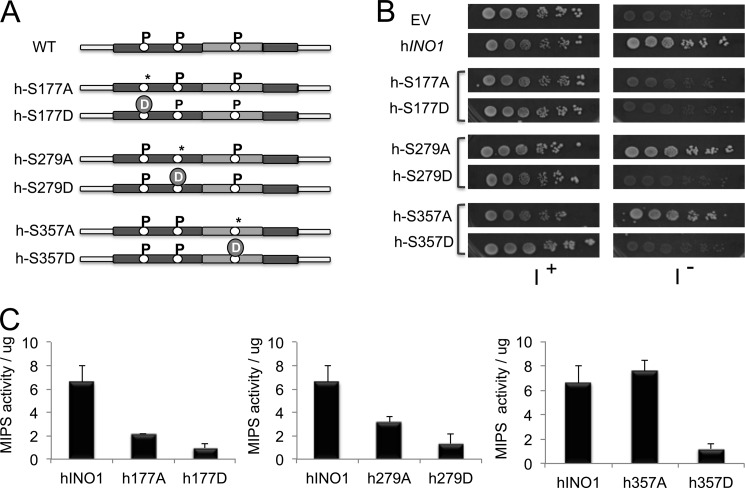
We previously cloned and expressed a human cDNA encoding MIPS and showed that human MIPS (hMIPS) is functional in yeast (31). Amino acid sequence alignment of yeast MIPS and hMIPS showed that the three residues we have found to modulate yeast MIPS are conserved in the human enzyme (Fig. 4B and supplemental Fig. S2). These hMIPS residues are Ser-177 and Ser-279, both of which are in the NAD+-binding domain, and Ser-357, which lies in the catalytic domain (Fig. 4B). To determine if hMIPS is regulated by phosphorylation, the hINO1 gene was transferred from pRSETA-hINO1 (31) and cloned into the overexpression vector pRD015. Two independent mutations were created for each residue: Ser to Ala and Ser to Asp (Fig. 8A). All of the mutations were confirmed by sequencing.
FIGURE 8.
Effect of site mutations of human MIPS on growth and enzyme activity. A, schematic representation of the mutated phosphorylation sites in the human MIPS protein. *, phosphodeficient mutation; D, phosphomimetic mutation. B, rescue of ino1Δ cells on I− medium by human MIPS. ino1Δ cells transformed with the empty vector pRD015, vector carrying wild type human INO1 or vector carrying the indicated site mutations were precultured in Ura− I+ medium, washed twice in distilled water to remove inositol, and spotted in a 10-fold dilution series on Ura− medium in the presence or absence of 75 μm inositol and grown for 3 days at 30 °C. C, effect of site mutations on the activity of human MIPS. ino1Δ cells expressing the wild type (hINO1) or the indicated mutated MIPS were grown at 30 °C to the exponential phase in selective SM supplemented with 75 μm inositol. MIPS was purified and assayed for activity using the method of Barnett et al. (37), as described under “Experimental Procedures,” and activity per μg of protein is indicated. Values are the mean of at least two experiments ± S.E. (error bars).
To determine if any of the three conserved residues are important for the function of hMIPS, all constructs along with the controls were transformed into ino1Δ cells. Transformants were grown on selective media with or without inositol. The wild type hINO1 gene rescued growth on I− medium, indicating that hMIPS expressed from the pRD015 vector is functional in yeast (Fig. 8B). Mutations S177A and S177D did not support growth in the absence of inositol (Fig. 8B). Consistent with this observation, hMIPS purified from these mutants showed decreased activity (Fig. 8C), suggesting that a serine residue is required at this site for the activity of the enzyme. For residue Ser-279, the S279A mutation supported growth on I− medium, although enzyme activity was lower than wild type. The S279D mutation did not support growth on I− medium and caused a big decrease in activity, suggesting that phosphorylation of this residue inhibits hMIPS activity and, hence, the synthesis of inositol. The S357A mutation of the catalytic domain supported growth, but S357D did not. Consistent with this, S357A caused a slight increase in activity, whereas S357D caused a decrease in activity, suggesting that phosphorylation of Ser-357 inhibits hMIPS activity.
Similar to the yeast enzyme, dephosphorylation of hMIPS led to increased activity (data not shown). Taken together, hMIPS, similar to its yeast counterpart, is regulated by phosphorylation of at least three residues, all of which are inhibitory when phosphorylated.
The S184A/S374A Double Mutation Confers Growth Advantage and Resistance to VPA
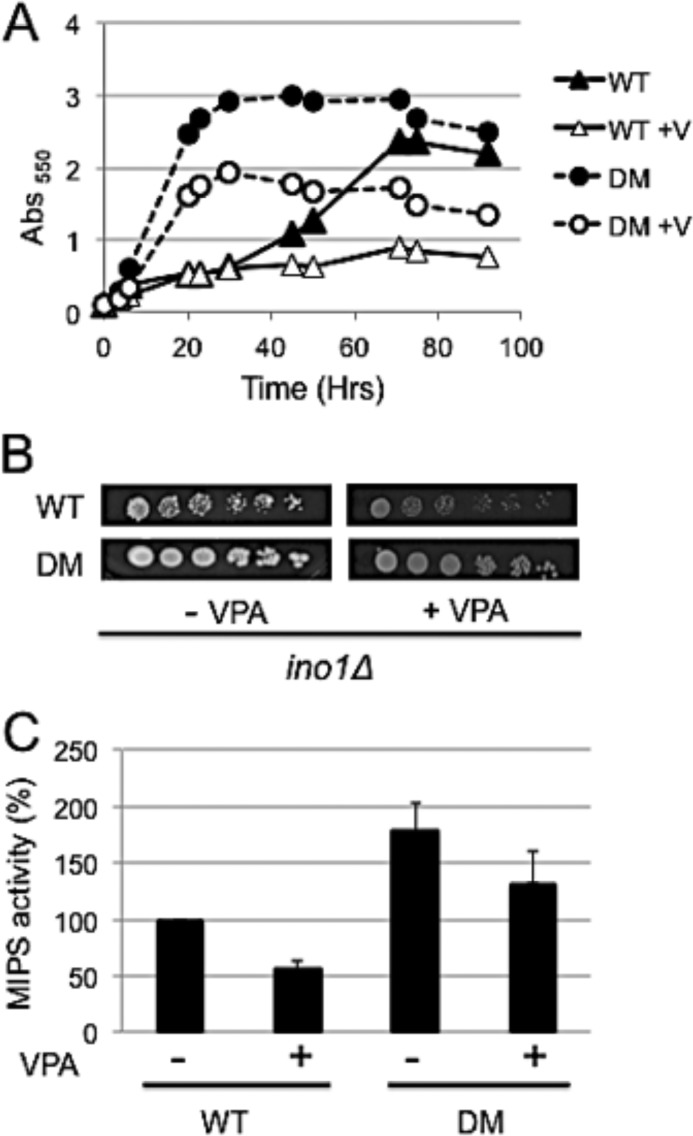
To determine the effect of loss of both Ser-184 and Ser-374 phosphosites, ino1Δ cells were transformed with a vector containing the yeast INO1 gene with both S184A and S374A mutations. As shown in Fig. 9A, cells carrying the S184A/S374A mutated INO1 (DM) exhibited a greatly increased growth rate and reduced lag phase compared with cells carrying the wild type INO1 gene (WT). Importantly, the double mutant exhibited significantly better growth in the presence of VPA (Fig. 9, A and B).
FIGURE 9.
The double mutation S184A/S374A (DM) confers a growth advantage, increases MIPS activity, and partially rescues sensitivity to VPA. A, ino1Δ cells carrying the low copy vector with the wild type INO1 (WT) or double mutant S184A/S374A were grown in Leu− medium in the presence or absence of 1 mm VPA (V) at 30 °C with constant shaking. Growth was monitored at A550. B, cells bearing the low copy vector with the wild type INO1 or the double mutation were precultured in Leu− I+ medium to the mid-logarithmic phase, harvested, washed twice, serially diluted, and spotted onto Leu− plates with or without 1 mm VPA and incubated at 30 °C. C, ino1Δ cells carrying the pRD vector with the wild type INO1 or double mutant S184A/S374A were grown to the mid-logarithmic phase, treated with VPA for 3 h, and then harvested. MIPS was purified, and activity was assayed using the method of Barnett et al. (37), as described under “Experimental Procedures.” Values are the mean of at least two experiments ± S.E. (error bars).
Consistent with increased growth conferred by the double mutation, the activity of the double mutant MIPS was almost twice that of wild type MIPS (Fig. 9C). Importantly, the decrease in activity in response to VPA was twice as much in wild type MIPS (50%) as in the double mutated MIPS (25%). Together, the data show that loss of the two phosphosites Ser-184 and Ser-374 leads to increased MIPS activity and decreased sensitivity to VPA.
DISCUSSION
The current study shows for the first time that MIPS is regulated at the post-translational level by phosphorylation. We report the following novel findings. 1) Yeast MIPS activity is regulated by phosphorylation of at least three residues. 2) Phosphorylation of the corresponding residues affects activity of human MIPS. 3) Eliminating the two inhibitory phosphosites confers resistance to VPA. These findings identify phosphorylation as a novel mechanism of regulation of inositol synthesis and suggest that VPA-mediated inositol depletion may result from phosphorylation of MIPS.
MIPS was identified as a phosphoprotein by phospholabeling and phosphoamino acid analysis (Fig. 3). Consistent with this finding, mass spectrometry identified five phosphosites in MIPS isolated from VPA-treated cells (Thr-48, Ser-177, Ser-184, Ser-296, and Ser-374) (Figs. 4 and 5 and supplemental Fig. S1). Mutation of three of the five sites modulated MIPS activity, including Ser-184, Ser-296, and Ser-374. Phosphomimetic mutations of these three sites decreased enzymatic activity. Among the three residues, only one phosphorylation-deficient mutation, S296A, decreased the activity of MIPS, indicating that this site is crucial for function.
The decreased activity of MIPS carrying either the S184D or S374D mutation suggests that phosphorylation of these two residues promotes conformational changes that alter the activity of the enzyme or block access of the substrate to its catalytic domain. These residues lie in two functionally critical domains. Residue Ser-184 is in the middle of the NAD+-binding domain, which encompasses residues 66–326, and is one of 14 residues that directly interact with NAD+ (17). The adenine portion of NAD+ specifically forms tight hydrogen bonds between N1 and Ser-184 (16). Structural studies suggest that the binding of NAD+ to the apoenzyme is a prerequisite for the orderly binding of the substrate glucose 6-phosphate to the active site (16, 38). Therefore, it is likely that NAD+ binding is disrupted when Ser-184 is phosphorylated, resulting in decreased activity of MIPS, which is dependent on NAD+ as a cofactor.
Residue Ser-374 lies within the catalytic domain of MIPS. For MIPS to complete its catalytic cycle, the active site folds and completely encapsulates the substrate in an extreme example of induced fit (16, 17, 38). We have previously shown that multisubstrate adducts that carry a phosphate group on their glucitol side are more potent inhibitors, suggesting that the presence of a phosphate group in the catalytic domain inhibits activity (39). Phosphorylation of Ser-374 may perturb access of the substrate to the catalytic domain or may destabilize the induced fit by creating steric hindrance, thus decreasing catalytic activity. Additionally, the catalytic domain is populated with hydrophobic residues (15, 17). The negative charge of the phosphate group may alter the electrostatic balance of these residues and thus cause further perturbation of enzyme activity.
Whereas the S184A and S374A mutations did not significantly affect activity, both S296A and S296D mutations decreased activity of the enzyme, suggesting that the presence of a serine residue at this location is critical. This residue is also in the NAD+-binding domain. Interestingly, Ser-296 is within a stretch of seven residues, all of which are completely conserved. It is possible that any distortion in this region destabilizes the protein structure or disrupts the interaction between monomers that is needed to form the tetrameric structure of the enzyme, leading to loss of activity.
Alignment of the amino acid sequences of hMIPS and yeast MIPS revealed a consensus of 49.3% (supplemental Fig. S2). Of the 533 amino acids of yeast MIPS, 263 residues are conserved. The highest level of conservation is in the catalytic domain (68%). The observation that the three regulatory sites identified in yeast MIPS are conserved in the human homolog was intriguing. The inability of hMIPS carrying the S357D mutation to rescue inositol auxotrophy of yeast ino1Δ cells (Fig. 8B) and the decreased activity of the enzyme (Fig. 8C) suggest that phosphorylation perturbs the active site or obstructs binding of the substrate. Similarly, phosphorylation of the human Ser-279 may perturb the activity of the enzyme as a consequence of altering its structure or obstructing the binding of NAD+. The decreased activity of both S177A and S177D of hMIPS suggests that a serine residue is critical at that position. The crystal structure of MIPS has been solved for S. cerevisiae (17, 40), Mycobacterium tuberculosis (41), and Archaeoglobus fulgidus (14). To our knowledge, the crystal structure of hMIPS has not been reported. The structural knowledge pertaining to the catalytic mechanism of MIPS is based largely on the S. cerevisiae and A. fulgidus models (14, 17). The catalytic and NAD+-binding domains of hMIPS are recognized based on sequence similarity with the yeast homolog. The current study is the first to shed light on the mechanism of regulation of hMIPS by characterizing the human protein.
Based on the prediction of NetPhos Yeast, the three identified residues Ser-184, Ser-296, and Ser-374 are potential phosphorylation sites for PKA, GSK3, and PKC, respectively. With regard to the human MIPS, human Ser-177 is predicted to be a PKA recognition site. In preliminary experiments, MIPS activity decreased when treated with PKC, suggesting that PKC may be one of the regulators of MIPS activity. GSK3 treatment increased MIPS activity. This is consistent with the slight decrease in activity we observed in the dephosphorylated double mutant, in which only the Ser-296 site remains to be phosphorylated. It also supports our earlier finding that the GSK-3 mutant shows decreased MIPS activity and increased sensitivity to VPA (42).
We have shown in a previous study that VPA causes a decrease in inositol 3-phosphate and myo-inositol (33), suggesting that VPA inhibits MIPS. Inhibition of MIPS by VPA was not observed in vitro (31), suggesting that VPA inhibition is indirect. Our current findings suggest that at least one mechanism whereby VPA causes inositol depletion may involve the phosphorylation of Ser-184 and Ser-374. First, phosphorylation of yeast MIPS is increased in cells grown in VPA (Fig. 3B). Second, the double mutation S184A/S374A partially rescues sensitivity of yeast cells to VPA (Fig. 9, A and B) and increases activity of the enzyme (Fig. 9C). We speculate that VPA triggers a signal(s) that culminates in the phosphorylation of MIPS.
It is possible that MIPS is regulated by phosphorylation of additional sites, which may not have been detected in the current study. The use of the GAL1-driven vector was essential for obtaining sufficient amounts of the enzyme for characterizing MIPS activity and for the MS analysis. However, overexpression may have perturbed the stoichiometric balance of phosphorylation, thus underestimating the importance of phosphorylation of some residues. It is also possible that overexpression may have mistargeted the protein and exposed it to non-physiological phosphorylation. Phosphorylation of some amino acids may be transient (rapidly reversible). It is also possible that phosphorylation may occur in a sequential manner, in which a phosphate at one residue may be required to activate a kinase that phosphorylates another residue(s). For example, phosphorylation of Ser-374 may facilitate the phosphorylation of Ser-184 because the increase in activity following dephosphorylation of a single mutant is almost half of that observed for the WT. Alternatively, phosphorylation may occur at different growth stages in response to different cues or signals.
Although the replacement of serine with alanine or aspartate creates phosphodeficient or phosphomimetic mutations, respectively, these mutations may not recapitulate the phosphorylated residues' functions. The mutations may alter the structural conformation of the enzyme, triggering changes in activity that are not necessarily reflective of a phosphosite. However, our findings are strongly supported by MS data, which unambiguously show that the residues identified in MIPS are phosphosites. The correlation between the growth experiments and enzyme activity demonstrates that MIPS activity is regulated by phosphorylation.
The work reported here identifies for the first time a novel mechanism of regulation of inositol biosynthesis. The knowledge that MIPS is regulated by phosphorylation will facilitate studies to identify signals that play a role in regulating this enzyme, which is crucial for maintaining inositol homeostasis.
Acknowledgment
We thank Dr. Marie Migaud for thoughtful comments and critique of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant DK 081367 (to M. L. G.). This work was also supported by Wayne State University Graduate Research Fellowship and Graduate Research Enhancement Funds (to R. M. D.). The Wayne State University Proteomics Core Facility is supported by National Institutes of Health Center Grants P30 ES06639 and P30 CA22453.

This article contains supplemental Table S1 and Figs. S1 and S2.
- UASINO
- inositol-sensitive upstream activating sequence
- MIPS
- myo-inositol-3-phosphate synthase
- hMIPS
- human myo-inositol-3-phosphate synthase
- VPA
- valproate
- SM
- synthetic medium.
REFERENCES
- 1. Di Paolo G., De Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 2. Falkenburger B. H., Jensen J. B., Dickson E. J., Suh B. C., Hille B. (2010) Phosphoinositides. Lipid regulators of membrane proteins. J. Physiol. 588, 3179–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gaspar M. L., Aregullin M. A., Jesch S. A., Henry S. A. (2006) Inositol induces a profound alteration in the pattern and rate of synthesis and turnover of membrane lipids in Saccharomyces cerevisiae. J. Biol. Chem. 281, 22773–22785 [DOI] [PubMed] [Google Scholar]
- 4. Garcia-Perez A., Burg M. B. (1991) Role of organic osmolytes in adaptation of renal cells to high osmolality. J. Membr. Biol. 119, 1–13 [DOI] [PubMed] [Google Scholar]
- 5. Bachhawat N., Mande S. C. (1999) Identification of the INO1 gene of Mycobacterium tuberculosis H37Rv reveals a novel class of inositol-1-phosphate synthase enzyme. J. Mol. Biol. 291, 531–536 [DOI] [PubMed] [Google Scholar]
- 6. Chen L., Zhou C., Yang H., Roberts M. F. (2000) Inositol-1-phosphate synthase from Archaeoglobus fulgidus is a class II aldolase. Biochemistry 39, 12415–12423 [DOI] [PubMed] [Google Scholar]
- 7. Lohia A., Hait N. C., Majumder A. L. (1999) l-myo-Inositol 1-phosphate synthase from Entamoeba histolytica. Mol. Biochem. Parasitol. 98, 67–79 [DOI] [PubMed] [Google Scholar]
- 8. Loewus M. W., Loewus F. (1971) The isolation and characterization of d-glucose 6-phosphate cycloaldolase (NAD-dependent) from Acer pseudoplatanus L. cell cultures. Its occurrence in plants. Plant Physiol. 48, 255–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adhikari J., Majumder A. L. (1988) l-myo-Inositol-1-phosphate synthase from mammalian brain. Partial purification and characterisation of the fetal and adult enzyme. Indian J. Biochem. Biophys. 25, 408–412 [PubMed] [Google Scholar]
- 10. Maeda T., Eisenberg F., Jr. (1980) Purification, structure, and catalytic properties of l-myo-Inositol-1-phosphate synthase from rat testis. J. Biol. Chem. 255, 8458–8464 [PubMed] [Google Scholar]
- 11. Dean-Johnson M., Henry S. A. (1989) Biosynthesis of inositol in yeast. Primary structure of myo-inositol-1-phosphate synthase (EC 5.5.1.4) and functional analysis of its structural gene, the INO1 locus. J. Biol. Chem. 264, 1274–1283 [PubMed] [Google Scholar]
- 12. Guan G., Dai P., Shechter I. (2003) cDNA cloning and gene expression analysis of human myo-inositol 1-phosphate synthase. Arch. Biochem. Biophys. 417, 251–259 [DOI] [PubMed] [Google Scholar]
- 13. Seelan R. S., Lakshmanan J., Casanova M. F., Parthasarathy R. N. (2009) Identification of myo-inositol-3-phosphate synthase isoforms. Characterization, expression, and putative role of a 16-kDa γc isoform. J. Biol. Chem. 284, 9443–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Neelon K., Roberts M. F., Stec B. (2011) Crystal structure of a trapped catalytic intermediate suggests that forced atomic proximity drives the catalysis of mIPS. Biophys. J. 101, 2816–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Geiger J. H., Jin X. (2006) The structure and mechanism of myo-inositol-1-phosphate synthase. Subcell. Biochem. 39, 157–180 [DOI] [PubMed] [Google Scholar]
- 16. Jin X., Geiger J. H. (2003) Structures of NAD+- and NADH-bound 1-l-myo-Inositol 1-phosphate synthase. Acta Crystallogr. D Biol. Crystallogr. 59, 1154–1164 [DOI] [PubMed] [Google Scholar]
- 17. Stein A. J., Geiger J. H. (2002) The crystal structure and mechanism of 1-l-myo-inositol-1-phosphate synthase. J. Biol. Chem. 277, 9484–9491 [DOI] [PubMed] [Google Scholar]
- 18. Majumder A. L., Chatterjee A., Ghosh Dastidar K., Majee M. (2003) Diversification and evolution of l-myo-inositol 1-phosphate synthase. FEBS Lett. 553, 3–10 [DOI] [PubMed] [Google Scholar]
- 19. Carman G. M., Henry S. A. (1999) Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 38, 361–399 [DOI] [PubMed] [Google Scholar]
- 20. Greenberg M. L., Lopes J. M. (1996) Genetic regulation of phospholipid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 60, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nunez L. R., Henry S. A. (2006) Regulation of 1D-myo-inositol-3-phosphate synthase in yeast. Subcell. Biochem. 39, 135–156 [DOI] [PubMed] [Google Scholar]
- 22. Henry S. A., Kohlwein S. D., Carman G. M. (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. White M. J., Hirsch J. P., Henry S. A. (1991) The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper. J. Biol. Chem. 266, 863–872 [PubMed] [Google Scholar]
- 24. Culbertson M. R., Henry S. A. (1975) Inositol-requiring mutants of Saccharomyces cerevisiae. Genetics 80, 23–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deranieh R. M., Greenberg M. L. (2009) Cellular consequences of inositol depletion. Biochem. Soc. Trans. 37, 1099–1103 [DOI] [PubMed] [Google Scholar]
- 26. Villa-García M. J., Choi M. S., Hinz F. I., Gaspar M. L., Jesch S. A., Henry S. A. (2011) Genome-wide screen for inositol auxotrophy in Saccharomyces cerevisiae implicates lipid metabolism in stress response signaling. Mol. Genet. Genomics 285, 125–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Belmaker R. H. (2004) Bipolar disorder. N. Engl. J. Med. 351, 476–486 [DOI] [PubMed] [Google Scholar]
- 28. McLaurin J., Franklin T., Chakrabartty A., Fraser P. E. (1998) Phosphatidylinositol and inositol involvement in Alzheimer amyloid-β fibril growth and arrest. J. Mol. Biol. 278, 183–194 [DOI] [PubMed] [Google Scholar]
- 29. Shimohama S., Tanino H., Sumida Y., Tsuda J., Fujimoto S. (1998) Alteration of myo-inositol monophosphatase in Alzheimer's disease brains. Neuroscience Lett. 245, 159–162 [DOI] [PubMed] [Google Scholar]
- 30. Nahorski S. R., Ragan C. I., Challiss R. A. (1991) Lithium and the phosphoinositide cycle. An example of uncompetitive inhibition and its pharmacological consequences. Trends Pharmacol. Sci. 12, 297–303 [DOI] [PubMed] [Google Scholar]
- 31. Ju S., Shaltiel G., Shamir A., Agam G., Greenberg M. L. (2004) Human 1-d-myo-Inositol-3-phosphate synthase is functional in yeast. J. Biol. Chem. 279, 21759–21765 [DOI] [PubMed] [Google Scholar]
- 32. Shaltiel G., Shamir A., Shapiro J., Ding D., Dalton E., Bialer M., Harwood A. J., Belmaker R. H., Greenberg M. L., Agam G. (2004) Valproate decreases inositol biosynthesis. Biol. Psychiatry 56, 868–874 [DOI] [PubMed] [Google Scholar]
- 33. Vaden D. L., Ding D., Peterson B., Greenberg M. L. (2001) Lithium and valproate decrease inositol mass and increase expression of the yeast INO1 and INO2 genes for inositol biosynthesis. J. Biol. Chem. 276, 15466–15471 [DOI] [PubMed] [Google Scholar]
- 34. Chang Y. F., Martin S. S., Baldwin E. P., Carman G. M. (2007) Phosphorylation of human CTP synthetase 1 by protein kinase C. Identification of Ser462 and Thr455 as major sites of phosphorylation. J. Biol. Chem. 282, 17613–17622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Choi H. S., Han G. S., Carman G. M. (2010) Phosphorylation of yeast phosphatidylserine synthase by protein kinase A. Identification of Ser46 and Ser47 as major sites of phosphorylation. J. Biol. Chem. 285, 11526–11536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Choi H. S., Su W. M., Morgan J. M., Han G. S., Xu Z., Karanasios E., Siniossoglou S., Carman G. M. (2011) Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae. Identification of Ser602, Thr723, and Ser744 as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem. 286, 1486–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barnett J. E., Brice R. E., Corina D. L. (1970) A colorimetric determination of inositol monophosphates as an assay for d-glucose 6-phosphate-1l-myoinositol 1-phosphate cyclase. Biochem. J. 119, 183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jin X., Foley K. M., Geiger J. H. (2004) The structure of the 1l-myo-inositol-1-phosphate synthase-NAD+-2-deoxy-d-glucitol 6-(E)-vinylhomophosphonate complex demands a revision of the enzyme mechanism. J. Biol. Chem. 279, 13889–13895 [DOI] [PubMed] [Google Scholar]
- 39. Deranieh R. M., Greenberg M. L., Le Calvez P. B., Mooney M. C., Migaud M. E. (2012) Probing myo-inositol 1-phosphate synthase with multisubstrate adducts. Org. Biomol. Chem. 10, 9601–9619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kniewel R., Buglino J. A., Shen V., Chadha T., Beckwith A., Lima C. D. (2002) Structural analysis of Saccharomyces cerevisiae myo-inositol phosphate synthase. J. Struct. Funct. Genomics 2, 129–134 [DOI] [PubMed] [Google Scholar]
- 41. Norman R. A., McAlister M. S., Murray-Rust J., Movahedzadeh F., Stoker N. G., McDonald N. Q. (2002) Crystal structure of inositol 1-phosphate synthase from Mycobacterium tuberculosis, a key enzyme in phosphatidylinositol synthesis. Structure 10, 393–402 [DOI] [PubMed] [Google Scholar]
- 42. Azab A. N., He Q., Ju S., Li G., Greenberg M. L. (2007) Glycogen synthase kinase-3 is required for optimal de novo synthesis of inositol. Mol. Microbiol. 63, 1248–1258 [DOI] [PubMed] [Google Scholar]